Abstract
2,3-Butanediol (23BD) is a high-value chemical usually produced petrochemically but which can also be synthesized by some bacteria. To date, the best microbial 23BD production rates have been observed using pathogenic bacteria in fermentation systems that depend on sugars as the carbon and energy sources for product synthesis. Here we present evidence of 23BD production by three nonpathogenic acetogenic Clostridium species—Clostridium autoethanogenum, C. ljungdahlii, and C. ragsdalei—using carbon monoxide-containing industrial waste gases or syngas as the sole source of carbon and energy. Through an analysis of the C. ljungdahlii genome, the complete pathway from carbon monoxide to 23BD has been proposed. Homologues of the genes involved in this pathway were also confirmed for the other two species investigated. A gene expression study demonstrates a correlation between mRNA accumulation from 23BD biosynthetic genes and the onset of 23BD production, while a broader expression study of Wood-Ljungdahl pathway genes provides a transcription-level view of one of the oldest existing biochemical pathways.
INTRODUCTION
2,3-Butanediol (23BD) is a commodity chemical usually produced from oil. It can be used as a precursor in the manufacture of a range of chemical products, including the solvents methyl ethyl ketone (MEK), gamma-butyrolactone (GBL), and 1,3-butadiene (13, 55, 58). Commercially, the key downstream products of 23BD have a potential global market of around 32 million tons per annum, valued at approximately $43 billion in sales.
23BD is known to be produced by a range of sugar (or citrate)-fermenting microbes, including Bacillus amyloliquefaciens (13), Bacillus subtilis (58), Enterobacter aerogenes (11), Klebsiella pneumoniae (7), Klebsiella oxytoca (51), Lactococcus lactis (26), Paenibacillus polymyxa (14), and Serratia marcescens (60). In this study, we present evidence of 23BD production by 3 acetogenic members of the Clostridium genus (Clostridium autoethanogenum, Clostridium ljungdahlii, and Clostridium ragsdalei) using gases (carbon monoxide [CO] or hydrogen [H2] and CO) as the source of carbon and energy.
To date, acetogens have been reported to produce acetate, ethanol, butyrate, and butanol, all of which are synthesized directly from acetyl-coenzyme A (acetyl-CoA) (16, 34, 36). However, whether metabolites known to require pyruvate as a precursor for their synthesis can be produced in significant quantities by microbes using gases as opposed to carbohydrates as the source of metabolic carbon and energy has not yet been demonstrated. In this study, the production of the pyruvate-derived metabolites 23BD and lactate by acetogenic clostridia was investigated. The purpose of this study was 2-fold: to gain insight into the genetic control of carbon flow in acetogenic organisms and to gain insight into the potential versatility of such an approach as a platform for the production of a broader spectrum of fuel or chemical products.
The ability of microbes to use CO relies on the operation of the Wood-Ljungdahl metabolic pathway (Fig. 1 A). The biochemistry associated with this pathway has been described in a variety of excellent review articles (16, 17, 45, 56). CO and/or CO2 feeds the methyl (eastern) and carbonyl (western) branches of the pathway, resulting in the formation of acetyl-CoA, which serves as the central precursor for all catabolic and anabolic processes. In the methyl branch, CO or CO2 is fixed in a sequence of tetrahydrofolate (THF)- and cobalamin-dependent reactions into a methyl group, which is then combined with CO (used either directly or after enzymatic reduction of CO2) to form acetyl-CoA catalyzed by the CODH/ACS (carbon monoxide dehydrogenase/acetyl-CoA synthase) complex (Fig. 1A).
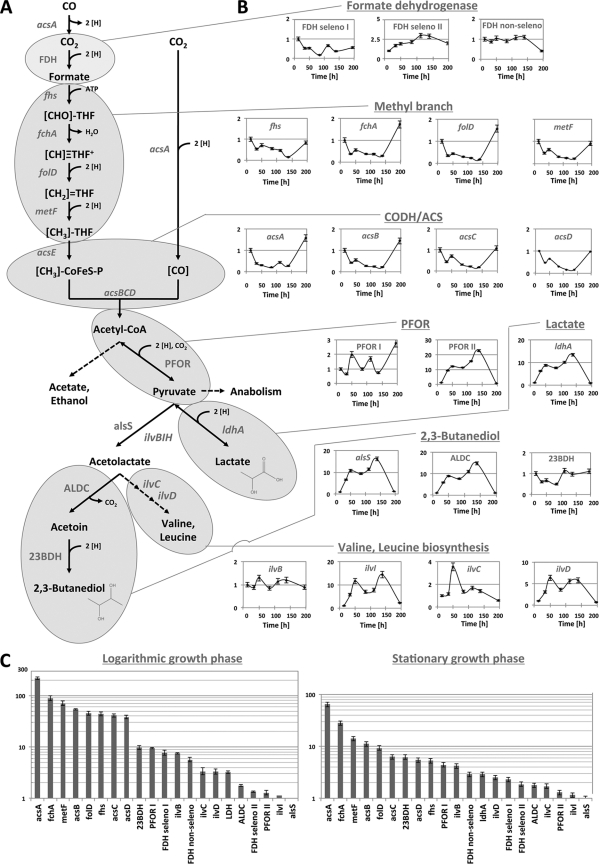
Fig. 1.
(A) Proposed pathway for production of 23BD and lactate from CO via the Wood-Ljungdahl pathway (16, 17, 45, 56). Single reactions shown do not represent stoichiometric fermentation balances. The graphs represent normalized mRNA levels over complete growth relative to those at the first time point (14 h) (B) and compared to the acetolactate synthase gene at the logarithmic (50 h) and stationary (139 h) growth phases (C). 23BDH, 2,3-butanediol dehydrogenase; acs, genes for CODH/ACS complex; acsA, CODH subunit gene; acsB, ACS subunit gene; acsC, corrinoid iron-sulfur protein large subunit gene; acsD, corrinoid iron-sulfur protein small subunit gene; acsE, methyltransferase subunit gene; ALDC, acetolactate decarboxylase; CFeSP, corrinoid iron-sulfur protein; fchA, formimido-tetrahydrofolate cyclodeaminase gene; FDH, formate dehydrogenase; fhs, formyl-tetrahydrofolate synthase gene; folD, bifunctional methylene-tetrahydrofolate dehydrogenase/formyl-tetrahydrofolate cyclohydrolase gene; ilvC, ketol acid reductoisomerase gene; ilvD, dihydroxy acid dehydratase gene; ilvIH, acetolactate synthase gene; metF, methylene-tetrahydrofolate reductase gene; THF, tetrahydrofolate.
Reducing equivalents required for metabolic processes are generated from either CO via the CODH enzyme or H2 via hydrogenase enzymes (33). In the absence of H2 in the input gas, the reducing equivalents are provided solely from CO via the biological water-gas shift reaction (CO + H2O → CO2 + 2 H+ + 2e−). If H2 is present in the gas, additional reducing equivalents are available (H2 → 2 H+ + 2e−), enabling a proportional increase in carbon assimilation. The use of CO (E0′ = −520 mV) and H2 (E0′ = −414 mV), which are not equivalent because of having significantly different redox potentials, allows a broad spectrum of resources to be considered for product synthesis (50).
The results presented herein show that the use of acetogens is a viable strategy for the production of 23BD. Furthermore, evidence for the biosynthetic pathway used for its synthesis from CO is given together with the results of a gene expression study examining the transcription of genes encoding biosynthetic enzymes from CO metabolism through to the biosynthesis of 23BD and lactate. Additionally, stereochemistry and toxicity analyses were undertaken to define the commercially important structural specificity and potential production limits of this molecule by gas fermentation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. autoethanogenum DSM 10061 and C. ljungdahlii DSM 13582 were obtained from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany), and C. ragsdalei ATCC BAA-622 was obtained from the American Type Culture Collection (Manassas, VA). All organisms were cultivated under strict anaerobic conditions (29) in modified PETC medium (ATCC medium 1754 with yeast extract and fructose omitted) at 30°C (C. ragsdalei) or 37°C (C. autoethanogenum and C. ljungdahlii). Heterotrophic growth was performed on 0.5% (wt/vol) fructose, while steel mill waste gas (composition, 44% CO, 32% N2, 22% CO2, and 2% H2; collected from a steel site in Glenbrook, New Zealand) was used for autotrophic growth at a pressure of 200 kPa.
Growth experiments were carried out in 100 ml medium, using 500-ml Schott Duran GL45 Pressure Plus bottles with butyl rubber stoppers. 23BD and lactate toxicity experiments were performed in 125-ml Bellco serum bottles with butyl rubber septa, using 50 ml medium containing increasing concentrations of 23BD (distilled from broth of C. autoethanogenum) and dl-lactic acid (Sigma-Aldrich, St. Louis, MO) (NaOH was used to compensate the pH of the medium after addition of lactic acid). All growth experiments were performed at least in triplicate. Growth was monitored by measuring the optical density at 600 nm (OD600), and metabolic end products were analyzed by high-performance liquid chromatography (HPLC).
Analytics.
Metabolite concentrations were routinely determined using an Agilent 1100 series HPLC system (Agilent Technologies, Santa Clara, CA) equipped with a refractive index detector (RID) operated at 35°C and an Alltech IOA-2000 organic acid column. The column was kept at 60°C. Slightly acidified water (0.005 M H2SO4) was used as the mobile phase, with a flow rate of 0.7 ml/min. To remove proteins and other cell residues, 400 μl of sample was mixed with 100 μl of 2% (wt/vol) 5-sulfosalicylic acid, and the samples were centrifuged at 14,000 × g for 3 min. The supernatant (10 μl) was then injected into the HPLC for analysis. Optical activity was measured at room temperature on a polartronic NH8 instrument (Schmidt+Haensch, Berlin, Germany) at 589 nm and 20°C. Pure chiral standards [d-(−)-23BD, l-(+)-23BD, and meso-23BD] were obtained from Sigma-Aldrich (St. Louis, MO). For 23BD identification, an Agilent gas chromatography-mass spectrometry (GC-MS) system with a quadrupole mass selective detector operated at 70 eV was used. A ZB-1701 column (30 m by 250 μm by 150-μm film thickness) (Phenomenex, Torrance, CA) with a 5-m guard column was used for all analyses.
Sequencing and analysis of genes and enzymes.
The genome sequence of C. ljungdahlii (33) was retrieved from GenBank (accession no. NC_014328), while sequences of C. autoethanogenum and C. ragsdalei were obtained by a whole-genome shotgun approach combined with bidirectional primer walking.
Genomic DNAs used as templates were isolated from overnight cultures (6). One hundred milliliters of culture was harvested by centrifugation (6,000 × g, 15 min, 4°C), washed with potassium phosphate buffer (10 mM; pH 7.5), and suspended in 1.9 ml STE buffer (50 mM Tris-HCl, 1 mM EDTA, 200 mM sucrose [pH 8.0]). This suspension was treated with 300 μl lysozyme (∼100,000 U) for 30 min at 37°C and with 280 μl of an SDS solution (10% [wt/vol]) for 10 min. RNA was digested by addition of 240 μl of an EDTA solution (0.5 M; pH 8), 20 μl Tris-HCl (1 M; pH 7.5), and 10 μl RNase A (50,000 U). Proteolysis was achieved by addition of 100 μl proteinase K (0.5 U) for 1 to 3 h at 37°C. Finally, 600 μl of sodium perchlorate (5 M) was added, followed by phenol-chloroform extraction and isopropanol precipitation. The purity and quantity of DNA were verified using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and a Qubit fluorometer (Invitrogen, Carlsbad, CA) as well as by gel electrophoresis.
Shotgun sequencing (39) was carried out using a 454 GS FLX Titanium instrument (Roche Applied Science, Indianapolis, IN). For C. autoethanogenum, 511,611 reads with a total length of 188,798,483 bp were obtained, and for C. ragsdalei, 430,028 reads totaling 150,049,168 bp were obtained, and the reads were assembled using the Newbler package (Roche Applied Science, Indianapolis, IN). Additionally, primer walking experiments using the Sanger method (47) were performed to confirm the identities of relevant sequences.
Genes and enzymes were predicted using the BLAST (3), COG (53), and TIGRFAM (25) databases. Motif scans were performed against the PROSITE (28) and Pfam (19) databases. T boxes were identified using the RFam database (22).
Gene expression studies.
To follow gene expression over time, duplicate samples were taken during every phase of growth of a C. autoethanogenum culture. The bacterial cell pellets (1.7 × 1010 to 2.5 × 1010 cells) were harvested by centrifugation (6,000 × g, 5 min, 4°C), snap-frozen in liquid nitrogen, and stored at −80°C until RNA extraction. Total RNA was isolated using a PureLink RNA minikit (Invitrogen, Carlsbad, CA) and then eluted in 100 μl of RNase-free water. After DNase I treatment (Roche Applied Science, Indianapolis, IN), the reverse transcription step was then carried out with a standardized amount of total RNA, using a SuperScript III reverse transcriptase kit (Invitrogen, Carlsbad, CA). RNA was checked using an Agilent Bioanalyzer 2100 instrument (Agilent Technologies, Santa Clara, CA) and by gel electrophoresis. A no-reverse-transcriptase control was performed for every primer pair.
All quantitative PCRs (qPCRs) were performed in duplicate, using a MyiQ single-color detection system (Bio-Rad Laboratories, Carlsbad, CA) and a total reaction volume of 15 μl with 25 ng of cDNA template, 67 nM (each) primers (see Table S2 in the supplemental material), and 1× iQ SYBR green supermix (Bio-Rad Laboratories, Carlsbad, CA). The reaction conditions were 95°C for 3 min followed by 40 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 30 s. For detection of primer dimerization or other artifacts of amplification, a melting curve analysis was performed immediately after completion of the qPCR (38 cycles of 58°C to 95°C at 1°C/s). All qPCR products were chosen to be between 110 and 140 bp.
Two housekeeping genes (guanylate kinase and formate tetrahydrofolate ligase genes) were included for each cDNA sample for normalization. Threshold cycle (CT) values are shown in the supplemental material. Derivation of relative gene expression was conducted using Relative Expression Software Tool (REST) 2008 version 2.0.7 (43), which uses the following formula to calculate relative expression: relative expression = concentration of gene of interest/geometric mean concentration of housekeeping genes, where concentration = AeCT (where Ae = amplification efficiency). Dilution series of cDNA spanning 4 log units were used to generate standard curves, and the resulting amplification efficiencies were used to calculate the concentration of mRNA.
Nucleotide sequence accession numbers.
The relevant sequences of C. autoethanogenum and C. ragsdalei were deposited in GenBank under accession numbers HQ876009 to HQ876032.
RESULTS
23BD production.
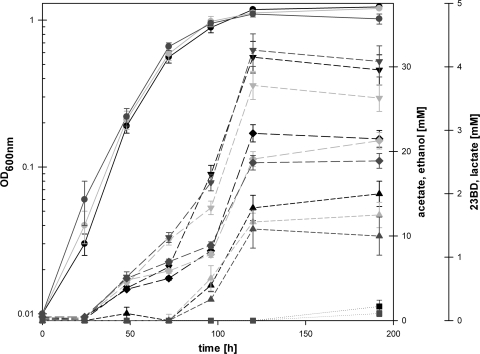
Growth and metabolic profiles of three closely related acetogenic species, C. autoethanogenum (1), C. ljungdahlii (52), and C. ragsdalei (27), on CO-containing steel mill waste gas as the sole energy and carbon source were compared in Schott bottle growth experiments (Fig. 2). Growth rates and maximum cell densities were found to be similar for all three species, although C. ragsdalei has a lower optimal growth temperature. No growth occurred when steel mill waste gas was omitted. As described in previous studies (1, 27, 52), acetate (28 to 32 mM) and ethanol (19 to 22 mM) were found to be the main metabolic end products. However, careful analysis also revealed synthesis of small amounts of 23BD (1.4 to 2 mM) and traces of lactate (0.1 to 0.2 mM) during the stationary growth phase for all three species (Fig. 2). This conclusion was independently confirmed by GC-MS analysis. The molar ratio of 23BD to ethanol was around 1:10, and that of 23BD to acetate was approximately 1:15. None of these products were detected in a medium control with steel mill waste gas in the headspace.
Fig. 2.
Growth experiments with C. autoethanogenum (black), C. ljungdahlii (light gray), and C. ragsdalei (dark gray) on steel mill waste gas (44% CO, 32% N2, 22% CO2, and 2% H2) as the sole carbon and energy source. Circles, biomass; triangles facing down, acetate; diamonds, ethanol; triangles facing up, 23BD; squares, lactate.
To investigate which of the different isomeric forms of 23BD [d-(−), l-(+), or meso] was produced, the substance was purified from cultures of C. autoethanogenum via distillation and further analyzed against chiral standards (55). HPLC chromatograms yielded two peaks, corresponding to 94% d-(−)-23BD and 6% meso-23BD. This was confirmed by measuring the optical activity of the sample, which returned an angle of rotation of −12.05°, compared to −12.93° for the pure d-(−)-standard (while the meso form is optically inactive).
Metabolic pathway for production of 23BD from CO.
To reconstruct the metabolic pathway for synthesis of 23BD from CO, similarities were sought with the well-documented enzymology of 23BD production in sugar-fermenting Enterobacteriaceae and Bacillus species as well as in the acetoin-producing organism Clostridium acetobutylicum (13, 30, 57). An analysis of the recently published C. ljungdahlii genome (33) revealed homologues of the genes encoding the three principle enzymes involved in 23BD production from the central intermediate pyruvate: acetolactate synthase, acetolactate decarboxylase, and 23BD dehydrogenase (see Fig. S1 in the supplemental material). The presence of these genes was also confirmed in C. autoethanogenum and C. ragsdalei by whole-genome shotgun sequencing and primer walking experiments. While these genes are usually clustered together in a common operon in Enterobacteriaceae and Bacillus species (10, 30, 46, 54, 57), they were found to be scattered over the genome in C. ljungdahlii (see Fig. S1), which is also the case for the acetolactate synthase and acetolactate decarboxylase genes of C. acetobutylicum.
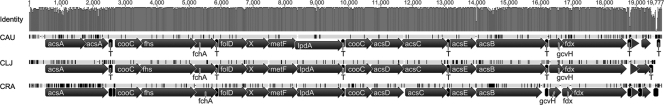
Synthesis of acetyl-CoA from CO via the Wood-Ljungdahl pathway has already been analyzed in detail for C. ljungdahlii (33). All genes (except for the three formate dehydrogenase genes, CLJU_c06990, CLJU_c08930, and CLJU_c20040) are arranged in a single cluster (CLJU_c37530-37670). Sequencing confirmed the same arrangement in C. autoethanogenum and C. ragsdalei (Fig. 3) but revealed differences in the nucleotide sequences of the genes in the three species. Most notable are variations in the acsA and acsB genes, encoding the catalytic domains of the key enzymes CODH and ACS, while the genes for the other CODH/ACS subunits and for the remaining enzymes of the methyl branch are highly conserved (Fig. 3). Conversion of acetyl-CoA and pyruvate is catalyzed by the action of a pyruvate:ferredoxin oxidoreductase (PFOR), whose gene is present in two copies in the genome of C. ljungdahlii (CLJU_c09340 and CLJU_c29340) that are also present in C. autoethanogenum and C. ragsdalei.
Fig. 3.
Arrangement and sequence identity of the Wood-Ljungdahl gene cluster in C. autoethanogenum (CAU), C. ljungdahlii (CLJ), and C. ragsdalei (CRA). The graph at the top shows the percentages of identity between all 3 sequences. Variations are marked in black or as gaps in the gray sequence bar above the annotations. Alignments were created using Geneious (Biomatters Ltd., Auckland, New Zealand). acs, genes for the CODH/ACS complex; acsA, CODH subunit gene; acsB, ACS subunit gene; acsC, corrinoid iron-sulfur protein large subunit gene; acsD, corrinoid iron-sulfur protein small subunit gene; acsE, methyltransferase subunit gene; cooC, gene for CODH accessory protein; fchA, formimido-tetrahydrofolate cyclodeaminase gene; fdx, ferredoxin gene; fhs, formyl-tetrahydrofolate synthase gene; folD, bifunctional methylene-tetrahydrofolate dehydrogenase/formyl-tetrahydrofolate cyclohydrolase gene; gcvH, gene for glycine cleavage system H protein; lpdA, gene for dihydrolipoamide dehydrogenase; metF, methylene-tetrahydrofolate reductase gene; T, predicted terminator; X, gene for conserved zinc finger protein of unknown function.
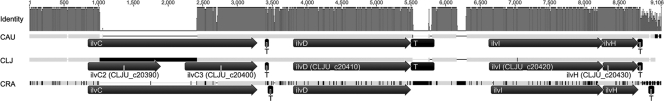
Initial conversion in the metabolic route from pyruvate to 23BD is catalyzed by an acetolactate synthase (EC 2.2.1.6), which links 2 molecules of pyruvate to α-acetolactate. This enzyme is found ubiquitously in nature, since acetolactate is also the precursor for biosynthesis of the branched-chain amino acids valine and leucine, but it exists in different forms. Accordingly, C. ljungdahlii possesses three different acetolactate synthase genes (33). Two of them, ilvB (CLJU_c32420) and ilvIH (CLJU_c20420-30), are likely to be involved in anabolism, while the third, alsS (CLJU_c38920), is predicted to encode a catabolic enzyme responsible for the formation of 23BD, with homology to the catabolic acetolactate synthase of C. acetobutylicum. Homologous genes (both anabolic and catabolic) were also found in C. autoethanogenum and C. ragsdalei. While the alsS gene was found to be exactly identical in all three strains, the genes involved in biosynthesis of branched-chain amino acids showed some variation. For example, the ilvB gene of C. autoethanogenum appears to be split into two open reading frames, in contrast to the case for both C. ljungdahlii and C. ragsdalei. This is due to a frameshift caused by a single base insertion, which was confirmed by repeated sequencing using different DNA preparations. The ilvIH genes of all three strains were found to be homologous, with slight differences in length, and in each case were located in a cluster containing other branched-chain amino acid biosynthesis genes (ilvC and ilvD, encoding a ketol acid reductoisomerase and a dihydroxy acid dehydratase, respectively). However, a difference between the three species investigated was found in the cluster of C. ljungdahlii (CLJU_c20390-430), which contains two identical copies of ilvC (possibly due to a gene doubling event), whereas only one ilvC gene is present in the ilvCDIH clusters of C. autoethanogenum and C. ragsdalei (Fig. 4). The genome of C. ljungdahlii also carries a third, orphan ilvC gene (CLJU_c15310) (see Fig. S1 in the supplemental material), which is also present in both C. autoethanogenum and C. ragsdalei.
Fig. 4.
Arrangement and identity of the ilvCDIH cluster in C. autoethanogenum (CAU), C. ljungdahlii (CLJ), and C. ragsdalei (CRA). The graph at the top shows the percentages of identity between all 3 sequences. Variations are marked in black or as gaps in the gray sequence bar above the annotations. Alignments were created using Geneious (Biomatters Ltd., Auckland, New Zealand). ilvC, ketol acid reductoisomerase gene; ilvD, dihydroxy acid dehydratase gene; ilvIH, acetolactate synthase gene; T, predicted terminator; T box, T box motif (RF00230).
In the second step, acetolactate is split into acetoin and CO2 by an acetolactate decarboxylase (EC 4.1.1.5). The respective gene (CLJU_c08380) was found to be highly conserved in all three strains as well as in the acetoin-producing organism C. acetobutylicum (see Table S3 in the supplemental material), while only minor homologies exist for sequences from other bacteria.
Acetoin is then finally reduced to 23BD by the action of a 23BD dehydrogenase (EC 1.1.1.4). The genome of C. ljungdahlii contains several dehydrogenase genes, some of whose products are likely to catalyze this reaction. The gene product of CLJU_c23220, for example, shows identity to (R,R)-butanediol dehydrogenases of several Bacillus strains known to produce 23BD, besides homology to several clostridial alcohol dehydrogenases of unknown function (see Table S3 in the supplemental material). A respective gene could also be identified in C. autoethanogenum and C. ragsdalei. The enzyme seems to be NADH and zinc dependent, since relevant binding domains could be identified. A search against the TIGRFAM database (25) revealed more potential candidate genes. A 23BD dehydrogenase/acetoin reductase motif (TIGR02415) was identified in three predicted short-chain dehydrogenase/reductase genes of C. ljungdahlii: CLJU_c14160 (E value = 9.1e−10), CLJU_c26580 (E value = 2.3e−06), and CLJU_c42160 (E value = 8.1e−50). However, the predicted structure indicates that all three genes are more likely to represent 3-oxoacyl-[acyl carrier protein] reductases involved in fatty acid biosynthesis, and CLJU_c14160 is also located in a respective fab gene cluster.
Gene expression studies.
Quantitative PCR was used to analyze the expression of the identified genes over time in a C. autoethanogenum culture (Fig. 1). The highest normalized mRNA levels were found for genes of the Wood-Ljungdahl gene cluster (Fig. 1C), which all had similar expression profiles (Fig. 1B). Among the three formate dehydrogenase genes, the gene encoding a selenocysteine-containing homologue of the CLJU_c06990 gene product yielded the highest normalized mRNA levels. While one PFOR gene (homologue of CLJU_c09340) was found to be expressed constitutively at a high level, the other PFOR gene (homologue of CLJU_c29340) was upregulated only at the end of growth (20-fold increase in normalized mRNA level).
Genes involved in synthesis of 23BD and lactate were found to be upregulated massively (15-fold increase in normalized mRNA level) during stationary growth phase, when the bulk of 23BD and lactate is produced. A slight upregulation was already observed during mid-/late exponential growth phase (after 50 h), which is shortly before the observed onset of 23BD production (at 75 h of growth). This slight difference in timing can be explained by the time needed to synthesize the active enzymes from the mRNAs as well as the limits in detection of produced 23BD. Only the putative 23BD dehydrogenase gene was expressed constitutively at a high normalized mRNA level over the whole period of growth. The expression of genes involved in the initial steps of branched-chain amino acid production was also analyzed. While expression of ilvB was found to be relatively stable, that of ilvIH, ilvC, and ilvD appears to be regulated.
Toxicity of 23BD.
To investigate whether 23BD has toxic effects on growth, cultures of C. autoethanogenum were challenged with increasing concentrations (0 to 200 g/liter) of previously distilled 23BD. Growth and acetate production ceased at concentrations of added 23BD of 40 to 50 g/liter (see Fig. S2 in the supplemental material).
DISCUSSION
Ethanol production from CO is considered to be a promising approach to low-carbon fuel production, and at least three companies, including Coskata, IneosBio, and LanzaTech, are seeking to develop the technology as a commercial process (35). The three most industrially relevant species, C. autoethanogenum (49), C. ljungdahlii (21), and C. ragsdalei (27), were compared in this study examining 23BD production from CO-rich off-gases produced by the steel industry. While only small variations in growth and ethanol production were observed, all three species were shown to produce small amounts of the four-carbon compound 23BD. To the best of our knowledge, 23BD production has not yet been reported as a native clostridial fermentation product. 23BD was coproduced with ethanol at a molar ratio of 1:10 (23BD to ethanol). Taking into account that for fermentation processes with C. ljungdahlii ethanol concentrations of over 50 g/liter were reported (44), production of 23BD from CO could become a viable industrial process.
To further develop this process as a strategy for 23BD production, it is important to identify the genes involved in synthesis of 23BD from CO in this system as well as to understand their regulation. Through sequence homology, we were able to identify a set of genes in the genome of C. ljungdahlii that are potentially involved in production of 23BD from pyruvate. The presence of these genes in C. autoethanogenum and C. ragsdalei was subsequently confirmed by sequencing. Quantitative PCR studies revealed that the expression of this gene set was generally highly upregulated during the culture stationary growth phase (Fig. 1B), which correlated well with the 23BD production observed (Fig. 2). The only exception was the putative 23BD dehydrogenase gene, which was found to be expressed constitutively at a much higher normalized mRNA level (Fig. 1C), suggesting that this gene might also be involved in other processes.
An analysis of the sequence for this putative enzyme revealed that it shows homology to (R,R)-specific 23BD dehydrogenases from several Bacillus strains (around 60% identity) known to produce 23BD (see Fig. S3 in the supplemental material). A knockout of this gene in Bacillus subtilis abolished production of 23BD and led to accumulation of the precursor acetoin (41). However, since very small amounts of 23BD were detected in very late stationary phase for this B. subtilis mutant strain (41), it was speculated that there is a second 23BD dehydrogenase which has yet to be identified. This might also be true for C. ljungdahlii, as further proteins with potential 23BD dehydrogenase motifs were identified.
A homologous enzyme from Clostridium beijerinckii (78% identity) was recently expressed in the acetoin-producing organism C. acetobutylicum (48), resulting in the production of d-(−)-23BD. This matches the results found for C. autoethanogenum, with 23BD samples comprising 96% d-(−)-23BD and 4% meso-23BD. The small amount of measured meso-23BD may be a result of the presence of l-(+)-acetoin, which is then converted by the (R,R)-23BD dehydrogenase into the meso compound (30, 55). All three isomeric forms of 23BD have already been reported as bacterial fermentation products, and synthesis of 2 enantiomers by one species is not uncommon (30, 37).
The stereochemistry of acetoin itself is a function of the acetolactate decarboxylase (30, 55), which converts acetolactate to acetoin. In C. acetobutylicum, which possesses the acetolactate decarboxylase most similar to the enzymes of C. auto-ethanogenum (74% identity), C. ljungdahlii, and C. ragsdalei, a ratio of d-(−)-acetoin to l-(+)-acetoin of 12:1 was found (48). No acetolactate decarboxylase gene was found in the genome of C. beijerinckii, which might explain why this strain is not able to produce 23BD, despite having a functional 23BD dehydrogenase (48). The function of this enzyme in C. beijerinckii is not known. In the presence of oxygen, acetolactate is also spontaneously converted to a diacetyl (13, 42), but given that the present system is anaerobic, it is unsurprising that no evidence for this reaction could be found.
Because acetolactate is also a precursor for biosynthesis of the branched-chain amino acids valine, leucine, and isoleucine, the biosynthetic genes associated with the production of these amino acids were also analyzed. These genes had generally higher normalized mRNA levels than the 23BD pathway genes (Fig. 1C). In Gram-positive bacteria, the regulation of genes involved in amino acid biosynthesis is often RNA dependent, via a riboswitch mechanism (24). Consistent with this, T box motifs (RF00230) were identified upstream of ilvB, ilvC, and ilvD (Fig. 4). Interestingly, two T boxes were found to be located upstream of the ivlIH genes, which showed the greatest degree of expression regulation (Fig. 1B). Expression in C. ljungdahlii and C. ragsdalei might differ from that in C. autoethanogenum, as C. ljungdahlii contains two copies of ilvC, while a predicted terminator region between ilvD and ilvIH is absent in C. ragsdalei (Fig. 4).
In addition to 23BD, traces of lactate were detected in cultures of C. autoethanogenum, C. ljungdahlii, and C. ragsdalei (Fig. 2). As with 23BD, lactate is produced from pyruvate by the action of a lactate dehydrogenase (see the supplemental material). This demonstrates the capability of acetogens to synthesize products which do not stem directly from acetyl-CoA (such as ethanol, acetate, butanol, and butyrate). The conversion of acetyl-CoA to pyruvate is enabled by the pyruvate:ferredoxin oxidoreductase enzyme coupled with CO oxidation by the CODH enzyme (Fig. 1A) (20). PFOR is widespread in anaerobic bacteria and some microaerophilic bacteria and archaea. Its catalytic properties have been studied extensively, but mainly in the oxidative direction (for experimental reasons), although in many organisms the physiological function is assumed to be the synthesis of pyruvate from acetyl-CoA and CO2 (20, 59). All three organisms possess two PFOR genes, but high normalized mRNA levels were detected only for the homologue of CLJU_c09340, which is thus hypothesized to be the primary PFOR gene expressed under auto-trophic growth conditions. Genes involved in electron transport from CO (E0′ = −520 mV) and H2 (E0′ = −414 mV) to ferredoxin (E0′ ≈ −400 mV) and NAD(P) (E0′ = −320 mV) and the coupling sites associated with the respective electron transport have yet to be investigated, since an understanding has only recently begun to emerge (8, 9, 33, 40).
Of all the genes investigated in this study, those of the Wood-Ljungdahl cluster yielded the highest normalized mRNA levels. This observation is not surprising given that these genes are directly involved in carbon uptake. Despite the large size of the cluster (20 kbp) and the presence of several predicted terminator structures (7 for C. autoethanogenum, 6 for C. ljungdahlii, and 9 for C. ragsdalei), all genes showed similar expression profiles (Fig. 1B), with the most expression directly after inoculation (presumably to synthesize the first new enzymes) and at the end of growth (potentially as a response to the depletion of carbon monoxide from the headspace).
The three formate dehydrogenase genes, which are located separately on the genome and whose products catalyze one of the initial steps in the Wood-Ljungdahl pathway, showed completely different expression patterns at significantly lower normalized mRNA levels (Fig. 1). This might indicate an important role in regulation of this pathway, although nothing is known about the enzyme activities and electron donor/acceptor specificities. Two of the formate dehydrogenases are predicted to be seleno-enzymes, which are generally considered to have superior catalytic efficiencies to those of their cysteine-containing analogues (31). Thus, two primer pairs were used for each gene, binding upstream and downstream of the selenocysteine insertion sequence (SECIS element) (18). In both cases, the primer pairs resulted in similar mRNA accumulation levels, which was expected since the medium contained selenite. Among the three formate dehydrogenase genes, the CLJU_c06990 gene homologue encoding a selenocysteine-containing variant showed the highest (slightly) expression level during exponential phase, while the gene encoding the cysteine-containing variant yielded the highest normalized mRNA levels at stationary phase (Fig. 1C).
Another important aspect for development of an industrial process is end product inhibition, which is one of the major historical limitations in industrial butanol fermentations, where solvent concentrations of 1.5 to 2% could not be exceeded (2, 4, 34). This has significant impacts on the efficiency and economics of separation and purification, as product removal becomes increasingly energy intense and expensive (38). 23BD seems to be far less toxic than butanol, as no significant growth inhibition was observed up to a concentration of around 4%. It should be noted that an experimental design with addition of large amounts of 23BD before inoculation does not entirely reflect the physiological situation in a fermentation culture, where the concentration of 23BD increases gradually over time. Therefore, the limit might be even higher for endogenously produced 23BD, as cells have the opportunity to adapt slowly to the accumulating 23BD stress, e.g., by changing the membrane composition and fluidity (4). Relative concentrations of other metabolites (ethanol, acetate, and lactate) were also found to have a negative influence on the observed toxicity threshold for 23BD. This in turn is consistent with the additive impact reported for numerous fermentation coproducts on the microbial toxicity of individual molecules, for example, the reported deleterious influence of acetic acid on ethanol toxicity or of butyric acid on butanol toxicity (2).
Lactate in particular was found to be toxic to C. autoethanogenum at relatively low concentrations (<0.5%), but it could also be metabolized to some extent by the bacterium and therefore might act as a carbon storage strategy for the cell (see Fig. S3 in the supplemental material). This function has also been reported for 23BD in other bacteria, such as B. subtilis and K. pneumoniae (57), but no evidence (either experimental or from in silico analysis) for 23BD uptake could be found in C. autoethanogenum, C. ljungdahlii, or C. ragsdalei. This is important from a process perspective, as reutilization of the desired product is unwanted. The main drivers of synthesis of 23BD and lactate in C. autoethanogenum, C. ljungdahlii, and C. ragsdalei are hypothesized to be the offload of reducing equivalents and the deacidification of pyruvic acid (pKa = 2.4) to the pH-neutral 23BD or lactic acid (pKa = 3.9), as speculated for other 23BD- and lactate-producing bacteria (30, 57). Accordingly, 23BD and lactate production starts at the transition to stationary phase, when the pH of the medium has already dropped due to the production of acetic acid and a potential surplus of reducing equivalents exists.
To date, the most efficient 23BD production strains, K. pneumonia, K. oxytoca, E. aerogenes, and S. marcescens (30), are all categorized by the World Health Organization (WHO) as risk group 2 species (pathogenic), thus making their use in the large-scale production of 23BD particularly challenging and costly (13). Furthermore, in every case, these organisms rely on the use of plant-derived carbohydrates as a feedstock of carbon and energy for product synthesis. In other words, they are restricted to the use of farmed (lignocellulosic biomass) or, in some cases, food (sugar or starch) resources for 23BD production (13, 55, 58). In contrast, the present demonstration of 23BD production from industrial waste gases by acetogens has two potentially important commercial implications. First, it decouples the biological production of 23BD from a reliance on farmed or food sugars as the primary carbon and energy sources for product synthesis. In this study, for example, we demonstrate the viable use of by-product gases from the steel industry for 23BD production. Second, the three species of acetogens described here for the production of 23BD are each risk group 1 (nonpathogenic) organisms. The use of risk group 1 organisms is greatly preferred because of the comparatively lower health and safety risks and associated culture containment and handling costs.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 17 June 2011.
REFERENCES
- 1. Abrini J., Naveau H., Nyns E. J. 1994. Clostridium autoethanogenum, sp. nov., an anaerobic bacterium that produces ethanol from carbon monoxide. Arch. Microbiol. 161:345–351 [Google Scholar]
- 2. Alsaker K. V., Paredes C., Papoutakis E. T. 2010. Metabolite stress and tolerance in the production of biofuels and chemicals: gene-expression based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnol. Bioeng. 105:1131–1147 [DOI] [PubMed] [Google Scholar]
- 3. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 4. Bear S. H., Blaschek H. P., Smith T. L. 1987. Effect of butanol challenge and temperature on lipid composition and membrane fluidity of butanol-tolerant Clostridium acetobutylicum. Appl. Environ. Microbiol. 53:2845–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6. Bertram J., Dürre P. 1989. Conjugal transfer and expression of streptococcal transposons in Clostridium acetobutylicum. Arch. Microbiol. 151:551–557 [Google Scholar]
- 7. Biebl H., Zeng A. P., Menzel K., Deckwer W. D. 1998. Glycerol fermentation to 1,3-propanediol and 2,3-butanediol by Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 50:24–29 [DOI] [PubMed] [Google Scholar]
- 8. Biegel E., Müller V. 2010. Bacterial Na+-translocating ferredoxin:NAD+ oxidoreductase. Proc. Natl. Acad. Sci. U. S. A. 107:18138–18142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Biegel E., Schmidt S., González J., Müller V. 2010. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell. Mol. Life Sci. 68:613–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blomqvist K., et al. 1993. Characterization of the genes of the 2,3-butanediol operons from Klebsiella terrigena and Enterobacter aerogenes. J. Bacteriol. 175:1392–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Byun T. G., Zeng A. P., Deckwer W. D. 1994. Reactor comparison and scale-up for the microaerobic production of 2,3-butanediol by Enterobacter aerogenes at constant oxygen transfer rate. Bioprocess Eng. 11:167–175 [Google Scholar]
- 12. Reference deleted.
- 13. Celińska E., Grayek W. 2009. Biotechnological production of 2,3-butanediol—current state and prospects. Biotechnol. Adv. 27:715–725 [DOI] [PubMed] [Google Scholar]
- 14. De Mas C., Jansen N. B., Tsao G. T. 1988. Production of optically active 2,3-butanediol by Bacillus polymyxa. Biotechnol. Bioeng. 31:366–377 [DOI] [PubMed] [Google Scholar]
- 15. Reference deleted.
- 16. Drake H. L., Küsel K., Matthies C. 2006. Acetogenic prokaryotes, p. 354–420 In Dworkin M., Rosenberg E., Schleifer K. H., Stackebrandt E. (ed.), The prokaryotes, 3rd ed., vol. 2 (ecophysiology and biochemistry) Springer, New York, NY [Google Scholar]
- 17. Drake H. L., Küsel K. 2005. Acetogenic clostridia, p. 719–746 In Dürre P. (ed.), Handbook on clostridia. CRC Press, Boca Raton, FL [Google Scholar]
- 18. Engelberg-Kulka H., Liu Z., Li C., Reches M. 2001. An extended Escherichia coli “selenocysteine insertion sequence” (SECIS) as a multifunctional RNA structure. Biofactors 14:61–68 [DOI] [PubMed] [Google Scholar]
- 19. Finn R. D., et al. 2010. The PFAM protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Furdui C., Ragsdale S. W. 2000. The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotropic growth by the Wood-Ljungdahl pathway. J. Biol. Chem. 37:28494–28499 [DOI] [PubMed] [Google Scholar]
- 21. Gaddy J. L., Clausen W. C. December 1992. Clostridium ljungdahlii, an anaerobic ethanol and acetate producing microorganism. U.S. patent 5173429.
- 22. Gardner P. P., et al. 2008. Rfam: updates to the RNA families database. Nucleic Acids Res. 37:D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reference deleted.
- 24. Green N. J., Grundy F. J., Henkin T. M. 2010. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 584:318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haft D. H., Selengut D. H., White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res. 31:371–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hugenholtz J., Starrenburg M. J. 1992. Diacetyl production by different strains of Lactococcus lactis subsp. lactis var. diacetylactis and Leuconostoc spp. Appl. Microbiol. Biotechnol. 38:17–22 [Google Scholar]
- 27. Huhnke R., Lewis R. S., Tanner R. S. March 2008. Isolation and characterization of novel clostridial species. WO patent 2008/028055.
- 28. Hulo N., et al. 2008. The 20 years of PROSITE. Nucleic Acids Res. 36:245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hungate R. E. 1969. A roll tube method for cultivation of strict anaerobes, p. 117–132 In Norris J. R., Ribbons D. W. (ed.), Methods in microbiology, vol. 3B. Academic Press, New York, NY [Google Scholar]
- 30. Ji X. J., Huang H., Ouyang P. K. 2011. Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnol. Adv. 29:351–364 [DOI] [PubMed] [Google Scholar]
- 31. Jones J. B., Stadtman T. C. 1981. Selenium-dependent and selenium-independent formate dehydrogenases of Methanococcus vannielii. Separation of the two forms and characterization of the purified selenium-independent form. J. Biol. Chem. 256:656–663 [PubMed] [Google Scholar]
- 32. Reference deleted.
- 33. Köpke M., et al. 2010. Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc. Natl. Acad. Sci. U. S. A. 107:13087–13092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Köpke M., Dürre P. 2010. Biochemical production of biobutanol, p. 221–257 In Luque R., Campelo J., Clark J. H. (ed.), Handbook of biofuel production—processes and technologies. Woodhead Publishing, Cambridge, United Kingdom [Google Scholar]
- 35. Köpke M., Mihalcea C., Bromley J. C., Simpson S. D. 2011. Fermentative production of ethanol from carbon monoxide. Curr. Opin. Biotechnol. 22:320–325 [DOI] [PubMed] [Google Scholar]
- 36. Liou J. S., Balkwill D. L., Drake G. R., Tanner R. S. 2005. Clostridium carboxidivorans sp. nov., a solvent producing clostridium isolated from an agricultural settling lagoon, and reclassification of the acetogen Clostridium scatologenes strain SL1 as Clostridium drakei sp. nov. Int. J. Syst. Evol. Microbiol. 55:2085–2091 [DOI] [PubMed] [Google Scholar]
- 37. Maddox I. S. 1996. Microbial production of 2,3-butanediol, p. 269–291 In Roehr M. (ed.), Biotechnology: products of primary metabolism, vol. 6. VCH, Weinheim, Germany [Google Scholar]
- 38. Madson P. W. 2003. Ethanol distillation: the fundamentals, p. 319–336 In Jaques K. A., Lyons T. P., Kelsall D. R. (ed.), The alcohol textbook, 4th ed. Nottingham University Press, Nottingham, United Kingdom [Google Scholar]
- 39. Margulies M., et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Müller V., Imkamp F., Biegel E., Schmidt S., Dilling S. 2008. Discovery of a ferredoxin:NAD+-oxidoreductase (Rnf) in Acetobacterium woodii: a novel potential coupling site in acetogens. Ann. N. Y. Acad. Sci. 1125:137–146 [DOI] [PubMed] [Google Scholar]
- 41. Nicholson W. L. 2008. The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanediol dehydrogenase. Appl. Environ. Microbiol. 74:6832–6838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park H. S., Xing R., Whitman W. B. 1995. Nonenzymatic acetolactate oxidation to diacetyl by flavin, nicotineamide and quinone coenzymes. Biochim. Biophys. Acta 14:366–370 [DOI] [PubMed] [Google Scholar]
- 43. Pfaffl M. W., Horgan G. W., Dempfle L. 2002. Relative expression software tool (REST) for group wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:E36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Phillips J. R., Klasson K. T., Clausen E. C., Gaddy J. L. 1993. Biological production of ethanol from coal synthesis gas. Appl. Biochem. Biotechnol. 39/40:559–571 [Google Scholar]
- 45. Ragsdale S. W. 2004. Life with carbon monoxide. Crit. Rev. Biochem. Mol. Biol. 39:165–195 [DOI] [PubMed] [Google Scholar]
- 46. Renna M. C., Najimudin N., Winik L. R., Zahler S. A. 1993. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J. Bacteriol. 175:3863–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sanger F., Nicklen S., Coulson A. R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Siemerink M. A., et al. 2011. d-2,3-Butanediol production due to heterologous expression of an acetoin reductase in Clostridium acetobutylicum. Appl. Environ. Microbiol. 77:2582–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Simpson S. D., Forster R. L. October 2007. Microbial fermentation of gaseous substrates to produce alcohols. WO patent 117151.
- 50. Sipma J., et al. 2006. Microbial CO conversions with applications in synthesis gas purification and bio-desulfurization. Crit. Rev. Biotechnol. 26:41–65 [DOI] [PubMed] [Google Scholar]
- 51. Syu M. J. 2001. Biological production of 2,3-butanediol. Appl. Microbiol. Biotechnol. 55:10–18 [DOI] [PubMed] [Google Scholar]
- 52. Tanner R. S., Miller L. M., Yang D. 1993. Clostridium ljungdahlii sp. nov., an acetogenic species in clostridial rRNA homology group I. Int. J. Syst. Bacteriol. 43:232–236 [DOI] [PubMed] [Google Scholar]
- 53. Tatusov R. L., et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ui S., Hosaka T., Watanabe K., Mimura A. 1998. Discovery of a new mechanism of 2,3-butanediol stereoisomer formation in Bacillus cereus YUF-4. J. Ferment. Bioeng. 85:79–83 [Google Scholar]
- 55. Voloch M., et al. 1985. 2,3-Butanediol, p. 933–947 In Moo-Young M. (ed.), Comprehensive biotechnology: the principles, applications and regulations of biotechnology in industry, agriculture and medicine. Pergamon Press, New York, NY [Google Scholar]
- 56. Wood H. G. 1992. Life with CO or CO2 and H2 as a source of carbon and energy. FASEB J. 5:156–163 [DOI] [PubMed] [Google Scholar]
- 57. Xiao Z., Xu P. 2007. Acetoin metabolism in bacteria. Crit. Rev. Biochem. Microbiol. 33:127–140 [DOI] [PubMed] [Google Scholar]
- 58. Xiu Z. L., Zeng A. P. 2008. Present state and perspectives of downstream processing of biologically produced 1,3-propanediol and 2,3-butanediol. Appl. Microbiol. Biotechnol. 78:917–926 [DOI] [PubMed] [Google Scholar]
- 59. Yoon K. S., Hille R., Hemann C., Tabita F. R. 1999. Rubredoxin from the green sulfur bacterium Chlorobium tepidum functions as an electron acceptor for pyruvate ferredoxin oxidoreductase. J. Biol. Chem. 274:29772–29778 [DOI] [PubMed] [Google Scholar]
- 60. Zhang L., et al. 2010. Microbial production of 2,3-butanediol by a mutagenized strain of Serratia marcescens H30. Bioresour. Technol. 101:1961–1967 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.