Abstract
The dissimilatory iron-reducing bacterium Shewanella oneidensis MR-1 is capable of using extracellular DNA (eDNA) as the sole source of carbon, phosphorus, and nitrogen. In addition, we recently demonstrated that S. oneidensis MR-1 requires eDNA as a structural component during all stages of biofilm formation. In this study, we characterize the roles of two Shewanella extracellular endonucleases, ExeS and ExeM. While ExeS is likely secreted into the medium, ExeM is predicted to remain associated with the cell envelope. Both exeM and exeS are highly expressed under phosphate-limited conditions. Mutants lacking exeS and/or exeM exhibit decreased eDNA degradation; however, the capability of S. oneidensis MR-1 to use DNA as the sole source of phosphorus is only affected in mutants lacking exeM. Neither of the two endonucleases alleviates toxic effects of increased eDNA concentrations. The deletion of exeM and/or exeS significantly affects biofilm formation of S. oneidensis MR-1 under static conditions, and expression of exeM and exeS drastically increases during static biofilm formation. Under hydrodynamic conditions, a deletion of exeM leads to altered biofilms that consist of densely packed structures which are covered by a thick layer of eDNA. Based on these results, we hypothesize that a major role of ExeS and, in particular, ExeM of S. oneidensis MR-1, is to degrade eDNA as a matrix component during biofilm formation to improve nutrient supply and to enable detachment.
INTRODUCTION
Extracellular DNA (eDNA) occurs in significant amounts in aquatic and terrestrial ecosystems (68). Concentrations vary among different environments, ranging from 2 μg g−1 in dry soil (49) to up to 20 mg g−1 organic matter in activated sludge from wastewater treatment plants (51). The pool of eDNA provides a reservoir for horizontal gene transfer (45, 68) and a valuable source of carbon, nitrogen, and, in particular, phosphorus (11, 12, 48, 53). Accordingly, it has been demonstrated that several bacteria have the capacity to use DNA as the sole nutrient source (17, 48, 53).
In biofilms, the predominant bacterial lifestyle in nature, the cells are embedded in a self-produced matrix, forming highly structured aggregates that are commonly attached to surfaces (60). This matrix, which in many cases comprises the majority of the communities' biomass, consists of different biopolymers. It has previously been reported that eDNA is abundant in bacterial biofilms (19, 51); in addition, polysaccharides and proteins are common components (18). Studies of Pseudomonas aeruginosa demonstrated that eDNA in the matrix is required for structural integrity during the early stages of biofilm formation (70). Since then, eDNA has been recognized as an important factor for cell adhesion and as a structural component of the biofilm matrix for a growing number of Gram-positive and Gram-negative species and mixed communities (15, 59). However, its exact role in biofilm formation remains to be elucidated. In addition to a structural and/or adhesive role in microbial biofilms, eDNA was demonstrated to have antimicrobial activity through chelation of ions required for stabilizing lipopolysaccharides and/or outer membranes (47). The release of DNA is thought to be mediated by cell lysis (37, 41, 55, 56, 63), active transport (25), or vesiculation (1, 70). In contrast, given the significance of eDNA as a structural component, potential nutrient, antimicrobial agent, and reservoir for gene transfer, surprisingly little is known about the role of eDNA degradation in biofilms. Recently, it was demonstrated that nuclease-mediated degradation of eDNA is required for normal biofilm formation. It is thought that the nuclease activity enables detachment of cells from Staphylococcus aureus and Neisseria gonorrhoeae biofilms (41, 58).
The dissimilatory iron-reducing gammaproteobacterium Shewanella oneidensis MR-1 adheres to surfaces and forms complex and dynamic surface-associated biofilms (2, 42, 62, 64–66). Biofilm formation is thought to positively impact the ability of Shewanella species to access insoluble alternative electron acceptors, such as ferric or manganese minerals (10, 24, 43). We have recently demonstrated that biofilm formation of S. oneidensis MR-1 strongly depends on eDNA as a structural component. The DNA is predominantly released by prophage-mediated lysis of a subpopulation of the cells. In the absence of eDNA, S. oneidensis MR-1 is severely reduced in surface adhesion and subsequent formation of three-dimensional structures. DNase I treatments of S. oneidensis MR-1 biofilms released large amounts of biomass (22). In addition, a recent study showed that Shewanella species can use eDNA as the sole source of phosphorus, carbon, and energy. Accordingly, nuclease activity was determined in S. oneidensis MR-1 cultures, and two putative extracellular nucleases were identified (27, 53). The activities and functions of both proteins are not yet known. However, with respect to the important role of eDNA as structural component, we hypothesized that these nucleases might also affect S. oneidensis MR-1 biofilm formation. In this study, we characterized whether the two predicted extracellular endonucleases are involved in degradation of eDNA by S. oneidensis MR-1. Furthermore, we determined whether both proteins affect the ability of this species to exploit eDNA as a nutrient, to tolerate large amounts of eDNA, or to form biofilms.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. Escherichia coli and S. oneidensis strains were routinely grown in LB medium at 37°C and 30°C, respectively. For plates, agar was added to a final concentration of 1.5% (wt/vol). For the conjugation strain E. coli WM3064, 2,6-diaminopimelic acid (DAP) was added to the medium to a final concentration of 300 μM.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotypea | Source or reference |
|---|---|---|
| Bacterial strains | ||
| Escherichia coli | ||
| DH5α λ pir | φ80dlacZΔM15 Δ(lacZYA-argF)U169recA1hsdR17 deoR thi-l supE44 gyrA96 relA1/λ pir | 44 |
| WM3064 | thrB1004pro thi rpsL hsdS lacZΔM15 RP4-1360 Δ(araBAD)567 ΔdapA1341::[erm pir (wt)] | W. Metcalf, University of Illinois, Urbana |
| Shewanella oneidensis | ||
| MR-1 | S. oneidensis MR-1 wild type | 67 |
| S198 | MR-1, tagged with EGFP in a mini-Tn7 construct; Cmr | This work |
| S988 | MR-1 ΔexeM; deletion of gene SO_1066 | This work |
| S1015 | MR-1 ΔexeM, tagged with EGFP in a mini-Tn7 construct; Cmr | This work |
| S1223 | Wild type (reintegration of ΔexeM) | This work |
| S989 | MR-1 ΔexeS; deletion of gene SO_1844 | This work |
| S1016 | MR-1 ΔexeS, tagged with EGFP in a mini-Tn7 construct; Cmr | This work |
| S1212 | Wild type (reintegration of ΔexeS) | This work |
| S1034 | MR-1 ΔexeM ΔexeS | This work |
| S1044 | MR-1 ΔexeM ΔexeS, tagged with EGFP in a mini-Tn7 construct; Cmr | This work |
| Plasmids | ||
| pBBR1-MCS5 | oriTmobRK2oriRlacZα, cloning vector; Gmr | 36 |
| pBBR1-MCS5-TT-RBS-lux | luxCDABE and terminators lambda T0 rrnB1 T1 cloned into pBBR1-MCS5 for plasmid-based transcriptional fusions; Gmr | This work |
| pBBR1-MCS5-TT-Pmot-RBS-lux | motAB promoter of S. oneidensis MR-1 cloned into pBBR1-MCS5-TT-RBS-lux; Gmr | This work |
| pNTPS-138-R6K | oriT ori-R6K sacB, suicide plasmid for generating in-frame deletions; Kmr | 38 |
| pNTPS-R6K-dexeM | Fragment for in-frame deletion of SO_1066 in pNTPS-R6K; Kmr | This work |
| pNTPS-R6K-KI-exeM | exeM in pNTPS-R6K for reintegration; Kmr | This work |
| pNTPS-R6K-dexeS | Fragment for in-frame deletion of SO_1844 in pNTPS-R6K; Kmr | This work |
| pNTPS-R6K-KI-exeS | exeS in pNTPS-R6K for reintegration; Kmr | This work |
| pTNS2 | ori-R6K; encodes the TnsABC + D-specific transposition pathway; Apr | 9 |
| pUC18-mini-Tn7T-Gm-lux | ColE1 replicon, mini-Tn7; luxCDABE Apr Gmr | 9 |
| pUC18-R6KT-miniTn7T-egfp | NotI-egfp-Cmr-NotI fragment from pBK-miniTn7-gfp3 in pUC18-R6KT-miniTn7T | 22 |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance.
Anaerobic growth was assayed in LB medium adjusted to pH 7.5, containing 40 mM lactate and 20 mM fumarate as described previously (38). For growth assays, S. oneidensis strains were cultivated in modified M1 minimal medium (53), containing 30 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 30 mM NaCl, 3 mM MgCl2, 1.34 mM KCl, 6.8 μM CaCl2, and 10 ml of a 100× mineral solution used for 4M medium, containing 6.72 mM Na2EDTA, 5.66 mM H3BO3, 0.54 mM FeCl2, 0.5 mM CoCl2, 0.5 mM NiCl2, 0.39 mM Na2MO4, 0.15 mM Na2SeO4, 0.13 mM MnSO4, 0.1 mM ZnSO4, and 0.02 mM CuSO4 (20). Either 40 mM lactate, 0.86 mM NaH2PO4, 28 mM NH4Cl, or DNA was used as the source of carbon, phosphorus, and nitrogen, respectively. All growth experiments were performed using DNA from salmon sperm (Sigma, Deisenhofen, Germany).
If necessary, media were supplemented with 6 μg ml−1 chloramphenicol and/or 30 μg ml−1 kanamycin. Biofilms of S. oneidensis were cultivated in LM medium (52) without antibiotics containing 0.5 mM lactate. DNase I (Serva Electrophoresis GmbH, Heidelberg, Germany) was used at a concentration of 30 μg ml−1 in medium supplemented with 5 mM MgCl2. DDAO [7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one)] was used at a concentration of 4 μM to stain extracellular DNA in biofilms grown under hydrodynamic conditions.
Vector and strain construction.
Molecular methods were carried out according to standard protocols (57) or according to the manufacturer's instructions. Kits for the isolation of plasmids and the purification of PCR products were purchased from HISS Diagnostics GmbH (Freiburg, Germany). Enzymes were purchased from New England BioLabs (Frankfurt, Germany) and Fermentas (St. Leon-Rot, Germany). The strains and plasmids used in this study are summarized in Table 1.
Markerless in-frame deletion mutants of S. oneidensis MR-1 were constructed essentially as previously reported (38) using the suicide vector pNTPS-138-R6K and appropriate primer pairs, as summarized in Table S1 in the supplemental material. Complementation of the mutants was carried out by reintegration of the deleted fragment into the appropriate deletion strain using the same protocol as for the deletion. After complementation, all strains retained the wild-type phenotype.
For biofilm studies, S. oneidensis MR-1 strains constitutively expressing the green fluorescent protein (GFP) gene gfp were constructed by using a modified Tn7 delivery system as reported earlier (22). Briefly, the plasmid pUC18-R6KT-miniTn7T-egfp coding for enhanced GFP (EGFP) was used for tagging S. oneidensis strains by three-parental mating from the DAP auxotroph E. coli WM3064 and E. coli strain WM3064 harboring the helper plasmid pTNS2. Comparative growth and biofilm experiments ensured that no detrimental effects based on fluorescent protein expression occurred.
For the construction of constitutively luminescent S. oneidensis MR-1 strains, the luxCDABE operon of Photorhabdus luminescens and a transcriptional terminator cassette were amplified from pUC18-mini-Tn7T-Gm-lux and cloned into the broad-host-range vector pBBR1-MCS5, using KpnI and SacI/XhoI, respectively, yielding vector pBBR1-MCS5-TT-RBS-lux. Lux expression was conferred by cloning the constitutively transcribed S. oneidensis MR-1 motAB promoter upstream of the lux operon, using PstI and BamHI. The resulting vector, pBBR1-MCS5-TT-Pmot-RBS-lux, was introduced into the appropriate strains by electroporation or conjugation.
Cultivation of S. oneidensis MR-1 biofilms. (i) Static conditions.
Biofilm cultivation in polystyrene microtiter plates (Sarstedt, Newton, NC) was carried out essentially as previously described (65). Briefly, freshly diluted overnight cultures of S. oneidensis MR-1 strains (1:35 in LM medium) were transferred to wells of polystyrene microtiter plates (170 μl per well) and incubated for the indicated times at 30°C. Prior to processing, the density of the planktonic population in the wells was determined at 600 nm. Subsequently, 10 μl crystal violet (0.5% [wt/vol]) was added to the wells followed by 10 min of incubation. The wells were washed with 200 μl distilled water, and the remaining surface-attached biomass was quantified indirectly by the solubilization of retained crystal violet by the addition of 200 μl ethanol (96% [wt/vol]) followed by measurements of the absorbance at 570 nm using an Infinite M200 plate reader (Tecan, Switzerland). The relative degree of surface attachment was normalized to that of the wild type. The assay was repeated in at least three independent experiments.
(ii) Hydrodynamic conditions.
Biofilms were cultivated at room temperature in LM medium in custom-made three-channel flow cells with individual channel dimensions of 1 by 4 by 40 mm. Microscope coverslips (Roth, Germany) were used as a colonization surface and glued onto the channels with silicone (Sista-Henkel, Germany). Assembly, sterilization, and inoculation of the flow system were performed essentially as previously described (65). Analyses were carried out in triplicate in at least two independent experiments. For DDAO staining, the flow was briefly arrested and the dye was added to the medium in the bubble trap. This process took no longer than 1 min, and control channels ensured that the short arrest did not affect biofilm development. The biofilm cells were incubated with the DDAO for 1 h. Microscopic visualization was performed at defined spots close to the inflow before and after the treatment.
Microscopy and image acquisition.
Microscopic visualization of biofilms and image acquisition were conducted using an inverted Leica TCS SP5 confocal laser scanning microscope (Leica Microsystems, Wetzlar, Germany) equipped with ×10/0.3 Plan-Neofluar and ×63/1.2 W C-Apochromate objectives. The flow chambers were mounted on the microscopic stage without interrupting the flow. Live imaging of the cells was enabled by using strains that were constitutively expressing gfp; however, biofilms were not further incubated after DDAO staining. To display biofilm images, confocal laser scanning microscopy (CLSM) image stacks were processed using the IMARIS software package (Bitplane AG, Zürich, Switzerland) and Adobe Photoshop. For the quantification of the surface coverage, the image of the confocal plane displaying the cell layer attached to surface was selected. The amount of surface-attached biomass was determined by the amount of green pixels (cells) in relation to that of the background (black) using Adobe Photoshop CS2. For each data point, at least four individual images from at least two independent experiments were analyzed.
Quantification of extracellular DNA.
The quantification of extracellular DNA in the biofilm supernatant was carried out as previously described (22). Briefly, the supernatants of statically grown S. oneidensis MR-1 biofilms were collected after 1, 4, and 24 h of incubation and filter sterilized. One hundred microliters of a 1:200 dilution of PicoGreen fluorescent dye (Invitrogen/Molecular Probes, Darmstadt, Germany) was added to biofilm supernatant, and the DNA release was immediately measured fluorometrically at an excitation wavelength of 485 nm and an emission wavelength of 535 nm using a Tecan Infinite M200 reader (Tecan, Switzerland). The concentration of extracellular DNA was then calculated using DNA reference standards prepared in culture medium.
DNA degradation assay.
Nuclease activity was determined in a DNA degradation assay essentially as described earlier (53). Overnight cultures of S. oneidensis MR-1 strains grown in LB medium were diluted to an optical density at 600 nm (OD600) of 0.05 and incubated in LB or 4M medium until cells reached an OD600 of 1.9. Afterwards, 230-μl aliquots of washed cell culture and filter-sterilized supernatants, respectively, were mixed with 1,006 bp or 3,140 bp of PCR-amplified DNA fragments at a final concentration of 5 μg ml−1. The samples were incubated at 30°C. Every 15 min, 25-μl aliquots were removed and analyzed by agarose gel electrophoresis. The absence of DNA on the gel was assumed to be an indicator for complete DNA digestion. The assay was repeated in at least two independent experiments.
Extraction of total RNA from S. oneidensis MR-1.
RNA was extracted from static biofilm cultures incubated in petri dishes. To this end, overnight cultures of S. oneidensis MR-1 strains grown in LM medium were diluted to an OD600 of 0.01 and transferred into petri dishes for static biofilm formation. At appropriate time points, the medium supernatant was collected and the surface-associated cells were collected in 1:10 stop solution (5% [vol/vol] phenol, 95% [vol/vol] ethanol [pH 7.4]) via scraping prior to RNA extraction. Cells from biofilm supernatants and planktonic cultures were harvested by centrifugation (15 min at 4,600 × g and 4°C). Anaerobic cultures for RNA extraction were grown up to an OD600 of 0.5 before harvesting.
Harvested cells were washed with 2 ml AE buffer (20 mM sodium acetate, 1 mM EDTA) and resuspended in 600 μl AE buffer. Subsequently, 900 μl of hot phenol (preheated to 60°C in a water bath) and 10 μl of 25% (wt/vol) SDS were added to the solution, which was then incubated at 60°C for 10 min with occasional inversion and finally cooled on ice. After centrifugation (10 min, 13,000 × g), the aqueous phase was transferred to a Phase Lock gel tube (light, 2 ml; 5 PRIME GmbH, Hamburg, Germany) and further supplemented with 900 μl 60°C hot phenol and 62.5 μl 2 M sodium acetate solution (pH 5.2) followed by another 10 min of centrifugation (13,000 × g). These steps were repeated without adding sodium acetate until no interphase was visible. A total of 2.5 volumes of ice-cold ethanol (96% [vol/vol]) was used to precipitate the RNA in the samples by incubation at −80°C for at least 2 h. After centrifugation at 4°C for at least 30 min, the RNA was washed with ice-cold ethanol (70% [vol/vol]). After removal of the supernatant, the RNA precipitate was dried at room temperature for approximately 1 h and then dissolved in 100 μl diethyl pyrocarbonate (DEPC)-treated water. Contaminating DNA was removed using the Turbo DNA-free kit (Applied Biosystems, Darmstadt, Germany). RNA quality was determined by agarose gel electrophoresis.
Quantitative RT-PCR (qRT-PCR).
For quantitative reverse transcription-PCR (qRT-PCR), extracted total RNA was applied as a template for random-primed first-strand cDNA synthesis by using Bioscript reverse transcriptase (Bioline, Luckenwalde, Germany) according to the manufacturer's instructions. The cDNA was used as a template for quantitative PCR (real-time 7300 PCR machine; Applied Biosystems, Darmstadt, Germany) by using the Sybr green detection system (Applied Biosystems, Darmstadt, Germany). The signals were standardized to recA, with the cycle threshold (CT) determined automatically by the Real Time 7300 PCR software (Applied Biosystems), and the total number of cycles was set to 40. Samples were assayed in duplicate. The efficiency of each primer pair was determined using four different concentrations of S. oneidensis MR-1 chromosomal DNA (10, 1.0, 0.1, and 0.01 ng liter−1) as a template in quantitative PCRs.
Growth inhibition and killing assays.
Killing assays were performed as previously described (29, 47). Briefly, overnight cultures of wild-type and mutant strains carrying pBBR1-MCS5-TT-Pmot-lux were diluted to an OD600 of 0.05 in LB medium and were grown to an OD600 of 1.9. Luminescence of 180 μl cells was measured using a Tecan Infinite M200 plate reader (Tecan, Switzerland). Subsequently, 20 μl herring sperm DNA dissolved in LB medium was directly added to the wells to yield the appropriate end concentration and mixed well. Subsequently, the luminescence of the cells was monitored over time as a measure of viability. Each measurement was performed in duplicate, and the experiments were repeated at least two times. In addition, cells were removed 30 min after addition of eDNA and diluted in 1:10 steps, and 10 μl of each deletion was spotted onto an LB plate.
RESULTS
Identification of two extracellular endonucleases in S. oneidensis MR-1.
Previous bioinformatic analysis of S. oneidensis MR-1 revealed two genes, SO_1066 and SO_1844, which encode putative extracellular endonucleases. Both genes are followed by a Rho-independent transcriptional terminator structure and are likely to be transcribed monocistronically. SO_1844 is 2,847 bp in length and encodes a protein of 948 amino acids with a predicted molecular mass of 101 kDa. The protein has a predicted N-terminal signal sequence and is assumed to be transported in a Sec-dependent manner. Consistently, SO_1844 has so far only been identified in S. oneidensis MR-1 culture supernatants (53). SO_1066 is 2,616 bp in length, and the deduced protein of 871 amino acids has a predicted molecular mass of 93.7 kDa. SO_1066 has an N-terminal signal sequence and, in addition, a putative transmembrane domain at the C terminus which could function as a membrane anchor. In accordance with the predicted localization, SO_1066 was found to be associated with the cell envelope in S. oneidensis MR-1 (6, 61). Thus, we will refer to the proteins encoded by SO_1066 and SO_1844 as ExeM (extracellular endonuclease, membrane associated) and ExeS (extracellular endonuclease, secreted), respectively.
To determine whether ExeM and ExeS are active nucleases, we performed DNA degradation assays with S. oneidensis strains lacking one or both enzymes. To this end, we introduced in-frame deletions in the corresponding genes resulting in single mutants (ΔexeM and ΔexeS) and a double mutant (ΔexeM ΔexeS). We then determined the nuclease activity of filter-sterilized supernatants derived from exponentially growing cultures of the appropriate strains in LB medium (Fig. 1). In the supernatant of a wild-type culture, degradation of a defined PCR-derived DNA fragment of 1,006 bp was observed after 15 min, and degradation was completed after 45 min, while no nuclease activity was observed in plain LB medium. In contrast to the wild type, nuclease activity was significantly decreased in the supernatant of the mutant strain lacking ExeS. DNA degradation was visible after 30 min and was completed after 75 min. A mutant lacking ExeM exhibited even less nuclease activity, and nondegraded DNA fragments of the original size were still present in the supernatant after 75 min of incubation. The DNA degradation pattern of the double mutant equaled that of the ΔexeM mutant. Degradation of a larger 3,140-bp DNA fragment occurred significantly faster (Fig. 1); however, the relative phenotype of the mutants remained consistent. When a DNA fragment was directly added to cultures of the wild-type and mutant strains, a degradation pattern similar to that of the corresponding supernatants occurred (see Fig. S1 in the supplemental material). In a parallel analysis, we also determined the nuclease activity of the wild type and the nuclease mutants in mineral medium. The DNA degradation patterns equaled those obtained in LB medium (data not shown). Together, the results indicated that both ExeM and ExeS are active nucleases and that ExeM, which is predicted to be membrane associated, is also present in cell-free culture supernatants. Since significant DNA degradation occurs in the absence of ExeS and ExeM, S. oneidensis MR-1 likely possesses one or more additional proteins with nucleolytic activity.
Fig. 1.
DNA degradation by ExeM and ExeS in medium supernatants. Filter-sterilized supernatants of the S. oneidensis MR-1 wild-type strain and nuclease mutants were mixed with PCR-amplified Shewanella 1,006-bp (A) and 3,140-bp (B) DNA fragments. At the indicated times, aliquots of the samples were removed and analyzed by agarose gel electrophoresis. In the control lane, the PCR fragment was added to plain LB medium.
ExeM contributes to utilization of eDNA as the sole source of phosphorus.
Having established that ExeM and ExeS are active nucleases, we determined whether both enzymes are required to utilize DNA as the source of phosphorus. To this end, we tested growth of the wild type and the corresponding mutants (ΔexeM, ΔexeS, and ΔexeM ΔexeS) in mineral medium supplemented with 0.5 g liter−1 (0.05%) DNA in the absence of alternative phosphorus sources (Fig. 2). By quantitative RT-PCR (qRT-PCR), we determined that both exeM and exeS are highly upregulated under these conditions (see Fig. 5A). The wild type grew with DNA as the sole source of phosphorus; however, a significantly prolonged lag phase was observed (∼24 h). Notably, the exeS mutant grew comparably well with DNA, like the wild type, strongly indicating that ExeS is not required for utilization of eDNA. In contrast, the ΔexeM mutant had a significantly reduced growth rate with DNA as the phosphate source, and accordingly, the ΔexeM ΔexeS double deletion mutant exhibited a ΔexeM growth phenotype. Growth on DNA as the phosphate source was not totally abolished in these mutants. In contrast to phosphate limitation, almost no growth occurred with wild-type and mutant strains when eDNA was the sole source of nitrogen or carbon (data not shown). Based on these results, we concluded that ExeM contributes to utilizing eDNA as source of phosphate. As it was previously established that ExeM and ExeS are not the only proteins with nucleolytic activity in the supernatant, another nuclease might be involved in this process. In contrast, ExeS appears not to be required to use DNA as a nutrient in growing cultures and probably has a different role in S. oneidensis MR-1.
Fig. 2.
Aerobic growth of S. oneidensis MR-1 strains with DNA as the sole source of phosphorus. Growth of the wild-type (WT; circles) strain, the ΔexeM (triangles) and ΔexeS (squares) single mutant strains, and the ΔexeM ΔexeS (diamonds) double-mutant strain was followed for 60 h in mineral medium supplemented with either 0.86 mM NaH2PO4 (dashed lines), salmon sperm DNA (0.5 g liter−1; solid lines), or no source of phosphorus (dotted lines). The error bars represent the standard deviation.
Fig. 5.
Expression of exeM and exeS genes in S. oneidensis MR-1. (A) Transcriptional levels of exeM and exeS during exponential growth on DNA as the sole source of phosphate as determined by qRT-PCR. The bars display exeM and exeS expression during growth on DNA relative to expression levels during growth on NaH2PO4. (B) Differences in exeM and exeS transcriptional levels of surface-attached cells compared to those of planktonic cells during static biofilm formation. The transcription levels of exeM and exeS were determined by qRT-PCR 0.25, 1, and 24 h after attachment. Displayed are the transcription levels of the nuclease genes in surface-attached cells relative to transcription levels of cells in the supernatant. (C) Transcriptional levels of exeM and exeS under anaerobic conditions relative to aerobic conditions in planktonic cultures as quantified by qRT-PCR. All values are means of three replicates. Error bars display the standard deviations.
ExeM and ExeS do not significantly contribute to the tolerance toward elevated eDNA concentrations under planktonic conditions.
Since the matrix of S. oneidensis MR-1 biofilms was demonstrated to contain significant amounts of eDNA, we determined whether ExeM and ExeS could alleviate potential toxic effects of DNA. To this end, we constructed wild-type and mutant strains constitutively expressing the lux operon of Photorabdus luminescens and recorded the loss of luminescence by the S. oneidensis wild type and nuclease mutants upon addition of DNA to exponentially growing cultures. Under these conditions, significant inactivation of the wild type was observed upon addition of ≥1% DNA to the growth medium. Almost instant cell death occurred upon addition of 2% DNA (Fig. 3A), and the complete loss of viability due to cell lysis was confirmed by plating and microscopic observation (data not shown). Notably, we never observed a significant difference in the detrimental effect of DNA addition to cultures of the wild type or strains lacking exeM, exeS, or both at any of the concentrations tested (Fig. 3B). When sublethal concentrations of DNA were added to growing cultures, the growth rate was significantly decreased for a period of time before normal growth resumed. However, also under those conditions, no difference in growth rates occurred between the wild-type and mutant strains lacking ExeM, ExeS, or both (Fig. 3C). The analysis of exeM and exeS transcriptional levels by qRT-PCR 30 min after addition of eDNA revealed that expression of neither gene was significantly induced by the presence of DNA (data not shown). These results demonstrated that S. oneidensis MR-1 is susceptible to elevated levels of eDNA; however, under planktonic conditions, ExeM and ExeS do not protect the cells from these detrimental effects.
Fig. 3.
Detrimental effect of eDNA. (A) Exponentially growing LB cultures of S. oneidensis MR-1 harboring pBBR1-MCS5-TT-Pmot-RBS-lux were supplemented with DNA in the concentrations indicated, and the luminescence (counts per second [cps]) was measured over time. The data presented display luminescence relative to the untreated control. (B) Exponentially growing LB cultures of the S. oneidensis MR-1 wild-type strain and nuclease mutants harboring pBBR1-MCS5-TT-Pmot-RBS-lux were supplemented with eDNA at a concentration of 1.5%, and luminescence was measured over time. (C) Growth of S. oneidensis MR-1 wild type and ΔexeM ΔexeS mutant cultures in LB medium in the presence and absence of 0.25% DNA. All error bars display the standard deviation.
ExeM and ExeS affect biofilm formation.
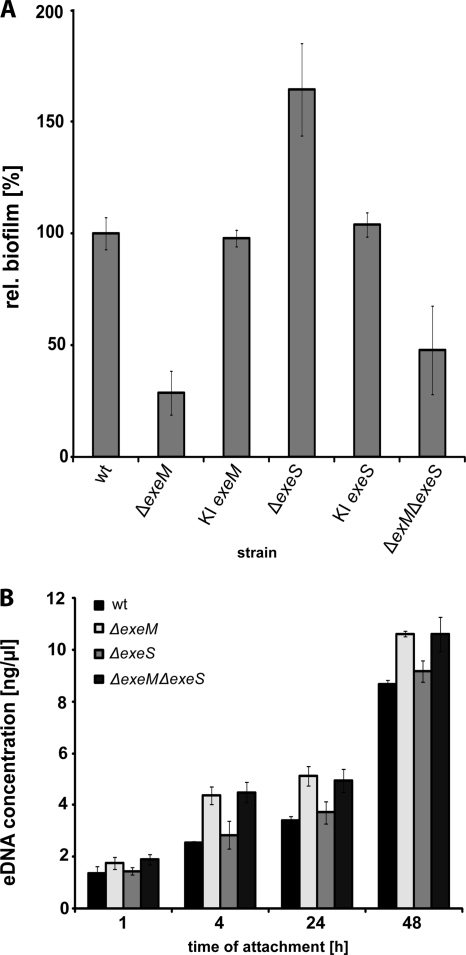
Since eDNA is a major factor in mediating cell-cell and cell-surface interactions in S. oneidensis MR-1, we next analyzed whether ExeM and/or ExeS contribute to biofilm formation of this species. Biofilm formation of the wild-type and mutant strains was characterized in static microtiter plate assays and in a hydrodynamic flow chamber system. When grown in microtiter plates (Fig. 4A), a ΔexeS mutant accumulated substantially more (164%) surface-attached biomass than the wild type. In contrast, the ΔexeM mutant displayed decreased biofilm formation (28% compared to that of the wild type). The ΔexeM ΔexeS double mutant formed less (48%) biofilm than wild-type cells. We also determined whether the absence of ExeM and ExeS results in a lower concentration of eDNA in the supernatant of static biofilm cultures (Fig. 4). After 4, 24, and 48 h of incubation, the amount of eDNA in the supernatants of the exeM mutant and the double mutant was significantly higher (∼15 to 30%), respectively, than that in supernatants obtained from the wild type and ΔexeS mutants, indicating the nucleolytic activity of ExeM. After 48 h of incubation, the concentration of eDNA in the supernatant reached up to 10 ng μl−1.
Fig. 4.
Role of ExeM and ExeS in S. oneidensis MR-1 static biofilm formation. (A) Biofilm formation of the S. oneidensis MR-1 wild-type strain and ΔexeM, ΔexeS, and ΔexeM ΔexeS nuclease mutant strains, as well as the corresponding complemented mutants (KI-) in microtiter plates. The strains were incubated in LM medium for 24 h. The surface-associated biomass was quantified using a crystal violet assay. The values are means of three replicates, and the standard deviations are displayed by error bars. (B) Amount of eDNA in the supernatant of static biofilm cultures of the wild type and the indicated nuclease mutants 1, 4, 24, and 48 h after surface attachment. The error bars display the standard deviation.
Due to the pronounced biofilm phenotype of exeM and exeS mutants in static biofilm cultures, we hypothesized that expression of exeM and/or exeS might be induced under these conditions. To analyze the regulation patterns of the two genes during biofilm formation under static conditions, we harvested cells from the supernatant and surface-associated cells at different time points during biofilm formation. RNA was prepared from the cells, and the exeM and exeS transcript levels were quantified by qRT-PCR. Both genes displayed highly increased transcript levels in surface-associated cells compared to cells from the supernatant already early during biofilm formation. One hour after surface attachment, exeM was upregulated by a factor of 7.9 and exeS by a factor of ∼20. Similar values were determined after 24 h of biofilm development (Fig. 5B). Thus, growth under static biofilm conditions strongly induces expression of exeM and exeS and might indicate that the cells undergo phosphate starvation under such conditions. Previous studies of S. oneidensis MR-1 biofilms have identified molecular oxygen levels as a major signal involved in biofilm formation (66). Based on these studies, we hypothesized that, in addition to phosphate limitation, expression of exeM and exeS might also be directly or indirectly controlled by oxygen levels. During exponential anaerobic growth, exeM was upregulated by a factor of about 2.9 (Fig. 5C). In contrast, expression of exeS decreased by a factor of 4.4 during anaerobic growth. Thus, oxygen levels might contribute to the upregulation of exeM during biofilm formation, while the drastically increased expression levels of exeS are independent from oxygen levels.
To analyze the biofilm formation in the flow chamber system via confocal laser scanning microscopy (CLSM), we constructed corresponding mutant strains constitutively expressing gfp. Under hydrodynamic conditions in the flow chamber, no visible phenotypic change occurred in the ΔexeS mutant (Fig. 6). Similar to the wild type, the mutant covered the surface after 24 h (94.73% ± 2.0% and 91.72% ± 7.0% surface coverage for the wild type and ΔexeS mutant, respectively), and distinct three-dimensional structures were formed after 48 h. However, under these conditions, the ΔexeM mutant had a distinct phenotype in biofilm formation. While the initial attachment was unaffected, ΔexeM mutants were not able to cover the surface after 24 h (44.95% ± 6.33% coverage versus 94.73% ± 2.0% for the wild type), but instead formed smaller, densely packed microcolonies. After 48 h, towering three-dimensional structures were formed. Notably, few cells of the ΔexeM mutant were observed that were not associated with the biofilm. As in the static microtiter plate system, the biofilm phenotype of the ΔexeM ΔexeS double mutant resembled that of the ΔexeM single mutant. To visualize the amount of eDNA in biofilms grown under hydrodynamic conditions, we applied DDAO staining (Fig. 6). eDNA was present in both wild-type and ΔexeS mutant biofilms in similar amounts to those previously described (22). However, a striking difference occurred in ΔexeM and ΔexeM ΔexeS mutant biofilms, which were covered by a thick layer of eDNA. Together, the results demonstrated that ExeM and ExeS affect biofilm formation. While a significant phenotype of a ΔexeS mutant only occurred under static conditions, the loss of ExeM results in altered biofilm formation under both static and hydrodynamic conditions, and large amounts of eDNA accumulated in the matrix.
Fig. 6.
Role of ExeM and ExeS in S. oneidensis MR-1 biofilm formation under hydrodynamic conditions. Biofilm formation of GFP-tagged wild-type and nuclease mutant strains under hydrodynamic conditions was monitored by CLSM after 1, 4, 24, and 48 h of attachment. After 48 h of incubation, eDNA was visualized by DDAO staining (red). The lateral edge of each micrograph equals 250 μm.
DISCUSSION
Earlier studies have identified substantial concentrations of eDNA in marine sediments (13, 14), an environment from which numerous Shewanella species have been isolated (26). The eDNA is assumed to provide a major source of phosphorus in marine sediments, and significant nuclease activities have been determined in these environments (11, 12). Accordingly, a recent study demonstrated that Shewanella species are capable of eDNA degradation and that eDNA is a particularly valuable source of phosphorus (53). Significant nuclease activity occurred in Shewanella cultures, indicating a role for extracellular nucleases in concert with phosphatases in exploiting DNA as a nutrient, as has recently been demonstrated for Pseudomonas aeruginosa (48). Two putative extracellular endonucleases were previously identified in Shewanella oneidensis MR-1 (53), and both are present in all Shewanella species sequenced so far. Our study provides evidence that these two nucleases, now designated ExeM and ExeS, are contributing to eDNA degradation. Both are highly upregulated under phosphate-limited conditions, as has been observed for other species, such as Corynebacterium glutamicum (31), Bacillus licheniformis (30, 69), and P. aeruginosa (48). However, the ability of S. oneidensis to utilize DNA as a source of phosphate was not abolished in the absence fof ExeM and not affected after loss of ExeS under all conditions tested, which is likely due to the fact that S. oneidensis produces one or more additional nucleases. This remarkable abundance of nucleolytic activity underlines the significance of eDNA for Shewanella species. In addition, our study strongly suggests that ExeS and, in particular, ExeM have functions beyond exploiting DNA as a nutrient.
ExeS is assumed to be transported via the cytoplasmic membrane, and the protein has so far only been identified in cell-free supernatants (53). In contrast, two studies addressing the membrane proteome of S. oneidensis MR-1 have provided evidence that ExeM is associated with the cell envelope. However, it is not yet clear whether ExeM is an outer membrane protein (61) or whether the protein is localized to the cytoplasmic membrane (6). The latter would be rather inconsistent with a role in degradation of an extracellular compound such as eDNA unless it is first transported through the outer membrane. Although ExeM has been proposed to be associated with the cell envelope, our study indicates that ExeM-dependent nuclease activity occurs in the cell-free supernatant. If ExeM is located on the outside of the outer membrane, the protein might be occasionally released into the supernatant. In addition, we have shown that S. oneidensis MR-1 undergoes cell lysis (22), and ExeM might also be transported by or attached to vesicles produced by Shewanella (23). Notably, DNA uptake systems involved in natural transformation are thought to involve membrane-associated nuclease activity (7, 8). In addition, a recent study provides evidence that an extracellular nuclease, Dns, affects natural transformation in Vibrio cholerae (3). This nuclease is expressed in a cell-density-dependent fashion and prevents transformation by degradation of eDNA at low cell densities. ExeM and/or ExeS may have a similar role in S. oneidensis MR-1; however, it remains to be demonstrated whether this species is naturally competent and whether the two nucleases are involved in or affect DNA uptake. Bioinformatic analysis readily identifies numerous genes/gene clusters that have most likely been acquired by S. oneidensis MR-1 through lateral gene transfer, such as a second set of flagellar stators, motAB (52). Moreover, this species harbors an integron integrase system (16), indicating active uptake of eDNA in S. oneidensis MR-1.
While ExeM is involved in utilizing eDNA as a source of phosphate, our study provided evidence that both ExeM and ExeS contribute to biofilm formation of S. oneidensis MR-1. The pronounced biofilm phenotype and the significant upregulation of the nucleases during surface-associated growth strongly suggest functional roles of ExeM and ExeS in the degradation of eDNA in the biofilm matrix. A recent study of Pseudomonas aeruginosa and other species has demonstrated that high concentrations of eDNA are lethal to bacterial cells (47). This effect is thought to be due to the chelating properties of DNA that result in a cation-limited environment leading to the perturbation of inner and outer membranes. Here, we provide evidence that S. oneidensis is susceptible to detrimental effects of eDNA in a similar fashion. Our previous studies indicated that, under hydrodynamic conditions, eDNA is not evenly distributed but is particularly abundant in the densely packed three-dimensional structures occurring after 24 h (22). We have shown that, under planktonic conditions, ExeM and ExeS do not provide short-term protection from toxic effects of eDNA. This is not surprising since smaller fragments of DNA probably can also provide chelating functions until being completely degraded. However, it is conceivable that ExeM and ExeS might help to counteract a gradual accumulation of eDNA in the matrix before it reaches inhibitory levels.
An important but not well understood stage occurring during biofilm formation is the detachment of cells from the community, which enables bacteria to leave under unfavorable conditions and contributes to biological dispersal and survival of the cells (32, 34). Previous studies on S. oneidensis MR-1 have shown that cells constantly detach from biofilms and that rapid detachment can be induced by a rapid drop in the molecular oxygen level (64, 66). To actively leave the biofilm, cells are required to modify the matrix that keeps them in the community. A number of matrix-degrading enzymes have been described for various species that contribute to detachment. Some target protein compounds of the matrix (4, 21, 39), while others degrade the polysaccharide components (5, 33). A recent study has shown that the staphylococcal thermonuclease (nuc) affects biofilm formation of Staphylococcus aureus (41). In the absence of nuc, S. aureus forms significantly thicker biofilms, indicating that this nuclease promotes biofilm dispersal. Similar observations were made for Neisseria gonorrhoeae (58). Similarly, with respect to the important role of eDNA as a structural matrix component in biofilms of S. oneidensis (22), we hypothesize that ExeM and ExeS are involved in detachment and dispersal of Shewanella cells from biofilms. ExeS is predicted to be released into the supernatant to degrade eDNA within the surface-associated community. Thus, a loss of ExeS would be expected to result in a more resistant biofilm matrix containing more biomass, as observed in the static microtiter plate assay. However, under hydrodynamic conditions, ExeS is probably removed by the medium flow before a concentration is reached that affects matrix formation. Accordingly, we did not observe a phenotype of exeS mutants in flow chamber-grown biofilms of S. oneidensis MR-1. In contrast to ExeS, ExeM is thought to be associated with the cell envelope. Hence, this nuclease would not be removed under hydrodynamic conditions and might degrade DNA in close proximity to the cell. Biofilms formed by exeM mutants in the hydrodynamic flow chamber exhibit tight cell-cell interactions and are covered by a thick layer of eDNA. In contrast, in the static microtiter plate assay this mutant displays less surface-associated biomass. A previous study has demonstrated that tight cell-cell interactions due to hyperpiliation cause a similar phenotype of S. oneidensis MR-1 (65). This finding indicates that detachment is an important prerequisite for normal biofilm formation of this species and suggests that a role of ExeM is to degrade eDNA to prevent the formation of too tight cell-cell interactions that would prevent detachment.
exeM and exeS were upregulated upon phosphate starvation in S. oneidensis MR-1. Interestingly, there are reports from other species that directly link the availability of phosphorus to biofilm formation. In Pseudomonas fluorescens Pf0-1, a low level of phosphate leads to activation of the Pho regulon and increased production of RapA (46). RapA is a diesterase that degrades the signaling molecule c-di-GMP. Generally, a low level of c-di-GMP is associated with a transition from a sessile to planktonic lifestyle in bacteria (28). More specifically, the degradation of c-di-GMP by RapA is thought to inhibit the secretion of a large adhesin, LapA, which is required for P. fluorescens biofilm formation (46). Similarly, activation of the Pho system positively regulates motility and decreases biofilm formation in Vibrio cholerae (54). Biofilm formation is also severely impaired in Proteus mirabilis mutants lacking the high-affinity transporter Pst (50). Thus, for a number of species, phosphate limitation appears to favor the transition from the biofilm to the planktonic lifestyle. Accordingly, production of a nuclease that degrades the matrix component eDNA under those conditions not only might improve the acquisition of nutrients but also would facilitate detachment from the community. It will be interesting to determine whether Shewanella biofilms also depend on phosphate in a similar fashion and whether growth in biofilms might lead to rapid depletion in phosphate. Thus, further studies will address how regulation of ExeM and ExeS occurs and whether they are specifically produced or activated, e.g., during induced biofilm detachment.
Taken together, we have demonstrated here that ExeM and ExeS are required for normal biofilm formation of S. oneidensis MR-1. We hypothesize that this effect is due to degradation of eDNA as a matrix component, enabling detachment from biofilms. Since Shewanella species are not confined to extreme habitats, it is likely that they commonly occur in mixed-species rather than in monospecies communities (35, 40). In contrast to many other extracellular polymeric substance (EPS) compounds, eDNA represents a universal matrix component and is required for interaction of mixed-species biofilms (15). Thus, the production of extracellular nucleases might be an effective means to enable detachment from such multispecies biofilms. Given the wide distribution of eDNA as a structural component of bacterial biofilms, we expect that similar systems are required for biofilm formation and dispersal of other species.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Penelope Higgs, Martin Thanbichler, and Chris van der Does for critical reading of the manuscript.
The study was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG; TH831/3-1) and the Max-Planck-Gesellschaft.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Allesen-Holm M., et al. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114–1128 [DOI] [PubMed] [Google Scholar]
- 2. Bagge D., Hjelm M., Johansen C., Huber I., Gram L. 2001. Shewanella putrefaciens adhesion and biofilm formation on food processing surfaces. Appl. Environ. Microbiol. 67:2319–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blokesch M., Schoolnik G. K. 2008. The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae. J. Bacteriol. 190:7232–7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boles B. R., Horswill A. R. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyd A., Chakrabarty A. M. 1994. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 60:2355–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown R. N., Romine M. F., Schepmoes A. A., Smith R. D., Lipton M. S. 2010. Mapping the subcellular proteome of Shewanella oneidensis MR-1 using sarkosyl-based fractionation and LC-MS/MS protein identification. J. Proteome Res. 9:4454–4463 [DOI] [PubMed] [Google Scholar]
- 7. Burton B., Dubnau D. 2010. Membrane-associated DNA transport machines. Cold Spring Harbor Perspect. Biol. 2:a000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen I., Christie P. J., Dubnau D. 2005. The ins and outs of DNA transfer in bacteria. Science 310:1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choi K. H., et al. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443–448 [DOI] [PubMed] [Google Scholar]
- 10. Das A., Caccavo F., Jr 2000. Dissimilatory Fe(III) oxide reduction by Shewanella alga BrY requires adhesion. Curr. Microbiol. 40:344–347 [DOI] [PubMed] [Google Scholar]
- 11. Dell'Anno A., Corinaldesi C. 2004. Degradation and turnover of extracellular DNA in marine sediments: ecological and methodological considerations. Appl. Environ. Microbiol. 70:4384–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dell'Anno A., Danovaro R. 2005. Extracellular DNA plays a key role in deep-sea ecosystem functioning. Science 309:2179. [DOI] [PubMed] [Google Scholar]
- 13. Dell'Anno A., Fabiano M., Duineveld G. C. A., Kok A., Danovaro R. 1998. Nucleic acid (DNA, RNA) quantification and RNA/DNA ratio determination in marine sediments: comparison of spectrophotometric, fluorometric, and high-performance liquid chromatography methods and estimation of detrital DNA. Appl. Environ. Microbiol. 64:3238–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dell'Anno A., Fabiano M., Mei M. L., Danovaro R. 1999. Pelagic-benthic coupling of nucleic acids in an abyssal location of the northeastern Atlantic Ocean. Appl. Environ. Microbiol. 65:4451–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dominiak D. M., Nielsen J. L., Nielsen P. H. 2011. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ. Microbiol. 13:710–721 [DOI] [PubMed] [Google Scholar]
- 16. Drouin F., Melancon J., Roy P. H. 2002. The IntI-like tyrosine recombinase of Shewanella oneidensis is active as an integron integrase. J. Bacteriol. 184:1811–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Finkel S. E., Kolter R. 2001. DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol. 183:6288–6293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flemming H. C., Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633 [DOI] [PubMed] [Google Scholar]
- 19. Frølund B., Palmgren R., Keiding K., Nielsen P. H. 1996. Extraction of extracellular polymers from activated sludge using a cation exchange resin. Water Res. 30:1749–1758 [Google Scholar]
- 20. Gescher J. S., Cordova C. D., Spormann A. M. 2008. Dissimilatory iron reduction in Escherichia coli: identification of CymA of Shewanella oneidensis and NapC of Escherichia coli as ferric reductases. Mol. Microbiol. 68:706–719 [DOI] [PubMed] [Google Scholar]
- 21. Gjermansen M., Nilsson M., Yang L., Tolker-Nielsen T. 2010. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol. Microbiol. 75:815–826 [DOI] [PubMed] [Google Scholar]
- 22. Gödeke J., Paul K., Lassak J., Thormann K. M. 2011. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J. 5:613–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gorby Y., et al. 2008. Redox-reactive membrane vesicles produced by Shewanella. Geobiology 6:232–421 [DOI] [PubMed] [Google Scholar]
- 24. Gorby Y. A., et al. 2006. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. U. S. A. 103:11358–11363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hamilton H. L., Dominguez N. M., Schwartz K. J., Hackett K. T., Dillard J. P. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 55:1704–1721 [DOI] [PubMed] [Google Scholar]
- 26. Hau H. H., Gralnick J. A. 2007. Ecology and biotechnology of the genus Shewanella. Annu. Rev. Microbiol. 61:237–258 [DOI] [PubMed] [Google Scholar]
- 27. Heidelberg J. F., et al. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118–1123 [DOI] [PubMed] [Google Scholar]
- 28. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 29. Hilpert K., Hancock R. E. 2007. Use of luminescent bacteria for rapid screening and characterization of short cationic antimicrobial peptides synthesized on cellulose using peptide array technology. Nat. Protoc. 2:1652–1660 [DOI] [PubMed] [Google Scholar]
- 30. Hoi le T., et al. 2006. The phosphate-starvation response of Bacillus licheniformis. Proteomics 6:3582–3601 [DOI] [PubMed] [Google Scholar]
- 31. Ishige T., Krause M., Bott M., Wendisch V. F., Sahm H. 2003. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 185:4519–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaplan J. B. 2010. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 89:205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaplan J. B., Ragunath C., Ramasubbu N., Fine D. H. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J. Bacteriol. 185:4693–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karatan E., Watnick P. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73:310–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kobayashi T., et al. 2008. Phylogenetic and enzymatic diversity of deep subseafloor aerobic microorganisms in organics- and methane-rich sediments off Shimokita Peninsula. Extremophiles 12:519–527 [DOI] [PubMed] [Google Scholar]
- 36. Kovach M. E., et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 37. Lappann M., et al. 2010. A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis. Mol. Microbiol. 75:1355–1371 [DOI] [PubMed] [Google Scholar]
- 38. Lassak J., Henche A. L., Binnenkade L., Thormann K. M. 2010. ArcS, the cognate sensor kinase in an atypical Arc system of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 76:3263–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee S. F., Li Y. H., Bowden G. H. 1996. Detachment of Streptococcus mutans biofilm cells by an endogenous enzymatic activity. Infect. Immun. 64:1035–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu W., et al. 2010. Geochip-based functional gene analysis of anodophilic communities in microbial electrolysis cells under different operational modes. Environ. Sci. Technol. 44:7729–7735 [DOI] [PubMed] [Google Scholar]
- 41. Mann E. E., et al. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4:e5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McLean J. S., et al. 2008. Investigations of structure and metabolism within Shewanella oneidensis MR-1 biofilms. J. Microbiol. Methods 74:47–56 [DOI] [PubMed] [Google Scholar]
- 43. McLean J. S., et al. 2010. Quantification of electron transfer rates to a solid phase electron acceptor through the stages of biofilm formation from single cells to multicellular communities. Environ. Sci. Technol. 44:2721–2727 [DOI] [PubMed] [Google Scholar]
- 44. Miller V. L., Mekalanos J. J. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Molin S., Tolker-Nielsen T. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14:255–261 [DOI] [PubMed] [Google Scholar]
- 46. Monds R. D., Newell P. D., Gross R. H., O'Toole G. A. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63:656–679 [DOI] [PubMed] [Google Scholar]
- 47. Mulcahy H., Charron-Mazenod L., Lewenza S. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 4:e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mulcahy H., Charron-Mazenod L., Lewenza S. 2010. Pseudomonas aeruginosa produces an extracellular DNase that is required for utilization of DNA as a nutrient source. Environ. Microbiol. 12:1621–1629 [DOI] [PubMed] [Google Scholar]
- 49. Niemeyer J., Gessler F. 2002. Determination of free DNA in soils. J. Plant Nutr. Soil Sci. 165:121–124 [Google Scholar]
- 50. O'May G. A., et al. 2009. The high-affinity phosphate transporter Pst in Proteus mirabilis HI4320 and its importance in biofilm formation. Microbiology 155:1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Palmgren R., Nielsen P. H. 1996. Accumulation of DNA in the exopolymeric matrix of activated sludge and bacterial cultures. Water Sci. Technol. 34:233–240 [Google Scholar]
- 52. Paulick A., et al. 2009. Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol. Microbiol. 71:836–850 [DOI] [PubMed] [Google Scholar]
- 53. Pinchuk G. E., et al. 2008. Utilization of DNA as a sole source of phosphorus, carbon, and energy by Shewanella spp.: ecological and physiological implications for dissimilatory metal reduction. Appl. Environ. Microbiol. 74:1198–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pratt J. T., McDonough E., Camilli A. 2009. PhoB regulates motility, biofilms, and cyclic di-GMP in Vibrio cholerae. J. Bacteriol. 191:6632–6642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qin Z., et al. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153:2083–2092 [DOI] [PubMed] [Google Scholar]
- 56. Rice K. C., et al. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 104:8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sambrook K., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 58. Steichen C. T., Cho C., Shao J. Q., Apicella M. A. 2011. The Neisseria gonorrhoeae biofilm matrix contains DNA and an endogenous nuclease controls its incorporation. Infect. Immun. 79:1504–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Steinberger R. E., Holden P. A. 2005. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ. Microbiol. 71:5404–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stoodley P., Sauer K., Davies D. G., Costerton J. W. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187–209 [DOI] [PubMed] [Google Scholar]
- 61. Tang X., et al. 2007. Profiling the membrane proteome of Shewanella oneidensis MR-1 with new affinity labeling probes. J. Proteome Res. 6:724–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Teal T. K., Lies D. P., Wold B. J., Newman D. K. 2006. Spatiometabolic stratification of Shewanella oneidensis biofilms. Appl. Environ. Microbiol. 72:7324–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Thomas V. C., Thurlow L. R., Boyle D., Hancock L. E. 2008. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 190:5690–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thormann K. M., et al. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 188:2681–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Thormann K. M., Saville R. M., Shukla S., Pelletier D. A., Spormann A. M. 2004. Initial phases of biofilm formation in Shewanella oneidensis MR-1. J. Bacteriol. 186:8096–8104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Thormann K. M., Saville R. M., Shukla S., Spormann A. M. 2005. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 187:1014–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Venkateswaran K., et al. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 2:705–724 [DOI] [PubMed] [Google Scholar]
- 68. Vlassov V. V., Laktionov P. P., Rykova E. Y. 2007. Extracellular nucleic acids. Bioessays 29:654–667 [DOI] [PubMed] [Google Scholar]
- 69. Voigt B., et al. 2006. The extracellular proteome of Bacillus licheniformis grown in different media and under different nutrient starvation conditions. Proteomics 6:268–281 [DOI] [PubMed] [Google Scholar]
- 70. Whitchurch C. B., Tolker-Nielsen T., Ragas P. C., Mattick J. S. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.