Abstract
Rhizobium sp. strain NGR234 is a unique alphaproteobacterium (order Rhizobiales) that forms nitrogen-fixing nodules with more legumes than any other microsymbiont. Since we have previously described the complete genome sequence of NGR234, we now report on a genome-wide functional analysis of the genes and enzymes involved in autoinducer I hydrolysis in this microbe. Altogether we identified five cosmid clones that repeatedly gave a positive result in our function-based approach for the detection of autoinducer I hydrolase genes. Of these five cosmid clones, two were located on pNGR234b and three were on cNGR234. Subcloning and in vitro mutagenesis in combination with BLAST analyses identified the corresponding open reading frames (ORFs) of all cosmid clones: dlhR, qsdR1, qsdR2, aldR, and hitR-hydR. Analyses of recombinant DlhR and QsdR1 proteins by using high-performance liquid chromatography-mass spectrometry (HPLC-MS) demonstrate that these enzymes function as acyl homoserine lactone (AHL) lactonases. Furthermore, we showed that these enzymes inhibited biofilm formation and other quorum-sensing-dependent processes in Pseudomonas aeruginosa, Chromobacterium violaceum, and Agrobacterium tumefaciens. Finally, our experimental data suggest that competitive colonization of roots in the rhizospheres of cowpea plants is affected by DlhR and QsdR1.
INTRODUCTION
Quorum sensing (QS) is a cell density-dependent system of gene regulation in prokaryotes. Through the accumulation of bacterially produced signaling molecules (autoinducers), the bacterial population is able to sense increases in cell density and alter gene expression accordingly (55). This allows coordinated expression of genes at the population level which are most effective at higher cell densities, such as those for pathogenicity, biofilm formation, production of extracellular proteins, and others. For an excellent review on interspecies signaling, see reference 41. Many quorum-sensing mechanisms involve N-acyl homoserine lactones (N-AHLs) in Gram-negative bacteria. While the general mechanisms of the synthesis of N-AHLs are well understood, it is quite intriguing that only a limited number of proteins that interfere with these bacterial quorum-sensing molecules via enzymatic hydrolysis are known. Three main types of enzymes act on autoinducer I (AI-I) molecules. (i) Lactonases, which hydrolyze the lactone ring moiety in a reversible way, form the major class of enzymes identified to date (11, 34, 35). More recently several novel lactonases have been reported: AidH (28), AiiM (54), and BpiB05 (3). (ii) Acylases are known to interfere with the autoinducer I-like molecules by cleaving the acyl side chain off the homoserine lactone moiety. Acylases have been identified in a variety of microorganisms, such as Comamonas spp. (50), Pseudomonas aeruginosa (20), Pseudomonas syringae (42), Ralstonia spp. (24), Rhodococcus erythropolis (49), Shewanella spp. (29), and Streptomyces spp. (31). (iii) Oxidoreductases are enzymes that target the acyl side chain by oxidative or reducing activities and thus catalyze the chemical modification of N-AHLs and not their degradation. To date only two such enzymes with oxidoreductase activity have been discovered. A P-450/NADPH-P450 reductase has been isolated from Bacillus megaterium and characterized in detail (6, 7). The respective AHL-oxidizing enzyme was designated CYP102A1, and it was able to hydrolyze both AHLs and fatty acids with various chain lengths. In addition, Uroz and colleagues reported the presence of another enzyme in crude cell extracts of R. erythropolis W2 (49). They demonstrated that the 3-oxo substituent of 3-oxo-C14-homoserine lactone (3-oxo-C14-HSL) was reduced to yield the corresponding derivative, 3-hydroxy-C14-HSL, and this was also observed for 3-oxo-C8-HSL, 3-oxo-C10-HSL, and 3-oxo-C12-HSL. The autoinducer was thereby inactivated.
Rhizobium sp. strain NGR234 (referred to here as NGR234) is able to nodulate more than 120 genera of legumes (33). The complete genome analysis identified two loci linked to the synthesis of autoinducer I molecules. TraI synthesizes an acyl-HSL that is probably N-3-oxooctanoyl-l-homoserine lactone (17), and ngrI probably encodes a coumestrol derivative of the autoinducer I type molecules (40).
In this study, we demonstrate that NGR234 carries a surprisingly large number of functional genes involved in the degradation of N-AHLs. Using a recently published function-based screening protocol (39), we identified at least five loci (three on the chromosome and two on pNGR234b) that are actively involved in N-AHL degradation or modification. Overexpression, purification, and biochemical characterization were carried out for two of the five proteins and showed that one was similar to a metal-dependent β-lactamase while the other one resembled a bacterial dienelactone hydrolase. The remaining three loci, identified to be responsible for quorum-quenching (QQ) or AI-I-modifying activity, encode a metal-dependent β-lactamase, an acetaldehyde dehydrogenase, and a putative histidine triad protein linked with a predicted NUDIX hydrolase. Furthermore, we were able to show that extra copies of the dlhR and qsdR1 genes strongly affect plant root colonization.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Microbiological materials used in the present work are listed in Table 1. Escherichia coli strains and Pseudomonas aeruginosa PAO1 were grown at 37°C in Luria-Bertani medium (37) supplemented with appropriate antibiotics. NGR234 was cultivated in yeast extract-mannitol medium (YEMA) (52) or TY medium (37) at 30°C. Agrobacterium tumefaciens NTL4 (14), carrying a traI::lacZ promoter fusion, was grown in LB or AT medium (46) containing 0.5% glucose per liter at 28°C. Chromobacterium violaceum CV026 was cultivated in LB or TY medium at 28°C.
Table 1.
Bacterial strains, plasmids, and constructs used in this study
| Strain or construct | Description | Reference(s) or source |
|---|---|---|
| EPI100-T1 phage T1-resistant E. coli | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74recA1 endA1 araD139 Δ(ara leu)7697galU galK λ−rpsL nupG | Epicentre Technologies, Madison, WI |
| E. coli XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1lac [F′ proABlacIqZΔM15 Tn10 (Tetr)] | Stratagene, La Jolla, CA |
| E. coli BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm(DE3) | Novagen, Darmstadt, Germany |
| Rhizobium sp. strain NGR234 | Wild-type New Guinea isolate; Rifr | 47 |
| Pseudomonas aeruginosa PAO1 | Wild-type strain of P. aeruginosa; Ampr | 19 |
| Agrobacterium tumefaciens NTL4(pCF218)(pCF372) | Reporter strain for AHL detection; traI::lacZ Tetr Spr | 14, 15, 25 |
| Chromobacterium violaceum CV026 | Reporter strain for autoinducer I; mini-Tn5 in cviI | 27 |
| pWEB-TNC | Cosmid cloning vector derived from pWE15; linearized with SmaI; ColE1; cos site; T7 promoter; Ampr Cmr | Epicentre Technologies, Madison, WI |
| pWEB-TNC-A5 | pWEB-TNC with a 40.4-kb insert from NGR234 | This study |
| pWEB-TNC-B2 | pWEB-TNC with a 34.0-kb insert from NGR234 | This study |
| pWEB-TNC-B9 | pWEB-TNC with a 42.0-kb insert from NGR234 | This study |
| pWEB-TNC-C6 | pWEB-TNC with a 37.7-kb insert from NGR234 | This study |
| pWEB-TNC-G2 | pWEB-TNC with a 33.3-kb insert from NGR234 | This study |
| pET21a | E. coli His6-tagged expression vector; Ampr | Novagen, Darmstadt, Germany |
| pET21a::dlhR | pET21a containing dlhR gene cloned into NdeI and HindIII restriction sites | This study |
| pET21a::qsdR1 | pET21a containing qsdR1 gene cloned into NdeI and XhoI restriction sites | This study |
| pET21a::gtfU | pET21a containing gtfU gene cloned into NdeI and HindIII restriction sites; served as control | This study |
| pBBR1MCS | Broad-host-range vector; low copy; Cmr | 22 |
| pBBR1MCS-5 | Broad-host-range vector; low copy; Gmr | 22 |
| pBBR1MCS::Ptaq::His-dlhR | pBBR1MCS containing the His6 tag-dlhR fragment amplified by T7 promoter/T7 terminator primer from pET21a::dlhR | This study |
| pBBR1MCS::Ptaq::His-qsdR1 | pBBR1MCS containing the His6 tag-qsdR1 fragment amplified by T7 promoter/T7 terminator primer from pET21a::qsdR1 | This study |
Unless otherwise specified, media were supplemented with antibiotics, as required, at the following final concentrations: for E. coli containing pET21a and pWEB-TNC cosmid clones and P. aeruginosa PAO1 cultures, ampicillin at 100 μg/ml; for A. tumefaciens NTL4 cultures, spectinomycin at 50 μg/ml and tetracycline at 4.5 μg/ml; for E. coli and NGR234 containing pBBR1MCS and related constructs, chloramphenicol at 50 μg/ml; for E. coli and NGR234 containing pBBR1MCS-5, gentamicin at 10 μg/ml; and for NGR234 cultures, rifampin at 25 μg/ml.
Transformation and electroporation procedures.
Plasmid or cosmid transformation in E. coli XL1-Blue and E. coli BL21(DE3) was done following standard heat shock and electroporation protocols (37). NGR234 was transformed by electroporation using a protocol for Rhizobium leguminosarum (16) with minor modifications.
NGR234 cosmid library construction.
NGR234 was grown overnight in 30 ml of TY medium with rifampin. The genomic DNA of NGR234 was isolated with the AquaPure kit (Bio-Rad Laboratories, Hercules, CA). For the construction of the NGR234 genomic cosmid library, the pWEB-TNC cosmid cloning kit (Epicentre Biotechnologies, Madison, WI) was used, with the provided protocol modified as follows. Shearing of genomic DNA was accomplished by partial digestion with Bsp143I (Sau3AI), and the end repair reaction mixture was dialyzed against water for 2 h. The ligation products were packaged using Gigapack III Gold packaging extract (Stratagene, La Jolla, CA) as recommended by the manufacturer and recovered by transfection into EPI100-T1 phage-resistant E. coli cells (referred to here as EPI100). The cells were spread on LB agar medium with ampicillin and incubated overnight, and colonies were transferred into 96-well microtiter plates containing 150 μl liquid LB medium with ampicillin and allowed to grow overnight. Microtiter plates were stored at −70°C after addition of 50 μl of 86% glycerol. A total of 603 cosmid clones were generated.
Screening of the NGR234 genomic library for N-AHL-degrading clones and their genetic analysis.
For the identification of cosmid clones capable of inactivating AHLs or of blocking AHL receptors/promoters, the biosensor strain A. tumefaciens NTL4 was used as previously described (3, 39, 56). This strain carries a traI::lacZ reporter and does not synthesize autoinducer, and it therefore is capable of reporting the presence of traI-inducing metabolites. Monitoring of a possible altered P. aeruginosa PAO1 motility phenotype triggered by positive tested cosmid clones and recombinant proteins was accomplished by motility assays as previously described (3, 39). The 33- to 42-kb large inserts of positive tested cosmid clones were end sequenced using M13_for and T7 promoter primers (see Table S1 in the supplemental material). For the detection of open reading frames (ORFs) involved in quorum-sensing inhibition, either subcloning, in vitro transposon mutagenesis, or direct cloning was employed. For subcloning, either EcoRI (Fermentas, St-Leon-Rot, Germany) fragments of cosmid clones were ligated with T4 DNA ligase (Promega, Mannheim, Germany) into EcoRI-linearized pTZ19R vector or Taq polymerase-amplified genes of interest were ligated into pDrive cloning vector (Qiagen [Hilden, Germany] PCR cloning kit) using the primer pairs indicated in Table S1 in the supplemental material and subjected further analyses. For in vitro transposon mutagenesis, the EZ-Tn5 <KAN-2> insertion kit (Epicentre Biotechnologies, Madison, WI) was used according to the manufacturer's protocol. ORFs were amplified using the primer pairs indicated in Table S1 in the supplemental material and directly cloned into pET21a. The amplicon and the vector were digested with NdeI and HindIII for dlhR or with XhoI for qsdR1 and ligated directionally, yielding pET21a::dlhR and pET21a::qsdR1, respectively. All clones obtained by subcloning, transposon mutagenesis, or direct cloning were assayed by A. tumefaciens NTL4 screening and motility assays with P. aeruginosa PAO1 as described above. For sequencing of plasmids and constructs, the T7 promoter/T3 promoter/T7 terminator/M13_20/M13_rev primers were used. Automated sequencing was performed with an ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA), based on the Sanger technique. Nucleotide and amino acid sequence comparisons were accomplished using the BLAST program (1) and the publically available databases SwissProt, GenBank, KEGG, and ProSite. Multiple-sequence alignments were performed using BioEdit.
Purification of His-tagged proteins.
For preparation of purified His6-tagged proteins, constructs pET21a::dlhR and pET21a::qsdR1 were transformed into E. coli BL21(DE3) cells. Precultures (50 ml) were cultivated in LB medium with ampicillin overnight at 37°C. The precultures were used to inoculate 1-liter LB main cultures, where the optical density at 600 nm (OD600) was adjusted to 0.1. The expression cultures were grown at 37°C to an OD600 of 0.6 to 0.8, expression was induced by addition of 100 μM IPTG (isopropyl-β-d-1-thiogalactopyranoside), and the main cultures were incubated overnight at 17°C with shaking at 140 rpm. Cells were harvested by centrifugation at 10,000 rpm and 4°C for 20 min and resuspended in 1× LEW buffer (Macherey-Nagel, Dueren, Germany) prior to disruption through ultrasonication (3 times for 10 min with 2-min cooling breaks). The lysate was centrifuged at 13,000 rpm and 4°C for 30 min, and the obtained supernatant was then purified using Protino Ni-TED 2000 packed columns (Macherey-Nagel, Dueren, Germany) following the manufacturer's protocol. To remove imidazole from eluted proteins, the eluted proteins were dialyzed against a 200-fold volume of 100 mM potassium phosphate buffer with pH values varied according to the protein being purified (for dlhR, pH 7.5, and for qsdR1, pH 7.0). The protein purities and molecular masses were determined by SDS-gel electrophoresis.
β-Galactosidase activity assay using reporter strain A. tumefaciens NTL4.
o-Nitrophenyl-β-d-galactopyranoside (ONPG) tests were performed as previously described (39) with minor modifications; 5 μl of a 10−8 M solution of 3-oxo-C8-HSL was added to 1 to 100 μl of purified protein extracts (400 μg/ml) and incubated at 30°C in 100 mM potassium phosphate buffer at pH 7.0.
HPLC-MS analysis.
High-pressure liquid chromatography-mass spectrometry (HPLC-MS) tests were done as previously published (3, 39) with minor modifications as follows. For chemical analysis, a 3-oxo-C8-HSL stock solution was prepared in dimethyl sulfoxide (DMSO) (0.2 mg 3-oxo-C8-HSL/μl DMSO). Protein extracts were purified from E. coli BL21(DE3) expressing dlhR and qsdR1 genes as described above. Protein amounts of 0.005 to 0.3 mg/ml were mixed with 10 μl of the 3-oxo-C8-HSL stock solution in 100 mM potassium phosphate buffer (for dlhR, pH 7.5, and for qsdR1, pH 7.0). After incubation for 20 h at 30°C, the mixtures were extracted twice with 1 volume ethyl acetate, and the combined organic layers were concentrated in vacuo. For a detailed description of the subsequent HPLC-MS analysis, see references 10 and 39.
Rhizosphere colonization experiments.
Rhizosphere colonization tests were done as previously described (45). Cowpea seeds (Vigna unguiculata subsp. unguiculata) were treated for 15 min in 70% ethanol and rinsed three times with sterile double-deionized water. These sterilized seeds were placed on 0.5× TY agar (lacking CaCl2) and germinated at 30°C. After 24 h, germinating seeds with no visible contamination were transferred into sterile 50-ml Falcon tubes (Fisher Scientific GmbH, Schwerte, Germany) filled with ∼10 g autoclaved vermiculite and 5 ml 0.25× Hoagland solution (18). NGR234 inocula consisted of NGR234 cells harboring either pBBR1MCS-5 (referred to here as pBBR-5) as a control or constructs pBBR1MCS::Ptaq::His-dlhR and pBBR1MCS::Ptaq::His-qsdR1 (referred to here as pBBR::dlhR/pBBR::qsdR1). NGR234 inocula were grown in 250 ml YEMA medium with appropriate antibiotics, and cell densities were determined by OD600 and adjusted in fresh medium to either 103 cells/ml or 105 cells/ml. One hundred microliters of these bacterial suspensions was used to inoculate the 24-h-old seedlings. Bacterial solutions of the constructs and the control strain were used in combination (1:1) or as single-strain inocula. The germinated and inoculated seedlings were incubated in a light- and humidity-controlled incubator (Rumed Rubarth Apparate GmbH, Laatzen, Germany) under controlled conditions (day/night: 24/19°C, 16/8 h, and 50% relative humidity) for 4 days. Roots were harvested after this time period, shaken to remove vermiculite, cut into segments, and placed in 1.5-ml Eppendorf tubes (Eppendorf AG, Hamburg, Germany) containing 1 ml of distilled water with 0.01% Tween 20. Appropriate dilutions were plated on YEMA agar containing Congo red and the required antibiotics.
RESULTS
Sequence-based identification of QQ genes in NGR234 that are involved in AHL degradation.
In order to identify possible genes and proteins interfering with autoinducer I (AI-I) molecules, we performed a detailed analysis of the NGR234 genome data (40). The results of these searches are summarized in Table 2. A total of 18 possible QQ genes were identified by careful BLAST analyses and searches for conserved motifs. Only five of these 18 ORFs were similar to previously identified autoinducer hydrolase genes. The observed identities for the known QQ genes ranged from 24% to 34% (Table 2). Eleven of these genes were grouped as metal-dependent hydrolases, including seven proteins belonging to the metallo-β-lactamase family. The products of four ORFs were annotated as putative β-lactamase family proteins and one as a metal-dependent phosphohydrolase. Altogether, 13 of these proteins shared a conserved zinc-binding motif of previously published AHLases. This “HxHx∼DH” pattern is found in several groups of metallohydrolases, including metallo-β-lactamases, glyoxalases II, and arylsulfatases, and is probably essential for AHL-degrading activity (12). Furthermore, two ORFs were similar to those for dienelactone hydrolases. In addition to the ORFs given in Table 2, we identified five other proteins encoded within the NGR234 genome sequence which may be involved in QQ processes. These proteins were annotated as putative amidohydrolases and are designated NGR_c01920, NGR_c18520, NGR_c27330, NGR_c28580, and NGR_c33720 (data not shown). Additionally, we identified 14 genes and ORFs (see Table S2 in the supplemental material) that encoded possible hydrolases with no clear function assigned.
Table 2.
Possible and verified QQ loci identified in the NGR234 genome
| ORFa | Possible and/or annotated function | Conserved HxHx∼DH motif | Size of protein (no. of amino acids) | Identity/similarity (%) to published QQ proteinb |
|---|---|---|---|---|
| NGR_c10650 | Metallo-β-lactamase family protein | + | 556 | |
| NGR_b16870 | Putative metallo-β-lactamase family protein QsdR1 | + | 321 | Aii2 (24/39) |
| NGR_b15850 | Metallo-β-lactamase family protein | + | 312 | AhlK (34/44) |
| NGR_b12260 | Metallo-β-lactamase family protein | + | 277 | |
| NGR_c22830 | Putative β-lactamase family protein | + | 214 | |
| NGR_c27960 | Metal-dependent hydrolase | − | 280 | |
| NGR_c05660 | Metallo-β-lactamase family protein | + | 304 | |
| NGR_c03760 | Metallo-β-lactamase family protein | + | 383 | Aii2 (27/44) |
| NGR_c05950 | Putative β-lactamase family protein | + | 254 | AhlD (30/47) |
| NGR_c16020 | Putative β-lactamase family protein QsdR2 | + | 279 | |
| NGR_c00430 | Putative metal-dependent hydrolase | − | 174 | |
| NGR_c08460 | Metal-dependent phosphohydrolase | − | 219 | |
| NGR_c06480 | Metal-dependent hydrolase | + | 289 | QlcA (27/49) |
| NGR_c32270 | Putative β-lactamase family protein | + | 336 | |
| NGR_c17430 | Metallo-β-lactamase family protein | + | 306 | |
| NGR_c10940 | Metal-dependent hydrolase | + | 235 | |
| NGR_b22150 | Dienelactone hydrolase-like protein DlhR | − | 358 | |
| NGR_c03800 | Dienelactone hydrolase-like protein | − | 292 | |
| NGR_c23150c | Acetaldehyde dehydrogenase AldR | − | 503 | |
| NGR_c35560 | Predicted NUDIX hydrolase (putative) | − | 346 | |
| NGR_c35570 | Histidine triad protein, hitR-hydR | − | 202 |
ORF identification numbers for NGR234 were derived from the GenBank entries U00090, CP000874, and CP001389. Genes/ORFs in bold were verified by functional searches in this work.
Identity and similarity values were obtained using the NCBI database. Only identities and similarities to the best hit are given: Aii2 from an uncultured Bacillus sp. (accession no. CAD44268), AhlK from Klebsiella pneumonia (accession no. AAO47340), AhlD from Arthrobacter sp. strain IBN110 (accession no. AAP57766), and QlcA from uncultured Acidobacteria, cosmid p2H8 (accession no. ABV58973).
The gene/ORF could be identified only by functional searches and was not found in the initial sequence-based searches.
Function-based identification of QQ loci in NGR234 involved in AHL degradation.
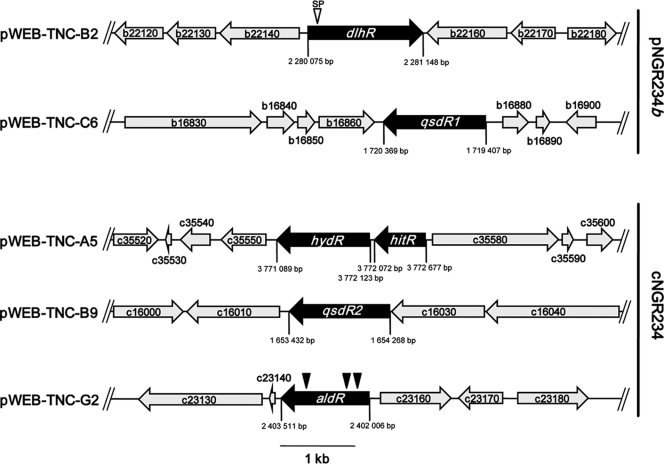
Besides the sequence-based screening, we used a function-based approach for QQ genes to ensure identification of all potential genes involved in quenching of the bacterial autoinducer I signaling molecule. To functionally identify the genes and ORFs that were involved in QQ in NGR234 in vivo, we employed a previously published function-based screening technology (39). For this purpose we constructed an NGR234 cosmid clone library with insert sizes ranging from 25 to 42 kb. In the initial screen, 603 cosmid clones were tested directly in the EPI100 host using the above-mentioned screening protocol, including the A. tumefaciens NTL4 biosensor strain carrying a traI::lacZ reporter gene and ATsoft agar supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and 3-oxo-C8-HSL. This screening was repeated at least three times. Accumulated data from several repetitions of the function-based screening identified five clones that consistently gave a positive result. All five clones reproducibly stayed colorless in the biosensor assay and displayed a deviant phenotype compared to negative controls. At this stage the five cosmid clones were end sequenced and compared with the complete NGR234 genome sequence. All clones carried inserts ranging from 33 to 42 kb; three of the cosmid clones were mapped on cNGR234 (i.e., pWEB-TNC-A5, pWEB-TNC-B9, and pWEB-TNC-G2) and two on pNGR234b (pWEB-TNC-B2 and pWEB-TNC-C6) (Fig. 1). The insert sizes, location on the chromosome or on pNGR234b, and number of ORFs located on these cosmids are given in Table S3 in the supplemental material.
Fig. 1.
Identification of NGR234 AHL-degrading cosmid clones. Partial physical maps of the identified cosmid clones pWEB-TNC-B2, -C6, -A5, -B9, and -G2 carrying the putative genes dlhR, qsdR1, hitR-hydR, qsdR2, and aldR are shown. Cosmid clones pWEB-TNC-B2 and -C6 were mapped on pNGR234b, whereas cosmids pWEB-TNC-A5, -B9, and -G2 were located on the bacterial chromosome. Black arrows indicate the ORFs that were involved in QQ. Gray-shaded arrows indicate flanking ORFs that were not linked to the QQ phenotypes. ORFs are designated using ORF identification numbers from GenBank entries CP000874 and CP001389 (prefix NGR_∼). The scale bar represents 1 kb. A possible secretion signal (SP) in dlhR is indicated by an inverse open triangle. Positions of in vitro transposon insertions in the identified QQ genes are indicated by filled and inverse triangles.
Crude cell extracts of cosmid clones affect motility, biofilm formation, and pyocyanin production in P. aeruginosa PAO1.
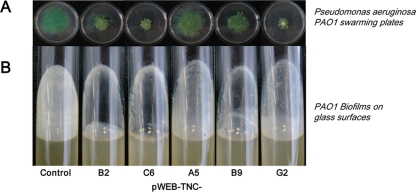
For the further verification of the influence of the isolated cosmids on QS-dependent processes in other Gram-negative bacteria, we used P. aeruginosa PAO1 motility assays, as motility such as swarming/swimming or biofilm formation is QS dependent in PAO1 (21, 43, 53). Crude cell extracts were obtained from E. coli XL1-Blue cells harboring the cosmids pWEB-TNC-A5, -B2, -B9, -C6, and -G2. Crude cell extracts (5 to 100 μg) of cosmids were added to autoclaved and cooled swarming agar and inoculated with P. aeruginosa PAO1. The swarming ability of the PAO1 isolate was strongly altered after incubation for 16 h with crude cell extracts of cosmid clones. All five clones could significantly reduce the swarming motility of PAO1 and additionally alter the phenotypic appearance of colonies on the plates. The control, which was E. coli XL1-Blue carrying a randomly chosen negatively tested cosmid, was treated like the other five cosmids and incubated with PAO1. However, PAO1 was not affected in its swarming capabilities by the control (Fig. 2A). Further tests measuring the ability of PAO1 to form biofilms on glass surfaces suggested that the crude cell extracts of the cosmids pWEB-TNC-A5, -B2, -B9, -C6, and -G2 influenced biofilm formation. For this test, 5 to 100 μg of crude cell extracts of the cosmids and the control was added to exponentially growing cells of PAO1. In all test tubes supplemented with protein extracts from the five cosmid clones, a less developed biofilm formation was visible; no such effect was observed with the controls (Fig. 2B). Together these data further suggested that crude cell extracts of the isolated cosmid clones interfered with bacterial QS activities. Furthermore, crude cell extracts of these cosmid clones significantly reduced pyocyanin production in PAO1 (data not shown). The blue compound pyocyanin produced by P. aeruginosa PAO1 is also regulated by QS-dependent processes (38).
Fig. 2.
P. aeruginosa PAO1 motility and biofilm assays. (A) Altered swarming motility of P. aeruginosa PAO1 on swarming plates supplemented with crude cell extracts of cosmids pWEB-TNC-B2, -C6, -A5, -B9, and -G2. Plates were supplemented with 5 to 100 μg of crude cell extracts of E. coli harboring the QQ cosmids. The plates were inoculated with 1 × 108 cells of P. aeruginosa PAO1 and incubated overnight. (B) Biofilm assays using sterile test tubes filled with LB medium and supplemented with 5 to 100 μg of crude cell extracts of cosmid clones. The tubes were inoculated with 1 × 108 cells of P. aeruginosa PAO1 and incubated at 37°C and 140 rpm for 6 to 8 h. Controls in panels A and B were either E. coli cells or E. coli harboring a negatively tested pWEB-TNC cosmid clone inoculated with PAO1. For overview purposes, only one control is displayed.
Identification of possible QQ ORFs and similarities to known QQ proteins.
For the identification of the genes responsible for the observed phenotypes in the A. tumefaciens NTL4 bioassay, a detailed analysis of the insert sequences of these cosmid clones in combination with in vitro transposon mutagenesis and subcloning was performed. Transposon mutagenesis was accomplished for pWEB-TNC-G2, subcloning of random EcoRI restriction fragments of cosmids into pTZ19R vector was carried out for pWEB-TNC-B2, and specific subclones of PCR-amplified genes of interest were established for pWEB-TNC-C6, -A5, and -B9 in the pDrive or pTZ19R cloning vector. In the latter cases only the genes of interest and no additional DNA was cloned. All subclones constructed from the pWEB-TNC-A5, B2, -B9, and -C6 cosmids or mutants obtained from pWEB-TNC-G2 were analyzed by functional assays using A. tumefaciens NTL4 and the C. violaceum reporter strain CV026. Together these tests identified the dlhR gene on cosmid pWEB-TNC-B2 on a 4.4-kb EcoRI fragment, the qsdR1 gene on pWEB-TNC-C6, the aldR gene on pWEB-TNC-G2, and the qsdR2 gene on pWEB-TNC-B9 as responsible for the observed phenotypes. On pWEB-TNC-A5, the hitR-hydR locus was associated with the observed phenotype. The identified genes and their flanking regions are depicted in Fig. 1 and indicated in Table 2. These data suggested that NGR234 probably carries at least five gene loci involved in modification or possible degradation of the AI-I signaling molecules.
The NGR234 ORF NGR_b16870 (960 bp) encodes a 321-amino-acid protein designated QsdR1 (for quorum sensing signal degrading enzyme from Rhizobium sp. strain NGR234). The translational start codon of QsdR1 is preceded by a possible ribosome-binding site AGAGGA, and possible −10 as well as −n sequences were identified according to previously reported rhizobial consensus promoter sequences (26). QsdR1 originating from cosmid pWEB-TNC-C6 is highly similar to a hypothetical protein from Rhizobium etli (RetlG_22662), with an E value of 2e−124, and to a hypothetical protein of Agrobacterium tumefaciens (Atu4307), with an E value of 1e−121. The protein belongs to the hydrolases of the β-lactamase superfamily and is grouped in class B of these enzymes, requiring a bivalent metal ion (Zn2+) for activity (8) (Table 2). Although an NCBI database search uncovered high homologies only to rather conserved hypothetical proteins with β-lactamase domains, a direct comparison with known representatives of AHLases confirmed that the qsdR1 product is highly similar to the Aii2 hydrolase from an uncultured Bacillus sp. (5). Additionally, BLAST searches indicated two conserved regions among the QsdR1 sequence and several known AHLases, such as AiiA (Table 3). The first short sequence is 141HMHMDHIG148 and the second is 236TGGHTPGH243.
Table 3.
Biochemically characterized QQ hydrolases in microorganisms and metagenomes
| Protein | Source or strain | Protein family | Conserved motif | Reference |
|---|---|---|---|---|
| AiiA | Bacillus sp. strain 240B1 | Metallo-β-lactamase superfamily | HxHx∼DH | 12 |
| AiiB/AttM | Agrobacterium tumefaciens strain C58 | Metallo-β-lactamase superfamily | HxHx∼DH | 5 |
| AhlD | Arthrobacter sp. strain IBN110 | Metallo-β-lactamase superfamily | HxHx∼DH | 32 |
| AhlK | Klebsiella pneumoniae | Metallo-β-lactamase superfamily | HxHx∼DH | 32 |
| QlcA | Soil metagenome | Metallo-β-lactamase superfamily | HxHx∼DH | 36 |
| QsdR1 | Rhizobium sp. strain NGR234 | Metallo-β-lactamase superfamily | HxHx∼DH | This study |
| AidH | Ochrobactrum anthropi ATCC 49188 | α/β-Hydrolase fold family | GX-Nuc-XG | 28 |
| AiiM | Microbacterium testeceum StLB037 | α/β-Hydrolase fold family | NDa | 54 |
| BpiB04 | Soil metagenome | Glycosyl hydrolase family | ND | 39 |
| BpiB07 | Soil metagenome | Dienelactone hydrolase family | ND | 39 |
| DlhR | Rhizobium sp. strain NGR234 | Dienelactone hydrolase family | GSD(L) | This study |
| BpiB01 | Soil metagenome | Hypothetical protein | ND | 39 |
| BpiB05 | Soil metagenome | Hypothetical protein | ND | 3 |
| QsdA | Rhodococcus erythropolis W2 | PTE superfamily | PTE domain | 51 |
ND, none detected.
The ORF NGR_c16020 (834 bp) encodes QsdR2, a 279-amino-acid protein which was also assigned to the β-lactamase superfamily. Prior to the putative ATG start codon, an AGGAGA ribosome-binding site as well as possible −10 and −35 promoter sequences could be identified according to published rhizobial consensus sequences (26). QsdR2 showed a very high similarity to a hypothetical protein from Sinorhizobium meliloti 1021 (SMc01194), with an E value of 3e−146, and to a β-lactamase domain-containing protein of Sinorhizobium medicae WSM419 (E value, 4e−146). QsdR2 shares a short conserved motif within the AHLases but is otherwise not similar to AHLases. The identified pattern is 83HAHADH89.
A detailed amino acid analysis of ORF NGR_b22150 (1,071 bp), encoding DlhR (for dienelactone hydrolase from Rhizobium sp. strain NGR234), revealed that this gene product was most similar to a hypothetical protein (Atu0247) from Agrobacterium tumefaciens strain C58. The observed similarity was 75% and the identity was 58%, with an E value of 3e−89 (Table 2). The deduced amino acid sequence of DlhR contained a multidomain COG4188 spanning nearly the whole protein and a smaller Pfam03403 domain structure. COG4188 represents a predicted dienelactone hydrolase domain, whereas Pfam03403 is involved in a subfamily of phospholipases A2, which are responsible for inactivation of platelet-activating factor through cleavage of an acetyl group. In addition, a conserved GSD(L) motif containing the active-site serine residue typical of GDSL esterases (family II of lipolytic enzymes) was identified (2). Further analysis of the upstream sequence of the predicted ATG start codon of dlhR revealed a possible ribosome-binding site as well as both promoter (−10/−35) regions. Additionally, the prediction of the presence and location of a signal peptide (SignalP 3.0 server, http://www.cbs.dtu.dk/services/SignalP/) on dlhR uncovered a 99% probability for a cleavage site between amino acids 22 and 23 (beginning with methionine). The predicted signal peptide on dlhR is MIPSHVPAALALAVAFAAPCHAF.
ORF NGR_c23150 (1,506 bp) encodes a putative acetaldehyde dehydrogenase, a 503-amino-acid protein, which we designated AldR (for acetaldehyde dehydrogenase from Rhizobium sp. strain NGR234). Similarities to proteins present in Sinorhizobium meliloti 1021, Sinorhizobium medicae WSM419, Rhizobium etli CFN42, Rhizobium leguminosarum bv. trifolii WSM2304, and Mesorhizobium loti MAFF303099 could be observed, with a maximum identity up to 100% and E values of 0.0 (Table 2).
Cosmid clone pWEB-TNC-A5 carries two ORFs that appeared to be involved in QQ. ORF NGR_c35570 (606 bp), encoding HitR (for histidine triad protein from Rhizobium sp. strain NGR234), and NGR_c35560, encoding HydR (for NUDIX hydrolase from Rhizobium sp. strain NGR234), might be responsible for the QQ phenotype. Gene hitR is predicted to encode a protein with 202 amino acids which was highly similar to homologous proteins in Sinorhizobium meliloti 1021 (E value, 1e−104). The hydR gene (1,035 bp) is predicted to encode a protein with 346 amino acids. The highest observed similarity of HydR was to a potential NUDIX hydrolase of Sinorhizobium medicae WSM419 and Sinorhizobium meliloti 1021, with E values of 0.0. Although we have no experimental evidence, it is highly possible that the two genes form an operon (Fig. 1).
We have conclusively identified three proteins (QsdR1, QsdR2, and DlhR) that are at least weakly similar to previously known QQ proteins. Furthermore, we have identified one protein (AldR) and one locus (hitR-hydR) that have not been reported to be involved in QS-modifying activities. All these findings are in line with the data derived from our sequence-based analyses (Table 2), where qsdR1, qsdR2, dlhR, and hitR-hydR were already identified. Furthermore, we extend the list of potential QQ genes by the aldR gene, which was identified only by function-based searches.
Heterologously expressed DlhR and QsdR1 quench QS-dependent processes in A. tumefaciens.
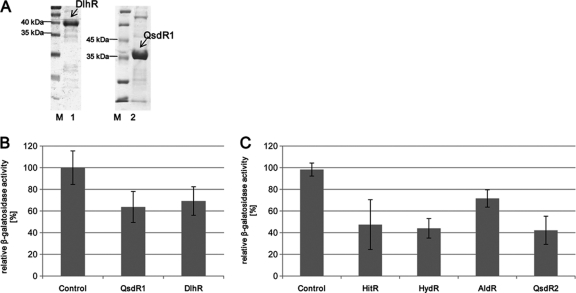
To verify the QQ activities of several of the identified proteins by more sophisticated methods, we focused on the enzyme candidates on pNGR234b. Thus, we expressed the dlhR and qsdR1 genes in E. coli and purified the corresponding recombinant His6-tagged proteins. For heterologous expression, the dlhR and qsdR1 genes were cloned directionally into pET21a using primers with inserted restriction sites (see Table S1 in the supplemental material) and overexpressed as described above. The estimated molecular masses of the His6-tagged and purified proteins of DlhR and QsdR1 (Fig. 3A) were in accordance with the theoretical molecular masses of 39.3 kDa (DlhR) and 36.0 kDa (QsdR1).
Fig. 3.
Purification of recombinant DlhR and QsdR1 and β-galactosidase assay using the same QQ proteins and test with pDrive clones of hitR, hydR, aldR, and qsdR2. (A) SDS-15% PAGE of protein extracts of E. coli BL21(DE3) cells expressing pET21a::dlhR and pET21a::qsdR1. Lanes: M, molecular mass markers; 1, purified His6-tagged DlhR at 39.3 kDa; 2, purified His6-tagged QsdR1 at 36.0 kDa. (B) β-Galactosidase activity in A. tumefaciens NTL4 cells after addition of 3-oxo-C8-HSL and 6 μg of either DlhR or QsdR1. (C) β-Galactosidase activity in A. tumefaciens NTL4 cells after addition of 3-oxo-C8-HSL and crude cell extracts of pDrive::hitR (HitR), pDrive::hydR (HydR), pDrive::aldR (AldR), and pDrive::qsdR2 (QsdR2). In panels B and C, KPO4 buffer and crude NTL4 extracts served as controls. Data are means of at least three independent measurements. Error bars indicate simple standard deviations.
The purified proteins were investigated for their QQ impact using A. tumefaciens NTL4. The initial screening employed for detection of AHL-degrading cosmids within the NGR234 library was also used for verifying the hydrolytic activities of the recombinant and purified DlhR and QsdR1 proteins. Different concentrations of both proteins incubated with 3-oxo-C8-HSL solution (4.1 × 10−6 M) and applied on ATsoft agar containing A. tumefaciens NTL4 caused a colorless appearance of the agar, indicating their effect on the QS response in A. tumefaciens NTL4 (data not shown).
Additionally, β-galactosidase assays combining hydrolytic cleavage of ONPG, A. tumefaciens NTL4, and our proteins confirmed these findings. Prior to incubation with NTL4 overnight, 5 μl of 3-oxo-C8-HSL (4.1 × 10−8 M) was mixed with recombinant DlhR or QsdR1 protein extracts (4 to 400 μg/μl) and incubated for 1.5 h at 30°C. After this short incubation time, the levels of detected AHLs were significantly lower than those of the controls. In general, we could detect less than 70% of the added 3-oxo-C8-HSL (Fig. 3B) after incubation with DlhR. The β-galactosidase assay with 10 to 20 μg of recombinant and purified QsdR1 revealed an even more pronounced 3-oxo-C8-HSL degradation (Fig. 3B). On average, the β-galactosidase assays suggested that the recombinant protein degraded 50% of the added 3-oxo-C8-HSL. Consequently, these tests confirmed the ability of recombinant DlhR and QsdR1 to degrade 3-oxo-C8-HSL. Further tests using crude cell extracts of pDrive clones harboring the hitR, hydR, aldR, and qsdR2 clones verified their hydrolytic activities in ONPG assays as well (Fig. 3C).
Uncovering the cleavage mechanism of DlhR and QsdR1 proteins by HPLC-MS analysis.
To verify AHL degradation by DlhR and QsdR1 and to detect the cleavage mechanisms by which the N-AHLs are inactivated (lactonolysis or amidolysis), we performed HPLC-MS analysis. Therefore, 10 μl of 3-oxo-C8-HSL was incubated with 0.005 to 0.3 mg/ml purified protein in 100 mM potassium phosphate buffer, pH 7.0. After incubation and extraction, reaction products were analyzed by HPLC-MS-diode array detection (DAD). Spontaneous degradation of 3-oxo-C8-HSL was evaluated in control experiments with a purified glycosyl transferase (GtfU) subjected to the same concentrations and conditions.
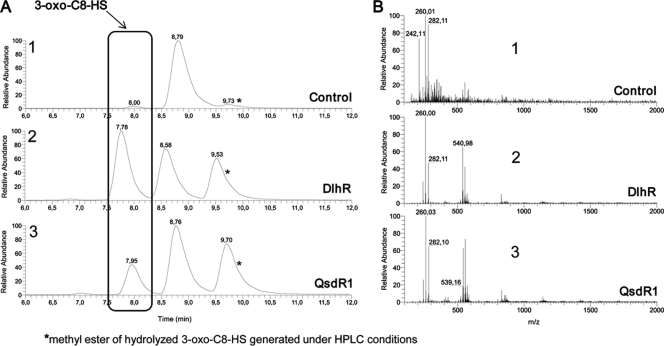
Repeated measurements with both proteins indicated that the underlying mechanism of degradation was a hydrolysis of the lactone ring of 3-oxo-C8-HSL. Determined by HPLC analysis followed by mass spectrometry, the enzymatic degradation resulted in a mixture of 3-oxo-C8-HS with a retention time (Rt) of ∼7.9 min and a methyl ester of hydrolyzed 3-oxo-C8-HS with an Rt of ∼9.6 min, generated only under given HPLC conditions. Peaks detected at an Rt of ∼8.6 min display the unhydrolyzed 3-oxo-C8-HSL form (Fig. 4).
Fig. 4.
HPLC-MS analysis of recombinant DlhR and QsdR1. (A) HPLC-UV spectra recorded at 252 nm for 3-oxo-C8-HSL after incubation with GtfU as a control, DlhR, and QsdR1. (B) Respective mass spectra recorded for samples treated with either control protein GtfU, DlhR, or QsdR1. (A1) The HPLC-UV chromatogram depicts the control incubated with 3-oxo-C8-HSL, displaying only one distinct peak at an Rt of 8.8 min representing 3-oxo-C8-HSL. No lactone hydrolysis could be observed. (B1) The mass spectrum of the GtfU control (at an Rt of 8.0 min) shows an (M + H)+ ion at an m/z of 242.1 as significant for this compound. An (M + H)+ ion at an m/z of 260.01 and an (M + Na)+ ion at an m/z of 282.1 were also detected owing to spontaneous degradation of 3-oxo-C8-HSL. (A2) HPLC-UV chromatogram for 3-oxo-C8-HSL incubated with DlhR, showing the peak at an Rt of 7.8 min for the cleavage product 3-oxo-C8-HS (opened lactone ring). The peak at an Rt of 8.6 min indicates the unhydrolyzed 3-oxo-C8-HSL, and an irrelevant by-product (a methyl ester), which is generated only under HPLC conditions, was detected at an Rt of 9.6 min. (B2) The corresponding mass spectrum shows three characteristic ions: an (M + H)+ ion at an m/z of 260.0, an (M + Na)+ ion at an m/z of 282.1, and a (2M + Na)+ ion at an m/z of 540.2. (A3) HPLC-UV chromatogram of 3-oxo-C8-HSL hydrolyzed by QsdR1. The cleavage product, an opened lactone ring form, was detected at an Rt of 8.0 min; consequently, QsdR1 was able to enzymatically degrade 3-oxo-C8-HSL. Both peaks for the nonhydrolyzed form of 3-oxo-C8-HSL as well as the methyl ester were recorded for QsdR1 as well. (B3) The mass spectrum as already given for DlhR showed the characteristic ions [an (M + H)+ ion, an (M + Na)+ ion, and a (2M + Na)+ ion at the same m/z].
3-Oxo-C8-HSL incubated with control protein GtfU resulted in the detection of almost exclusively the nonhydrolyzed form of 3-oxo-C8-HSL. Figure 4A, panel 1, shows the HPLC-UV spectrum at 252 nm for the control, displaying only one distinct peak at an Rt of 8.8 min representing 3-oxo-C8-HSL. A peak showing the relative abundance of the hydrolyzed form was under the detection limit; consequently, no significant lactone hydrolyzation linked to the control could be detected. The mass spectrum of the control (at an Rt of 8.0 min) showed an (M + H)+ ion at an m/z of 242.1 that was significant for this compound. An (M + H)+ ion at an m/z of 260.01 and an (M + Na)+ ion at an m/z of 282.1 were also detected owing to spontaneous degradation of 3-oxo-C8-HSL (Fig. 4B, panel 1). Both proteins incubated with 3-oxo-C8-HSL were able to hydrolyze the lactone ring of this N-AHL. HPLC-UV spectra detected at 252 nm for DlhR and QsdR1 showed almost identical retention times for the cleavage product 3-oxo-C8-HS with the opened lactone ring. Peaks were detected at an Rt of 7.8 min for DlhR and at an Rt of 8.0 min for QsdR1 (Fig. 4A, panels 2 and 3). The relative abundance of the 3-oxo-C8-HS compound found for both proteins was considerably higher than that for the control, thus showing a good enzymatic degradation of 3-oxo-C8-HSL by our proteins. Mass spectra for the proteins showed a characteristic (M + H)+ ion at an m/z of 260.0, an (M + Na)+ ion at an m/z of 282.1, and a (2M + Na)+ ion at an m/z of 540.2 (Fig. 4B, panels 2 and 3).
In summary, the control incubated under identical conditions as for DlhR and QsdR1 with 3-oxo-C8-HSL did not produce a peak at an Rt of ∼7.9 min, which is characteristic for the cleavage form, 3-oxo-C8-HS. The HPLC-UV as well as mass spectral data for both recombinant proteins confirmed an enzymatic activity, giving evidence that DlhR and QsdR1 act as true lactonases in NGR234.
DlhR and QsdR1 interfere with rhizosphere colonization of cowpea roots.
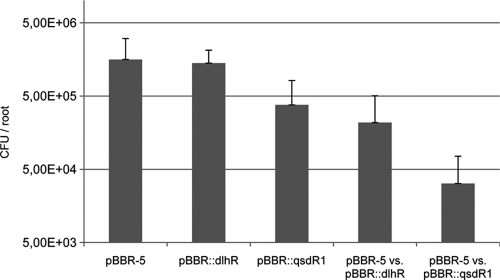
We speculated that DlhR and QsdR1 would be important for plant rhizosphere colonization. In order to test this hypothesis, we constructed two strains carrying extra copies of the two genes in the vector pBBR1MCS. NGR234 cells containing pBBR1MCS constructs expressed the dlhR and qsdR1 genes under the control of the Ptaq promoter. Constructs were verified by sequencing the inserts. We then carried out root colonization tests on cowpea roots by inoculating each mutant individually and in competition with the control strain (NGR234 harboring pBBR1MCS-5). The plants were harvested after 4 days and root colonization analyzed as described in Materials and Methods. Under our experimental conditions, NGR234 grew and colonized the root most rapidly at between 3 and 6 days after inoculation (data not shown), and these tests indicated that the control strain was most efficient in root colonization in all tested combinations (Fig. 5). While the strain carrying an extra copy of the dlhR gene was in general only slightly affected in its capability to colonize the root surface, the strain carrying the qsdR1 gene was significantly affected in its rhizosphere colonization capabilities. Additional competition experiments confirmed these findings (Fig. 5). In these tests the strains carrying extra copies of the dlhR or the qsdR1 gene were outnumbered by the control strain and colonized the rhizospheres at a statistically significantly (P ≤ 0.01) lower level. In fact, the qsdR1 strain was outnumbered by at least an order of magnitude by the control strain when coinoculated into the rhizosphere at equal numbers (CFU/ml), and the strain carrying extra copies of dlhR was outnumbered by a factor of 5 to 7. From these tests we concluded that both genes contribute to rhizosphere colonization fitness of NGR234 and are possibly involved in the degradation of plant-derived or microbial autoinducer molecules in the rhizosphere.
Fig. 5.
Growth of NGR234 in the cowpea rhizosphere. Bacteria inoculated on day 0 were quantified in the inocula by measuring the CFU. After 4 days, bacteria were recovered from developed roots. Data represent mean numbers (CFU) of bacteria per root and standard deviations from a minimum of five replicates. From left to right: control pBBR-5 (NGR234 harboring the empty pBBR1MCS- 5), pBBR::dlhR (NGR234 with an extra copy of dlhR in pBBR1MCS), pBBR::qsdR1 (NGR234 with an extra copy of qsdR1 in pBBR1MCS), pBBR-5 versus pBBR::dlhR (competition experiments with NGR234 carrying control pBBR-5 versus NGR234 carrying an extra copy of dlhR), and pBBR-5 versus pBBR::qsdR1 (competition experiments with NGR234 carrying control pBBR-5 versus NGR234 carrying an extra copy of qsdR1). The last two bars indicate the recovered bacteria carrying the extra copies of the corresponding QQ genes (i.e., dlhR and qsdR1). Error bars indicate simple standard deviations.
DISCUSSION
In this paper we describe the isolation of five NGR234-derived genes and loci that interfere with AI-I signaling molecules, and we have partially biochemically characterized two of the proteins. The NGR234-derived QQ clones were initially identified because they repeatedly interfered with quorum sensing in an A. tumefaciens NTL4-based bioassay using a traI::lacZ promoter fusion. The genes and loci identified and linked to the QS inhibitory phenotypes were designated dlhR, qsdR1, qsdR2, aldR, and hitR-hydR. All these genes, when expressed, resulted in strongly reduced motility and biofilm phenotypes in P. aeruginosa PAO1 (Fig. 2A and B). With respect to the reduced biofilm formation and motility induced through the expression of the NGR234-derived QQ genes, we hypothesized that these phenotypes were a result of the N-AHL degradation caused by the identified proteins. In fact, the results presented in Fig. 3B strongly supported that hypothesis, and further data using more sophisticated analytical technologies confirmed these findings for the recombinant and purified DlhR and QsdR1 proteins (Fig. 4). These results are all in line with the reports on enzymatic degradation of N-AHLs. Enzymatic degradation of the quorum-sensing signal molecules is an established method of quorum quenching and has been reviewed recently (11, 34, 35). Three main types of microbial enzymes that act on the N-AHLs are known: (i) oxidoreductases, (ii) acylases, and (iii) lactonases. Besides oxidoreductases, which have been uncovered in Gram-positive bacteria (6, 7, 49), oxidases have been identified in algae, acting on the 3-oxo group (4). Acylases were found in organisms such as Comamonas (50), P. aeruginosa (20), P. syringae (42), Ralstonia spp. (24), R. erythropolis (49), Shewanella (29), and Streptomyces (31). Aminoacylases cleave the lactone ring off the fatty acids, whereas lactonases, which constitute the largest group of AHL-degrading enzymes, hydrolyze the lactone ring in a reversible way. Today, only a small number of different functionally verified classes of lactonase proteins that efficiently hydrolyze the lactone ring in N-AHLs are known. The experimentally verified and biochemically characterized lactonases are summarized in Table 3. The lactonases are grouped into several clusters according to their overall similarity and the original microbe in which the first gene of the cluster was identified. The first known clusters are designated AiiA, AttM, and PON1-3 (13). Recently six other types of lactonases (i.e., QsdA [51], BpiB01, BpiB04, and BpiB07 [39]; AiiM [54], and AidH [28]) have been identified and extend the diversity of lactonase family proteins. These proteins are probably each members of novel N-AHL lactonase families. Zn2+-dependent lactonases have been described for Bacillus (5, 9, 23, 48), Agrobacterium (5), Rhodococcus (30), Streptomyces (31), Arthrobacter (32), Pseudomonas (44), and Klebsiella (32). The novel lactonase derived from Rhodococcus, with the gene designated qsdA, forms a new family within the metal-dependent lactonases (51). Furthermore, we have recently identified four novel lactone-hydrolyzing enzymes isolated from the metagenome (3, 39). Within this framework it is noteworthy that we have not only identified three lactonases (DlhR, QsdR1, and QsdR2) in NGR234, but we also identified two completely novel genes in NGR234 that had not been linked to QQ in earlier studies. These are the aldR gene and the hitR-hydR locus. Although we have not characterized all the respective proteins in detail, our data strongly suggest that they are involved in autoinducer degradation or modification in the sense that P. aeruginosa PAO1 responds with reduced motility and other QS-dependent phenotypes.
The richness of ORFs linked to QQ activity in NGR234 probably suggests that this is an important feature, which is possibly needed during rhizosphere colonization and during growth in free soil. This hypothesis is strongly supported by our rhizosphere colonization tests using NGR234 mutant strains carrying extra copies of the dlhR and the qsdR1 genes (Fig. 5). These strains were in general less effective during root colonization than a wild-type control strain carrying an empty vector.
A detailed genome comparison of several of the sequenced rhizobial genomes suggests that rhizobial isolates appear to commonly have several QQ genes encoded in their genomes, and those appear to be present on the chromosomes or the larger megaplasmids but not on the symbiotic plasmids (Table 4). A comparison of the genomes revealed the presence of four of the NGR234 QQ genes in the S. meliloti SM1021 genome. SM1021, however, lacks the dlhR locus. Also, dlhR as well as the qsdR1 locus are missing in M. loti MAFF303099. Furthermore, an analysis of the nearly complete USDA257 genome revealed that besides the dlhR gene, all NGR234 QQ genes were present in this broad-host microbe as well (unpublished data from our lab). A more detailed analysis of the presence and absence of the NGR234 QQ genes in other rhizobia is given in Table 4.
Table 4.
NGR234 QQ genes in related and completed rhizobial genomes
| NGR234 QQ locus | Homologue/orthologue ina: |
||||
|---|---|---|---|---|---|
| Rleg3841 | SM1021 | Bjap110 | Retli42 | ML99 | |
| dlhR (NGR_b22150) | RRZL01754 | ND | ND | RRET01614 | ND |
| qsdR1 (NGR_b16870) | RRZL02285 | RSM05795 | RBRJ02791 | RRET00556 | ND |
| qsdR2 (NGR_c16020) | RRZL02453 | RSM02893 | RBRJ04514 | RRET02102 | RMLO00322 |
| aldR (NGR_c23150) | RRZL04239 | RSM03645 | RBRJ04784 | RRET03644 | RMLO05206 |
| hitR-hydR (NGR_c35560/c35570) | RRZL00137/RRZL00138 | RSM01522/RSM01523 | RBRJ08122/RBRJ08123 | RRET00118/RRET00119 | RMLO04260/RMLO04261 |
Homologous/orthologous genes are indicated by the corresponding gene/ORF number. Data for the analysis were extracted from the respective genome projects and the corresponding GenBank files: NGR234, Rhizobium sp. strain NGR234 (CP000874 and CP001389); Rleg3841, Rhizobium leguminosarum bv. viciae 3841 (AM236080 to AM236086); SM1021, Sinorhizobium meliloti strain 1021 (AL591786); Bjap110, Bradyrhizobium japonicum strain USDA110 (BA000040); Retli42, Rhizobium etli strain CFN42 (AF311739); ML99, Mesorhizobium loti strain MAFF303099 (AP003017). ND, not detected.
The results from this study probably have two main implications. First, NGR234 and perhaps many other rhizosphere-associated organisms appear to have a surprisingly large number of different ways to degrade or modify autoinducers present in their environment. Second, data from this study emphasize the ecological importance of QQ during root colonization.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the BMBF-Kompetenznetzwerk “Biotech GenoMik+” and GenoMik-Transfer.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Arpigny J. L., Jaeger K. E. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343:177–183 [PMC free article] [PubMed] [Google Scholar]
- 3. Bijtenhoorn P., et al. 2011. BpiB05, a novel metagenome-derived hydrolase acting on N-acylhomoserine lactones. J. Biotechnol. doi:10.1016/j.jbiotec.2010.12.016 [DOI] [PubMed] [Google Scholar]
- 4. Borchardt S. A., et al. 2001. Reaction of acylated homoserine lactone bacterial signaling molecules with oxidized halogen antimicrobials. Appl. Environ. Microbiol. 67:3174–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carlier A., et al. 2003. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-acyl homoserine lactonase activity. Appl. Environ. Microbiol. 69:4989–4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chowdhary P. K., et al. 2007. Bacillus megaterium CYP102A1 oxidation of acyl homoserine lactones and acyl homoserines. Biochemistry 46:14429–14437 [DOI] [PubMed] [Google Scholar]
- 7. Chowdhary P. K., Stewart L., Lopez C., Haines D. C. 2008. A single mutation in P450BM-3 enhances acyl homoserine lactone:acyl homoserine substrate binding selectivity nearly 250-fold. J. Biotechnol. 135:374–376 [DOI] [PubMed] [Google Scholar]
- 8. Daiyasu H., Osaka K., Ishino Y., Toh H. 2001. Expansion of the zinc metallo-hydrolase family of the beta-lactamase fold. FEBS Lett. 503:1–6 [DOI] [PubMed] [Google Scholar]
- 9. Dong Y. H., Gusti A. R., Zhang Q., Xu J. L., Zhang L. H. 2002. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl. Environ. Microbiol. 68:1754–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dong Y. H., et al. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813–817 [DOI] [PubMed] [Google Scholar]
- 11. Dong Y. H., Wang L. Y., Zhang L. H. 2007. Quorum-quenching microbial infections: mechanisms and implications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362:1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dong Y. H., Xu J. L., Li X. Z., Zhang L. H. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. U. S. A. 97:3526–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dong Y. H., Zhang L. H. 2005. Quorum sensing and quorum-quenching enzymes. J. Microbiol. 43(Spec. No.): 101–109 [PubMed] [Google Scholar]
- 14. Fuqua W. C., Winans S. C. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fuqua W. C., Winans S. C. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garg B., Dogra R. C., Sharma P. K. 1999. High-efficiency transformation of Rhizobium leguminosarum by electroporation. Appl. Environ. Microbiol. 65:2802–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He X., et al. 2003. Quorum sensing in Rhizobium sp. strain NGR234 regulates conjugal transfer (tra) gene expression and influences growth rate. J. Bacteriol. 185:809–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoagland D. R., Arnon D. I. 1950. The water-culture method for growing plants without soil. California Agricultural Experiment Station Circular 1-32. [Google Scholar]
- 19. Holloway B. W., Krishnapillai V., Morgan A. F. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang J. J., Petersen A., Whiteley M., Leadbetter J. R. 2006. Identification of QuiP, the product of gene PA1032, as the second acyl-homoserine lactone acylase of Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 72:1190–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kohler T., Curty L. K., Barja F., van Delden C., Pechere J. C. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kovach M. E., et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 23. Lee S. J., et al. 2002. Genes encoding the N-acyl homoserine lactone-degrading enzyme are widespread in many subspecies of Bacillus thuringiensis. Appl. Environ. Microbiol. 68:3919–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin Y. H., et al. 2003. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 47:849–860 [DOI] [PubMed] [Google Scholar]
- 25. Luo Z. Q., Clemente T. E., Farrand S. K. 2001. Construction of a derivative of Agrobacterium tumefaciens C58 that does not mutate to tetracycline resistance. Mol. Plant Microbe Interact. 14:98–103 [DOI] [PubMed] [Google Scholar]
- 26. MacLellan S. R., MacLean A. M., Finan T. M. 2006. Promoter prediction in the rhizobia. Microbiology 152:1751–1763 [DOI] [PubMed] [Google Scholar]
- 27. McClean K. H., et al. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703–3711 [DOI] [PubMed] [Google Scholar]
- 28. Mei G. Y., Yan X. X., Turak A., Luo Z. Q., Zhang L. Q. 2010. AidH, an alpha/beta-hydrolase fold family member from an Ochrobactrum sp. strain, is a novel N-acylhomoserine lactonase. Appl. Environ. Microbiol. 76:4933–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morohoshi T., Nakazawa S., Ebata A., Kato N., Ikeda T. 2008. Identification and characterization of N-acylhomoserine lactone-acylase from the fish intestinal Shewanella sp. strain MIB015. Biosci. Biotechnol. Biochem. 72:1887–1893 [DOI] [PubMed] [Google Scholar]
- 30. Park S. Y., et al. 2006. N-Acylhomoserine lactonase producing Rhodococcus spp. with different AHL-degrading activities. FEMS Microbiol. Lett. 261:102–108 [DOI] [PubMed] [Google Scholar]
- 31. Park S. Y., et al. 2005. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl. Environ. Microbiol. 71:2632–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park S. Y., et al. 2003. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 149:1541–1550 [DOI] [PubMed] [Google Scholar]
- 33. Pueppke S. G., Broughton W. J. 1999. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol. Plant Microbe Interact. 12:293–318 [DOI] [PubMed] [Google Scholar]
- 34. Rasmussen T. B., Givskov M. 2006. Quorum-sensing inhibitors as anti-pathogenic drugs. Int. J. Med. Microbiol. 296:149–161 [DOI] [PubMed] [Google Scholar]
- 35. Rasmussen T. B., Givskov M. 2006. Quorum sensing inhibitors: a bargain of effects. Microbiology 152:895–904 [DOI] [PubMed] [Google Scholar]
- 36. Riaz K., et al. 2008. A metagenomic analysis of soil bacteria extends the diversity of quorum-quenching lactonases. Environ. Microbiol. 10:560–570 [DOI] [PubMed] [Google Scholar]
- 37. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 38. Schaber J. A., et al. 2004. Analysis of quorum sensing-deficient clinical isolates of Pseudomonas aeruginosa. J. Med. Microbiol. 53:841–853 [DOI] [PubMed] [Google Scholar]
- 39. Schipper C., et al. 2009. Metagenome-derived clones encoding two novel lactonase family proteins involved in biofilm inhibition in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 75:224–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmeisser C., et al. 2009. Rhizobium sp. strain NGR234 possesses a remarkable number of secretion systems. Appl. Environ. Microbiol. 75:4035–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shank E. A., Kolter R. 2009. New developments in microbial interspecies signaling. Curr. Opin. Microbiol. 12:205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shepherd R. W., Lindow S. E. 2009. Two dissimilar N-acyl-homoserine lactone acylases of Pseudomonas syringae influence colony and biofilm morphology. Appl. Environ. Microbiol. 75:45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shrout J. D., et al. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264–1277 [DOI] [PubMed] [Google Scholar]
- 44. Sio C. F., et al. 2006. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect. Immun. 74:1673–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Streit W. R., Joseph C. M., Phillips D. A. 1996. Biotin and other water-soluble vitamins are key growth factors for alfalfa root colonization by Rhizobium meliloti 1021. Mol. Plant Microbe Interact. 9:330–338 [DOI] [PubMed] [Google Scholar]
- 46. Tempe J., Petit A., Holsters M., Montagu M., Schell J. 1977. Thermosensitive step associated with transfer of the Ti plasmid during conjugation: possible relation to transformation in crown gall. Proc. Natl. Acad. Sci. U. S. A. 74:2848–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trinick M. J. 1980. Relationships amongst the fast-growing rhizobia of Lablab purpureus, Leucaena leucocephala, Mimosa spp., Acacia farnesiana and Sesbania grandiflora and their affinities with other rhizobial groups. J. Appl. Microbiol. 49:39–53 [Google Scholar]
- 48. Ulrich R. L. 2004. Quorum quenching: enzymatic disruption of N-acylhomoserine lactone-mediated bacterial communication in Burkholderia thailandensis. Appl. Environ. Microbiol. 70:6173–6180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Uroz S., et al. 2005. N-Acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology 151:3313–3322 [DOI] [PubMed] [Google Scholar]
- 50. Uroz S., et al. 2007. N-Acyl homoserine lactones are degraded via an amidolytic activity in Comamonas sp. strain D1. Arch. Microbiol. 187:249–256 [DOI] [PubMed] [Google Scholar]
- 51. Uroz S., et al. 2008. A Rhodococcus qsdA-encoded enzyme defines a novel class of large-spectrum quorum-quenching lactonases. Appl. Environ. Microbiol. 74:1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vincent J. M. 1970. A manual for the practical study of root-nodule bacteria. Blackwell Scientific, Oxford, United Kingdom [Google Scholar]
- 53. Wagner V. E., Li L. L., Isabella V. M., Iglewski B. H. 2007. Analysis of the hierarchy of quorum-sensing regulation in Pseudomonas aeruginosa. Anal. Bioanal Chem. 387:469–479 [DOI] [PubMed] [Google Scholar]
- 54. Wang W. Z., Morohoshi T., Ikenoya M., Someya N., Ikeda T. 2010. AiiM, a novel class of N-acylhomoserine lactonase from the leaf-associated bacterium Microbacterium testaceum. Appl. Environ. Microbiol. 76:2524–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Waters C. M., Bassler B. L. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319–346 [DOI] [PubMed] [Google Scholar]
- 56. Zhu J., et al. 1998. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 180:5398–5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.