Abstract
Dehalococcoides spp. are an industrially relevant group of Chloroflexi bacteria capable of reductively dechlorinating contaminants in groundwater environments. Existing Dehalococcoides genomes revealed a high level of sequence identity within this group, including 98 to 100% 16S rRNA sequence identity between strains with diverse substrate specificities. Common molecular techniques for identification of microbial populations are often not applicable for distinguishing Dehalococcoides strains. Here we describe an oligonucleotide microarray probe set designed based on clustered Dehalococcoides genes from five different sources (strain DET195, CBDB1, BAV1, and VS genomes and the KB-1 metagenome). This “pangenome” probe set provides coverage of core Dehalococcoides genes as well as strain-specific genes while optimizing the potential for hybridization to closely related, previously unknown Dehalococcoides strains. The pangenome probe set was compared to probe sets designed independently for each of the five Dehalococcoides strains. The pangenome probe set demonstrated better predictability and higher detection of Dehalococcoides genes than strain-specific probe sets on nontarget strains with <99% average nucleotide identity. An in silico analysis of the expected probe hybridization against the recently released Dehalococcoides strain GT genome and additional KB-1 metagenome sequence data indicated that the pangenome probe set performs more robustly than the combined strain-specific probe sets in the detection of genes not included in the original design. The pangenome probe set represents a highly specific, universal tool for the detection and characterization of Dehalococcoides from contaminated sites. It has the potential to become a common platform for Dehalococcoides-focused research, allowing meaningful comparisons between microarray experiments regardless of the strain examined.
INTRODUCTION
The genus Dehalococcoides contains obligate anaerobes capable of reductively dechlorinating a variety of common groundwater contaminants (1, 12, 18, 31). The utility of Dehalococcoides in bioremediation of chlorinated-solvent-contaminated sites has lead to the development of Dehalococcoides-containing cultures utilized as industrial tools (12, 29), methods for identifying and tracking Dehalococcoides growth and dechlorination (26, 45, 54), and methods for identifying the novel metabolism associated with the dechlorination reaction (2, 27, 30, 40). The Dehalococcoides appear to be globally distributed; they have been isolated from sites in North America (e.g., strains DET195 [37], MB [7], and FL2 [19]) and Europe (e.g., strain CBDB1 [3]) and identified at many contaminated sites and more remote environments (1, 15, 22). Genome sequences currently exist for five strains of Dehalococcoides, strain DET195 (48), strain CBDB1 (31), strain BAV1 (38), strain VS (38), and strain GT (http://genome.jgi-psf.org/deh_g/deh_g.home.html). Several more genome sequences are in progress from mixed-culture metagenomes, including from the KB-1 enrichment consortium, which contains at least two Dehalococcoides strains (12). Comparative genomics has revealed that the Dehalococcoides spp. share a core genome having high synteny and conservation of nucleotide identity, with two regions of high plasticity (HPRs) where large genomic rearrangements and gene variation occur (38).
The Dehalococcoides spp. exhibit a wide range of substrate specificities on halogenated compounds (12, 24, 37, 52). The reductive dehalogenases are the enzyme family capable of catalyzing the removal of chlorine ions from a substrate (23, 47). From sequenced genomes, it is clear that each strain contains a unique subset of reductive dehalogenases, explaining their different substrate specificities (38).
The close conservation of the Dehalococcoides genomes outside the HPRs is highlighted by their 16S rRNA identities, which range from 98 to 100% identical for all known strains (Fig. 1), a lack of variation discordant with the diverse substrate specificities exhibited by the different strains. Quantitative PCR of Dehalococcoides 16S is a common tool used within consulting companies for identification of Dehalococcoides at sites (41, 46), but it is not useful for strain examinations or separate tracking of native and augmented organisms. Quantitative PCR assays for several of the reductive dehalogenases with known functions have been developed for assessing a site's potential for dechlorination (8, 45), but these generate information for, at best, a few genes.
Fig. 1.
Maximum likelihood phylogenetic tree from a 16S rRNA gene alignment, including the 5 Dehalococcoides strains involved in probe design experiments, the in silico tester strain, Dehalococcoides strain GT, and the nearest sequenced relative to the Dehalococcoides group, Dehalogenimonas lykanthroporepellens strain BL-DC-9. Numbers on nodes refer to bipartition support from 100 bootstrap replicates. The 16S sequence alignment was generated using the Geneious muscle global alignment plug-in (11, 13), with the phylogeny and bootstrap replicates generated using RAxML version 7.0.3 under the GTR-GAMMA model of sequence evolution (50).
There is an identified need for common tools and techniques to allow better comparisons across Dehalococcoides strains as well as for tools robust for use with pure strains, mixed bacterial enrichment cultures, and environmental samples. In particular, the ability to detect and identify native Dehalococcoides at a contaminated site and to track specific strains of bioaugmented Dehalococcoides during the course of a biodegradation treatment is a recognized need.
Oligonucleotide arrays have been used to examine intraspecies genomic variation for a number of well-characterized genera, including Saccharomyces (17, 60), and for Escherichia coli (57, 58). There are several currently available arrays for examining Dehalococcoides. PhyloChip, a 16S rRNA-based microbial diagnostic microarray (6, 10, 63), allows distinguishing of Dehalococcoides from other genera but does not allow strain differentiation (10). The GeoChip, a functional gene array focused on biogeochemical functions (20, 21, 53), allows identification of key functional genes in different Dehalococcoides strains, along with other geochemically important bacterial processes (20, 21, 44). More specifically for the Dehalococcoides, a whole-genome-tiled microarray exists for strain 195 (25), which has been used to extensively examine that strain's metabolic profile under different treatments (28) and to examine a mixed community containing two different Dehalococcoides strains (56). Recently, an oligonucleotide array was developed covering all genes from four sequenced Dehalococcoides genomes, with cross-hybridization of probes stringently prevented (32). This pangenus array design works well for the four Dehalococcoides genomes it represents but will only provide useful data for unsequenced Dehalococcoides genomes exhibiting a high sequence similarity to the design genomes.
Here we describe and validate an option for Dehalococcoides array probe design that increases the utility of the array for diverging strains. Oligonucleotide probes are designed for groups of orthologous genes, such that the probe hybridizes to conserved regions within the genes and, hence, has a higher likelihood of matching an as-yet-unsequenced ortholog from a novel strain. The program ProDesign (14) implements this approach using gene clusters defined by sequence similarity, generating the best probe set for a given set of clusters while optimizing probe melting temperatures and minimizing cross-hybridization of probes to nontarget genes.
Probes were designed by ProDesign for clusters of conserved genes from the Dehalococcoides spp. for which genomic sequence information was available (strains 195, VS, CBDB1, BAV1, and KB-1). The clusters of orthologous genes were defined such that some probes allow universal detection of Dehalococcoides, while others provide strain-specific differentiation. The functionality of this pangenome probe set was compared to oligonucleotide probe sets designed for the five Dehalococcoides strains individually by the Agilent eArray platform in comparative genomic hybridizations. The comparison between the pangenome probe set (designed by ProDesign) and the strain-specific probe sets (designed by Agilent's eArray) was extended to an in silico examination of the coverage of the genes of Dehalococcoides strain GT, whose genome was not available at the time of probe design. The pangenome probe set provided a higher expected coverage of strain GT's genes than a combined data set of all of the single genome probes, highlighting the advantage of designing probes for clustered homologs.
MATERIALS AND METHODS
KB-1 metagenome sequence.
DNA was extracted from the KB-1 enrichment culture using a cetyltrimethylammonium bromide (CTAB) protocol (59), with volumes scaled up for higher yield as described in the alternate protocol, omitting subsequent cesium chloride gradient centrifugation steps. Clone libraries with 8-kb and 3-kb inserts were created by the Joint Genome Institute (JGI) using their in-house protocols (http://www.jgi.doe.gov/sequencing/protocols), and end sequencing was conducted using an AB13730xl Sanger sequencing machine. The metagenome was sequenced in two stages: an initial 10 Mb of sequence, which was available at the time of the pangenome probe design, and a subsequent final 95-Mb sequence, which is publically available (http://genome.jgi-psf.org/aqukb/aqukb.home.html).
Identification and clustering of the Dehalococcoides genes.
All coding genes from the four available Dehalococcoides genomes (strains 195, VS, CBDB1, and BAV1 [RefSeq accession no. NC_002936.3, NC_013552.1, NC_007356.1, NC_009455.1, respectively]) were combined into a database. Dehalococcoides genes from the KB-1 metagenome were identified by a BLAST search (5) of the initial 10 Mb of the KB-1 metagenome sequence against this database of Dehalococcoides genes. Metagenome sequences whose BLAST hits had E values of ≤1 × 10−5 and percent identities of >90% were aligned with the EMBOSS Water program (49) to generate full-length gene alignments within the longer contig sequences. The 1,146 sequences identified were then translated, and sequences truncated by stop codons were removed, leaving a final set of 933 KB-1 gene sequences. These sequences represent high-confidence Dehalococcoides genes and were included in the panarray probe design to ensure that the probe design was based on the full breadth of available nucleotide diversity within the Dehalococcoides genus.
The final gene set comprised 6,812 sequences. These genes were clustered at the nucleotide level using cd-hit-est (33–35) at sequence identity thresholds of 80 to 95% identity (ID) at 1% intervals, with an alignment coverage control (cd-hit-est with flag-aL) of 80%.
Pangenome probe design.
The program ProDesign/OpSelector (14) was used to generate probes for the clustered Dehalococcoides genes. Clusters were reverse complemented, and the full duplicated set of clusters was used in a separate round of ProDesign to allow multiple probes to be designed to the same cluster. Several parameter conditions were tested, with the final set as follows: seed weight of 12, seed span of 24, probe length of 50 to 60 nucleotides (nt), and a final melting temperature (Tm) of 89.5°C. To prevent nonspecific hybridization, probes were tested against a set of available genomes from soil bacteria and close relatives to organisms present in dechlorinating enrichment cultures, and cross-hybridizing probes were redesigned.
For probe design based on the prediction of hybridization, clustering homologs based on a threshold of sequence identity is preferable to best-reciprocal BLAST match approaches meant to identify and cluster orthologs (as has been used for other Dehalococcoides comparisons [4, 38]). A sequence identity of 95% was chosen as the optimal clustering threshold which maximized the ProDesign probe coverage of the clusters. This resulted in 4,232 clusters, of which 3,857 had at least one probe designed (91.1%). The clustering at 95% gave high relative numbers of core gene clusters (defined here as a cluster containing a sequence from all five genomes, in which the KB-1 data are considered a “genome”) and clusters containing genes from subsets of the five genomes, compared to clusters containing a sequence from a single genome. The use of reverse-complemented sequences in a second round of ProDesign probe design provided additional coverage (i.e., coverage of a cluster not previously having a sense-strand probe designed to it) of 405 clusters, representing 418 genes (6.1% of total genes). Any probe to a reverse-complemented cluster was subsequently returned to an antisense sequence for array printing. The final pangenome probe set coverage statistics are presented in Table 1. The distribution of the five genomes within clusters is depicted as a Venn diagram in Fig. S1 in the supplemental material. The probe sequences are presented in Table S1 in the supplemental material.
Table 1.
Summary of probe sets included in the array design
| Probe set name/genome covered | Design program | No. of target genes | No. of target clustersa | No. of probes | Coverage (%) | No. of probes/targets | Mean probe length (no. of nt) | Mean probe Tm (°C) | No. of probes with x-hyb potentialb |
|---|---|---|---|---|---|---|---|---|---|
| PanDhc (all 5) | ProDesign with clustering | 6,812 | 4,232 | 5,514 (to 5,410 genes) | 79.4 | 1 or 2 | 49 | 89.5 | 0 |
| BAV1 | Agilent eArray | 1,371 | N/A | 1,356 | 98.9 | 1 | 60 | 80 | 2 |
| CBDB1 | Agilent eArray | 1,458 | N/A | 1,456 | 99.9 | 1 | 60 | 80 | 3 |
| DET195 | Agilent eArray | 1,580 | N/A | 1,510 | 95.6 | 1 | 60 | 80 | 3 |
| KB-1 | Agilent eArray | 933 | N/A | 931 | 99.8 | 1 | 60 | 80 | 4 |
| VS | Agilent eArray | 1,470 | N/A | 1,459 | 99.3 | 1 | 60 | 80 | 20 |
N/A, not applicable.
x-hyb, cross-hybridization.
Dehalococcoides strain-specific probe design.
The Agilent eArray system (with all default parameters for bacterial genomes) was used to design probes for the complete gene complement of each individual genome sequence (or partial genome sequence, in the case of KB-1). The statistical properties of these probe sets are presented in Table 1, and their sequences are listed in Table S1 in the supplemental material.
The final array design was built on a 4 × 44K Agilent oligonucleotide array. It contains triplicate copies of the 5 strain-specific probe sets and the pangenome probe set generated by ProDesign/OpSelector, with any remaining spot filled by random selection of pangenome probes.
DNA microarray template preparation, hybridization, and signal processing.
Pure strain DNA for Dehalococcoides strains VS, CBDB1, and BAV1 was generously provided by Alfred Spormann (Stanford University), Lorenz Adrian (Helmholtz Centre for Environmental Research [UFZ]), and Frank Löffler (Georgia Institute of Technology), respectively. Mixed-culture DNA from the D2 batch reactor (43) containing Dehalococcoides strain DET195 was generously provided by Ruth Richardson (Cornell University). KB-1 DNA was extracted in-house from 50 ml of the KB-1 enrichment culture using the MoBio UltraClean soil DNA isolation kit, according to the manufacturer's directions.
Two 4 × 44K oligonucleotide array slides were ordered from Agilent, providing a total of 8 arrays and a total of 16 possible DNA samples using a 2-dye system. The design of the test array DNA hybridizations is depicted in Table S2 in the supplemental material.
Amplification and labeling of DNA, as well as hybridization, washing, scanning, and quantification of arrays, were done by the University Health Networks Microarray Center (UHN). For array hybridization, 50 ng of each Dehalococcoides DNA sample was amplified and chemically labeled according to the Agilent WGA+ULS (version 3) protocol for comparative genome hybridizations (CGHs). Equal quantities (200 ng) of amplified and labeled DNA were spotted on each array. Hybridization was conducted at 65°C with shaking at 20 rpm for 20 h. Quantification of array intensities was done with a G2565C DNA scanner, and intensities were analyzed using the Agilent Feature Extraction software version 10.5. Raw intensity values and background-subtracted spot intensities were generated following all Agilent Feature Extraction steps in the CGH_105_Dec08 Agilent protocol. For all subsequent analyses, the background-subtracted spot intensities are used as the prenormalization values.
Array normalization.
The intensities of the red channel duplicate samples were compared to determine if there was significant between-array error. For the Dehalococcoides strain CBDB1 DNA samples, one array replicate showed significant noise within its signal intensities. Upon examination of the KB-1 (green) intensities of the same array, a similar trend was observed, indicating that this array had not hybridized comparably to the other arrays. Subsequently, these array data were excluded from the test set.
For the remaining 7 arrays, total array signal intensities were calculated for the KB-1 replicate samples (green channel), and the average total intensity was determined. For each array, both the red and green channel intensities were scaled by the factor normalizing the total green channel intensity to the average total intensity. For the 7 KB-1 replicate arrays, probe intensity values were taken as the trimmed mean of the 7 scaled values (average of the values lying within the interval of the mean ± 3 times the standard deviation of all seven values). For the duplicate red channel samples (VS, DET195, and BAV1), the average intensity of the duplicates was taken. Probes with absolute duplicate pairwise differences outside the mean plus 3 times the standard deviation were flagged as poorly performing duplicates. For the single array with a Dehalococcoides strain CBDB1 DNA sample, probe values were kept as the scaled values from total intensity normalization.
Individual probes were spotted on the array with a minimum of 3 replicates. Following array duplicate merging, as described above, single-probe intensity values were determined by taking the average of the probe replicates within an array. Previously flagged probes from the duplicate averaging were excluded. Replicate probe pairs whose pairwise difference fell outside the mean ± 3 times the standard deviation were also excluded, and the final averaged probe value was taken from all remaining, reliable probe values.
Expected hybridization patterns.
In order to determine the fraction of probes hybridizing correctly, the expected behavior of each probe was determined bioinformatically.
For the pangenome probes, probes were expected to hybridize to DNA from Dehalococcoides strains if a gene from that strain was present within the cluster to which the probe was designed. For the eArray-designed strain-specific probes, probes designed for a specific genome were universally expected to hybridize to that strain's DNA sample.
To determine the expected nontarget strain cross-hybridization, each probe was blasted against the five Dehalococcoides genomes using BLASTn, with a drop-off value for gapped alignments of 150, a nucleotide mismatch penalty of −1, a word size of 7, and filtering for repeated sequences implemented (conditions for short query sequences adapted from reference 34) (5, 42). For each eArray-designed 60-mer, this yielded a BLAST score of nucleotide identity between 0 and 60. A BLAST score ratio (BSR) was calculated for each probe using the BLAST score of that probe against a nonspecific genome, divided by the BLAST score of the probe against its specific genome (score = 60). The expected cross-hybridization patterns for BSR thresholds between 83% and 95%, at 1% intervals, were determined for the strain-specific probes as well as for the pangenome probe set.
Signal threshold determination.
The effects of signal threshold for determining positive hybridization on probe performance were examined for each set of expected hybridization patterns.
Probe sensitivity, specificity, accuracy, and Fβ score were calculated for each data set as follows: sensitivity was defined as the observed true positives (TP) divided by the expected positives (TP + false negatives [FN]), specificity was calculated as the observed true negatives (TN) divided by the total number of expected negatives (TN + false positives [FP]), accuracy was defined as the observed true positives divided by the total observed positives [TP/(TP + FP)], and the Fβ score was defined as [(1 + β2) × accuracy × sensitivity]/(β2 × accuracy + sensitivity), where β is equal to 0.1 (chosen to weight toward a lower false-positive rate) (55). These parameters of probe performance were examined for intensity values of 1,000 to 1 × 106 over intervals of 1,000. From these calculations, it was determined that a threshold fluorescence value between 1 × 104 and 3 × 104 would yield consistently high probe performance statistics across all examined data sets.
A second method for determining threshold intensity values was adapted from Oh et al. (42). Here, instead of using a single reference genome with designed probes and several tester strain DNA samples, any pair of Dehalococcoides genomes with lower than 90% average nucleotide identity (ANI) was utilized as a reference-tester pair (and vice versa). The ANI for the five strains was determined by taking the average of the nucleotide identity across full gene alignments for all reciprocal best BLAST matches for genes between two genomes (see Table S3 in the supplemental material).
For each combination of tester genome and reference genome, the BLAST score of the reference genome probes against the tester genome and the BLAST score ratio (BLAST score of the probe against the tester genome/BLAST score of the probe against the reference genome) were determined. The average signal intensity for each BLAST score was calculated, as well as the log of the average signal intensity ratio between the tester and reference genomes.
Plots of the average signal intensity versus the BLAST score for the tester genome and the log(average hybridization intensity ratio) versus the BLAST score ratio for the tester/reference genome were made for each permutation of the tester and reference genomes (12 permutations in total) (see Fig. S2 in the supplemental material). The calculations for each pair were merged into one data set, and plots were generated for the entire data set as a whole (see Fig. S3 in the supplemental material). The point of inflection on Fig. S3B occurs at a BLAST score ratio of 81 to 85%, corresponding to an optimal threshold of 1.17 × 104 to 1.78 × 104, which agrees with the observed optimal thresholds seen using the more conventional sensitivity and accuracy measures described above.
From these threshold determination trials, a threshold of 1.46 × 104 normalized fluorescence intensity was chosen, whereby a probe with fluorescence above this was considered ON (positive) and below this was considered OFF (negative). This threshold value represents the best agreement between the different threshold determinations utilized.
RESULTS AND DISCUSSION
Nontarget genome probe performance examination.
In order to determine the most accurate expected hybridization pattern for the strain-specific probes against nontarget genomes, sensitivity, specificity, accuracy, and the Fβ score were calculated for each possible expected hybridization pattern for BSR thresholds of 83 to 95%. From this, the optimal BSR for accurate prediction of probe cross-hybridization for the strain-specific probes on nontarget genomes was determined to be 83% (see Table S4 in the supplemental material). This finding is in keeping with the recently published determination that a BSR of 83% marks the beginning of meaningful biological hybridization between strains of bacteria (42).
The sensitivities, specificities, and Fβ scores for strain-specific probes and pangenome probes were compared (Fig. 2). When the expected hybridization was based on a BSR of ≥83%, the performances of the strain-specific and pangenome probes were comparable, with the pangenome probe set under its original design parameters showing slightly lower Fβ scores. Overall, the three data sets did not have significantly different performance values. This is interesting to note for the arrays with KB-1 and DET195 DNA, as these samples came from mixed cultures. Their equivalently high performance compared to that of arrays hybridized with pure strain DNA indicates that the probe sets are robust to more complex samples.
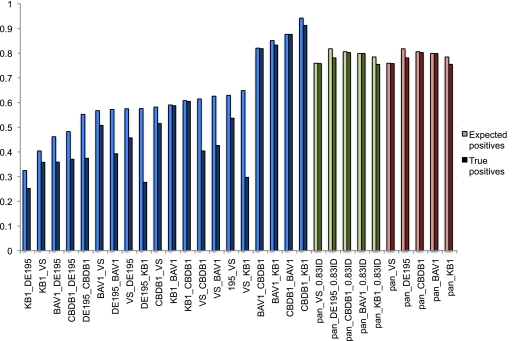
Fig. 2.
Sensitivity, specificity, and accuracy measures for the various data sets, in which column names are in the following format: probe set_DNA sample hybridized. Pale bars indicate sensitivity [TP/(TP + FN)], medium-intensity bars indicate specificity [TN/(TN + FP)], and dark bars indicate accuracy [TP/(TP + FP)]. Probe hybridization predictions (positive or negative) were based on either an 83% BSR (strain-specific probes in blue; pangenome probes in green) or, for the pangenome probes, gene presence within cd-hit-est clusters, as designed (in red).
The more telling comparison was the proportion of genes within a genome that are covered by the various probe sets (i.e., are predicted to have positive hybridization signals in the presence of that strain's DNA). For both the proportion of genes with predicted positive probes [(TP + FN)/total number of genes in genome] and the actual proportion of genes with observed positive signals (TP/total number genes in genome), the pangenome probes provided significantly higher gene coverage of any genome pair with lower than 99% ANI (Fig. 3). For Dehalococcoides, this means that strain-specific probes designed to “Pinellas” group genomes (strains CBDB1, BAV1, and KB-1 [9]) perform well in hybridizations against other Pinellas strains but that members of any other known group will have significantly lower coverage, as will Pinellas strains on probes designed for non-Pinellas strains. As an example, for examination of strain BAV1 DNA using the strain-specific probe set for strain DET195, only 57% of the gene complement is expected to be represented, and in reality, only 37% of strain BAV1 genes are detected using the strain DET195 probes. In comparison, the pangenome probe set predicts coverage of 80% or 86% of the BAV1 genes (predicted hybridization based on a BSR of ≥83% or cluster design, respectively) and provides >99% detection of the predicted genes, resulting in a coverage of 80% and 86%. It was not expected for the strain-specific probes to function equivalently well compared to the pangenome probe set, but the size of the discrepancy between the two methods was surprising. Dehalococcoides spp. are a closely related group, and a previous study has utilized a strain-specific microarray to examine a nontarget Dehalococcoides sp. (56), so it was anticipated that the strain-specific probe sets would have higher levels of detection of nontarget strains.
Fig. 3.
Proportion of genes per genome covered by probe sets, in which column names are in the following format: probe set_DNA sample hybridized. Light bars indicate the proportion of genes predicted to be detected, while dark bars indicate the actual proportion of genes detected. Predicted probe hybridization was based on either an 83% BSR (strain-specific probes in blue; pangenome probes in green) or, for the pangenome probes, gene presence within cd-hit-est clusters, as designed (in red). Detected positives were based on a normalized fluorescence signal threshold of 1.46 × 104. Pangenome probe set data in which statistical measures were based on an expected hybridization pattern are presented in red.
Pangenome probe set proteomic coverage.
The clustering approach allowed ProDesign to find probes which covered a large percentage of clusters (91%); however, the percentage of genes in the data set covered by a probe was only 79%. The low sequence coverage from the pangenome probe set was concerning, so a proteomic examination of the pathways covered by the probe set was undertaken. The complete gene complement of the 5 Dehalococcoides genomes was run through the KEGG Automatic Annotation Server (KAAS) (http://www.genome.jp/tools/kaas) for assignment of KEGG Orthology (KO) numbers to applicable genes. For complete genomes, a bidirectional best hit search was utilized, while single-directional best hit searches were undertaken for the sets of genes covered or not covered by the pangenome probe sets. The search database used included the default KAAS prokaryotic genomes with Geobacter metallireducens and Dehalococcoides strains 195, CBDB1, and VS added to the set. Table S5 in the supplemental material provides a list of the Dehalococcoides genes not represented on the pangenome array. From these, it is clear that while a certain proportion of Dehalococcoides genes are not represented within the pangenome probe set, many of the genes lacking coverage are from the ribosome complex (12.3%) or are hypothetical proteins (42.4%), meaning that coverage of the known metabolic proteins from Dehalococcoides is sufficient to allow examination of the Dehalococcoides response to environmental perturbations. Probes for all genes not covered by the pangenome probe set are present in the strain-specific probe sets. A combination of the pangenome probe set and the subset of strain-specific probes for genes not otherwise covered represents a functional array design for 100% detection of known Dehalococcoides genes.
Reductive dehalogenase homologous gene coverage.
The functional genes for reductive dechlorination of contaminant substrates are the reductive dehalogenases. The five-genome data set contained 105 reductive dehalogenase (RDH) homologous sequences annotated and available at the time of the array design: 17 from strain 195, 32 from strain CBDB1, 10 from strain BAV1 (draft genome), 36 from strain VS (draft genome), and 10 from the KB-1 metagenome.
The detection profile of the RDH genes further illustrates the utility of a pangenome approach. The combined 5-strain-specific probe sets provide 100% detection of the 105 RDH genes. The pangenome probe set provides detection of 98% of the reductive dehalogenases in the 105-gene data set (see Table S6 in the supplemental material for cluster descriptions). There are no core RDH genes, defined here as a cluster containing a gene from each representative genome under the 95% ID clustering conditions. However, there are several clusters containing genes from more than one genome, indicating that these RDHs are conserved across different strains to a degree that allows effective design of non-strain-specific probes. The single-genome strain-specific probe sets provide significantly lower coverage when treated separately, ranging from 20 to 61% observed coverage under an 83% BSR threshold of hybridization (Table 2). This is as expected but highlights the advantage of a pangenome approach for detection of this diverse and highly distributed gene family.
Table 2.
Predicted and observed detection of the reductive dehalogenases from the 5-genome data set under various expected hybridization conditions
| Probe set | Detection (%) |
|
|---|---|---|
| Predicted | Observed | |
| Panprobe (as designed) | 98.1 | 98.1 |
| Panprobe (by 83% BSR) | 98.1 | 98.1 |
| Strain specific (as designed) | ||
| All | 100.0 | 99.0 |
| DET195 | 16.2 | 16.2 |
| VS | 34.3 | 34.3 |
| BAV1 | 9.5 | 9.5 |
| CBDB1 | 30.5 | 30.5 |
| KB-1 spp. | 9.5 | 8.6 |
| Strain specific (by 83% BSR) | ||
| All | 100.0 | 99.0 |
| DET195 | 38.1 | 30.5 |
| VS | 70.5 | 20.0 |
| BAV1 | 21.9 | 61.0 |
| CBDB1 | 66.7 | 51.4 |
| KB-1 spp. | 35.2 | 31.4 |
Of the 105 RDH sequences, 4 represent genes of known function on specific substrates, as follows: tceA in strain DET195 (36), bvcA in strain BAV1 (30), vcrA in strain VS (40), and cbrA in strain CBDB1 (2). Both the pangenome probes and the strain-specific probes perform accurately for these genes, detecting them in the genomes where they are present. Interestingly, both the strain VS probe specific for vcrA and the pangenome probe designed to this single-gene cluster show “false-positive” hybridization for the KB-1 DNA sample. The KB-1 “genome” used to generate these probe sets did not contain a vcrA homolog, as the KB-1 genes were determined from a partial metagenome sequence. However, further metagenomic sequencing of the KB-1 culture has shown the presence of a vcrA homologous gene with extremely high sequence conservation to the VS vcrA used here for probe design. Both the pangenome probe designed to the vcrA-containing cluster and the strain-specific probe designed to the VS vcrA gene are a perfect match to the KB-1 vcrA gene sequence, confirming that the “false-positive” hybridization of these VS-specific probes to the KB-1 mixed-culture genomic DNA represents a true detection of a gene not encompassed in the array design. This is an interesting example whereby a researcher utilizing the strain-specific probe sets designed for strain 195, BAV1, or CBDB1 would not have detected this gene within KB-1, while utilization of a pangenome probe set (either combined strain specific or pangenome) does allow identification of the industrially relevant vcrA gene in the KB-1 consortium.
Testing the probe sets' performance on novel available Chloroflexi genomes.
The availability of the complete 95-Mb KB-1 consortium metagenome sequence provided an opportunity to examine the pangenome probe set's ability to detect genes from a novel Dehalococcoides species for which it was not implicitly designed but which were present on DNA utilized in the hybridization experiments. The partial metagenome available at the time of probe design comprised 10 Mb and contained 933 identified Dehalococcoides genes. A total of 681 additional putative Dehalococcoides genes were identified from the completed 95-Mb KB-1 metagenome. All probes (pangenome and strain specific) were blasted against these additional Dehalococcoides strain KB-1 genes under the same conditions as those described in Materials and Methods. A BLAST score ratio of 83% was set as a hypothetical cutoff for expected hybridization, based on the true hybridization data from the arrays. The pangenome probe set was also examined with a hypothetical cutoff identity of 95%, which approximates the conditions utilized during pangenome probe design. The actual detection of these genes under hybridization of the arrays with KB-1 DNA was examined using the previously determined signal threshold of 1.46 × 104. The proportion of the new KB-1 genes predicted to be covered by the probe sets and the proportion actually detected are presented in Table 3. From this, it can be seen that the pangenome probe set performs better than any one single-strain-specific probe set in detecting the additional KB-1 genes. While, when combined, the strain-specific probes do provide a higher level of coverage, they are subject to a higher level of chance in terms of their performance against previously unknown gene sequences. This is illustrated in Fig. 4, showing a partial alignment of the ribosomal small subunit S16 genes from all five Dehalococcoides considered here. The pangenome probe was designed to a cluster containing the four genes available at the time (from strains CBDB1, BAV1, VS, and DET195). It is clear from the alignment that the pangenome probe was designed by ProDesign to avoid an area of strain divergence within this gene, while each strain-specific probe incorporates this region, lowering their sensitivity to other nontarget strains. To whit, due to a high number of mismatches near the 3′ end of the VS probe, it failed to detect the highly homologous KB-1 S16 gene. This is a clear example of the benefits of clustering prior to probe design in order to allow more universal detection of Dehalococcoides.
Table 3.
Predicted coverage of all Dehalococcoides strain GT and D. lykanthroporepellens strain BL-DC-9 genes at an 83% BSR threshold for hybridizationa
| Probe set (program) | Predicted coverage (%) |
Detection (%) of strain KB-1 (681 added genes) | ||
|---|---|---|---|---|
| Strain GT (1,417 genes) | Strain BL-DC-9 (1,659 genes) | Strain KB-1 (681 added genes) | ||
| BAV1 (eArray) | 55.9 | 0.42 | 40.2 | 38.3 |
| CBDB1 (eArray) | 62.2 | 0.18 | 48.8 | 45.6 |
| DET195 (eArray) | 39.2 | 0.06 | 30.1 | 14.8 |
| VS (eArray) | 45.6 | 0.30 | 32.5 | 15.5 |
| KB-1 (eArray) | 40.6 | 0.06 | N/A | N/A |
| All strain-specific probes (eArray) | 77.7 | 1.02 | 71.8 (62.4) | 63.3 (49.2) |
| Pangenome set (ProDesign) | 83.1 | 0.66 | 60.3 | 70.9 |
Predicted and actual coverage for the Dehalococcoides genes identified from metagenome sequencing of the KB-1 consortium after probe design was completed. Predicted coverage is based on an 83% BSR threshold for existing probes on the novel strains' genes. For newly added Dehalococcoides KB-1 genes, parentheses indicate the detection level using a 95% BSR threshold, which approximates the original ProDesign conditions. Detection of the additional Dehalococcoides KB-1 genes was based on probes exhibiting fluorescence above the threshold implemented throughout (1.46 × 104). N/A, not applicable.
Fig. 4.
A partial nucleotide alignment (bases 76 to 187) of the ribosomal large subunit S16 genes from Dehalococcoides strains CBDB1, BAV1, DET195, and KB-1. Bases are colored according to sequence similarity across the alignment. The Dehalococcoides KB-1 sequence was obtained from further metagenome sequencing post-array design. The pangenome probe designed to the ribosomal protein S16-containing cluster and the strain-specific probes designed to individual ribosomal S16 genes are included in the alignment. The strain-specific probe for strain VS's ribosomal S16 gene failed to detect the KB-1 ribosomal protein S16 gene. All other probe/genome hybridization combinations were detected.
In order to test the ability of the pangenome probe set to detect and examine a novel Dehalococcoides complete gene complement, an in silico hybridization against the newly published Dehalococcoides strain GT genome (51; http://genome.jgi-psf.org/deh_g/deh_g.home.html) was carried out as described above for the new KB-1 genes. The number of strain GT genes predicted to be covered by the six probe sets is presented in Table 3. From this raw comparison, it can be seen that the pangenome probes provide significantly higher coverage, even compared to strain-specific probes for the Pinellas group strains. From the actual hybridization data, it was clear that the pangenome probe sets have a much more consistent ratio of detected to predicted positive probes. From this, it is likely that the comparison for strain GT in Table 3 should actually be more distinct, as the strain-specific probe sets have been shown to perform more poorly than predicted. Even using the in silico numbers alone, it is clear that the pangenome probe set provides improved detection of Dehalococcoides strain GT. Of the 123 genes covered solely by the strain-specific probe sets, over 47% are annotated as hypothetical, with a further 15% annotated as ribosome subunit proteins. Conversely, of the 199 GT genes that the pangenome probes uniquely detect, only 12% correspond to hypothetical proteins, while 11 of the 20 total reductive dehalogenase homologous genes are included in this set, meaning that the combined strain-specific probes will miss over half of strain GT's functionally important genes involved in respiration of chlorinated solvents.
A similar exercise was conducted using the Dehalogenimonas lykanthroporepellens strain BL-DC-9 genome (39; http://genome.jgi-psf.org/dehly/dehly.home.html), which is the nearest neighbor to the Dehalococcoides group for which sequence information is currently available (Fig. 1). This in silico prediction of probe performance gave very little expected hybridization of D. lykanthroporepellens strain BL-DC-9 to either the pangenome probes (∼1% of genes detected) or to the strain-specific probe sets (Table 3). This indicates that the probe sets are highly specific for Dehalococcoides, which, while this may limit their scope in detecting novel organisms with less than 83% sequence identity, also indicates that the array is highly specific to Dehalococcoides.
The in silico exercises utilizing Dehalococcoides strain GT and D. lykanthroporepellens strain BL-DC-9 show proof of the utility, flexibility, and specificity of the pangenome probes compared to those of probe sets designed to a single Dehalococcoides genome. This is due to the nature of the probe sets' design; the pangenome probe sets were designed to conserved regions of Dehalococcoides genes from clusters and, hence, should be more likely to match to other, previously unknown Dehalococcoides strains' genes.
Conclusions.
The Dehalococcoides pangenome probe set developed here represents a universal platform for the analysis of this industrially relevant genus. The probe set is highly specific to Dehalococcoides: it is robust to cross-hybridization from environmental bacteria, including the closest known relative to Dehalococcoides spp., the Dehalogenimonas spp. In addition, in silico comparisons utilizing the Dehalococcoides strain GT genome indicate that the pangenome probe set detects a larger proportion of a novel Dehalococcoides strain's genes than the set of combined strain-specific probes. Newly available Dehalococcoides genes from the KB-1 consortium allowed confirmation that probe design to clusters of highly similar genes increases the likelihood that a probe is designed to a conserved region of a gene, strengthening the universal detection of Dehalococcoides by this probe set.
A known weakness of this probe set is the lowered coverage of Dehalococcoides protein genes (∼80% represented in clusters with probes). Future work with this platform will be geared toward optimizing coverage of Dehalococcoides genomes while still maintaining the clustering advantages of this design. An immediately available alternative is the use of a subset of the strain-specific probes to complement the pangenome probe set to fill in the remaining 20% of missing genes. This would allow 100% coverage of known Dehalococcoides genes and would be acceptable for use with any of the currently sequenced strains. Additionally, this probe design method is flexible and can be applied to larger Dehalococcoides genomic data sets as sequencing information comes available, allowing this probe set to evolve with the growing knowledge base. An additional note is that here we examined probe performance with a constant amount of DNA utilized across all samples, chosen to avoid any expected detection limits (62). For RNA expression studies or environmental sample testing, a thorough determination of DNA concentration and signal intensity relationships will be required to assess the limits of detection for this probe set (16, 61).
Several different array-based methods for bacterial identification and genomic comparisons exist. Current general microbial detection microarrays do not provide strain differentiation of Dehalococcoides (PhyloChip and others [10, 63]) or do not provide complete coverage of the Dehalococcoides gene complement (e.g., GeoChip [20, 53]). As described here and elsewhere (25), Dehalococcoides strain-specific probe sets provide complete coverage of a strain of interest's gene complement but can only be used for partial examinations of nontarget strains (results above; see also reference 56). A pangenome or pangenus array approach provides the ability to compare novel strains to the known genomes in a more complete fashion and allows examination of hypothetical genes outside the known functional cannon. The existing Dehalococcoides pangenus Affymetrix array (32) and the combined strain-specific eArray-designed probe sets described here represent a straightforward design targeting each individual gene in the combined set. In contrast, the pangenome probe set designed with ProDesign is based on clustered genes such that probes are designed to highly conserved regions of similar genes. The pangenome probe set thus represents a flexible tool that can be applied to laboratory research, allowing multiple labs to work with a common platform regardless of which specific Dehalococcoides strain (sequenced or unsequenced) they are cultivating. The use of a common platform will facilitate collaboration between research groups, and it is possible that the use of identical probe sequences to examine different Dehalococcoides strains will provide higher consistency across different laboratories' experimental data. The pangenome probe set can also be utilized for the detection and identification of Dehalococcoides at contaminated sites or, as has become increasingly of interest, at pristine sites where dechlorinating organisms have not yet been exposed to human pollution. The sensitivity and specificity of a microarray paired with the universality of Dehalococcoides detection demonstrated by the ProDesign pangenome probe set provide a powerful tool for examining the global distribution and metabolic capacity of Dehalococcoides.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Joint Genome Institute for sequencing the KB-1 metagenome and other Dehalococcoides genomes. We also acknowledge the University Health Network microarray center (Toronto, Ontario, Canada) for helping with labeling and hybridization.
ProDesign development was funded in part by the Ontario Ministry of Health and Long-Term Care. This project was funded by the Government of Canada through Genome Canada and the Ontario Genomics Institute (grant 2009-OGI-ABC-1405), the Natural Sciences and Engineering Research Council of Canada (NSERC), and the U.S. Department of Defense Strategic Environmental Research Defense Program (SERDP). E.R.T. holds a Canada Research Chair in Analytical Genomics. L.A.H. was supported by an NSERC CGS-D scholarship.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Adrian L. 2009. ERC-group microflex: microbiology of Dehalococcoides-like Chloroflexi. Rev. Environ. Sci. Biotechnol. 8:1569–1705 [Google Scholar]
- 2. Adrian L., Rahnenfuhrer J., Gobom J., Holscher T. 2007. Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl. Environ. Microbiol. 73:7717–7724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adrian L., Szewzyk U., Wecke J., Gorisch H. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580–583 [DOI] [PubMed] [Google Scholar]
- 4. Ahsanul Islam M., Edwards E. A., Mahadevan R. 2010. Characterizing the metabolism of Dehalococcoides with a constraint-based model. PLoS Comput. Biol. 6:e1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 6. Bodrossy L., Sessitsch A. 2004. Oligonucleotide microarrays in microbial diagnostics. Curr. Opin. Microbiol. 7:245–254 [DOI] [PubMed] [Google Scholar]
- 7. Cheng D., He J. 2009. Isolation and characterization of “Dehalococcoides” sp. strain MB, which dechlorinates tetrachloroethene to trans-1,2-dichloroethene. Appl. Environ. Microbiol. 75:5910–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cupples A. M. 2008. Real-time PCR quantification of Dehalococcoides populations: methods and applications. J. Microbiol. Methods 72:1–11 [DOI] [PubMed] [Google Scholar]
- 9. Cupples A. M., Spormann A. M., McCarty P. L. 2004. Comparative evaluation of chloroethene dechlorination to ethene by Dehalococcoides-like microorganisms. Environ. Sci. Technol. 38:4768–4774 [DOI] [PubMed] [Google Scholar]
- 10. DeSantis T. Z., et al. 2007. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb. Ecol. 53:371–383 [DOI] [PubMed] [Google Scholar]
- 11. Drummond A. J., et al. 2010. Geneious version 5.0. Geneious, Auckland, New Zealand [Google Scholar]
- 12. Duhamel M., Mo K., Edwards E. A. 2004. Characterization of a highly enriched dehalococcoides-containing culture that grows on vinyl chloride and trichloroethene. Appl. Environ. Microbiol. 70:5538–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng S., Tillier E. R. 2007. A fast and flexible approach to oligonucleotide probe design for genomes and gene families. Bioinformatics 23:1195–1202 [DOI] [PubMed] [Google Scholar]
- 15. Fennell D. E., Carroll A. B., Gossett J. M., Zinder S. H. 2001. Assessment of indigenous reductive dechlorinating potential at a TCE-contaminated site using microcosms, PCR analysis, and site data. Environ. Sci. Technol. 35:1830–1839 [DOI] [PubMed] [Google Scholar]
- 16. Gao H., et al. 2007. Microarray-based analysis of microbial community RNAs by whole-community RNA amplification. Appl. Environ. Microbiol. 73:563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gresham D., et al. 2006. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science 311:1932–1936 [DOI] [PubMed] [Google Scholar]
- 18. Grostern A., Edwards E. A. 2006. Growth of Dehalobacter and Dehalococcoides spp. during degradation of chlorinated ethanes. Appl. Environ. Microbiol. 72:428–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He J., Sung Y., Krajmalnik-Brown R., Ritalahti K. M., Löffler F. E. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 7:1442–1450 [DOI] [PubMed] [Google Scholar]
- 20. He Z., et al. 2010. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 4:1167–1179 [DOI] [PubMed] [Google Scholar]
- 21. He Z., et al. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 1:67–77 [DOI] [PubMed] [Google Scholar]
- 22. Hendrickson E. R., et al. 2002. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 68:485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holliger C., Wohlfarth G., Diekert G. 1998. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22:383–398 [Google Scholar]
- 24. Jayachandran G., Gorisch H., Adrian L. 2003. Dehalorespiration with hexachlorobenzene and pentachlorobenzene by Dehalococcoides sp. strain CBDB1. Arch. Microbiol. 180:411–416 [DOI] [PubMed] [Google Scholar]
- 25. Johnson D. R., et al. 2008. Temporal transcriptomic microarray analysis of “Dehalococcoides ethenogenes” strain 195 during the transition into stationary phase. Appl. Environ. Microbiol. 74:2864–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnson D. R., Lee P. K., Holmes V. F., Alvarez-Cohen L. 2005. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl. Environ. Microbiol. 71:3866–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson D. R., Lee P. K., Holmes V. F., Fortin A. C., Alvarez-Cohen L. 2005. Transcriptional expression of the tceA gene in a Dehalococcoides-containing microbial enrichment. Appl. Environ. Microbiol. 71:7145–7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johnson D. R., Nemir A., Andersen G. L., Zinder S. H., Alvarez-Cohen L. 2009. Transcriptomic microarray analysis of corrinoid responsive genes in Dehalococcoides ethenogenes strain 195. FEMS Microbiol. Lett. 294:198–206 [DOI] [PubMed] [Google Scholar]
- 29. Jones E. J. P., Voytek M. A., Lorah M. M., Kirshtein J. D. 2006. Characterization of a microbial consortium capable of rapid and simultaneous dechlorination of 1,1,2,2-tetrachloroethane and chlorinated ethane and ethene intermediates. Bioremediat. J. 10:153–168 [Google Scholar]
- 30. Krajmalnik-Brown R., et al. 2004. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl. Environ. Microbiol. 70:6347–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kube M., et al. 2005. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23:1269–1273 [DOI] [PubMed] [Google Scholar]
- 32. Lee P. K., et al. 2011. Comparative genomics of two newly isolated Dehalococcoides strains and an enrichment using a genus microarray. ISME J. 5:1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li W., Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659 [DOI] [PubMed] [Google Scholar]
- 34. Li W., Jaroszewski L., Godzik A. 2001. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 17:282–283 [DOI] [PubMed] [Google Scholar]
- 35. Li W., Jaroszewski L., Godzik A. 2002. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 18:77–82 [DOI] [PubMed] [Google Scholar]
- 36. Magnuson J. K., Romine M. F., Burris D. R., Kingsley M. T. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141–5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maymo-Gatell X., Chien Y., Gossett J. M., Zinder S. H. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568–1571 [DOI] [PubMed] [Google Scholar]
- 38. McMurdie P. J., et al. 2009. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 5:e1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moe W. M., Yan J., Nobre M. F., da Costa M. S., Rainey F. A. 2009. Dehalogenimonas lykanthroporepellens gen. nov., sp. nov., a reductively dehalogenating bacterium isolated from chlorinated solvent-contaminated groundwater. Int. J. Syst. Evol. Microbiol. 59:2692–2697 [DOI] [PubMed] [Google Scholar]
- 40. Muller J. A., et al. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70:4880–4888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nishimura M., et al. 2008. Detection and identification of Dehalococcoides species responsible for in situ dechlorination of trichloroethene to ethene enhanced by hydrogen-releasing compounds. Biotechnol. Appl. Biochem. 51:1–7 [DOI] [PubMed] [Google Scholar]
- 42. Oh S., Yoder-Himes D. R., Tiedje J., Park J., Konstantinidis K. T. 2010. Evaluating the performance of oligonucleotide microarrays for bacterial strains with increasing genetic divergence from the reference strain. Appl. Environ. Microbiol. 76:2980–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rahm B. G., Richardson R. E. 2008. Correlation of respiratory gene expression levels and pseudo-steady-state PCE respiration rates in Dehalococcoides ethenogenes. Environ. Sci. Technol. 42:416–421 [DOI] [PubMed] [Google Scholar]
- 44. Rhee S. K., et al. 2004. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 70:4303–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ritalahti K. M., et al. 2006. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 72:2765–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schmidt K. R., Stoll C., Tiehm A. 2006. Evaluation of 16S-PCR detection of Dehalococcoides at two chloroethene-contaminated sites. Water Sci. Technol. 6:129–136 [Google Scholar]
- 47. Schumacher W., Holliger C., Zehnder A. J., Hagen W. R. 1997. Redox chemistry of cobalamin and iron-sulfur cofactors in the tetrachloroethene reductase of Dehalobacter restrictus. FEBS Lett. 409:421–425 [DOI] [PubMed] [Google Scholar]
- 48. Seshadri R., et al. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105–108 [DOI] [PubMed] [Google Scholar]
- 49. Smith T. F., Waterman M. S. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147:195–197 [DOI] [PubMed] [Google Scholar]
- 50. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 51. Sung Y., Ritalahti K. M., Apkarian R. P., Löffler F. E. 2006. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl. Environ. Microbiol. 72:1980–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Taş N., Van Eekert M. H. A., De Vos W. M., Smidt H. 2010. The little bacteria that can—diversity, genomics and ecophysiology of ‘Dehalococcoides’ spp. in contaminated environments. Microb. Biotechnol. 3:389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tiquia S. M., et al. 2004. Evaluation of 50-mer oligonucleotide arrays for detecting microbial populations in environmental samples. Biotechniques 36:664–670 [DOI] [PubMed] [Google Scholar]
- 54. Van Raemdonck H., Maes A., Ossieur W., Verthe K., Boon N. 2006. Real time PCR quantification in groundwater of the dehalorespiring Desulfitobacterium dichloroeliminans strain DCA1. J. Microbiol. Methods 67:294–303 [DOI] [PubMed] [Google Scholar]
- 55. van Rijsbergen C. V. 1979. Information retrieval, 2nd ed. Butterworth, London, England [Google Scholar]
- 56. West K. A., et al. 2008. Comparative genomics of “Dehalococcoides ethenogenes” 195 and an enrichment culture containing unsequenced “Dehalococcoides” strains. Appl. Environ. Microbiol. 74:3533–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Willenbrock H., Hallin P. F., Wassenaar T. M., Ussery D. W. 2007. Characterization of probiotic Escherichia coli isolates with a novel pan-genome microarray. Genome Biol. 8:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Willenbrock H., et al. 2006. Design of a seven-genome Escherichia coli microarray for comparative genomic profiling. J. Bacteriol. 188:7713–7721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wilson K. 2001. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. Chapter 2:Unit 2.4 [DOI] [PubMed] [Google Scholar]
- 60. Winzeler E. A., et al. 2003. Genetic diversity in yeast assessed with whole-genome oligonucleotide arrays. Genetics 163:79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu L., Liu X., Schadt C. W., Zhou J. 2006. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl. Environ. Microbiol. 72:4931–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu L., et al. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhou J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288–294 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.