Abstract
Noroviruses (NoV) in 78 wastewater samples from Luxembourg were quantified, cloned, and sequenced in 2008-2009. The concentrations of NoV genogroup II and the relative occurrences of certain genotypes changed significantly during the winter season. NoV genogroup I was frequently detected by real-time reverse transcription-PCR (RT-PCR), albeit at 30-fold lower concentrations than for genogroup II, hampering attempts to assess overall genetic diversity by the cloning/sequencing approach.
TEXT
Noroviruses (NoV) have been identified as a major etiologic agent of acute gastroenteritis. Wastewater is considered the primary source of environmental contamination, and the NoV genome can be detected in surface waters considerable distances downstream from wastewater discharge (21, 22) and in groundwaters (6). NoV outbreaks have been associated with the consumption of fecally contaminated recreational waters (8) and drinking waters (14, 15, 18). Since human NoVs cannot be cultured (5), detection relies mainly on molecular techniques. Based on the phylogenetic analysis of viral RNA-dependent RNA polymerase (RdRp) and the capsid protein gene sequences, five genogroups (GI to GV) are currently being recognized (26), while a sixth genogroup has been suggested (16). In humans, GI and GII are the most frequently detected genotypes. Our study compares two approaches to assess the presence of NoV genogroups in wastewaters, one using highly specific primers in real-time reverse transcription-PCR (RT-PCR) targeting the highly conserved ORF1-ORF2 junction and one using generic primers in a cloning/sequencing protocol targeting part of the RdRp gene located in ORF1.
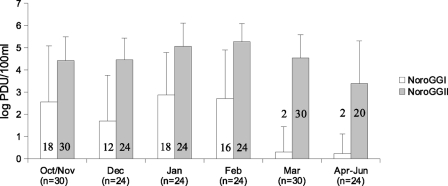
During the winter season of 2008-2009, a total of 78 samples were collected at the inlet of three main wastewater treatment plants of Luxembourg (from Schifflange, 90,000 equivalent inhabitants [EI]; from Pétange, 50,000 EI; and from Beggen, 300,000 EI). One hundred milliliters of wastewater was clarified by slow centrifugation (3,000 × g; 15 min). The supernatant was ultracentrifuged at 100,000 × g for 1 h 30, and the pellet was resuspended in 2 ml of Milli-Q water containing 0.01% of Tween 80. Viral RNA was extracted from 140 μl of concentrate using a QIAamp viral RNA minikit (Qiagen, Germany). A competitive internal RNA control (IC) was added at the end of the lysis step to assess the efficiency of both the extraction (excluding the lysis step) and the detection procedures, including the presence of real-time RT-PCR inhibitors (21). The IC was reverse transcribed and amplified using the same primers as the NoV GII assay. During amplification, the internal control was distinguished from NoV GII genomes by using a specific TaqMan probe (21). Five microliters of undiluted and 10×-diluted extracted RNA was analyzed for NoV GI and GII by real-time TaqMan one-step RT-PCR, targeting the ORF1-ORF2 junction and using the specific primers/probes JJV1NF, JJV1R, JJV1P, and RING-1b for GI and JJV2F, COG2R, and RING2-TP for GII (9, 10, 13, 21). Concentrations of the IC RNAs and NoV were determined independently for each extract, and no correction was applied (21). IC RNAs were detected in 76 (97%) undiluted and in all 78 (100%) diluted samples. According to the criteria of da Silva et al. (4), extraction and real-time RT-PCR efficiencies were considered “good” (IC recovery rate > 10%) in 62% of undiluted concentrates and in 97% of the diluted concentrates, suggesting that residual inhibitors could be partially overcome by dilution. NoV GI and GII were detected in 34 and 76 out of 78 samples, respectively. The overall concentrations differed significantly between undiluted samples positive for GI or GII (3.0 versus 4.5 log PCR detection units per 100 ml [PDU · 100 ml−1]; t test; P < 0.001). There was no statistically significant difference between NoV GI and GII concentrations between treatment plants (one-way analysis of variance [ANOVA]; P = 0.65 for GI and P = 0.28 for GII), but concentrations were found to differ significantly throughout the study period for GII (one-way ANOVA; P < 0.001) and to a lesser extent also for GI (one-way ANOVA; P = 0.017) (Fig. 1). The higher concentration of NoV GII than of GI, particularly during January-February, is most likely due to the synchronous high incidences of documented GII infections/outbreaks in the catchment population of the treatment plants (12) and/or higher viral loads of NoV GII in stool samples (3, 19).
Fig. 1.
Average concentration of norovirus GI and GII in raw wastewater samples. Values for negative samples were set to 0 for the averaging. Error bars indicate standard deviations. The numbers of samples are indicated in brackets for each month, while the numbers of positive samples are detailed in the graphic for each genogroup and each date.
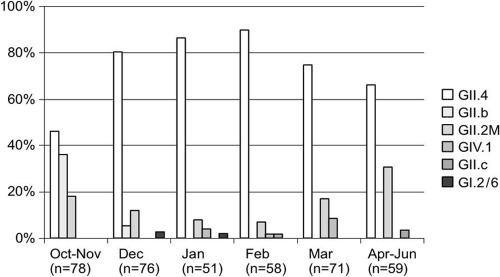
Five microliters of extracted RNA was amplified using the generic primers JV12y-JV13i targeting the RNA polymerase gene (25). RT-PCR products of appropriate size (327 bp) were purified (QIAquick PCR purification kit; Qiagen, Germany) and cloned into a plasmid vector using a Qiagen PCR cloning kit (Qiagen, Germany) according to the manufacturer's instructions. For each sample, between 5 and 10 colonies were selected for PCR, cloning, and sequencing. Isolated white colonies were picked up into 45 μl of DNase/RNase-free water and heated to 95°C for 5 min under agitation (400 rpm). After 1 min of centrifugation at 20,000 × g, the supernatant was tested by PCR using specific plasmid M13 forward and reverse primers. Amplified DNA was sequenced using a BigDye Terminator cycle sequencing ready reaction Kit (PE Applied Biosystems, Foster City, CA) and an ABI Prism 3130 genetic analyzer. A total of 393 clones were successfully sequenced, revealing 171 NoV variants. A neighbor-joining tree constructed with MEGA4 software from an alignment of 285 nucleotides of the partial RdRp gene indicated 8 distinct clusters supported by bootstrap values of >80% (Fig. 2). In each cluster, NoV genotypes were identified by comparison to the QuickTyping database. NoV GII.4 and GII.2 M represented 71.2% and 15.5% of all clones, respectively. Only 3 of 393 clones (0.8%), corresponding to 2 samples, were identified as GI. NoV diversities were similar in Pétange and Beggen, while neither GI nor GII.c could be detected in Schifflange (Table 1). The percentage of NoV GII.4 increased from October to February and decreased from February to June, while NoV GII.2 M showed the opposite pattern (Fig. 3). NoV GIV was detected from January to March, confirming its circulation in the water environment (11). Distinct viral sequences were recovered from the same sample, indicating the cocirculation of multiple NoV strains in the population. Similar observations have been reported before (1, 24), confirming that direct sequencing is likely to underestimate the diversity of NoV contained in wastewaters. GII.4 was identified in 71 samples out of 78 (91%) and represents the predominant lineage, which is consistent with previous studies (1, 7).
Fig. 2.
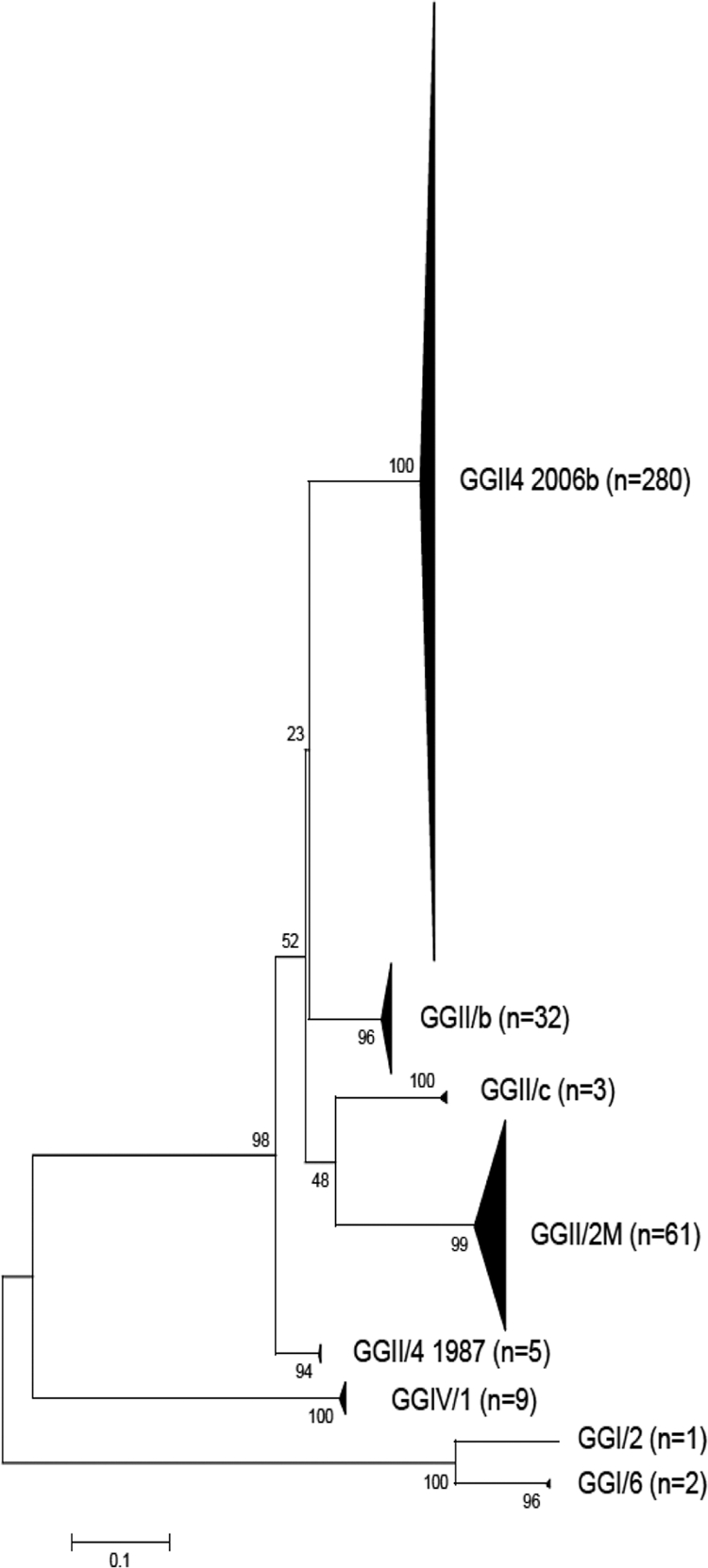
Neighbor-joining tree constructed from an alignment of the 393 partial RdRp sequences obtained in this study. NoV genotypes were identified by comparison to those in the QuickTyping database (http://www.noronet.nl/databases/).
Table 1.
Numbers and percentages of NoV genotypes by sampling site
| Plant | No. (%) of NoV genotypes at indicated site |
|||||
|---|---|---|---|---|---|---|
| GII.4 | GII.b | GII.2 M | GIV.1 | GII.c | GI.2/6 | |
| Pétange (n = 121) | 91 (75) | 11 (9.1) | 10 (8.3) | 7 (5.8) | 1 (0.8) | 1 (0.8) |
| Schifflange (n = 135) | 108 (80) | 16 (12) | 10 (7.4) | 1 (0.7) | 0 (0) | 0 (0) |
| Beggen (n = 137) | 86 (63) | 5 (3.6) | 41 (30) | 1 (0.7) | 2 (1.5) | 2 (1.5) |
Fig. 3.
Percentage of NoV genotypes by sampling date (for each month, genotypes are presented in the same order [GII.4, GII.b, GII.2 M, GIV.1, GII.c, and GI.2/6]; thus, the absence of a bar means that the corresponding genotype was not detected).
The recovery of a low number of GI clones compared to real-time RT-PCR results (2 versus 34 out of 78 samples) could be explained by a higher concentration of NoV GII than of GI (average of 1.5 log PDU · 100 ml−1, equivalent to a 30-fold difference). Specific primers targeting the ORF1-ORF2 junction were used for real-time RT-PCR while generic primers (JV12y-JV13i) targeting the RdRp gene in ORF1 (region A) were used for sequencing, as recommended for improved detection of NoV GI (20, 24, 25). Initial differences in concentrations combined with variability of primer affinity could lead to an underestimation of NoV GI in wastewaters. Also, it cannot be excluded that part of the NoV identified as GI when targeting the ORF1-ORF2 junction clusters with GII when the RdRp gene in ORF1 is considered, since recombination of NoVs is common (2). Despite saving material and time, our work clearly shows the limitations of using generic primers to determine NoV strain diversity. This limitation may be overcome by using more specific sets of primers, by increasing the number of clones, or by using “next-generation“ sequencing techniques (17). Samples of particular interest can be stored at −80°C and tested retrospectively when updated protocols become available (23).
Nucleotide sequence accession numbers.
The NoV sequence data can be found in GenBank under accession numbers FR668938 to FR669108.
Acknowledgments
This work was funded by the National Research Fund (FNR) from Luxembourg (SECAL Program, SENSORLUX project, FNR/SECAL/07/04).
We thank Laurent Solinhac (CRP-GL, Luxembourg) for his assistance in sequencing analysis, Nicolas Bonjean and Delphine Collard (CRP-GL, Luxembourg) for their technical assistance, and Romain Muno, Gianni Dipentima, and Guy Felten for their kind collaboration at the wastewater treatment plants of Pétange, Schifflange, and Beggen, respectively.
Footnotes
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Aw T. G., Gin K. Y., Ean Oon L. L., Chen E. X., Woo C. H. 2009. Prevalence and genotypes of human noroviruses in tropical urban surface waters and clinical samples in Singapore. Appl. Environ. Microbiol. 75:4984–4992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bull R. A., Tanaka M. M., White P. A. 2007. Norovirus recombination. J. Gen. Virol. 88:3347–3359 [DOI] [PubMed] [Google Scholar]
- 3. Chan M. C., et al. 2006. Fecal viral load and norovirus-associated gastroenteritis. Emerg. Infect. Dis. 12:1278–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. da Silva A. K., et al. 2007. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl. Environ. Microbiol. 73:7891–7897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duizer E., et al. 2004. Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 85:79–87 [DOI] [PubMed] [Google Scholar]
- 6. Gabrieli R., Maccari F., Ruta A., Pana A., Divizia M. 2009. Norovirus detection in groundwater. Food Environ. Virol. 1:92–96 [Google Scholar]
- 7. Gallimore C. I., Iturriza-Gomara M., Xerry J., Adigwe J., Gray J. J. 2007. Inter-seasonal diversity of norovirus genotypes: emergence and selection of virus variants. Arch. Virol. 152:1295–1303 [DOI] [PubMed] [Google Scholar]
- 8. Hoebe C. J., Vennema H., Roda Husman A. M., van Duynhoven Y. T. 2004. Norovirus outbreak among primary schoolchildren who had played in a recreational water fountain. J. Infect. Dis. 189:699–705 [DOI] [PubMed] [Google Scholar]
- 9. Jothikumar N., et al. 2005. Rapid and sensitive detection of noroviruses by using TaqMan-based one-step reverse transcription-PCR assays and application to naturally contaminated shellfish samples. Appl. Environ. Microbiol. 71:1870–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kageyama T., et al. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kitajima M., Haramoto E., Phanuwan C., Katayama H., Ohgaki S. 2009. Detection of genogroup IV norovirus in wastewater and river water in Japan. Lett. Appl. Microbiol. 49:655–658 [DOI] [PubMed] [Google Scholar]
- 12. Kremer J., et al. 2010. Genetic diversity of noroviruses from outbreaks, sporadic cases and wastewater in Luxembourg 2008-2009. Clin. Microbiol. Infect. [Epub ahead of print.] doi:10.1111/j.1469-0691.2010.03407.x [DOI] [PubMed] [Google Scholar]
- 13. Lyman W. H., et al. 2009. Prospective study of etiologic agents of acute gastroenteritis outbreaks in child care centers. J. Pediatr. 154:253–257 [DOI] [PubMed] [Google Scholar]
- 14. Maunula L. 2005. Norovirus outbreaks from drinking water. Emerg. Infect. Dis. 11:1716–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maunula L., et al. 2009. Enteric viruses in a large waterborne outbreak of acute gastroenteritis in Finland. Food Environ. Virol. 1:31–36 [Google Scholar]
- 16. Mesquita J. R., Barclay L., Nascimento M. S., Vinjé J. 2010. Novel norovirus in dogs with diarrhea. Emerg. Infect. Dis. 16:980–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakamura S., et al. 2009. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One 4:e4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nygard K., et al. 2003. Emerging genotype (GGIIb) of norovirus in drinking water, Sweden. Emerg. Infect. Dis. 9:1548–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ozawa K., Oka T., Takeda N., Hansman G. S. 2007. Norovirus infections in symptomatic and asymptomatic food handlers in Japan. J. Clin. Microbiol. 45:3996–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rutjes S. A., van den Berg H. H., Lodder W. J., Roda Husman A. M. 2006. Real-time detection of noroviruses in surface water by use of a broadly reactive nucleic acid sequence-based amplification assay. Appl. Environ. Microbiol. 72:5349–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Skraber S., Gantzer C., Helmi K., Hoffmann L., Cauchie H. M. 2009. Simultaneous concentration of enteric viruses and protozoan parasites: a protocol based on tangential flow filtration and adapted to large volumes of surface and drinking waters. Food Environ. Virol. 1:66–76 [Google Scholar]
- 22. Skraber S., Gassilloud B., Gantzer C. 2004. Comparison of coliforms and coliphages as tools for assessment of viral contamination in river water. Appl. Environ. Microbiol. 70:3644–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Skraber S., Italiaander R., Lodder W., Roda Husman A. M. 2005. Noroviruses in archival samples. Emerg. Infect. Dis. 11:489–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van den Berg H., Lodder W., van der Poel P. W., Vennema H., Roda Husman A. M. 2005. Genetic diversity of noroviruses in raw and treated sewage water. Res. Microbiol. 156:532–540 [DOI] [PubMed] [Google Scholar]
- 25. Vennema H., de Bruin E., Koopmans M. 2002. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase PCR. J. Clin. Virol. 25:233–235 [DOI] [PubMed] [Google Scholar]
- 26. Zheng D. P., et al. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312–323 [DOI] [PubMed] [Google Scholar]