Abstract
Twenty Lactococcus lactis strains with an L. lactis subsp. lactis phenotype isolated from five traditional cheeses made of raw milk with no added starters belonging to the L. lactis subsp. lactis and L. lactis subsp. cremoris genotypes (lactis and cremoris genotypes, respectively; 10 strains each) were subjected to a series of phenotypic and genetic typing methods, with the aims of determining their phylogenetic relationships and suitability as starters. Pulsed-field gel electrophoresis (PFGE) analysis of intact genomes digested with SalI and SmaI proved that all strains were different except for three isolates of the cremoris genotype, which showed identical PFGE profiles. Multilocus sequence typing (MLST) analysis using internal sequences of seven loci (namely, atpA, rpoA, pheS, pepN, bcaT, pepX, and 16S rRNA gene) revealed considerable intergenotype nucleotide polymorphism, although deduced amino acid changes were scarce. Analysis of the MLST data for the present strains and others from other dairy and nondairy sources showed that all of them clustered into the cremoris or lactis genotype group, by using both independent and combined gene sequences. These two groups of strains also showed distinctive carbohydrate fermentation and enzyme activity profiles, with the strains in the cremoris group showing broader profiles. However, the profiles of resistance/susceptibility to 16 antibiotics were very similar, showing no atypical resistance, except for tetracycline resistance in three identical cremoris genotype isolates. The numbers and concentrations of volatile compounds produced in milk by the strains belonging to these two groups were clearly different, with the cremoris genotype strains producing higher concentrations of more branched-chain, derived compounds. Together, the present results support the idea that the lactis and cremoris genotypes of phenotypic Lactococcus lactis subsp. lactis actually represent true subspecies. Some strains of the two subspecies in this study appear to be good starter candidates.
INTRODUCTION
Lactococcus lactis is a lactic acid bacterium (LAB) commonly dominant in milk and fermented dairy products. Not surprisingly, carefully selected strains of L. lactis are majority components of starter cultures for dairy fermentations (38). Worldwide, over 100 million metric tons of milk is transformed annually into dairy products using L. lactis starters, reflecting the industrial and thus economic importance of this organism (38). The growth of L. lactis in milk is associated with the rapid production of lactic acid, which provides flavor, assists in curd formation, prevents the growth of pathogenic and spoilage bacteria, and creates optimal biochemical conditions for ripening. Via their proteolytic and amino acid conversion pathways, lactococci further contribute to the final texture (moisture, softness) and flavor of dairy products (47). These functions determine the sensory quality, safety, and shelf life of fermented dairy products. L. lactis includes three subspecies (L. lactis subsp. cremoris, L. lactis subsp. lactis, and L. lactis subsp. hordniae) plus a diacetyl-forming biovariety (L. lactis subsp. lactis biovar diacetylactis). The lactose-negative L. lactis subsp. hordniae (45) has never been found in dairy products. L. lactis subsp. lactis is distinguished from L. lactis subsp. cremoris according to five phenotypic criteria: the ability to grow at 40°C, in 4% NaCl, and at pH 9.2, the ability to ferment maltose, and the capacity to deaminate arginine (25, 45), for all of which L. lactis subsp. cremoris strains are reported to be negative. In addition, L. lactis subsp. lactis biovar diacetylactis is distinguished by its ability to assimilate citrate, which is converted into diacetyl, a potent odorous compound.
Current dairy lactococcal starters are thought to be derived from a small number of well-adapted, genetically related lineages showing similar genetic profiles and phenotypic properties (24, 39, 55). Therefore, there is a great demand for new strains to solve technological problems such as insufficient acid production, frequent culture failure resulting from the attack of bacteriophages, and the development of undesirable flavors (28, 33, 37, 54). In addition, interest is spurred by the continuing search for strains that harbor unique flavor-forming activities (3) or that produce novel, broad-range antimicrobial agents (6). Strains with these properties might be of use in traditional fermentations but also might allow new processes to be developed.
For some cheese types, L. lactis subsp. cremoris strains are the preferred starter since their growth response in milk is better and because of the typical aroma profiles associated with them (42, 54). Although found in dairy environments, the natural niche of this subspecies remains elusive, and claims of having isolated novel L. lactis subsp. cremoris strains from milk and naturally fermented products are regarded as controversial (30, 33, 42, 54). Furthermore, distinction between L. lactis subsp. lactis and L. lactis subsp. cremoris is difficult since it is based on a set of phenotypic characteristics that may show strain-to-strain variation. Further, some strains of L. lactis showing an L. lactis subsp. lactis phenotype according to the classical distinction criteria have long been known to show a L. lactis subsp. cremoris genotype (cremoris genotype) (27). Recently, phenotypic L. lactis subsp. cremoris showing a L. lactis subsp. lactis genotype (lactis genotype) have also been reported (30, 52). Therefore, the L. lactis species has an unusual structure with two phenotypically distinct groups defining L. lactis subsp. lactis and L. lactis subsp. cremoris, which may belong to two distinct genotype groups (30, 36, 40, 52, 54). This makes the accurate identification of new isolates very difficult, yet this is a crucial first step in the development of new cultures. In addition, the phenotypic and genetic relationships between the subspecies of L. lactis and even within subspecies remain unclear.
As the use of molecular genetic techniques became universal, strains with an L. lactis subsp. cremoris genotype have been isolated from many sources, including vegetables and plants (28, 37) and milk and dairy products (11, 15, 18, 34, 43, 54). A few strains have also been isolated from Spanish traditional, starter-free cheeses made from raw milk (14, 21, 35).
This work reports a comparative phenotypic, genotypic, and technological characterization of 20 strains with an L. lactis subsp. lactis phenotype, 10 each belonging to the lactis and cremoris genotypes. To compare their properties and assess their functionality, the strains were subjected to genetic fingerprinting, carbohydrate fermentation tests, enzyme activity profiling, and antibiotic resistance-susceptibility assays. Growth and production of volatile compounds in milk were also examined. These studies allowed the molecular genetic and phenotypic profiles of the strains belonging to the two genotypes to be compared, to make comparisons with results in the literature, and to propose a new classification for the members of this species.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The bacteria studied were L. lactis subsp. lactis strains, 10 belonging to the lactis genotype and 10 belonging to the cremoris genotype. They were all isolated during the manufacturing and ripening stages of five traditional, Spanish cheeses made from raw milk without the deliberate addition of commercial starter cultures, which implies that the isolates were all wild strains. The cheese origins of the different strains are as follows: Cabrales (L. lactis subsp. lactis L39, 1AA59, 3AA15, 2BA36, and 4AA10 of the lactis genotype and L. lactis subsp. lactis 1AA23, 3AA9, 3AA11, 3AA23, and 4AA28 of the cremoris genotype), Peñamellera (L. lactis subsp. lactis 1A38 and 2A83 of the lactis genotype and L. lactis subsp. lactis 2A5, 2A22, and 2A27 of the cremoris genotype), Genestoso (L. lactis subsp. lactis GE-1 of the lactis genotype and L. lactis subsp. lactis GE2-14 of the cremoris genotype), and Casín (L. lactis subsp. lactis CAS3 and Q1-6 of the lactis genotype and L. lactis subsp. lactis LC44 of the cremoris genotype). These strains were previously identified by the sequencing of the 16S rRNA gene and comparison of the sequences against those in the GenBank and Ribosomal Database Project II databases (1).
L. lactis subsp. lactis CECT 185T (ATCC 19435T) (lactis genotype, L. lactis subsp. lactis phenotype), L. lactis subsp. cremoris CECT 967T (NCDO 607T) (cremoris genotype, L. lactis subsp. cremoris phenotype), L. lactis subsp. lactis MG 1363 (L. lactis subsp. lactis phenotype, cremoris genotype), and L. lactis subsp. lactis IL 1403 (ex-phenotype L. lactis subsp. lactis biovar diacetylactis, lactis genotype) strains were used as controls throughout this study. Unless otherwise stated, strains were grown statically in M17 (Scharlab, Barcelona, Spain) broth at 30°C for 18 to 24 h.
Molecular identification of strains.
The identifications of the isolates were verified by molecular methods, which included partial amplified rRNA gene restriction analysis (ARDRA), sequencing, and sequence comparison. For this, total genomic DNA was purified from overnight cultures using a GenElute bacterial genomic DNA kit (Sigma-Aldrich, St. Louis, MO) following the manufacturer's recommendations. Electrophoresis was performed in 1% agarose gels, and the DNA was stained with ethidium bromide (0.5 μg/ml) and photographed under UV light. ARDRA was performed after amplification of the 16S rRNA genes with the bacteria-specific universal primer 27F (S-D-Bact-0008-a-S-20; 5′-AGAGTTTGATCCTGGCTCAG-3′) and the bacteria/archaea-specific primer 1492R (S-*-Univ-1492R-b-A-21; 5′-GGTTACCTTGTTACGACTT-3′). Amplicons were purified using GenElute PCR clean-up columns (Sigma-Aldrich), digested with the restriction enzymes HaeIII and HhaI (Invitrogen Ltd., Paisley, United Kingdom), and electrophoresed as described above. Both strands of the amplicons were sequenced using both 27F and 1942R primers; the sequences were then aligned and compared to those in databases.
Restriction fragment length polymorphism (RFLP) typing by PFGE.
Intact genomic DNA from L. lactis strains was isolated and digested in agarose plugs as described by Howard et al. (26). Purified DNA was digested independently with 20 U of the restriction enzymes SmaI and SalI (Boehringer Mannheim, Mannheim, Germany) for 18 h at 37°C in the restriction buffer recommended by the manufacturer. DNA digests were separated in a contour-clamped homogeneous electric field (CHEF) in a CHEF-DRII apparatus (Bio-Rad, Richmond, CA). Low-range and bacteriophage lambda ladder pulsed-field gel electrophoresis (PFGE) markers were obtained from New England BioLabs (Ipswich, MA). Electrophoresis was carried out in 1% FastLane agarose gels (FMC Corporation, Philadelphia, PA) in 0.5× TBE (Tris-borate-EDTA) for 20 to 24 h at 14°C and 6 V cm−1. Pulse times ranged from 0.5 to 25 s for 12 h and from 25 to 50 s for 6 h for the SmaI digests and from 0.5 to 5 s for 12 h and from 5 to 30 s for 8 h for the SalI digests. Similarity clustering was performed with the MultiVariate Statistical Package (MVSP; Kovach Computing Services, Anglesey, Wales, United Kingdom) using the unweighted-pair group method with arithmetic averages and Sorensen's correlation.
MLST analysis.
DNA sequence analysis of 350 to 861 bp of intragenic regions of the genes encoding the ATP synthase alpha subunit (atpA), the phenylalanyl-tRNA synthase alpha subunit (pheS), the RNA polymerase alpha subunit (rpoA), the branched-chain aminotransferase (bcaT), the peptidase N (pepN), and the X-prolyl dipeptidyl aminopeptidase (pepX) was performed employing the oligonucleotides and PCR conditions reported by Rademaker et al. (40). For multilocus sequence typing (MLST) analysis, forward and reverse sequences were trimmed, aligned, and analyzed using MEGA (version 4) software (51). Sequences were then compared to one another, and the similarities of the patterns were analyzed by the neighbor-joining method.
Phenotypic characterization.
Phenotypic analysis of the strains was done in filter-sterilized medium, as follows. Growth at 40 and 45°C was tested in Elliker broth (Scharlau, Barcelona, Spain) and examined daily for up to 5 days. Similarly, growth in 4% and 6.5% NaCl and at pH 9.2 and 9.6 was assayed in Elliker broth at 30°C and checked daily for up to 5 days. To test for arginine hydrolase activity, strains were grown for 48 h at 30°C in an arginine broth composed of peptone (5%), tryptone (0.5%), yeast extract (0.5%), K2HPO4 (0.2%), l-arginine (0.5%), dextrose (0.05%), MgSO4 (250 mg/liter), and ascorbic acid (0.5 g/liter), pH 7.0. After incubation, cells were removed by centrifugation and 10 μl of the supernatant was mixed with a drop of Nessler's reagent (KI [5 g], HgCl2 [5 g], NaOH [4 g], and 100 ml of filter-sterilized H2O). Strains were recorded as negative or weakly or strongly positive, judged by the intensity of their orange coloration.
The carbohydrate fermentation profiles of the isolates and control strains were determined using the commercial PhenePlate system (Bactus, Stockholm, Sweden) as recommended by the supplier. Additionally, strains were examined using the API 20 Strep kit following the manufacturer's recommendations (bioMérieux, Montalieu-Vercieu, France).
In addition to the Voges-Proskauer test of the API 20 Strep system, acetoin (acetylmethylcarbinol) production was further analyzed in Clark and Lubs medium (casein and meat peptone [3.5 g/liter each], dextrose [5 g/liter], potassium phosphate [5 g/liter], pH 6.9) with incubation at 30°C for 72 h. To a 2.5-ml aliquot of the cultures, 0.6 ml of Barritt's reagent A (5% [wt/vol] α-naphthol in absolute ethanol) was added, followed by 0.2 ml of reagent B (40% [wt/vol] KOH in water). Reagents were mixed, and the contents of the tubes were left to settle for 10 min. Strains were recorded as negative or weakly or strongly positive, judged by the intensity of their red coloration.
Citrate assimilation is also included among the API 20 Strep tests. Citrate utilization was further assayed in Kempler and McKay medium (31) under anaerobic conditions in the dark at 30°C for 44 to 72 h.
Enzyme activities were measured using the commercial, semiquantitative API ZYM system (bioMérieux) following the manufacturer's recommendations. Sixty-five microliters of a cell suspension corresponding to McFarland standard 5 (spectrophotometric equivalent, 3 × 109 CFU ml−1) was inoculated into each well of the API ZYM strips, and the strips were incubated for 4 h at 30°C and developed as recommended.
MICs of antibiotics were determined by microdilution in VetMIC plates for LAB (National Veterinary Institute of Sweden, Uppsala, Sweden) containing 2-fold serial dilutions of 16 antibiotics. Colonies grown on LAB susceptibility test medium (LSM) (32) agar plates were suspended in 2 ml of sterile saline solution (Oxoid, Basingstoke, Hampshire, United Kingdom) to obtain a density corresponding to McFarland standard 1 (spectrophotometric equivalent, 3 × 108 CFU ml−1). The suspension was further diluted 1:1,000 with LSM (final cell concentration, 3 × 105 CFU ml−1). One hundred microliters of this inoculum was added to each well of the VetMIC plate, which was incubated at 28°C for 48 h. The MICs were defined as the lowest antibiotic concentration at which no visible growth was observed. The presence of tetracycline resistance genes was checked by PCR using the universal primers for genes encoding the ribosomal protection proteins DI (5′-GAYACICCIGGICAYRTIGAYTT-3′) and DII (5′-GCCCARWAIGGRTTIGGIGGIACYTC-3′) (10) and using the PCR conditions described by the latter authors. Amplicons were purified and sequenced, and the sequences were compared against those in GenBank.
Growth and acidification of milk.
Acid production was determined in UHT milk (Corporacion Alimentaria Peñasanta, S.A. [CAPSA], Siero, Spain). A 1% inoculum from an overnight M17 culture was washed in sterile water and used to inoculate the milk, which was then incubated at 22°C; samples were scored for clotting at 15, 18, and 24 h. The pH was measured at 24 h using a pH meter (Crison Instruments S.A., Barcelona, Spain). The appearance of the coagulum (whey drainage, curd firmness, presence of gas bubbles, curd breaking) was also recorded by visual inspection.
Production of volatile compounds in milk.
The volatile compounds produced in milk were determined after growth of the strains in 10 ml of UHT milk (CAPSA) at 30°C for 2, 5, and 21 days. Cultures were grown in screw-cap tubes with a rubber liner to prevent the escape of volatiles, supplied with 100 μl of internal standard (cyclohexanone, 0.36 mg ml−1), and stored at −80°C until analysis. The separation and quantification of volatile compounds were carried out by headspace (HS)/gas chromatography (GC)/mass spectrometry (MS) analysis using a combined system composed of the units G 1888 HS, 6890 GC, and 5975B MSD, respectively (Agilent Technologies, Wilmington, DE), equipped with an HP-Innovax capillary column (60 m by 0.25 μm; Agilent). Sample preparation and gas chromatographic separation were performed as described by Salazar et al. (44). Peaks were quantified as the relative total ionic count abundance with respect to the internal standard. The concentrations (μg/ml) of some volatile compounds (acetaldehyde, diacetyl, 2-propanone, acetic acid, 2-butanone, and ethanol) were calculated, using linear regression equations (R2 ≥ 0.99), from the standard curve obtained using five representative concentrations.
Nucleotide sequence accession numbers.
The partial sequences of the seven genes examined in the MLST analysis were deposited in the GenBank database under accession numbers JF297335 through JF297474.
RESULTS
Partial ARDRA with the restriction enzymes HaeIII and HhaI, followed by sequencing, unequivocally identified all 20 strains as belonging to L. lactis. As expected from previous in silico analyses, 10 showed the lactis and 10 showed the cremoris genotype. The digestion profiles of amplicons of both genotypes were identical with HaeIII, but they gave distinct banding patterns with HhaI. The ribosomal sequences of the strains showing a lactis genotype were mostly identical, matching the 16S rRNA sequence of the L. lactis subsp. lactis type strain (ATCC 19435T) and the 16S rRNA sequences of the sequenced strains L. lactis subsp. lactis IL 1403 and L. lactis subsp. lactis KF147, except for a single nucleotide change in the sequences of strains GE-1 and 2BA36 (adenine for guanine at positions 91 and 465 of the IL 1403 numbering, respectively). The sequences of the strains with the cremoris genotype differed in 10 positions from those of the lactis genotype: positions 70, 76, 82, 87, 91, 93, 95, 98, 183, and 195. Two distinct sequences were found among the strains with the cremoris genotypes. The sequences varied in a single nucleotide at position 183, corresponding to cytosine in six strains (2A5, 2A22, 2A27, 3AA9, 3AA23, and 4AA28) and to thymine in four strains (1AA23, 3AA11, GE2-14, and LC44).
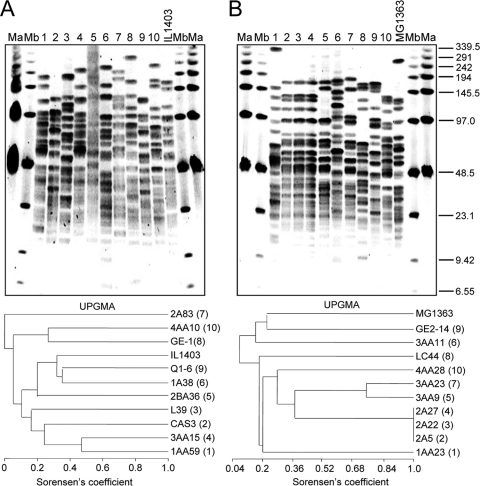
To assess the genetic diversity and relatedness of the strains, all were subjected to RFLP-PFGE analysis. L. lactis subsp. lactis MG 1363 (L. lactis subsp. lactis phenotype, cremoris genotype) and L. lactis subsp. lactis IL 1403 (ex-phenotype L. lactis subsp. lactis biovar diacetylactis, lactis genotype) were included as a control. Figure 1 shows the RFLP profiles obtained with the enzyme SalI and the clustering of the strains in terms of the Sorensen's coefficient of similarity. All L. lactis isolates of the lactis genotype were shown to be unrelated, as they shared a low similarity index. On the contrary, two isolates of the cremoris genotype from the same cheese sample (3AA9 and 3AA23) proved to be related (similarity index, 0.89). Moreover, three other cremoris genotype isolates (2A5, 2A22, and 2A27) from a single cheese batch showed identical digestion profiles, which indicates that all three could be replicates of the same strain. Similar PFGE results were obtained after digestion with SmaI (data not shown). However, the DNA of three strains (one belonging to the lactis genotype, GE-1, and two of the cremoris genotype, 3AA9 and 3AA23) was shown to be resistant to digestion with the last enzyme, which prevented a proper strain comparison. SmaI digestions allowed the estimation of the size of the chromosomes, which were shown to range approximately from 2,250 to 2,600 kbp for the lactis genotypes and 2,400 to 2,650 kbp for the cremoris genotypes. In conclusion, all 10 strains with the lactis genotype and at least 8 strains with the cremoris genotype could be considered different. In spite of this, isolates were all independently subjected to further genetic analyses and biochemical tests.
Fig. 1.
PFGE patterns of SalI-digested genomic DNA from L. lactis isolated from starter-free cheeses made of raw milk of lactis (A) and cremoris (B) genotypes. (A) Lanes 1 to 10, L. lactis genotype lactis 1AA59, CAS3, L39, 3AA15, 2BA36, 1A38, 2A83, GE-1, Q1-6, and 4AA10, respectively. (B) Lanes 11 to 20, L. lactis genotype cremoris 1AA23, 2A22, 2A27, 2A5, 3AA23, 3AA11, 3AA9, LC44, GE2-14, and 4AA28, respectively. IL 1403, L. lactis genotype lactis IL 1403; MG 1363, L. lactis genotype cremoris MG 1363. Lanes Ma and Mb, low-range and bacteriophage lambda ladder PFGE markers (New England BioLabs), respectively. Dendrograms of the similarity of the profiles of the strains in the respective panel expressed by the Sorensen's coefficient are shown below the panels. Clustering was performed by the unweighted-pair group method using arithmetic averages (UPGMA). Figures in parentheses after the strain code indicate the lane in the gels.
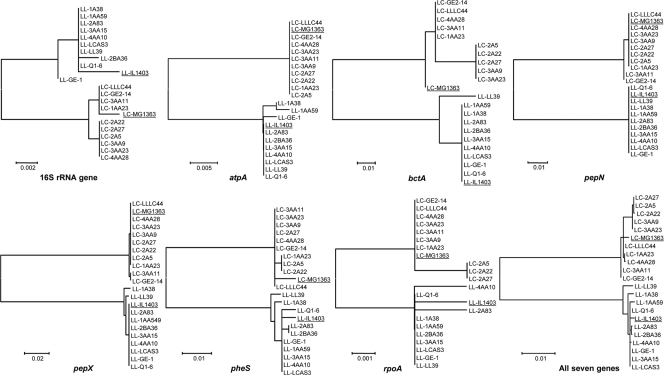
The genetic diversity of the lactis and cremoris genotype strains was further evaluated by MLST using partial nucleotide sequences of six genes, atpA, rpoA, pheS, pepN, bcaT, and pepX and the gene coding for 16S rRNA (Table 1). Amplicons and sequences from all these genes of all strains were obtained and aligned. Nucleotide positions of the sequences showing ambiguities were excluded from the analysis. Unique sequence types (STs) were obtained for every strain. After individual analysis of the seven genes and all genes together, two consistent and distinct clusters with similar topologies were obtained for the lactis and cremoris genotype strains (Fig. 2). The number of polymorphic sites varied strongly from gene to gene, ranging from 12 in 16S rRNA genes to 94 in pepX (Table 1). However, most of these nucleotide variations corresponded to nucleotide differences among strains of the lactis and cremoris genotypes, indicating that the actual number of alleles in the two species is much lower (from one to six; Table 1). Further, a majority of the substitutions were synonymous (they did not result in amino acid changes), as inferred from the low ratio of the number of nonsynonymous substitutions by the number of synonymous substitutions (dN/dS ratio; it was particularly low for the bcaT gene), which indicates conservation during evolution of amino acid sequences between members of the two subspecies. Together, the above results strongly suggest that the two clusters are composed of individual organisms showing a high degree of intracluster genetic similarity.
Table 1.
Genetic diversity at seven loci based on the nucleotide sequences used for MLST analysis and that of the 16S rRNA genes of 20 L. lactis subsp. lactis strains of both lactis and cremoris genotypes (10 each) used in this study
| Locus | Length (bp) |
G+C content (%) | No. of polymorphic sites | No. alleles by genotype |
dN/dS ratioa | |||
|---|---|---|---|---|---|---|---|---|
| Gene | Amplified fragment | Analyzed fragment | lactis | cremoris | ||||
| atpA | 1,503 | 1,141 | 861 | 42 | 49 | 4 | 1 | 0.30 |
| rpoA | 939 | 814 | 721 | 39 | 15 | 4 | 4 | 0.26 |
| pheS | 2,533 | 618 | 477 | 42 | 45 | 6 | 3 | 0.13 |
| bcaT | 1,047 | 493 | 350 | 42 | 50 | 3 | 2 | 0.02 |
| pepN | 1,023 | 482 | 473 | 37 | 46 | 1 | 2 | 0.10 |
| pepX | 2,269 | 602 | 508 | 39 | 94 | 4 | 2 | 0.20 |
| 16S rRNA gene | 1,548 | 1,465 | 605 | 49 | 12 | 3 | 2 | NAb |
The dN/dS ratio was calculated by dividing the number of nonsynonymous substitutions by the number of synonymous substitutions.
NA, not applicable.
Fig. 2.
Neighbor-joining cluster analysis of individual partial DNA sequences of the genes coding for 16S rRNA and the housekeeping protein-encoding genes atpA, rpoA, bacT, pepN, pepX, and pheS from 10 wild strains of L. lactis of the lactis genotype and 10 wild strains L. lactis of the cremoris genotype, as well as a seven-locus MLST analysis based on the composite data set for the seven genes. Bootstrap percentages (≥50) after 500 simulations are shown for single and composite sequence analyses. Sequences of the genome-sequenced L. lactis IL 1403 (lactis genotype) and MG 1363 (cremoris genotype) strains were used as a control and are underlined. LL and LC, L. lactis subsp. lactis having a lactis genotype and a cremoris genotype, respectively.
Since the same gene stretches as those reported by Rademaker et al. (40) were amplified and sequenced, the sequences obtained in the present work were trimmed in the same manner used by the latter authors. Sequences were then submitted to a recently developed MLST database for L. lactis (http://www-mlst.biotoul.fr/). Composite sequences of all seven genes were aligned with those of representative strains from the study of Rademaker et al (40). This allowed the relatedness of the present cheese isolates to be compared at the DNA level with those of lactococci from dairy and nondairy sources (Fig. 3). In the present study, most strains of both the lactis and cremoris genotypes clustered together, indicating greater similarity among themselves than to the dairy strains of Rademaker et al (40). Strains from different traditional cheeses grouped together, even though they were isolated from geographical areas more than 200 km apart, indicating that related STs are widespread. At the same time, some other strains from the same cheese batch clustered separately, suggesting the presence of unrelated STs. The cremoris genotype strains were split into two groups, one of which seems to be related to L. lactis subsp. lactis MG 1363 and some other strains with the L. lactis subsp. lactis phenotype and cremoris genotype (Fig. 3), and one of which is apparently closer to the true (phenotypic) L. lactis subsp. cremoris strains.
Fig. 3.
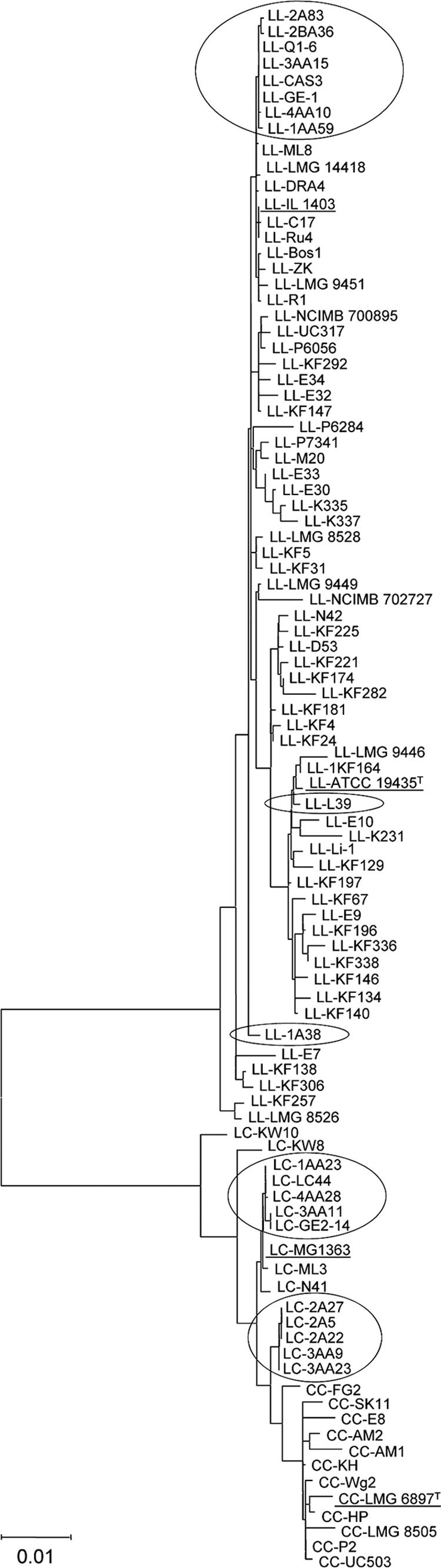
Diversity analysis of the wild L. lactis subsp. lactis strains of the lactis and cremoris genotypes studied in this work compared with L. lactis subsp. lactis and L. lactis subsp. cremoris strains of dairy (starter) and nondairy origin (40). Neighbor-joining cluster analysis of a seven-locus MLST analysis based on a composite data set of partial DNA sequences of the 16S rRNA, atpA, rpoA, bacT, pepN, pepX, and pheS genes. For sequence analysis, bootstrap percentages (≥50) after 500 simulations are shown. LL, LC, and CC, L. lactis subsp. lactis of the lactis genotype, the L. lactis subsp. lactis of the cremoris genotype, and L. lactis subsp. cremoris, respectively. The codes for the strains of this study appear enclosed in ovals on the diagram; type strains of both subspecies, L. lactis subsp. lactis ATCC 19435T (CECT 185T) and L. lactis subsp. cremoris LMG 6897T (CECT 967T), and L. lactis laboratory strains IL 1403 (genotype lactis) and MG 1363 (genotype cremoris) are underlined.
Table 2 summarizes the conventional tests used to distinguish the subspecies L. lactis subsp. lactis and L. lactis subsp. cremoris and the results obtained. All the strains studied hydrolyzed arginine and grew in 4% NaCl and at 40°C. All but four of the strains hydrolyzed hippurate, and all but the genotype cremoris type strain (L. lactis subsp. cremoris CECT 967T) grew at pH 9.2. In contrast, none of the strains used citrate, and single strains grew well at 45°C and in 6.5% NaCl. Strains did not give a positive reaction in the Voges-Proskauer test, except for a weak reaction of the L. lactis subsp. cremoris type strain, suggesting that none of the strains belonged to the biovar diacetylactis. Few phenotypic differences were seen between strains belonging to the lactis and cremoris genotypes; the most notable was growth at pH 9.6, for which all cremoris genotypes proved to be positive (although growth was weak), whereas only two strains of the lactis genotype were positive. In conclusion, according to the classical phenotypic criteria, the biochemical assays identified all 20 L. lactis cheese strains as belonging to the L. lactis subsp. lactis subspecies.
Table 2.
Phenotypic and biochemical properties of wild L. lactis subsp. lactis strains of lactis and cremoris genotypes isolated from starter-free, raw milk cheeses
| Genotype/straina | Presence of phenotypic property |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pH in milk, 22°C, 24 h | Hydrolysis of: |
Citrate utilization | Acetoin production | Growth at or in: |
|||||||
| Hippurate | Arginine | 40°C | 45°C | 4% NaCl | 6.5 NaCl | pH 9.2 | pH 9.6 | ||||
| Genotype lactis | |||||||||||
| 1AA59 | 4.19 | − | + | − | − | + | − | + | − | wb | − |
| CAS3 | 4.14 | + | + | − | − | + | w | + | + | + | − |
| L39 | 4.69 | + | + | − | − | + | − | + | − | + | w |
| 3AA15 | 4.14 | + | + | − | − | + | w | + | w | + | − |
| 2BA36 | 6.17 | + | + | − | − | + | + | + | − | + | − |
| 1A38 | 4.89 | + | + | − | − | + | − | + | w | + | + |
| 2A83 | 6.37 | + | + | − | − | + | − | + | − | + | − |
| GE-1 | 4.17 | + | + | − | − | + | − | + | − | + | − |
| Q1-6 | 4.78 | − | + | − | − | + | − | + | − | + | − |
| 4AA10 | 5.76 | − | + | − | − | + | − | + | − | w | − |
| CECT 185T | 4.24 | − | + | − | − | + | − | + | − | + | − |
| Genotype cremoris | |||||||||||
| 1AA23 | 4.24 | + | + | − | − | + | − | + | w | + | w |
| 2A5 | 4.30 | + | + | − | − | + | − | + | w | + | + |
| 2A22 | 4.29 | + | w | − | − | + | − | + | w | + | w |
| 2A27 | 4.27 | + | + | − | − | + | − | + | − | + | w |
| 3AA9 | 4.32 | w | w | − | − | + | − | + | − | + | w |
| 3AA11 | 4.23 | + | + | − | − | + | − | + | − | + | w |
| 3AA23 | 4.19 | − | + | − | − | + | − | + | − | + | + |
| LC44 | 4.23 | + | + | − | − | + | − | + | − | + | w |
| GE2-14 | 4.25 | + | + | − | − | + | − | + | − | + | w |
| 4AA28 | 4.25 | + | + | − | − | + | − | + | − | + | w |
| CECT 967T | 4.28 | + | − | − | w | − | − | − | − | − | − |
CECT 185T and CECT 976T are the L. lactis subsp. lactis and L. lactis subsp. cremoris type strains, respectively.
w, weak reaction or growth.
Carbohydrate fermentation profiles were analyzed by the combined use of the PhenePlate and API 20 Strep systems, in which the utilization of 51 sugar and polyalcohols was examined. The two systems always showed concordant results with the carbohydrates tested. Table 3 shows the combined results. Strain-to-strain variations were found among both the lactis and cremoris genotypes. However, the fermentation profiles shown by the strains of the cremoris genotype were wider owing to their use of l-arabinose, arbutin, glycerol, inosine, mannitol, starch, and d-xylose. Key sugars used for distinguishing strains belonging to the L. lactis subsp. lactis (positive) and L. lactis subsp. cremoris (negative) subspecies were in fact equally fermented by all the present strains (such as maltose and ribose) or were fermented by more strains of the cremoris genotype (mannitol).
Table 3.
Carbohydrate fermentation profiles of the wild L. lactis subsp. lactis strains of lactis and cremoris genotypes of this study
| Genotype/straina | Fermentation of carbohydrateb |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adonitol | Amigdalin | l-Arabinose | Arbutin | Gentibiose | Gluconate | Glycerol | Inosine | 5-keto-gluconate | Maltose | Mannitol | β-Methyl glucoside | Ribose | Salicine | Starch | Sucrose | d-Xylose | |
| Genotype lactis | |||||||||||||||||
| 1AA59 | − | − | − | − | − | − | − | − | − | + | − | − | + | + | w | + | − |
| CAS3 | − | + | − | − | + | − | − | w | − | + | + | + | + | + | + | + | − |
| L39 | − | − | − | − | − | − | − | − | − | + | − | − | + | + | − | − | − |
| 3AA15 | − | + | − | − | + | − | − | w | − | + | − | + | + | + | w | + | − |
| 2BA36 | − | + | − | − | + | − | − | + | − | + | w | + | + | − | + | + | − |
| 1A38 | w | − | − | − | + | w | − | − | − | + | − | + | + | + | w | w | − |
| 2A83 | − | − | − | − | + | − | − | − | − | + | − | + | + | + | w | − | − |
| GE-1 | − | + | − | w | + | − | + | − | − | + | w | + | + | + | w | + | − |
| Q1-6 | − | + | − | − | − | − | − | − | − | + | w | + | + | + | − | − | − |
| 4AA10 | − | + | − | w | + | − | w | − | − | + | − | + | + | + | w | + | − |
| CECT 185T | − | − | − | − | + | w | − | − | − | + | − | + | + | + | w | − | − |
| Genotype cremoris | |||||||||||||||||
| 1AA23 | − | + | w | − | + | − | w | w | − | + | + | + | + | + | + | + | + |
| 2A22 | − | + | w | w | + | − | w | w | − | + | + | + | + | + | + | + | + |
| 2A27 | − | + | − | w | + | − | − | + | − | + | + | + | + | + | + | + | + |
| 2A5 | − | + | − | w | + | − | − | w | − | + | + | + | + | + | + | + | + |
| 3AA23 | − | − | − | w | + | − | − | − | − | + | + | + | + | + | + | − | + |
| 3AA11 | − | − | + | w | w | − | w | w | w | + | + | w | + | w | + | + | − |
| 3AA9 | − | + | − | w | + | − | w | w | − | + | + | + | + | + | w | + | + |
| LC44 | − | + | − | − | + | − | w | w | − | + | w | + | − | + | + | − | − |
| GE2-14 | − | − | − | − | − | − | − | − | − | + | w | − | + | + | w | − | − |
| 4AA28 | − | − | + | − | + | − | − | − | − | + | w | − | + | + | w | − | − |
| CECT 967T | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
CECT 185T and CECT 976T are the L. lactis subsp. lactis and L. lactis subsp. cremoris type strains, respectively.
All strains fermented glucose, galactose, lactose, and lactulose, and all but L. lactis subps. cremoris CECT 967T fermented cellobiose and trehalose. None of the strains fermented d-arabitol, deoxyribose, doxyglucose, dulcitol, d-fucose, l-fucose, fumarate, galacturonic-lactone, glycogen, inositol, inulin, malinate, malonate, maltitol, mannonic acid lactone, melbionate, melezitose, melibiose, ornithine, palatinose, pyruvate, raffinose, rhamnose, sorbitol, sorbose, tagatose, l-tartrate, or urea under the assay conditions. w, weak reaction.
To further compare the biochemical properties of the strains, all were subjected to phenotypic profiling for enzyme activities using the API ZYM and API 20 Strep systems and for antibiotic resistance-susceptibility via determination of the MICs for 16 antibiotics using the VetMIC system. The results are shown in Tables 4 and 5, respectively. Only 10 of the 20 enzymes whose activities were assayed with the two kits were positive for the strains studied. Activities showed high variability among the strains and between genotypes (Table 4). The enzyme profiles of the cremoris genotype were usually greater and/or the level of activity was higher than those of the lactis genotype; these showed moderate esterase (C4) activity and high esterase-lipase (C8), acid phosphatase, naphthol-AS-BI-phosphohydrolase, and α-glucosidase activities. In contrast β-galactosidase and β-glucosidase activities were usually stronger for the lactis genotype strains.
Table 4.
Enzymatic activities measured with the API ZYM and 20 Strep systems in wild L. lactis subsp. lactis strains of lactis and cremoris genotypes isolated from raw milk, starter-free cheeses
| Genotype/ straina | Enzymatic activityb |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Alkaline phosphatase | Esterase (C4) | Esterase lipase (C8) | Pyrrolidonyl- arylamidase | Leucine arylamidase | Acid phosphatase | Naphthol-AS-BI- phosphohydrolase | β-Galactosidase | α-Glucosidase | β-Glucosidase | |
| Genotype lactis | ||||||||||
| 1AA59 | 2.5 | 0 | 5 | 30 | 10 | 2.5 | 0 | 40 | 2.5 | 0 |
| CAS3 | 2.5 | 0 | 2.5 | 2.5 | 10 | 15 | 2.5 | 10 | 0 | 30 |
| L39 | 2.5 | 0 | 5 | 30 | 15 | 20 | 10 | 2.5 | 15 | 40 |
| 3AA15 | 2.5 | 0 | 2.5 | 30 | 5 | 0 | 2.5 | 40 | 2.5 | 0 |
| 2BA36 | 2.5 | 0 | 5 | 20 | 5 | 5 | 5 | 40 | 5 | 0 |
| 1A38 | 2.5 | 0 | 5 | 2.5 | 20 | 10 | 2.5 | 0 | 0 | 40 |
| 2A83 | 2.5 | 0 | 5 | 5 | 5 | 5 | 5 | 0 | 0 | 0 |
| GE-1 | 0 | 0 | 5 | 20 | 40 | 15 | 2.5 | 20 | 0 | 40 |
| Q1-6 | 2.5 | 0 | 5 | 30 | 20 | 10 | 5 | 2.5 | 0 | 40 |
| 4AA10 | 2.5 | 0 | 5 | 20 | 10 | 10 | 2.5 | 0 | 0 | 15 |
| CECT 185T | 2.5 | 0 | 5 | 20 | 30 | 20 | 5 | 2.5 | 40 | 40 |
| Genotype cremoris | ||||||||||
| 1AA23 | 2.5 | 2.5 | 5 | 20 | 10 | 30 | 10 | 2.5 | 10 | 0 |
| 2A22 | 2.5 | 5 | 10 | 30 | 10 | 40 | 15 | 2.5 | 20 | 20 |
| 2A27 | 2.5 | 5 | 10 | 30 | 10 | 40 | 15 | 0 | 10 | 10 |
| 2A5 | 2.5 | 5 | 10 | 30 | 10 | 35 | 15 | 0 | 20 | 20 |
| 3AA23 | 2.5 | 0 | 5 | 20 | 20 | 40 | 20 | 0 | 40 | 40 |
| 3AA11 | 2.5 | 10 | 10 | 30 | 5 | 20 | 15 | 10 | 15 | 0 |
| 3AA9 | 2.5 | 5 | 10 | 30 | 10 | 30 | 15 | 0 | 15 | 20 |
| LC44 | 2.5 | 2.5 | 5 | 20 | 40 | 40 | 20 | 2.5 | 20 | 20 |
| GE2-14 | 2.5 | 5 | 5 | 30 | 5 | 5 | 5 | 10 | 5 | 0 |
| 4AA28 | 2.5 | 2.5 | 2.5 | 5 | 5 | 30 | 20 | 2.5 | 10 | 10 |
| CECT 967T | 0 | 2.5 | 5 | 20 | 5 | 10 | 5 | 0 | 0 | 0 |
CECT 185T and CECT 976T are the L. lactis subsp. lactis and L. lactis subsp. cremoris type strains, respectively.
Units of activity are expressed as nanomoles of substrate hydrolyzed under the assay conditions. Valine arylamidase, cysteine arylamidase, lipase (C14), trypsin, α-quimiotrypsin, α-galactosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase activities were never detected.
Table 5.
MICs of 16 antibiotics to wild L. lactis subsp. lactis strains of lactis and cremoris genotypes
| Genotype/straina | MIC (μg/ml)b |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gm | Km | Sm | Nm | Tc | Em | Cl | Cm | Am | PG | Va | Vi | Lz | Tm | Ci | Ri | |
| Genotype lactis | ||||||||||||||||
| 1AA59 | 4 | 8 | 32 | 8 | 0.50 | 0.25 | 0.12 | 8 | 0.50 | 0.25 | 0.50 | 1 | 2 | >64 | 8 | 16 |
| CAS3 | 4 | 16 | 32 | 8 | 0.25 | 0.25 | 0.12 | 2 | 0.25 | 0.25 | 0.50 | 2 | 2 | >64 | 4 | 16 |
| L39 | 1 | 8 | 16 | 2 | 0.50 | 0.25 | 0.25 | 4 | 0.25 | 0.25 | 0.50 | 2 | 4 | >64 | 4 | 16 |
| 3AA15 | 2 | 16 | 16 | 4 | 0.50 | 0.25 | 0.12 | 4 | 0.25 | 0.25 | 0.25 | 1 | 2 | >64 | 4 | 16 |
| 2BA36 | 2 | 8 | 32 | 2 | 0.50 | 0.25 | 0.25 | 8 | 0.12 | 0.25 | 0.25 | 2 | 4 | >64 | 4 | 16 |
| 1A38 | 4 | 16 | 64 | 8 | 0.50 | 0.25 | 0.25 | 4 | 0.25 | 0.25 | 0.50 | 2 | 2 | >64 | 2 | 32 |
| 2A83 | 2 | 8 | 32 | 8 | 0.50 | 0.50 | 0.12 | 4 | 0.25 | 0.12 | 0.50 | 0.50 | 2 | >64 | 4 | 32 |
| GE-1 | 4 | 16 | 32 | 8 | 0.50 | 0.25 | 0.12 | 8 | 0.25 | 0.25 | 0.25 | 1 | 4 | >64 | 8 | 16 |
| Q1-6 | 2 | 8 | 16 | 4 | 0.50 | 0.25 | 0.12 | 4 | 0.12 | 0.12 | 0.50 | 1 | 2 | >64 | 4 | 16 |
| 4AA10 | 4 | 32 | 64 | 16 | 0.50 | 0.25 | 0.25 | 8 | 0.25 | 0.25 | 0.50 | 2 | 2 | >64 | 4 | 16 |
| CECT 185T | 16 | 32 | 64 | 32 | 0.50 | 0.25 | 0.12 | 8 | 0.50 | 0.50 | 0.50 | 8 | 4 | >64 | 8 | >64 |
| Genotype cremoris | ||||||||||||||||
| 1AA23 | 1 | 8 | 16 | 16 | 0.50 | 0.25 | 0.25 | 4 | 0.25 | 0.25 | 0.50 | 2 | 2 | >64 | 8 | 8 |
| 2A22 | 4 | 16 | 64 | 32 | 64 | 0.25 | 0.50 | 4 | 0.25 | 0.25 | 0.50 | 2 | 2 | >64 | 16 | 32 |
| 2A27 | 8 | 32 | 64 | 32 | 64 | 0.25 | 0.50 | 4 | 0.25 | 0.25 | 0.50 | 2 | 2 | >64 | 16 | 32 |
| 2A5 | 8 | 16 | 64 | 16 | 64 | 0.25 | 0.50 | 4 | 0.25 | 0.25 | 0.50 | 2 | 2 | >64 | 16 | 32 |
| 3AA23 | 16 | 64 | 128 | 64 | 0.50 | 0.25 | 0.50 | 16 | 0.25 | 0.25 | 0.50 | 2 | 2 | >64 | 8 | 8 |
| 3AA11 | 2 | 8 | 32 | 8 | 1 | 0.12 | 0.25 | 8 | 0.25 | 0.25 | 0.50 | 2 | 2 | >64 | 8 | 32 |
| 3AA9 | 16 | 64 | 128 | 64 | 0.50 | 0.25 | 0.50 | 4 | 0.25 | 0.25 | 0.50 | 2 | 2 | >64 | 8 | 8 |
| LC44 | 2 | 16 | 32 | 8 | 0.50 | 0.25 | 0.25 | 8 | 0.25 | 0.50 | 0.50 | 2 | 2 | >64 | 8 | 32 |
| GE2-14 | 8 | 32 | 64 | 8 | 0.50 | 0.12 | 0.25 | 8 | 0.25 | 0.25 | 0.50 | 2 | 2 | >64 | 4 | 32 |
| 4AA28 | 2 | 16 | 32 | 4 | 0.50 | 0.25 | 0.25 | 8 | 0.12 | 0.12 | 0.50 | 2 | 2 | >64 | 8 | 16 |
| CECT 967T | 0.50 | 2 | 2 | 1 | 0.50 | 0.25 | 0.12 | 8 | 0.25 | 0.25 | 0.50 | 1 | 2 | 64 | 4 | >64 |
CECT 185T and CECT 976T are the L. lactis subsp. lactis and L. lactis subsp. cremoris type strains, respectively.
Antibiotic abbreviations: Gm, gentamicin; Km, kanamycin; Sm, streptomycin; Nm, neomycin; Tc, tetracycline; Em, erythromycin; Cl, clindamycin; Cm, chloramphenicol; Am, ampicillin; PG, penicillin G; Va, vancomycin; Vi, virginiamycin; Lz, linezolid; Tm, trimethoprim; Ci, ciprofloxacin; Ri, rifampin.
Few differences, if any, were observed in terms of the antibiotic MIC profiles among the strains of the lactis and cremoris genotypes (Table 5). High MICs were observed in both the lactis and cremoris genotypes for antibiotics to which lactococci have been reported to be intrinsically resistant (aminoglycosides, trimethoprim, and rifampin) (2). As an exception, the three identical strains of the cremoris genotype, 2A5, 2A22, and 2A27, showed atypical tetracycline MICs, compatible with the presence of dedicated resistance mechanisms. Following standard gene amplification, sequencing, and sequence analysis, all three strains were shown to harbor a tet(S) gene, which is thought to be responsible for strong resistance to this antibiotic.
Except for three lactis genotype strains (2BA36, 2A83, and 4AA10), all coagulated UHT milk at 22°C, reaching a final pH at 24 h ranging from 4.89 to 4.14 (Table 2). After 48 h in milk at 30°C, 11 volatile compounds were detected by HS/GC/MS, of which four were quantified (Table 6). The repeatability of this analysis was high; the coefficients of variation for the different volatile compounds and strains varied from 1 to 8%. Large strain-to-strain variations in either absolute or relative abundance of most of the volatile compounds were observed in strains of both the lactis and cremoris genotypes, especially with respect to the production of acetaldehyde, ethanol, and aldehyde- and alcohol-derived compounds from the catabolism of branched-chain amino acids. The volatile compound profiles of the lactis and cremoris genotype strains were qualitatively and quantitatively different. The lactis genotype strains produced more diacetyl and marginal levels of acetoin, while the cremoris genotype strains produced higher levels of acetaldehyde and all the known amino acid-derived flavor compounds, especially 2-methyl propanal, 3-methyl butanal, and 2- and 3-methyl butanol. Prolonged incubation of milk for 5 and 21 days did not show a significant difference either in the volatile profiles of the strains or in the abundance of the volatile compounds detected (data not shown).
Table 6.
Absolute or relative abundance of volatile compounds produced in UHT milk at 30°C for 48 h by the wild L. lactis subsp. lactis strains of the lactis and cremoris genotypes detected by HS/GC/MSa
| Genotype/strainb | Volatile compound concn (μg/ml) |
Relative abundancec |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acetaldehyde | 2-Propanone | Ethanol | Diacetyl | Acetic acid | Acetoin | Methanethiol | 2-Methyl propanal | 2-Methyl propanol | 2-Methyl butanal | 3-Methyl butanal | 2- and 3-Methyl butanol | |
| Genotype lactis | ||||||||||||
| 1AA59 | 6.41 | 11.47 | 802.33 | 7.31 | —d | 0.06 | 0.88 | 3.60 | — | 3.19 | 15.49 | 8.10 |
| CAS3 | 5.35 | 13.89 | 863.77 | 3.71 | — | — | 0.87 | — | — | — | — | — |
| L39 | 6.06 | 13.55 | 586.84 | — | — | — | 0.79 | 6.30 | 3.35 | 2.99 | 32.89 | 34.71 |
| 3AA15 | 5.60 | 13.87 | 865.09 | 8.35 | — | 0.06 | 0.91 | 4.42 | 0.88 | 3.06 | 15.64 | 8.71 |
| 2BA36 | 5.88 | 12.05 | 650.70 | 9.27 | — | 0.08 | 0.87 | 0.95 | — | 0.66 | 6.34 | 5.19 |
| 1A38 | 27.57 | 9.03 | 140.84 | 2.55 | — | 0.19 | — | — | — | — | — | — |
| 2A83 | 6.41 | 11.47 | 802.33 | 7.31 | — | 0.05 | 0.96 | 0.28 | — | — | 0.98 | 4.97 |
| GE-1 | 6.38 | 13.05 | 667.91 | 6.57 | — | — | 0.75 | — | — | — | — | — |
| Q1-6 | 1.55 | 9.93 | 153.24 | 4.31 | — | 0.43 | — | — | — | — | — | — |
| 4AA10 | 20.89 | 8.63 | 1,239.09 | 3.71 | — | — | 0.87 | — | — | — | — | — |
| CECT 185T | 5.26 | 1.38 | 238.9 | — | — | — | — | — | 0.35 | — | — | 2.48 |
| Genotype cremoris | ||||||||||||
| 1AA23 | 33.71 | 15.05 | 731.74 | 0.70 | — | — | — | — | — | — | — | — |
| 2A22 | 22.33 | 12.25 | 1,146.28 | 4.80 | — | — | 0.85 | 14.33 | 5.75 | 3.25 | 84.50 | 36.13 |
| 2A27 | 21.89 | 11.50 | 1,140.95 | 4.78 | — | — | 0.78 | 14.49 | 6.42 | 3.23 | 88.33 | 39.87 |
| 2A5 | 23.89 | 14.58 | 1,287.38 | 5.52 | — | — | 0.81 | 15.13 | 5.91 | 3.53 | 87.65 | 37.10 |
| 3AA23 | 17.91 | 9.75 | 1,170.95 | 4.84 | — | — | 0.46 | 19.76 | 9.10 | 2.79 | 89.76 | 53.99 |
| 3AA11 | 26.93 | 10.83 | 747.33 | 3.70 | — | 0.07 | 0.25 | 32.71 | 6.24 | 35.82 | 108.76 | 70.72 |
| 3AA9 | 79.56 | 10.76 | 823.03 | 2.40 | — | — | 0.38 | 17.38 | 9.76 | 0.95 | 58.39 | 66.17 |
| LC44 | 25.00 | 9.31 | 402.08 | — | — | — | 0.26 | — | — | — | — | — |
| GE2-14 | 33.81 | 12.46 | 679.72 | 2.56 | — | — | 0.24 | 35.74 | 6.06 | 35.09 | 113.76 | 83.26 |
| 4AA28 | 25.95 | 15.31 | 746.83 | — | — | — | 0.70 | 8.27 | 0.97 | 2.69 | 55.39 | 17.88 |
| CECT 967T | 2.25 | 0.87 | 77.50 | — | — | — | — | — | — | — | — | — |
Average results of duplicate analyses are shown.
CECT 185T and CECT 976T are the L. lactis subsp. lactis and L. lactis subsp. cremoris type strains, respectively.
Relative abundance compared to an internal control (cyclohexanone, 0.36 mg/ml).
—, not detected.
DISCUSSION
Traditional cheeses and nondairy fermented products are considered a potential source of new L. lactis strains with novel properties that might be able to replace or complement currently used dairy starters (29, 30, 37, 54). Additionally, wild strains are a source of phenotypic and genetic variability (13, 48) that might be used through genetic engineering to enhance the activity and performance of current starter strains (55). Phenotypically, the 20 strains of this study belonged to L. lactis subsp. lactis, which agrees well with the species identification of wild strains from milk (11, 18, 33) and traditional cheeses (14, 19, 34), as does the low percentage of cremoris genotypes.
In the past, the identification of Lactococcus species and subspecies was based entirely on phenotypic tests, primarily because species and subspecies are defined by their phenotypes (20). Phenotypic assays are sometimes ambiguous, can provide different results over the growth phase, and are dependent on culture conditions (19, 42, 52, 54). However, since the 1990s, identification has relied mostly on molecular genetic analyses (22, 42, 49), fuelled by the development of simple PCR-based methods (19, 36, 56). The use of molecular genetic techniques alone, however, has introduced some additional confusion into the taxonomy of L. lactis, complicating the unusual structure of this species (30, 40). Molecular genetic techniques allowed the recovery of strains with a cremoris genotype from different sources, including milk, dairy products, and plant material. It is not clear whether these new cremoris isolates have biochemical properties similar to those of isolates used as starters in the dairy industry. In fact, the phenotypic and technological characterization of cremoris genotype isolates and the comparison of their properties with those of recognized starter strains of both the lactis and cremoris subspecies have only rarely been undertaken (18, 37, 43, 54). Such studies are critical, however, for the selection of the most suitable strains for each application. Replacing the unreliable, traditional phenotypic tests with other phenotypic assays and molecular genetic techniques such as those used in the present work could help to discover new traits for distinguishing between L. lactis subspecies.
The typing results obtained in this study by PFGE agreed well with those previously obtained by random amplification of polymorphic DNA (RAPD) and repetitive extragenic palindromic (REP) techniques (1). In general, cluster analysis of macrorestriction patterns by PFGE showed less similarity between the strains than that obtained by PCR-based typing methods. PFGE is a powerful means of assessing genetic relationships for bacteria due to its larger genome coverage (greater than 90%) than other typing techniques (23). Cluster analysis of the typing results and sequencing of the 16S rRNA genes and housekeeping genes consistently provided two clear-cut clusters—formed separately by the strains of the lactis and cremoris genotypes—with only low-level similarity to one another. Similar results have been reported with other typing (11, 21, 37, 40) and sequencing (17, 40, 49, 54) techniques. The more robust MLST technique, which gathers together several gene sequences, produced strongly separated lactis and cremoris genotypes in deeply branched trees, as reported by other authors (39, 40, 50). Two major genomic lineages for the lactis and cremoris genotypes have also been recently recognized using pangenomic DNA array hybridization, determining the presence or absence of 4,571 gene orthologs (7). Proteomic analysis of the ribosomal proteins by matrix-assisted laser desorption ionization-time of flight mass spectrometry has provided similar results (52). All this would seem to indicate that, irrespective of their phenotype, the lactis and cremoris lineages are phylogenetically related but that they have long been on separate evolutionary paths. In fact, on the basis of 16S rRNA gene divergence (less than 0.8%), the deviation of the lactis and cremoris genotypes has been estimated to have occurred some 17 million years ago (9). Though it is difficult to infer divergence times for the different evolutionary steps, recent independent MLST analyses have confirmed an early separation of the lactis and cremoris genotypes (39, 40).
Despite the similarity of the lactis and cremoris genotypes shown in the present phenotypic assays, their member strains showed distinguishable carbohydrate utilization and enzyme activity profiles. Surprisingly, the strains with a cremoris genotype showed greater fermentation and enzyme activity profiles, even though true L. lactis subsp. cremoris strains are reported to have extremely reduced fermentation and enzyme activity profiles (20, 25, 45). The ability of L. lactis to ferment carbohydrates is thought to be related to the degree of adaptation of the strains to the dairy environment (30), independent of genotype. The genome sequence of two plant-associated L. lactis subsp. lactis strains shows the largest number of genes in the carbohydrate metabolism and transport category (46); therefore, they ferment more carbohydrates than any of the three sequenced dairy strains (MG 1363, IL 1403, and SK 11). Furthermore, the genome of L. lactis subsp. lactis MG 1363 (cremoris genotype) has been shown to carry more genes in these two categories (57) than that of L. lactis subsp. lactis IL 1403 (lactis genotype) (8); consequently, it can utilize more sugars. Chromosome size has been shown to vary widely among L. lactis strains of both subspecies (30, 39). However, the origin of the L. lactis strains has been shown to correlate with chromosome size, particularly for the phenotypic L. lactis subsp. cremoris strains, which have smaller chromosomes than the L. lactis subsp. lactis strains of both the lactis and cremoris genotypes (30). Chromosome sizes in this study fell within the normal range for L. lactis strains of either the lactis and cremoris genotype (30, 39). Adaptation of L. lactis to grow in milk is thought to have occurred by gene decay and acquisition of key traits (30, 46, 57), most probably under the high selective pressure imposed by cheese- and butter-making technologies. The analysis of the implicated genes and the whole-genome sequences of more strains might give clues about the current phylogenetic position of the lactis and cremoris genotypes and the evolutionary processes that gave rise to these dairy-adapted starter strains.
In the present work, acid production in milk was variable among the strains of the two genotypes; a few strains even failed to coagulate the provided UHT milk after 24 h of incubation at 22°C. In general, the cremoris genotypes usually caused the pH to decrease more than the lactis genotypes, as reported elsewhere (37, 54).
Wild and nondairy L. lactis strains have occasionally been associated with off-flavor production (3, 54), which correlates with the formation of large amounts of volatile compounds via the degradation of branched-chain amino acids (Leu, Ile, Val). These compounds have a very low taste threshold and have been connected with malty and burnt notes in dairy products (47). However, selected wild strains or combinations of wild and starter strains have been shown to enhance the typical flavors and to increase ripening indices (4, 5). Moreover, the same volatile compounds that caused the off flavors mentioned above seem to be involved in the desired strong flavors of cheeses made from raw milk (12); these flavors are strongly fostered in traditional cheese varieties, particularly those with protected designation of origin (PDO) status. Starter candidates might therefore be selected among wild L. lactis strains to ensure the production of intensely flavored cheeses.
In conclusion, the overall phenotypic and genotypic relatedness of the strains belonging to the lactis and cremoris genotypes suggests that they should be considered members of the same species, as their properties meet the criteria presently used in the species concept for prokaryotes (41). However, despite their similarity, the lactis and cremoris genotypes consistently cluster separately when investigated with simple molecular genetic techniques, suggesting that they should be considered true subspecies, a possibility contemplated in the species definition referred to above (41) and on the more recent recommendations for the taxonomy of prokaryotes (53). Separate or combined simple matching-based cluster analysis of the phenotypic traits analyzed in the present study (carbohydrate fermentation, enzyme activities, production of volatile compounds) consistently gave the same two well-separated clusters as the molecular genetic techniques (data not shown). Thus, in disagreement with some other authors (16, 40), irrespective of their phenotype, all the cremoris genotypes should be considered to belong to the cremoris subspecies.
Analysis of more L. lactis collections, including representative strains of all subspecies and biovars from different environments, using state-of-the-art high-throughput phenotypic (Biolog, cheese models) and molecular genetic (genome sequencing, microarray hybridization, comparative genomic hybridization) screening techniques, should further help in assessing the diversity of the lactococci. These studies should be aimed at correlating genomic makeup and phenotypic traits with industrial performance, which has ultimately to be assessed by carefully controlled trials using defined mixtures of phenotypic and genotypic strains of both subspecies.
ACKNOWLEDGMENTS
This research was supported by a project from the Spanish Ministry of Science and Innovation (MICINN) (AGL2007-61869-ALI). E. Fernández and A. Alegría were awarded a scholarship of the FPI program from MICINN (BES-2008-002031) and the Severo Ochoa program from FICYT (BP08-053), respectively. S. Delgado was supported by a research contract of MICINN under the Juan de la Cierva program (JCI-2008-02391).
P. Le Bourgeois, Université Paul Sabatier de Toulouse, Toulouse, France, is acknowledged for the critical reading of the manuscript.
Footnotes
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Alegría A., Delgado S., Roces C., López B., Mayo B. 2010. Bacteriocins produced by wild Lactococcus lactis strains isolated from traditional, starter-free cheeses made of raw milk. Int. J. Food Microbiol. 143:61–66 [DOI] [PubMed] [Google Scholar]
- 2. Ammor M. S., Flórez A. B., Mayo B. 2007. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 24:559–570 [DOI] [PubMed] [Google Scholar]
- 3. Ayad E. H. E., Verheul A., de Jong C., Wouters J. T. M., Smit G. 1999. Flavour forming abilities and amino acid requirements of Lactococcus lactis strains isolated from artisanal and non-dairy origin. Int. Dairy J. 9:725–735 [Google Scholar]
- 4. Ayad E. H. E., Verheul A., Wouters J. T. M., Smit G. 2000. Application of wild starter cultures for flavour development in pilot plant cheese making. Int. Dairy J. 10:169–179 [Google Scholar]
- 5. Ayad E. H. E., Verheul A., Engels W. J., Wouters J. T. M., Smit G. 2001. Enhanced flavour formation by combination of selected lactococci from industrial and artisanal origin with focus on completion of a metabolic pathway. J. Appl. Microbiol. 90:59–67 [DOI] [PubMed] [Google Scholar]
- 6. Ayad E. H. E., Verheul A., Wouters J. T. M., Smit G. 2002. Antimicrobial-producing wild lactococci isolated from artisanal and non-dairy origins. Int. Dairy J. 12:145–150 [Google Scholar]
- 7. Bayjanov J. R., et al. 2009. PanCGH: a genotype-calling algorithm for pangenome CGH data. Bioinformatics 25:309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bolotin A., et al. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bolotin A., Quinquis B., Sorokin A., Ehrlich D. S. 2004. Recent genetic transfer between Lactococcus lactis and enterobacteria. J. Bacteriol. 186:6671–6677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clermont D., Chesneau O., de Cespedes G., Horaud T. 1997. New tetracycline resistance determinants coding for ribosomal protection in streptococci and nucleotide sequence of tet(T) isolated from Streptococcus pyogenes A498. Antimicrob. Agents Chemother. 41:112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Corroler D., Mangin I., Desmasures N., Guéguen M. 1998. An ecological study of lactococci isolated from raw milk in the Camembert cheese registered designation of origin area. Appl. Environ. Microbiol. 64:4729–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Curioni P. M. G., Bosset J. O. 2002. Key odorants in various cheese types as determined by gas chromatography-olfactometry. Int. Dairy J. 12:959–984 [Google Scholar]
- 13. de la Plaza M., Fernández de Palencia P., Peláez C., Requena T. 2004. Biochemical and molecular characterization of alpha-ketoisovalerate decarboxylase, an enzyme involved in the formation of aldehydes from amino acids by Lactococcus lactis. FEMS Microbiol. Lett. 238:367–374 [DOI] [PubMed] [Google Scholar]
- 14. Delgado S., Mayo B. 2004. Phenotypic and genetic diversity of Lactococcus lactis and Enterococcus spp. strains isolated from northern Spain starter-free farmhouse cheeses. Int. J. Food Microbiol. 90:309–319 [DOI] [PubMed] [Google Scholar]
- 15. Dolci P., et al. 2008. Microbial dynamics of Calstelmagno PDO, a traditional Italian cheese, with focus on lactic acid bacteria ecology. Int. J. Food Microbiol. 122:302–311 [DOI] [PubMed] [Google Scholar]
- 16. Felis G. E., Molenaar D., Dellaglio F., van Hylckama Vlieg J. E. 2007. Dichotomy in post-genomic microbiology. Nat. Biotechnol. 25:848–849 [DOI] [PubMed] [Google Scholar]
- 17. Flórez A. B., de los Reyes-Gavilán C. G., Wind A., Mayo B., Margolles A. A. 2006. Ubiquity and diversity of multidrug resistance genes in Lactococcus lactis strains isolated between 1936 and 1995. FEMS Microbiol. Lett. 263:21–25 [DOI] [PubMed] [Google Scholar]
- 18. Franciosi E., Settanni L., Cavaza A., Poznanski E. 2008. Biodiversity and technological potential of wild lactic acid bacteria from raw cow's milk. Int. Dairy J. 19:3–11 [Google Scholar]
- 19. Garde S., Babin M., Gaya P., Núñez M., Medina M. 1999. PCR amplification of the gene acmA differentiates Lactococcus lactis subsp. lactis and L. lactis subsp. cremoris. Appl. Environ. Microbiol. 65:5151–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garvie E., Farrow J. A. E. 1982. Streptococcus lactis subsp. cremoris (Orla-Jensen) comb. nov. and Streptococcus lactis subsp. diacetylactis (Matuszewski et al.) nom. rev., comb. nov. Int. J. Syst. Bacteriol. 32:453–455 [Google Scholar]
- 21. Gaya P., Babín M., Medina M., Núñez M. 1999. Diversity among lactococci isolates from ewes' raw milk and cheeses. J. Appl. Microbiol. 87:849–855 [DOI] [PubMed] [Google Scholar]
- 22. Godon J.-J., Delorme C., Ehrlich S. D., Renault P. 1992. Divergence of genomic sequences between Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. cremoris. Appl. Environ. Microbiol. 58:4045–4047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goering R. V. 2010. Pulsed field gel electrophoresis: a review of application and interpretation in the molecular epidemiology of infectious disease. Infect. Genet. Evol. 10:866–875 [DOI] [PubMed] [Google Scholar]
- 24. Hansen E. B. 2002. Commercial bacterial starter cultures for fermented foods of the future. Int. J. Food Microbiol. 78:119–131 [DOI] [PubMed] [Google Scholar]
- 25. Holt J. G. 1994. Gram positive cocci, p. 527–558 In Holt J. G., Krieg N. R., Sneath P. H. A., Staley J. T., Williams S. T. (ed.), Bergey's manual of determinative bacteriology, 9th ed. Williams & Wilkins, Baltimore, MD [Google Scholar]
- 26. Howard P. J., Harsono K. D., Luchansky J. B. 1992. Differentiation of Listeria monocytogenes, Listeria innocua, Listeria ivanovii, and Listeria seeligeri by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 58:709–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jarvis A. W., Jarvis B. D. W. 1981. DNA homology among lactic streptococci. Appl. Environ. Microbiol. 41:77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kelly W., Davey G. P., Ward L. J. H. 1998. Characterization of lactococci isolated from minimally processed fresh fruit and vegetables. Int. J. Food Microbiol. 45:85–92 [DOI] [PubMed] [Google Scholar]
- 29. Kelly W., Ward L. J. H. 2002. Genotypic vs. phenotypic biodiversity in Lactococcus lactis. Microbiology 148:3332–3333 [DOI] [PubMed] [Google Scholar]
- 30. Kelly W. J., Ward L. J. H., Leahy S. C. 2010. Chromosomal diversity in Lactococcus lactis and the origin of dairy starter cultures. Genome Biol. Evol. 2:729–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kempler G. M., McKay L. L. 1981. Biochemistry and genetics of citrate utilization in Streptococcus lactis subsp. diacetylactis. J. Dairy Sci. 64:1527–1539 [Google Scholar]
- 32. Klare I., et al. 2005. Evaluation of new broth media for microdilution antibiotic susceptibility testing of lactobacilli, pediococci, lactococci, and bifidobacteria. Appl. Environ. Microbiol. 71:8982–8986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klijn N., Weerkamp A. H., de Vos W. M. 1995. Detection and characterization of lactose-utilizing Lactococcus spp. in natural ecosystems. Appl. Environ. Microbiol. 61:788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mannu L., Paba A., Pes M., Scintu M. F. 2000. Genotypic and phenotypic heterogeneity among lactococci isolated from traditional pecorino sardo cheese. J. Appl. Microbiol. 89:191–197 [DOI] [PubMed] [Google Scholar]
- 35. Nieto-Arribas P., Seseña S., Poveda J. M., Palop M. L., Cabezas L. 2009. Genotypic and technological characterization of Lactococcus lactis isolates involved in processing of artisanal Manchego cheese. J. Appl. Microbiol. 107:1505–1517 [DOI] [PubMed] [Google Scholar]
- 36. Nomura M., Kobayashi M., Okamoto T. 2002. Rapid PCR-based method which can determine both phenotype and genotype of Lactococcus lactis subspecies. Appl. Environ. Microbiol. 68:2209–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nomura M., Kobayashi M., Narita T., Kimoto-Nira H., Okamoto T. 2006. Phenotypic and molecular characterization of Lactococcus lactis from milk and plants. J. Appl. Microbiol. 101:396–405 [DOI] [PubMed] [Google Scholar]
- 38. Parente E., Cogan T. M. 2004. Starter cultures: general aspects, p. 123–147 In Fox P. O. (ed.), Cheese: chemistry, physics and microbiology, 3rd ed. Elsevier, Oxford, United Kingdom [Google Scholar]
- 39. Passerini D., et al. 2011. Genes but not genomes reveal bacterial domestication of Lactococcus lactis. PLoS One 5:e15306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rademaker J. L. W., et al. 2007. Diversity analysis of dairy and non-dairy Lactococcus lactis isolates, using a novel multilocus sequence analysis scheme and (GTG)5-PCR fingerprinting. Appl. Environ. Microbiol. 73:7128–7137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roselló-Mora R., Amman R. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39–67 [DOI] [PubMed] [Google Scholar]
- 42. Salama M. S., Sandine W. E., Giovannoni S. J. 1993. Isolation of Lactococcus lactis subsp. cremoris from nature by colony hybridization with rRNA probes. Appl. Environ. Microbiol. 59:3941–3945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Salama M. S., Musafija-Jeknic T., Sandine W. E., Giovannoni S. J. 1995. An ecological study of lactic acid bacteria: isolation of new strains of Lactococcus lactis including Lactococcus lactis subspecies cremoris. J. Dairy Sci. 78:1004–1017 [Google Scholar]
- 44. Salazar N., Gueimonde M., Hernández-Barranco A. M., Ruas-Madiedo P., de los Reyes-Gavilán C. G. 2008. Exopolysaccharides produced by intestinal Bifidobacterium strains act as fermentable substrates for human intestinal bacteria. Appl. Environ. Microbiol. 74:4737–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schleifer K. H., et al. 1985. Transfer of Streptococcus lactis and related streptococci to the genus Lactococcus gen. nov. Syst. Appl. Microbiol. 6:183–195 [Google Scholar]
- 46. Siezen R. J., et al. 2008. Genome-scale genotype-phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl. Environ. Microbiol. 74:424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smit G., Smit B. A., Engels W. J. M. 2005. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 29:591–610 [DOI] [PubMed] [Google Scholar]
- 48. Smit B. A., et al. 2005. Identification, cloning, and characterization of a Lactococcus lactis branched-chain alpha-keto acid decarboxylase involved in flavor formation. Appl. Environ. Microbiol. 71:303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Swindell S. R., Griffin H. G., Gasson M. J. 1994. Cloning, sequencing and comparison of three lactococcal l-lactate dehydrogenase genes. Microbiology 140:1301–1305 [DOI] [PubMed] [Google Scholar]
- 50. Taïbi A., Dabour N., Lamoureux M., Roy D., Lapointe G. 2010. Evaluation of the genetic polymorphism among Lactococcus lactis subsp. cremoris strains using comparative genomic hybridization and multilocus sequence analysis. Int. J. Food Microbiol. 144:20–28 [DOI] [PubMed] [Google Scholar]
- 51. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 52. Tanigawa K., Kawabata H., Watanabe K. 2010. Identification and typing of Lactococcus lactis by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 76:4055–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tindall B. J., Rosselló-Móra R., Busse H. J., Ludwig W., Kämpfer P. 2010. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 60:249–266 [DOI] [PubMed] [Google Scholar]
- 54. Urbach E., Daniels B., Salama M., Sandine W. E., Giovannoni S. J. 1997. The ldh phylogeny for environmental isolates of Lactococcus lactis is consistent with rRNA genotypes but not with phenotypes. Appl. Environ. Microbiol. 63:694–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. van Hylckama Vlieg J. E., et al. 2006. Natural diversity and adaptive responses of Lactococcus lactis. Curr. Opin. Biotechnol. 17:183–190 [DOI] [PubMed] [Google Scholar]
- 56. Ward L. J., Brown J. C., Davey G. P. 1998. Two methods for the genetic differentiation of Lactococcus lactis ssp. lactis and cremoris based on differences in the 16S rRNA gene sequence. FEMS Microbiol. Lett. 166:15–20 [DOI] [PubMed] [Google Scholar]
- 57. Wegmann U., et al. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]