Abstract
Noroviruses and rotaviruses from a gastroenteritis outbreak affecting >300 people near Garda Lake (Northern Italy) in 2009 were investigated. Characterization of viruses from 40 patient stool samples and 5 environmental samples identified three distinct rotavirus and five norovirus genotypes; two of the latter were detected in both patient and environmental samples.
TEXT
Norovirus is a major cause of sporadic and epidemic gastroenteritis worldwide, affecting children and adults (24). Five norovirus genogroups are known, among which GI, GII, and GIV affect humans and GIII and GV infect bovines and mice, respectively. Genogroups I and II encompass at least 29 genotypes (3, 21, 34). Group A rotavirus remains the main cause of severe diarrhea in children and an occasional cause in adults (8). Based on genetic characterization of outer-capsid protein VP7 (11) and VP4 (10), at least 23 G genotypes and 32 P serotypes are recognized (1, 6, 9, 20, 30), several of which infect both humans and animals (8). In June 2009, a gastroenteritis outbreak started in a Garda Lake hotel (Northern Italy) (26), and several hundred cases were reported to local public health authorities in the following 4 weeks. Waterborne transmission was suspected, involving the lake water consumed by municipal communities following physicochemical treatment (26). Microbiological analysis of 36 stool samples from 299 cases investigated confirmed the presence of bacterial and viral agents, also detected in environmental samples (26).
The present study provides detailed molecular characterization of viruses involved in the gastroenteritis outbreak to clarify the possible source of infection.
Thirty-six stool samples were obtained from patients enrolled as cases in the epidemiological study (26). Four samples collected 3 weeks later were also examined.
RNA was extracted from 160 μl of stool suspensions (10%) by using a QIAamp viral RNA minikit (Qiagen, Hilden, Germany). Three microliters of total RNA were screened for human norovirus and rotavirus by reverse transcription (RT)-PCR (SuperScript III one-step RT-PCR system with Platinum Taq; Invitrogen, Carlsbad, CA), followed by seminested PCR for rotavirus. For norovirus, primers G1SKF/G1SKR or G2SKF/G2SKR (0.5 μM) were used, amplifying a 338-bp open reading frame 2 (ORF2) (capsid) fragment of genogroup I or II, respectively (15). A confirmatory RT-PCR targeting ORF1 (RNA-dependent RNA polymerase [RdRp], 327 bp) was also performed using primers JV12/JV13 (0.5 μM) (31). Reactions were performed following the manufacturer's instructions, with annealing temperatures of 52°C and 40°C for ORF2 and ORF1, respectively.
For rotavirus G and P typing, RT-PCR protocols described previously (10, 11) were used. Amplification of the VP7 (881 bp) and VP4 (663 bp) regions was performed with 0.5 μM VP7F/VP7R and VP4F/VP4R primers (13, 14) with an annealing temperature of 52°C. The RT-PCR products (1:100) were used as templates for seminested multiplex PCR using mixtures of primers (50 nM, with annealing temperatures of 42°C for G typing and 45°C for P typing) specific for typical and emerging human G and P genotypes (11, 14, 25). Detection of bovine rotavirus and GIII noroviruses in environmental samples was performed by RT-PCR as described previously (7, 23, 29). Amplicons were detected on GelRed (Biotium, Inc., Hayward, CA)-stained 1.5% agarose gel. Sequence analysis of amplified norovirus diagnostic DNA was performed by Macrogen, Inc., Seoul, South Korea. Sequences were compared with the NCBI and FBVE (http://www.rivm.nl/mpf/norovirus/typingtool) databases. A phylogenetic tree was constructed using BioNumerics (version 6.1; Applied Maths, Kortrijk, Belgium). Environmental samples were examined as follows: sample A, water (500 ml) and sediment at the lake water intake point; sample B, water (500 ml) from a private swimming pool; sample C, filter sediment; and sample D, untreated lake water from upstream of the treatment plant. Water was prefiltered through glass wool (18), filtered through nitrocellulose membranes, eluted, and subjected to organic flocculation (33).
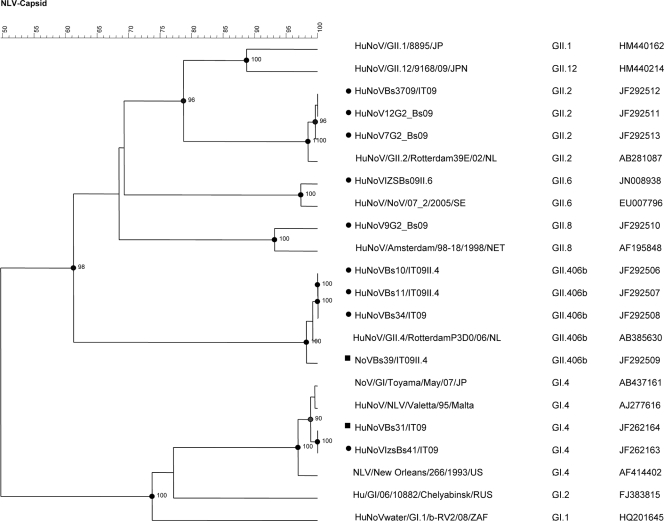
Thirty-four cases (85%), including 24 previously diagnosed cases (26), were positive for norovirus by one or more RT-PCR methods. Nineteen samples (47.5%) were positive for rotavirus, of which 17 also contained norovirus. Thirteen samples were positive for GI norovirus, 11 for GII, and 10 for both GI and GII. Sequence analysis was performed for 22 norovirus samples (14 GI and 8 GII) from 18 subjects (Table 1), revealing five different strains (Fig. 1). Most noroviruses (14/22) belonged to genotype GI.4 (GenBank sequence accession number JF262163, 100% nucleotide identity). Four different GII genotypes were also identified: three strains were GII.2 (GenBank sequence accession numbers JF292511 to JF292513, 100% nucleotide identity between each other), one was GII.8 (GenBank sequence accession number JF292510), one was GII.6 (GenBank sequence accession number JN008938), and three were GII.42006b (GenBank sequence accession numbers JF292506 to JF292508), which is a variant of the prototype Lordsdale strain (2, 28).
Table 1.
Genotypes of norovirus and rotavirus strains from clinical and environmental samples taken during a gastroenteritis outbreak in Italy in 2009
| Type of sampleg | No. of norovirus strains of indicated genotype (% nucleotide identity)a |
No. of rotavirus strains of indicated genotypeb |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GI.4c (83-97.0) | GI.4 (99.0) | GII.6 (97.0) | GII.8 (98.0) | GII.4v06 (99.0) | GII.2 (98.0) | Ratio to totald | G2P[4] | G3P[6]e | G1P[8] | G1P[4] | Ratio to totald | |
| Stool | 14 | 1 | 1 | 3 | 3 | 22/34 | 11 | 7 | 1 | 19/19 | ||
| Water | 1 | 1/1 | 1 | 1/1 | ||||||||
| Sediment | 1f | 1f | 2/2 | 1 | 1/1 | |||||||
Genotyping was performed by sequencing. Percent sequence identity was determined by BLAST.
Genotyping was performed by multiplex PCR.
Genotyping was based on ORF1 diagnostic fragment sequence.
Number of genotyped strains/total number of strains detected.
P[6] is a rare genotype in humans and is possibly of animal origin.
These two strains were detected in a single sample.
Forty clinical and 5 environmental samples were investigated.
Fig. 1.
Phylogenetic tree based on 300-nucleotide capsid fragment (ORF2) of Italian strains of human (•) and environmental (▪) origin and other norovirus strains. Tree constructed by using Bionumerics version 6.5 using UPGMA (unweighted-pair group method using average linkages) bootstrap values of >90% is shown. Genotypes and GenBank accession numbers are indicated. NoV, norovirus; HuNoV, human norovirus; NLV, Norwalk-like virus.
Three different G types were detected among 19 rotavirus-positive samples, namely, G2 (11 cases), G3 (7 cases), and G1 (1 cases), showing P[4], P[6], or P[8] genotypes, respectively. These results were confirmed by sequence analysis (GenBank sequence accession numbers JN001846 to JN001850) and suggested that at least three different rotavirus strains, of genotypes G1P[8], G2P[4], and G3P[6], were involved. Although the first two are common strains affecting humans, strain G3P[6] bears a possibly animal-origin genotype, P[6] (Table 1).
Two environmental samples were positive for both GI and GII norovirus and for rotavirus (Table 1). ORF2 sequence analysis of the lake water (sample D) showed a GI.4 norovirus (GenBank sequence accession number JF262164), and both G1 and P[4] rotavirus. The latter were also identified in sediment (sample A), together with norovirus GI.4 (ORF1, GenBank sequence accession number JF297568) and GII.4v2006b (ORF2, GenBank sequence accession number JF292509). Environmental samples were investigated for bovine rotavirus and norovirus strains but none were found, weakening the hypothesis of possible farm waste release into the lake.
The numerous norovirus (five) and rotavirus (three) genotypes involved and identical genomic sequences between patients suggest a common infection source, like human stools or sewage-contaminated water. Its identification in most cases suggests norovirus as the main cause of the outbreak. Despite the higher number of GII norovirus types identified, the single GI norovirus infected the greatest proportion of subjects. Only 3 of 18 sequence-confirmed subjects harbored the globally predominant strain GII.4v2006b (2, 28), associated with most European outbreaks (17). However, GI noroviruses are frequently reported in surface waters (19, 22), and the GI.4 identified in Italy might represent an emerging strain. The occurrence of G2P[4] rotavirus in most patients as opposed to only one case infected with the predominant G1P[8] is equally surprising.
The detection of human norovirus and rotavirus in lake water and sediment at the intake point near the water treatment plant is suggestive of human fecal contamination. The occurrence of the same norovirus (GI.4) in both stool samples and environmental samples, together with the large number (17) of norovirus/rotavirus mixed infections, suggests that lake water may indeed have transmitted the infections. In particular, the GI.4 norovirus detected showed 100% nucleotide identity in all samples (Fig. 1). The presence of the uncommon G1P[4] human strain in lake samples cannot be conclusively proved, as G1 and P[4] genes may have been amplified from distinct viruses, such as G1P[8] and G2P[4], identified in patients.
In principle, a water origin of the outbreak remains tentative, since the environmental contamination revealed might have followed the outbreak. No virus was detected in water from a home swimming pool, indicating that the viral concentration was below detection limits, at least during sampling. However, the association of norovirus with waterborne outbreaks (4, 5, 12, 17, 32) is undisputed, and acceptable hyperchlorination limits may be insufficient in cases of abnormal viral contamination (16, 27).
Acknowledgments
This study was partially supported by the following grants from the Ministry of Health, Italy: “Creazione di una rete di laboratori virologici di riferimento (Noronet-Italia) per il controllo delle epidemie di gastroenterite da Norovirus in Italia,” CCM 2009, and “Sviluppo di un sistema diagnostico per la rivelazione di agenti di zoonosi batterici e virali e dei loro fattori di patogenicità da applicare nella filiera di produzione del suino,” Ricerca Finalizzata 2006-MSRF0106 Programma straordinario.
Footnotes
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Abe M., et al. 2009. Molecular epidemiology of rotaviruses among healthy calves in Japan: isolation of a novel bovine rotavirus bearing new P and G genotypes. Virus Res. 144:250–257 [DOI] [PubMed] [Google Scholar]
- 2. Allen D. J., Gray J. J., Gallimore C. I., Xerry J., Iturriza-Gómara M. 2008. Analysis of amino acid variation in the P2 domain of the GII-4 norovirus VP1 protein reveals putative variant-specific epitopes. PLoS One 3:e1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ando T., Noel J. S., Fankhauser R. L. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181:336–348 [DOI] [PubMed] [Google Scholar]
- 4. Blackburn B. G., et al. 2004. Surveillance for waterborne-disease outbreaks associated with drinking water—United States, 2001-2002. MMWR Surveill. Summ. 53:23–45 [PubMed] [Google Scholar]
- 5. Boccia D., et al. 2002. Waterborne outbreak of Norwalk-like virus gastroenteritis at a tourist resort, Italy. Emerg. Infect. Dis. 8:563–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Collins P. J., Martella V., Buonavoglia C., O'Shea H. 2010. Identification of a G2-like porcine rotavirus bearing a novel VP4 type, P[32]. Vet. Res. 41:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Di Bartolo I., Ponterio E., Monini M., Ruggeri F. M. 2011. A pilot survey of bovine norovirus in Northern Italy. Vet. Rec. doi:10.1136/vr.d2625 [DOI] [PubMed]
- 8. Estes M. K., Kapikian A. Z. 2007. Rotaviruses, p. 1917–1974 In Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus E. E. (ed.), Fields virology , 5th ed. Lippincott, Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 9. Fukai K., et al. 2007. Molecular characterization of a novel bovine group A rotavirus. Vet. Microbiol. 123:217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gentsch J. R., et al. 1992. Identification of group A rotavirus gene 4 types by PCR. J. Clin. Microbiol. 30:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gouvea V., et al. 1990. PCR amplification and typing of rotavirus nucleic acid from stools. J. Clin. Microbiol. 28:276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hafliger D., Hubner P., Luthy J. 2000. Outbreak of viral gastroenteritis due to sewage-contaminated drinking water. Int. J. Food Microbiol. 54:123–126 [DOI] [PubMed] [Google Scholar]
- 13. Iturriza-Gomara M., Kang G., Gray J. J. 2004. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J. Clin. Virol. 31:259–265 [DOI] [PubMed] [Google Scholar]
- 14. Iturriza-Gomara M., Green J., Brown D. W., Desselberger U., Gray J. J. 2000. Diversity within the VP4 gene of rotavirus P[8] strains: implications for reverse transcription-PCR genotyping. J. Clin. Microbiol. 38:898–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kojima S., et al. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 100:107–114 [DOI] [PubMed] [Google Scholar]
- 16. Koopmans M., Duizer E. 2004. Foodborne viruses: an emerging problem. Int. J. Food Microbiol. 90:23–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kroneman A., et al. 2008. Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the Foodborne Viruses in Europe network from 1 July 2001 to 30 June 2006. J. Clin. Microbiol. 46:2959–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lambertini E., et al. 2008. Concentration of enteroviruses, adenoviruses, and noroviruses from drinking water by use of glass wool filters. Appl. Environ. Microbiol. 74:2990–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lysén M., et al. 2009. Genetic diversity among food-borne and waterborne norovirus strains causing outbreaks in Sweden. J. Clin. Microbiol. 47:2411–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martella V., et al. 2007. Identification of group A porcine rotavirus strains bearing a novel VP4 (P) genotype in Italian swine herds. J. Clin. Microbiol. 45:577–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martella V., et al. 2008. Detection and molecular characterization of a canine norovirus. Emerg. Infect. Dis. 14:1306–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maunula L., Miettinen I. T., von Bonsdorff C. H. 2005. Norovirus outbreaks from drinking water. Emerg. Infect. Dis. 11:1716–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Monini M., et al. 2008. Molecular characterization of bovine rotavirus strains circulating in northern Italy, 2003-2005. Vet. Microbiol. 129:384–389 [DOI] [PubMed] [Google Scholar]
- 24. Patel M. M., Hall A. J., Vinjé J., Parashar U. D. 2009. Noroviruses: a comprehensive review. J. Clin. Virol. 44:1–8 [DOI] [PubMed] [Google Scholar]
- 25. Samajdar S., et al. 2006. Changing pattern of human group A rotaviruses: emergence of G12 as an important pathogen among children in eastern India. J. Clin. Virol. 36:183–188 [DOI] [PubMed] [Google Scholar]
- 26. Scarcella C., et al. 2009. An outbreak of viral gastroenteritis linked to municipal water supply, Lombardy, Italy, June 2009. Euro Surveill. 14(29):pii19274. [DOI] [PubMed] [Google Scholar]
- 27. Shin G. A., Sobsey M. D. 2008. Inactivation of norovirus by chlorine disinfection of water. Water Res. 42:4562–4568 [DOI] [PubMed] [Google Scholar]
- 28. Siebenga J., Kroneman A., Vennema H., Duizer E., Koopmans M. 2008. Food-borne Viruses in Europe network. Food-borne Viruses in Europe network report: the norovirus GII. 4 2006b (for US named Minerva-like, for Japan Kobe034-like, for UK V6) variant now dominant in early seasonal surveillance. Euro Surveill. 13(2):pii8009. [PubMed] [Google Scholar]
- 29. Smiley J. R., Hoet A. E., Tråvén M., Tsunemitsu H., Saif L. J. 2003. Reverse transcription-PCR assays for detection of bovine enteric caliciviruses (BEC) and analysis of the genetic relationships among BEC and human caliciviruses. J. Clin. Microbiol. 41:3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ursu K., et al. 2009. Molecular analysis of the VP7 gene of pheasant rotaviruses identifies a new genotype, designated G23. Arch. Virol. 154:1365–1369 [DOI] [PubMed] [Google Scholar]
- 31. Vinjé J., Koopmans M. P. 1996. Molecular detection and epidemiology of small round-structured viruses in outbreaks of gastroenteritis in the Netherlands. J. Infect. Dis. 174:610–615 [DOI] [PubMed] [Google Scholar]
- 32. Werber D., et al. 2009. Massive outbreak of viral gastroenteritis associated with consumption of municipal drinking water in a European capital city. Epidemiol. Infect. 137:1713–1720 [DOI] [PubMed] [Google Scholar]
- 33. Wyn-Jones A. P., Pallin R., Dedoussis C., Shore J., Sellwood J. 2000. The detection of small round-structured viruses in water and environmental materials. J. Virol. Methods 87:99–107 [DOI] [PubMed] [Google Scholar]
- 34. Zheng D. P., et al. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312–323 [DOI] [PubMed] [Google Scholar]