Abstract
Phage metagenomes isolated from wastewater over a 12-month period were analyzed. The results suggested that various strains of Proteobacteria, Bacteroidetes, and other phyla are likely to participate in transduction. The patterns of 16S rRNA sequences found in phage metagenomes did not follow changes in the total bacterial community.
TEXT
Bacteriophages are key biological entities that control the size of bacterial populations, establish lysogeny (temperate phages), and introduce horizontal gene transfer via the transduction processes. Phages have been shown to exist in most environments, including wastewater (9), and transduce various genetic markers, including antibiotic resistance (3). Several studies point out that the bacterial rRNA gene can be packed into generalized transducing phage particles (2, 5, 13). We recently showed that an analysis of 16S rRNA gene sequences in the phage metagenome can be successfully used to study bacteria included in horizontal gene transfer mediated by transduction (4). In the current study, we analyzed 16S rRNA genes present in bacteriophage-derived (PH) and total (TOT) bacterial metagenomes isolated from wastewater over a 1-year period in order to assess the likely extent of transductional transfer in this environment.
Water from an oxidation tank of a municipal wastewater treatment plant located in Belfast (Northern Ireland) was analyzed. Samples (15 liters) were collected in August 2009 (A09), September 2009 (S09), January 2010 (J10), March 2010 (M10), and July 2010 (Jul10). Isolation and purification of phage particles was carried according to the protocol described by Thurber et al. (17) with modifications as described in our previous work (4). The removal of bacterial DNA from phage preparation was achieved as described previously (1, 4). All DNA manipulations were performed using standard techniques (12). PCRs were carried out using universal 16S rRNA gene primers, and 1.4-kbp fragments were obtained from both total bacterial and phage metagenomic DNA preparations. By using these fragments, eight 16S rRNA gene libraries were constructed in the pJET1.2 vector (Fermentas). These libraries were analyzed as described before (4), and the summary of the analysis is presented in Table 1. Sequences from libraries PH A09 and TOT A09 were reported previously (4), and 16S rRNA sequences from libraries PH M10, PH J10, PH Jul10, TOT M10, TOT J10, and TOT Jul10 were deposited in the EMBL database. Various phylogenetically diverse species of bacteria were detected in wastewater, corresponding to previously published results (6, 14, 18). Phage-borne 16S rRNA genes were found for most bacterial groups, excluding Nitrospirae, Planctomycetes, and Gemmatimonadetes (Table 1). It is notable that phage-derived 16S rRNA genes were found for groups that seem to be present in the minority in the wastewater environment (e.g., Firmicutes and Cyanobacteria, which are not detected in total bacterial metagenome libraries) (Table 1). However, corresponding species were always detected in the total community by using PCR with species-specific primers (results not presented). Phage-derived sequences of Alpha-, Beta-, and Gammaproteobacteria were present in all samples taken over a 12-month period, whereas Cyanobacteria sequences were found in the July 2010 sample only. Actinobacteria sequences were not detected in July 2010 phage samples, although Actinobacteria were present in significant numbers in the total metagenome obtained from the same sample (Table 1). These results indicate that during the 1-year period, transduction in wastewater was likely to encompass a significant part of the inhabiting bacteria; however, it did not correlate directly with dominant bacterial groups inhabiting this environment.
Table 1.
Enumeration of 16S rRNA gene sequences representing various bacterial phyla detected in metagenomic libraries
| Phylum and class | No. of analyzed clones in 16S rRNA gene library (no. of unique sequences)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| TOT |
PH |
|||||||
| A09 | J10 | M10 | Jul10 | A09 | J10 | M10 | Jul10 | |
| Proteobacteria | ||||||||
| Alphaproteobacteria | 11 (3) | 0 | 3 (3) | 8 (7) | 28 (3) | 9 (2) | 8 (3) | 2 (2) |
| Betaproteobacteria | 24 (4) | 16 (2) | 31 (8) | 1 (1) | 16 (2) | 14 (2) | 6 (3) | 3 (2) |
| Gammaproteobacteria | 0 | 6 (2) | 2 (2) | 2 (1) | 29 (4) | 7 (1) | 15 (3) | 17 (5) |
| Deltaproteobacteria | 3 (2) | 1 | 4 (2) | 8 (3) | 0 | 1 | 0 | 1 |
| Actinobacteria | 41 (5) | 4 (1) | 1 | 23 (5) | 7 (1) | 0 | 0 | 0 |
| Firmicutes | 0 | 0 | 0 | 0 | 14 (1) | 2 (1) | 0 | 3 (2) |
| Nitrospirae | 1 | 0 | 0 | 1 (1) | 0 | 0 | 0 | 0 |
| Planctomycetes | 6 (2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Bacteroidetes | 0 | 21 (2) | 23 (3) | 5 (2) | 0 | 14 (2) | 4 (2) | 1 (1) |
| Chloroflexi | 0 | 0 | 0 | 3 (2) | 0 | 4 (2) | 0 | 2 (2) |
| Cyanobacteria | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 23 (1) |
| Gemmatimonadetes | 0 | 0 | 0 | 1 (1) | 0 | 0 | 0 | 0 |
Numbers of the sequenced 16S rRNA genes are presented; numbers of unique sequences are given in parentheses. PH, phase-derived library; TOT, total bacterial library; A09, August 2009; J10, January 2010; M10, March 2010; Jul10, July 2010.
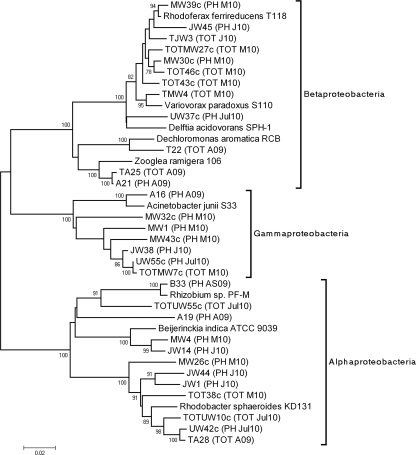
Phylogenetic analyses of the 16S rRNA gene sequences were conducted with MEGA software version 4 (MEGA4) (15). Trees were generated by neighbor joining (11) using the maximum composite likelihood model (16). Phylogenies were also evaluated by the maximum parsimony method and the unweighted pair group method with arithmetic mean (UPGMA) and found to be similar. Figure 1 shows the phylogenetic tree for Proteobacteria species found in TOT and PH libraries. It is clear that in Alpha- and Betaproteobacteria in particular, a number of diverse strains likely participate in transductional gene transfer. A number of highly homologues sequences were also observed for various metagenomes analyzed over the 12-month period. Examples of these are TA28 and UW42c (Alphaproteobacteria), which were detected in the total bacterial metagenome in August 2009 (TOT A09) as well as in the phage metagenome in July 2010 (PH Jul10), and TOTMW7c (TOT M10) and UW55c (PH Jul10) (Fig. 1).
Fig. 1.
Phylogenetic analysis of 16S rRNA gene sequences of Proteobacteria from the oxidation tank of a wastewater treatment plant. The phylogenetic tree was inferred using the neighbor-joining method (11). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. Phylogenetic analyses were conducted with MEGA4 (15). Sequences from phage (PH) and total (TOT) metagenome libraries are labeled as follows: August 2009 (A09), August-September 2009 (AS09), January 2010 (J10), March 2010 (M10), and July 2010 (Jul10). Only one sequence from each metagenome is presented when the 16S rRNA gene sequence identity was higher than 99%. 16S rRNA gene sequences of these clones were aligned with representatives of Alphaproteobacteria (Rhizobium sp. PF-M, GenBank accession number DQ202284; Beijerinckia indica ATCC 9039, NC_010581; Rhodobacter sphaeroides KD131, NC_011958), Betaproteobacteria (Dechloromonas aromatica RCB, NC_007298; Variovorax paradoxus S110, NC_012791; Delftia acidovorans SPH-1, NC_010002; Rhodoferax ferrireducens T118, NC_007908; Zooglea ramigera 106, NR_026130), Gammaproteobacteria (Acinetobacter junii S33, AB101444), Bacteroidetes (Flavobacterium johnsoniae UW101, NC_009441), Chloroflexi (Dehalococcoides sp. GT, NC_013890), and Actinobacteria (Ilumatobacter fluminis YM22-133, AB360343; Acidimicrobium ferrooxidans DSM10331, NC_013124). The bar at the bottom of the figure shows estimated sequence divergence.
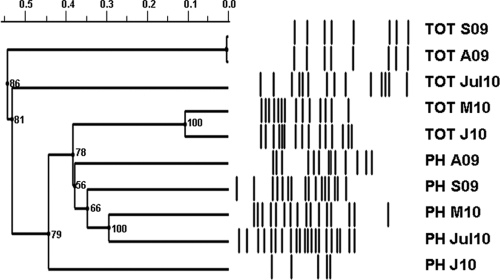
To further analyze the changes in 16S rRNA gene patterns in the wastewater environment, denaturing gradient gel electrophoresis (DGGE) analyses were performed with a CBS Scientific system. Polyacrylamide (6%) gradient gels were formed as described previously (7, 8) and a urea gradient range of 20 to 100% (100% denaturant contained 7 M urea and 40% deionized formamide) was used. DGGE profiles were analyzed using various approaches to quantify the similarity between lanes, including the Jaccard coefficient, the Dice similarity, and the Ochiai coefficient, which minimizes the effect of different numbers of bands in various lanes. Analysis was performed using the Phoretix 1D v10.3 (TotalLab Ltd.) software package. The results of the DGGE analyses are presented in Fig. 2. It is notable that bands representing 16S rRNA gene patterns in phage metagenomes isolated in August and September 2009 and in March and July 2010 are clustered together and distinct from those from total metagenomic libraries. The number of phage-derived 16S rRNA gene sequences from January 2010 seem to be lower than in other samples, although the number of bands in the total bacterial metagenome was higher in the same sample and the pattern was similar to that of March 2010. Although total bacterial profiles for August and September 2009 samples are very similar, profiles of phage metagenomes showed more pronounced differences.
Fig. 2.
DGGE analysis of 16S rRNA gene sequences from wastewater samples isolated from 2009 to 2010. Designation of samples is the same as in Fig. 1. Samples were analyzed as described in the text. DGGE images were digitalized using the Phoretix 1D v10.3 (TotalLab Ltd.) software package. Cluster analysis of 16S rRNA DGGE profiles was performed using the UPGMA clustering and Ochiai coefficient-based similarities with cophenetic correlation coefficients shown at the nodes. The pairwise comparison is based on 22 matching bands.
In this work, we chose a typical municipal wastewater treatment plant to analyze changes in phage-borne 16S rRNA gene patterns in a complex bacterial community over a 1-year period. We were exploiting the facts that generalized transducing phages can pack any region of the host's genome (2) and that activated sludge phage metagenomes indeed contain large amounts of bacterial sequences (10). By using a similar culture-independent approach, it was previously demonstrated that Aeromonas and Acinetobacter species appear to participate in transduction in wastewater (13) and that a number of distinctive bacterial strains belonging to several bacterial taxa apparently have transducing phages (4). Here, analysis of the 16S rRNA patterns over a 12-month period demonstrated that a large number of phylogenetically distant bacterial strains are very likely to participate in transduction in the wastewater environment. Closely related sequences are found in metagenomic samples taken in different periods. It was observed that changes in the 16S rRNA gene patterns found in phage metagenomes over the analyzed period did not mirror the changes in the total bacterial community of the studied environment. It is important to note that other mechanisms of gene exchange (i.e., conjugation and possibly transformation) may play important roles in the wastewater environment. An analysis of the relative significance of these types of horizontal gene transfer will be essential for understanding the functioning of microbial communities.
Nucleotide sequence accession numbers.
16S rRNA sequences from libraries PH M10, PH J10, PH Jul10, TOT M10, TOT J10, and TOT Jul10 were deposited in the EMBL database under accession numbers FR774592 to FR774635.
Acknowledgments
This work was supported by the QUESTOR Centre (Queens University of Belfast).
We thank Ciaran Prunty for expert help in collecting samples and Giovanna E. Felis, Verona University, for expert help in phylogenetic analysis.
Footnotes
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Allander T., Emerson S. U., Engle R. E., Purcell R. H., Bukh J. 2001. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. U. S. A. 98:11609–11614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beumer A., Robinson J. B. 2005. A broad-host range, generalized transducing phage (SN-T) aquires 16S rRNA genes from different genera of bacteria. Appl. Environ. Microbiol. 71:8301–8304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Colomer-Lluch M., Jofre J., Muniesa M. 2011. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS One 6:e17549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Del Casale A., Flanagan P. V., Larkin M. J., Allen C. C. R., Kulakov L. A. 2011. Analysis of transduction in wastewater bacterial populations by targeting the phage-derived 16S rRNA gene sequences. FEMS Microbiol. Ecol. 76:100–108 [DOI] [PubMed] [Google Scholar]
- 5. Ghosh D., et al. 2008. Prevalence of lysogeny among soil bacteria and presence of 16S rRNA and trzN genes in viral-community DNA. Appl. Environ. Microbiol. 74:495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miura Y., et al. 2007. Bacterial community structures in MBRs treating municipal wastewater: relationship between community stability and reactor performance. Water Res. 41:627–637 [DOI] [PubMed] [Google Scholar]
- 7. Muyzer G., de Waal E. C., Uitterlinden A. G. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of PCR-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Myers R. M., Fischer S. G., Lerman L. S., Maniatis T. 1985. Nearly all single base substitutions in DNA fragments joined to a GC-clamp can be detected by denaturing gradient gel electrophoresis. Nucleic Acids Res. 13:3131–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Otawa K., et al. 2007. Abundance, diversity, and dynamics of viruses on microorganisms in activated sludge processes. Microb. Ecol. 53:143–152 [DOI] [PubMed] [Google Scholar]
- 10. Parsley L. C., et al. 2010. Census of the viral metagenome within an activated sludge microbial assemblage. Appl. Environ. Microbiol. 76:2673–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 12. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 13. Sander M., Schmieger H. 2001. Method for host-independent detection of generalized transducing bacteriophages in natural habitats. Appl. Environ. Microbiol. 67:1490–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Snaidr J., Amann R., Huber I., Ludwig W., Schleifer K.-H. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 16. Tamura K., Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512–526 [DOI] [PubMed] [Google Scholar]
- 17. Thurber R. V., Haynes M., Breitbart M., Wegley L., Rohwer F. 2009. Laboratory procedures to generate viral metagenomes. Nat. Protoc. 4:470–483 [DOI] [PubMed] [Google Scholar]
- 18. Wagner M., Loy A. 2002. Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotechnol. 13:218–227 [DOI] [PubMed] [Google Scholar]