Abstract
The genetic diversity of 35 Bacillus sphaericus strains was analyzed by a newly developed multilocus sequence typing (MLST) scheme, toxin gene pool survey, and mosquito bioassay. The results demonstrated that strains assigned to the same sequence type (ST) had the same occurrence of toxin genes. Further sequence analysis revealed that toxic strains presented a nearly clonal population structure, whereas nontoxic strains had a high level of heterogeneity and were significantly distinct from toxic strains.
TEXT
Bacillus sphaericus is an aerobic, mesophilic, spore-forming, and Gram-positive bacterium that is commonly isolated from soil. Some strains are toxic toward mosquito larvae and have been widely used as biocontrol agents for disease-transmitting mosquitoes (18). The larvicidal activity of B. sphaericus mainly originates from the binary toxins (Bin proteins), which are produced during sporulation and form the major toxic components in commercial B. sphaericus strains. Other identified toxin proteins, such as Mtx1, Mtx2, and Mtx3, are produced during vegetative growth but play a minor role in toxicity because of their low production and quick degradation by B. sphaericus proteinases (23). Recently, a new two-component set of Cry toxins, Cry48Aa and Cry49Aa, were characterized in B. sphaericus strain IAB59, and a purified equimolar mixture of both proteins exhibited high target specificity against Culex quinquefasciatus (10, 11).
The evolutionary model and systematic classification of B. sphaericus is a continual source of debate. Several typing methods, including DNA homology analysis (13), serotyping (2), multilocus enzyme electrophoresis (MLEE) (25), 16S rRNA gene analysis (16, 17), and mosquitocidal toxin gene detection by PCR (8), have been used for the identification of toxic B. sphaericus. Based on the results of DNA-DNA hybridization experiments, isolates of B. sphaericus can be divided into five DNA groups, and mosquitocidal B. sphaericus isolates were all found within group IIA (13). Serotyping revealed that pathogenic B. sphaericus strains can be grouped into 9 (H1, H2, H3, H5, H6, H9, H25, H26, and H48) of 49 serotypes (2). Furthermore, the 16S rRNA gene sequences of toxic strains were essentially identical. Likewise, a PCR-based method for classification according to the toxin genes provides very limited information and is insufficient to distinguish B. sphaericus as a single species. Thus, the evolutionary relationship between toxic and nontoxic strains has yet to be clearly established. In order to characterize the isolates and explore the population structure of B. sphaericus, a multilocus sequence typing (MLST) scheme was employed for the analysis of 35 B. sphaericus strains for the first time (Table 1) by sequencing six chromosomally encoded housekeeping genes, adk (adenylate kinase), ccpA (catabolite control protein A), pycA (pyruvate carboxylase), glyA (serine hydroxylmethyl transferase), glcK (glucose 6-phosphate kinase), and glpF (glycerol uptake facilitator protein).
Table 1.
B. sphaericus strains used in this study and their characteristics
| Strain | Origin | ST | Allelic profilea | PCR result forb: |
Serotypec | Toxicityd | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| binA | binB | mtx1 | Mtx2 | Mtx3 | cry48Aa | cry49Aa | ||||||
| C3-41 | China | 1 | 1, 2, 6, 6, 5, 1 | + | + | + | + | + | − | − | H5 | H |
| 2362 | USA | 1 | 1, 2, 6, 6, 5, 1 | + | + | + | + | + | − | − | H5 | H |
| 1593 | Indonesia | 1 | 1, 2, 6, 6, 5, 1 | + | + | + | + | + | − | − | H5 | H |
| 2317-2 | USA | 1 | 1, 2, 6, 6, 5, 1 | + | + | + | + | + | − | − | H5 | H |
| Bs44-4 | China | 1 | 1, 2, 6, 6, 5, 1 | + | + | + | + | + | − | − | H5 | H |
| Bs77-5 | China | 1 | 1, 2, 6, 6, 5, 1 | + | + | + | + | + | − | − | H5 | H |
| Bs90-2 | China | 1 | 1, 2, 6, 6, 5, 1 | + | + | + | + | + | − | − | H5 | M |
| Bs91-5 | China | 1 | 1, 2, 6, 6, 5, 1 | + | + | + | + | + | − | − | H5 | H |
| Bs150-3 | China | 1 | 1, 2, 6, 6, 5, 1 | + | + | + | + | + | − | − | H5 | M |
| Bs356-11 | China | 1 | 1, 2, 6, 6, 5, 1 | + | + | + | + | + | − | − | H5 | H |
| Bs436-8 | China | 1 | 1, 2, 6, 6, 5, 1 | + | + | + | + | + | − | − | H5 | H |
| S-35 | China | 1 | 1, 2, 6, 6, 5, 1 | + | + | + | + | + | − | − | H5 | H |
| S-70 | China | 1 | 1, 2, 6, 6, 5, 1 | + | + | + | + | + | − | − | H5 | H |
| S-128 | China | 1 | 1, 2, 6, 6, 5, 1 | + | + | + | + | + | − | − | H5 | H |
| 47-6b | China | 2 | 1, 6, 3, 7, 10, 4 | + | + | + | + | + | + | + | H6 | H |
| IAB59 | Ghana | 2 | 1, 6, 3, 7, 10, 4 | + | + | + | + | + | + | + | H6 | H |
| IAB763 | Ghana | 2 | 1, 6, 3, 7, 10, 4 | + | + | + | + | + | + | + | H6 | H |
| IAB769 | Ghana | 2 | 1, 6, 3, 7, 10, 4 | + | + | + | + | + | + | + | H6 | H |
| Bs208-6 | China | 3 | 4, 8, 9, 1, 8, 7 | − | − | − | − | − | − | − | ND | N |
| Bs225-2 | China | 3 | 4, 8, 9, 1, 8, 7 | − | − | − | − | − | − | − | ND | N |
| 2297 | USA | 4 | 6, 6, 6, 7, 2, 11 | + | + | + | + | + | − | − | H25 | H |
| IAB872 | Ghana | 5 | 1, 2, 6, 7, 4, 2 | + | + | + | + | + | − | − | H48 | H |
| IAB881 | Ghana | 6 | 1, 6, 6, 6, 11, 1 | + | + | − | − | + | − | − | H3 | H |
| Bs117-3 | China | 6 | 1, 6, 6, 6, 11, 1 | + | + | − | − | + | − | − | H5 | M |
| NRS1693 | USA | 7 | 7, 4, 2, 2, 7, 6 | − | − | − | + | − | − | − | H2 | N |
| 2314-2 | Thailand | 8 | 2, 5, 4, 4, 3, 5 | − | − | − | − | − | − | − | H9 | N |
| LP1-G | Singapore | 9 | 1, 2, 6, 3, 6, 8 | + | + | − | − | + | + | + | H3 | H |
| Bs-197 | India | 10 | 1, 6, 1, 7, 11, 4 | + | + | + | + | + | − | − | H1 | M |
| 2173 | USA | 11 | 1, 2, 6, 7, 11, 3 | − | − | − | − | + | + | + | H26 | M |
| Cok31 | Turkey | 12 | 6, 2, 6, 7, 11, 1 | − | − | + | + | + | − | − | H9 | L |
| SSII-1 | India | 13 | 6, 6, 6, 7, 11, 4 | − | − | + | + | + | − | − | H2 | L |
| Kellen Q | USA | 14 | 3, 2, 6, 3, 10, 2 | − | − | + | + | + | − | − | H1 | L |
| Dak614 | France | 15 | 8, 3, 7, 9, 9, 8 | − | − | − | − | − | − | − | H4 | N |
| SO7001 | France | 16 | 7, 1, 5, 8, 12, 9 | − | − | − | + | − | − | − | ND | N |
| Bs227-3 | China | 17 | 5, 7, 8, 5, 1, 10 | − | − | − | − | − | − | − | ND | N |
The number refers to the allele number, and allelic profile is given in the following order: adk, ccpA, pycA, glyA, glcK, glpF.
+, present; −, absent.
Serotype is as determined by de Barjac et al. (2). ND, not determined.
Effect on larvae of Culex quinquefasciatus. H, high toxicity (50% lethal concentration [LC50] ≤ 1 ng of fermentation broth per ml); M, moderate toxicity (LC50 ≈ 100 ng of fermentation broth per ml); N, nontoxic; L, low toxicity (LC50 ≥ 1 μg of fermentation broth per ml). Note that the number of nontoxic strains used for analysis is much less than the number of toxic strains. This is because although different degenerate PCR strategies have been tried (data not shown), for most of the nontoxic strains, the products of the six loci for MLST are not available simultaneously due to the genetic diversity.
The six MLST gene fragments of 35 strains were amplified and sequenced using primers designed based on the nucleotide sequences of these loci in B. sphaericus strain C3-41 (GenBank accession number NC_010382) (see Table S1 in the supplemental material). All the nucleotide sequences will be submitted to the MLST database (http://www.mlst.net/). Data are available from the authors on request. The sequences obtained for each gene were aligned without indels. Each unique sequence was assigned an arbitrary allele number, and the combination of allele numbers for all six loci of a given isolate was then assigned an arbitrary sequence type (ST). Descriptive analysis of the genetic variability at MLST loci was performed with the DNAsp package, version 5.10.00 (19). As shown in Table 2, the MLST gene fragments varied in length from 414 bp (glpF) to 654 bp (glcK), and the average GC contents of different gene fragments ranged from 37.4% (glcK) to 42.0% (pycA). The ratio of nonsynonymous to synonymous mutations (Ka/Ks) with a Jukes-Cantor correction was much less than one for all loci, from 0.0185 (ccpA) to 0.1196 (glcK), indicating the absence of a strong positive selection pressure and the suitability of these loci for genetic population studies. The genetic diversity of the six loci ranged from π = 0.0249 (adk) to π = 0.0715 (glcK), with the number of alleles ranging from 8 (adk) to 12 (glcK). The average number of alleles per locus (9.5) was higher than that observed by MLEE (4.0) (25). Altogether, 17 unique allelic profiles, or sequence types (STs), which were numbered sequentially, were observed in the 35 isolates (Table 1). All toxic strains belonging to the DNA IIA group and involving the nine serotypes could be assigned to 11 STs, indicating that the MLST method possessed a better discriminatory power.
Table 2.
Sequence variation at six loci
| Locus | Targeta | Fragment (gene)b size (bp) | No. of alleles | Avg GC content (%) | Ksc | Kad | Ka/Ks | πe | No. (%) of polymorphic sites |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Toxic | Nontoxic | |||||||||
| adk | Bsph_4593 | 531 (654) | 8 | 39.5 | 0.1013 | 0.0075 | 0.0740 | 0.0249 | 77 (14.5) | 2 (0.4) | 72 (13.6) |
| ccpA | Bsph_4200 | 579 (1,005) | 8 | 40.4 | 0.3192 | 0.0059 | 0.0185 | 0.0428 | 132 (22.8) | 1 (0.2) | 128 (21.6) |
| pycA | Bsph_1392 | 534 (3,435) | 9 | 42.0 | 0.3608 | 0.0086 | 0.0238 | 0.0480 | 133 (24.9) | 2 (0.4) | 128 (24.0) |
| glyA | Bsph_1005 | 552 (1,242) | 9 | 40.2 | 0.2419 | 0.0054 | 0.0223 | 0.0414 | 117 (21.2) | 3 (0.6) | 105 (19.0) |
| glcK | Bsph_1259 | 654 (909) | 12 | 37.4 | 0.3872 | 0.0463 | 0.1196 | 0.0715 | 250 (38.2) | 6 (0.9) | 235 (36.0) |
| glpF | Bsph_0241 | 414 (804) | 11 | 39.0 | 0.2488 | 0.0164 | 0.0659 | 0.0496 | 107 (25.8) | 7 (1.7)f | 104 (25.1) |
Values in parentheses represent the total length of the gene.
Ks, number of synonymous changes per synonymous site.
Ka, number of nonsynonymous changes per nonsynonymous site.
π, nucleotide diversity per site.
Toxic strain LP1-G shared an identical glpF gene fragment with nontoxic strain Dak614 and, therefore, was not included in the analysis.
For a better understanding of the relationship of allele diversity between toxic and nontoxic strains, the occurrence of toxin genes and mosquitocidal activities against C. quinquefasciatus larvae were surveyed. PCR assays with seven toxin genes, binA, binB, mtx1, mtx2, mtx3, cry48Aa, and cry49Aa, were performed for the 35 strains (see primers in Table S1 in the supplemental material). The bioassays were carried out with the method proposed by the World Health Organization (24), and four toxicity levels, high, moderate, low, and nontoxicity, were determined. It was observed that highly or moderately toxic B. sphaericus strains contained at least one of the two pairs of genes binA-binB and cry48Aa-cry49Aa, whereas low-toxicity strains possessed only three mtx genes and nontoxic strains contained only the mtx2 gene or no toxin genes at all. Remarkably, the strains assigned to the same ST harbored the same toxin genes (Table 1).
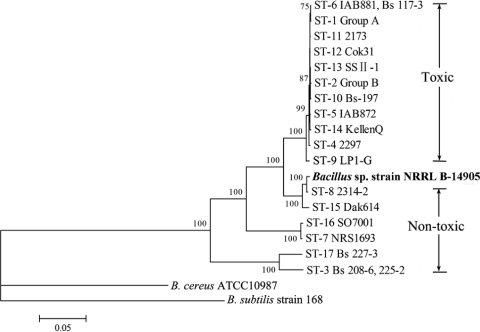
Using MEGA version 4.0 software (22), a neighbor-joining (NJ) phylogenetic tree based on the Kimura two-parameter model was constructed for the 3,264-bp concatenated sequences of six loci of B. sphaericus isolates (Fig. 1). The nucleotide sequences of the equivalent loci of B. cereus ATCC 10987, B. subtilis strain 168, and Bacillus sp. strain NRRL B-14905 (GenBank accession numbers NC_003909, NC_000964, and NZ_AAXV00000000) were included for comparison. The phylogenetic tree revealed that all toxic B. sphaericus strains were most closely related and fell into one distinct tight cluster despite their worldwide distribution, whereas nontoxic strains were more diverse and were grouped into several other clusters. NJ trees were also constructed individually for all six loci to examine the degrees of congruence between trees (see Fig. S1 in the supplemental material). Five trees based on adk, ccpA, pycA, glyA, and glcK showed similar topologies, while the tree derived from glpF showed minor differences. Specifically, ST9 (strain LP1-G) shared an identical glpF gene fragment with ST15 (strain Dak614). Previous studies showed that B. sphaericus and Bacillus sp. strain NRRL B-14905 have similar genotypic and phenotypic characteristics, including the inability to metabolize polysaccharides, suggesting that they should be reclassified into one genus (1, 20). In this study, the phylogenetic analysis revealed that B. sphaericus strains were closer to Bacillus sp. strain NRRL B-14905 than to B. cereus ATCC 10987 and Bacillus subtilis strain 168, which were positioned as outgroups on the phylogenetic tree (Fig. 1; also see Fig. S1 in the supplemental material).
Fig. 1.
NJ phylogenetic tree constructed from the concatenated sequences of six loci of B. sphaericus and three related Bacillus species. One definite toxic-specific cluster (Toxic) was identified. Group A and Group B refer to strains of the indicated ST listed in Table 1. B. cereus ATCC 10987 and B. subtilis strain 168 were used as outgroups. Bootstrap values above 70% are indicated. All branch lengths are drawn to scale.
To examine the effect of recombination on diversification of B. sphaericus population, split decomposition analysis with SplitsTree version 4.10 software (7) was performed. When the concatenated sequences were investigated, the split graph showed a networklike structure (see Fig. S2 in the supplemental material), and evidence of significant recombination was found (P = 0.044). However, the evidence of recombination disappeared when ST9 (strain LP1-G) was removed from the analysis. This could be explained by the investigation of horizontal gene transfer (HGT) for the concatenated sequences using the RDP3Beta40 package (15), which revealed only one HGT event (P ≤ 1.518 × 10−5). Specifically, the glpF gene fragment of ST9 (strain LP1-G) was involved in the HGT event, consistent with the incongruence between the glpF gene tree and the other five gene trees. Evidence for recombination was also tested by determining the standardized index of association, IsA (5), with the use of SRART version 2.0 software (9). The IsA values were significantly higher than 0 (IsA = 0.526, P < 0.001) for 35 B. sphaericus strains, indicating that alleles were in strong linkage disequilibrium. However, when taking STs as the unit, which excluded the pandemic isolates from the datasets, a lower value was observed (IsA = 0.241, P < 0.001), indicating the presence of a limited amount of recombination based on the nucleotide differences which did not completely destroy the linkage between alleles. This observation was in accordance with the results of RDP analysis, although the latter detected the HGT event caused by the migration of nucleotide fragments. Nevertheless, the split graphs should be interpreted carefully because of the low number of alleles obtained. Furthermore, the ClonalFrame version 1.1 package (4) was employed to evaluate the influences of recombination relative to those of mutation on sequence diversification: two complementary measures of recombination rate were calculated, ρ/θ (the relative frequencies of occurrence of recombination and mutation) and r/m (the relative effects of recombination and mutation). Five independent ClonalFrame runs were performed for 17 unique STs, with each consisting of 100,000 burn-in iterations and 200,000 sampling iterations, and the samples from five independent runs were then concatenated for further analysis. We found a mean value for ρ (the recombination rate times two) of 1.152. The frequency of occurrence of recombination relative to that of mutation (ρ/θ) was 0.007, and the effect of recombination relative to that of mutation (r/m) was 0.113 (see Table S2 in the supplemental material). This estimate of the frequency and effect of recombination indicated that recombination was relatively rare compared to the rates for most species, such as the B. cereus group (21), Campylobacter coli (14), and Listeria monocytogenes (3), and that mutations were largely responsible for the generation of sequence diversity in B. sphaericus. Overall, only a limited amount of recombination in the evolution of B. sphaericus might happen based on the analysis by several methods.
In conclusion, although the presence of a certain degree of recombination cannot be excluded, our results demonstrated that there was a nearly clonal population structure in toxic strains, as opposed to the otherwise broad diversity among the nontoxic B. sphaericus isolates. One question is, why is there relatively little sequence diversity in toxic strains compared to that in nontoxic B. sphaericus strains? The results of previous studies indicate that Bacillus anthracis is a recently emerging pathogen compared to Bacillus cereus. It has been suggested that B. anthracis has evolved as a monomorphic lineage because of strict coevolution involving both the chromosome and the two large plasmids pXO1 and pXO2 following acquisition of the virulence plasmids by HGT (12). Similarly, compared to mosquitocidal B. sphaericus, nontoxic B. sphaericus is likely to be a more ancient lineage and has undergone an evolutionary radiation in response to different selection pressures, in which sequence variation is enhanced by limited recombination. Until the latter period of evolution, some strains have acquired the toxic genes by coincident HGT events, such as binA-binB, which seemed to be spread by a transposon according to the bilateral sequence analysis from Hu et al. (6). Furthermore, mosquito-bacterium interactions or the coevolution of B. sphaericus with other bacteria (e.g., Bacillus thuringiensis) in mosquito breeding habitats may provide more clues for the evolution or acquisition of toxin genes. The combination of a B. sphaericus Bin-like protein (Cry49Aa) and a B. thuringiensis 3-domain Cry protein (Cry48Aa) in some B. sphaericus strains may provide an important argument for this speculation. Once they obtain the ability for toxicity to mosquitoes, these toxic strains expand quickly and form an independent population structure, in which sequence variation is mainly introduced by point mutations. However, more extensive studies are necessary to accurately assess the population structure of B. sphaericus, especially for nontoxic strains.
Supplementary Material
Acknowledgments
We are grateful to Quanxin Cai for his technical assistance and to Simon Rayner and Na Han for useful suggestions and critical reading.
The project was supported by grants from the Chinese Academy of Sciences (KSCX2-SW-301-10′KSCX2-SW-315), an NFSC grant (30800002), and a 973 grant (2009CB118902) from China.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 17 June 2011.
REFERENCES
- 1. Ahmed I., Yokota A., Yamazoe A., Fujiwara T. 2007. Proposal of Lysinibacillus boronitolerans gen. nov. sp. nov., and transfer of Bacillus fusiformis to Lysinibacillus fusiformis comb. nov. and Bacillus sphaericus to Lysinibacillus sphaericus comb. nov. Int. J. Syst. Evol. Microbiol. 57:1117–1125 [DOI] [PubMed] [Google Scholar]
- 2. de Barjac H., Larget-Thiéry I., Cosmao Dumanoir V., Ripouteau H. 1985. Serological classification of Bacillus sphaericus strains on the basis of toxicity to mosquito larvae. Appl. Microbiol. Biotechnol. 21:85–90 [Google Scholar]
- 3. den Bakker H. C., Didelot X., Fortes E. D., Nightingale K. K., Wiedmann M. 2008. Lineage specific recombination rates and microevolution in Listeria monocytogenes. BMC Evol. Biol. 8:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Didelot X., Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haubold B., Hudson R. R. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847–848 [DOI] [PubMed] [Google Scholar]
- 6. Hu X., et al. 2008. Complete genome sequence of the mosquitocidal bacterium Bacillus sphaericus C3-41 and comparison with those of closely related Bacillus species. J. Bacteriol. 190:2892–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huson D. H., Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 8. Jagtap S. C., Jagtap C. B., Kumar P., Srivastava R. B. 2009. Detection of Bacillus sphaericus mosquitocidal toxin genes by multiplex colony PCR. Can. J. Microbiol. 55:207–209 [DOI] [PubMed] [Google Scholar]
- 9. Jolley K. A., Feil E. J., Chan M. S., Maiden M. C. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230–1231 [DOI] [PubMed] [Google Scholar]
- 10. Jones G. W., et al. 2007. A new Cry toxin with a unique two component dependency from Bacillus sphaericus. FASEB J. 21:4112–4120 [DOI] [PubMed] [Google Scholar]
- 11. Jones G. W., Wirth M. C., Monnerat R. G., Berry C. 2008. The Cry48Aa-Cry49Aa binary toxin from Bacillus sphaericus exhibits highly restricted target specificity. Environ. Microbiol. 10:2418–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kolsto A. B., Tourasse N. J., Okstad O. A. 2009. What sets Bacillus anthracis apart from other Bacillus species? Annu. Rev. Microbiol. 63:451–476 [DOI] [PubMed] [Google Scholar]
- 13. Krych V. K., Johnson J. L., Yousten A. A. 1980. DNA homologies among strains of Bacillus sphaericus. Int. J. Syst. Bacteriol. 30:476–484 [Google Scholar]
- 14. Lang P., et al. 2010. Expanded multilocus sequence typing and comparative genomic hybridization of Campylobacter coli isolates from multiple hosts. Appl. Environ. Microbiol. 76:1913–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin D., Rybicki E. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563 [DOI] [PubMed] [Google Scholar]
- 16. Nakamura L. K. 2000. Phylogeny of Bacillus sphaericus-like organisms. Int. J. Syst. Evol. Microbiol. 50:1715–1722 [DOI] [PubMed] [Google Scholar]
- 17. Porwal S., Lal S., Cheema S., Kalia V. C. 2009. Phylogeny in aid of the present and novel microbial lineages: diversity in Bacillus. PLoS One 4:e4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Priest F. G. 1992. Biological control of mosquitoes and other biting flies by Bacillus sphaericus and Bacillus thuringiensis. J. Appl. Bacteriol. 72:357–369 [DOI] [PubMed] [Google Scholar]
- 19. Rozas J., Sanchez-DelBarrio J. C., Messeguer X., Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497 [DOI] [PubMed] [Google Scholar]
- 20. Siefert J. L., et al. 2000. Phylogeny of marine Bacillus isolates from the Gulf of Mexico. Curr. Microbiol. 41:84–88 [DOI] [PubMed] [Google Scholar]
- 21. Sorokin A., et al. 2006. Multiple-locus sequence typing analysis of Bacillus cereus and Bacillus thuringiensis reveals separate clustering and a distinct population structure of psychrotrophic strains. Appl. Environ. Microbiol. 72:1569–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 23. Wirth M. C., Yang Y., Walton W. E., Federici B. A., Berry C. 2007. Mtx toxins synergize Bacillus sphaericus and Cry11Aa against susceptible and insecticide-resistant Culex quinquefasciatus larvae. Appl. Environ. Microbiol. 73:6066–6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. World Health Organization, 1985. Informal consultation on the development of Bacillus sphaericus as microbial larvicide. TDR/BCV/SPHAERICUS/85.3.1-24 Word Health Organization, Geneva, Switzerland [Google Scholar]
- 25. Zahner V., Rabinovitch L., Cavados C. F. G., Momen H. 1994. Multilocus enzyme electrophoresis on agarose gel as an aid to the identification of entomopathogenic Bacillus sphaericus strains. J. Appl. Bacteriol. 76:327–335 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.