Abstract
Fourier transform infrared (FT-IR) spectroscopy and Raman spectroscopy were used to study the cell injury and inactivation of Campylobacter jejuni from exposure to antioxidants from garlic. C. jejuni was treated with various concentrations of garlic concentrate and garlic-derived organosulfur compounds in growth media and saline at 4, 22, and 35°C. The antimicrobial activities of the diallyl sulfides increased with the number of sulfur atoms (diallyl sulfide < diallyl disulfide < diallyl trisulfide). FT-IR spectroscopy confirmed that organosulfur compounds are responsible for the substantial antimicrobial activity of garlic, much greater than those of garlic phenolic compounds, as indicated by changes in the spectral features of proteins, lipids, and polysaccharides in the bacterial cell membranes. Confocal Raman microscopy (532-nm-gold-particle substrate) and Raman mapping of a single bacterium confirmed the intracellular uptake of sulfur and phenolic components. Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) were employed to verify cell damage. Principal-component analysis (PCA), discriminant function analysis (DFA), and soft independent modeling of class analogs (SIMCA) were performed, and results were cross validated to differentiate bacteria based upon the degree of cell injury. Partial least-squares regression (PLSR) was employed to quantify and predict actual numbers of healthy and injured bacterial cells remaining following treatment. PLSR-based loading plots were investigated to further verify the changes in the cell membrane of C. jejuni treated with organosulfur compounds. We demonstrated that bacterial injury and inactivation could be accurately investigated by complementary infrared and Raman spectroscopies using a chemical-based, “whole-organism fingerprint” with the aid of chemometrics and electron microscopy.
INTRODUCTION
Fruit and vegetable extracts can inhibit the growth of pathogenic and spoilage microorganisms in foods and are being evaluated as food preservatives, in part from consumer demands for fewer chemical food preservatives. Foods with high levels of phenolic compounds have received the greatest study, and their effectiveness as antimicrobial agents was validated (1, 2, 7, 11). For example, extracts from cranberries inhibited various pathogen growths (25, 55, 56, 57). The antimicrobial effect of garlic is attributed primarily to organosulfur compounds (19, 23, 24, 45, 47, 49, 52), such as allicin (5, 10, 14, 32), ajoene (38), and diallyl sulfides (41, 48, 59). However, others have demonstrated that the contribution of antimicrobial properties is also from phenolic compounds (6, 21, 31).
Vibrational spectroscopies (both infrared [IR] and Raman spectroscopies) are widely employed to characterize complicated biological systems based upon the chemical compositions of analytes (36, 37, 39). When infrared and Raman spectroscopies are coupled, a unique, wide-spectral fingerprint is generated across a wave number region (from 1,800 to 400 cm−1 for both IR and Raman spectroscopies) to identify and differentiate analytes, such as bacteria (20, 50). The major advantages of using vibrational spectroscopies to study bacteria are the high detection speed, reagentless operation, relatively precise identification of the components involved, and the ability to distinguish biological specimens, including bacteria, to the species and strain levels (42, 54). Bacteria can be identified within 6 h by Raman spectroscopy and 8 h by infrared spectroscopy (9, 22, 29). Recently, these spectroscopic methods have been intensively applied to study bacterial injury and inactivation by various treatments, such as bacteriocin (8), antibiotics (28, 33, 40), heat (3, 4), and sonication (27). Infrared spectroscopy is useful for bulk bacterial detection (the detection limit is ∼104 to 105 CFU/ml) (12), while Raman spectroscopy can potentially detect single bacterial cells (46), especially when a nanosubstrate is employed to signal enhancement, a technique called surface-enhanced Raman spectroscopy (SERS) (13).
In the current study, injury of Campylobacter jejuni was investigated. This food-borne pathogen was exposed to food components, specifically garlic concentrates and organosulfur compounds, at levels at which these components could be present in muscle-based foods, such as chicken and beef (59). C. jejuni can survive outside its host for an extended period, and it is now considered to be the most prevalent cause of bacterial food-borne illness in the world (43). C. jejuni-infected individuals experienced abdominal cramps, fever, and diarrhea accompanied by gross blood and leukocytes. Previous studies have examined the effectiveness of fruit extracts (53), plant essential oils and extracts (15, 26), and phenolic compounds (16, 17) for controlling the growth of C. jejuni. However, there are no reported studies involving Allium species, such as garlic.
To the best of our knowledge, this is the first paper combining infrared and Raman spectroscopies to study bacterial stress and injury. Rapid detection and quantification of sublethally injured bacterial cells are important for food safety since microorganisms can repair themselves if conditions are favorable and then start to grow in food products, presenting the potential for pathogenicity (58). These two vibrational spectroscopic techniques provide complementary biochemical information about bacterial cell composition for a more complete approach to the analysis of bacterial cellular response under environmental stress. The objective of this study was to combine spectroscopic methods of analysis with morphological tests to better understand the mechanism of bacterial injury resulting from exposure to garlic-derived organosulfur compounds.
MATERIALS AND METHODS
Chemicals and reagents.
Folin-Ciocalteu reagent, 2,2 diphyenyl-1-picrylhydrazyl (DPPH), 6-hydroxy-2,5,7,8-tetramethyl-2-carboxylic acid (Trolox), gallic acid, acetonitrile, methanol, and tetrahydrofuran were obtained from Sigma-Aldrich (St. Louis, MO). Sodium carbonate was purchased from J. T. Baker, Inc. (Phillipsburg, NJ). All chemicals used were high-performance liquid chromatography (HPLC) grade. Diallyl sulfide (purity, 98%) and crude diallyl disulfide (purity, 80%) were purchased from Sigma-Aldrich Chemical Co. (Milwaukee, WI). Diallyl disulfide was further purified by fractional distillation to a final purity greater than 98% as determined by HPLC (59). Diallyl trisulfide (purity, 97%) was purchased from Fisher Scientific (Pittsburgh, PA).
Reverse-phase high-performance liquid chromatography with a diode array detector (Agilent 1100 HPLC system; Palo Alto, CA) set at 240 nm was used to check the purity and stability of diallyl constituents used for this study. These compounds were analyzed with a Nova-Pak C18 column (4 μm, 4.6 by 150 mm; Waters Corporation, Milford, MA) and with a C18 guard column (5 μm, 3.9 by 20 mm; Waters Corporation, Milford, MA) using a mobile phase of 70:27:3 (acetonitrile-water-tetrahydrofuran [vol/vol/vol]) and a flow rate of 1 ml per min (41). The injection volume of the sample was 10 μl.
Culture preparation.
Four C. jejuni strains (ATCC 33291, ATCC 33560, ATCC 43430, and ATCC 49943) were obtained from Microbiologics, Inc. (St. Cloud, MN). Each bacterial strain was separately inoculated into 50 ml of campylobacter enrichment broth consisting of campylobacter nutrient broth no. 2 (CM0067; Oxoid, England) supplemented with campylobacter growth supplement (SR0232E; Oxoid, England). The C. jejuni broth specimens were then incubated in anaerobic jars at 42°C for 48 h under microaerophilic conditions (10% CO2, 5% O2, and 85% N2) using Pack-MicroAero (Mitsubishi Gas Chemical America, Inc., New York, NY). After 48 h of incubation at 42°C, 10 ml broth with each strain was transferred to individual 50-ml sterile centrifuge tubes. Cells were harvested by centrifugation at 5,000 rpm (Fisher AccuSpin model 400 benchtop centrifuge; Pittsburgh, PA) for 15 min at 22°C. To eliminate any effect of broth components and bacterial metabolites, the resultant pellets were resuspended in 10 ml of sterile 0.85% (wt/vol) saline solution and centrifuged again. After the second centrifugation, the pellets were suspended in 2% (wt/vol) buffer-peptone water (Difco), forming culture suspensions. Equal volumes of the suspensions containing one of the four strains were combined, forming a cocktail.
Media.
Bacterial viable counts were performed in duplicate by plating on Preston campylobacter nonselective medium. Preston campylobacter nonselective agar was prepared according to the manufacturer's instructions and consisted of campylobacter agar base (CM0689; Oxoid, England), campylobacter growth supplement (SR0232E; Oxoid, England), and lysed horse blood (Remel, Lenexa, KS). Nonselective agar plates were incubated at 42°C for 48 h under microaerophilic conditions (10% CO2, 5% O2, and 85% N2).
Antibacterial effects of garlic concentrate and garlic-derived organosulfur compounds on C. jejuni.
Fresh garlic was obtained from Global Farms Enterprises, Inc. (Los Angeles, CA), stored at room temperature (ca. 22°C), and used within 2 weeks. The peeled cloves were put into a juice extractor (Waring Pro) to produce fresh garlic juice under aseptic conditions. The raw garlic juice was immediately put into 50-ml conical sterile centrifuge tubes and centrifuged at 7,000 rpm (Fisher accuSpin model 400 benchtop centrifuge, Pittsburgh, PA) for 10 min at 22°C. The supernatant was recovered and filtered through a 10.0-μm-pore-size polycarbonate membrane (K99CP04700; GE Water & Process Technologies, Trevose, PA) and then through a 1-μm-pore-size polycarbonate membrane (K10CP04700; GE Water & Process Technologies, Trevose, PA) and, finally, through a 0.4-μm-pore-size polycarbonate membrane (K04CP04700; GE Water & Process Technologies, Trevose, PA) under vacuum to remove potential microbial contamination, generating the garlic concentrate. The whole process to make the garlic concentrate was completed within 20 min. Then, the garlic concentrate was stored at 4°C and protected to avoid light exposure. The concentrate was added into 0.85% (wt/vol) sterile saline water and/or nutrient broth no. 2 within 30 min to avoid loss of volatile organosulfur compounds.
Garlic concentrate was prepared at various concentrations (0, 6.25, 12.5, 25, 37.5, 50, 75, and 100 μl/ml) in 100 ml of sterilized 0.85% (wt/vol) saline water to study its bactericidal effect with limited nutrients and in 100 ml of nutrient broth no. 2 to investigate the inhibitory or suppressive effect of garlic concentrate. Each concentration of garlic extract was added to both saline water and broth and then inoculated with 1 ml of 7 log CFU/ml of a C. jejuni cocktail to achieve an initial inoculum level of approximately 5 log CFU/ml. Each inoculate was mixed well by vortexing and then incubated at 4, 22, and 35°C for 0, 1, 3, 5, 7, 10, and 24 h (saline water samples) and 0, 1, and 3 days (broth samples) microaerobically. At each sampling time, the samples were serially diluted with 2% (wt/vol) sterile buffer peptone water, and the appropriate dilution was spiral plated. After incubation at 42°C for 48 h, numbers of viable cells were determined.
Measurement of total phenolic content and total antioxidant capacity.
Total phenolic content and total antioxidant capacity were quantified because these two parameters are related to the antimicrobial activity of plant extracts (25, 31, 44). Two grams of chopped garlic was extracted with 15 ml methanol under conditions of magnetic stirring for 2 h. The extract was centrifuged at 4,000 rpm for 20 min, and the supernatant was filtered. The extraction procedure was repeated three times, and the supernatants were pooled together. The weight of dry matter of the extracts was determined to standardize the concentrations at 1 mg/ml.
The total phenolic content of each extract was determined in duplicate by the Folin-Ciocalteu procedures according to the method of Sun et al. (51), with minor changes. In brief, Folin-Ciocalteu reagent was diluted 10-fold with deionized water. Garlic extract (0.1 ml) was mixed with 0.75 ml of the diluted Folin-Ciocalteu reagent and incubated for 10 min at room temperature (ca. 22°C). Then, 0.75 ml of 2% (wt/vol) sodium carbonate solution was added. The mixture was allowed to stand in the dark (ca. 22°C) for 45 min before the absorbance at 765 nm was measured using an Ultrospec 4000 UV/visible-light spectrophotometer (Pharmacia Biotech, Cambridge, United Kingdom) against a blank containing deionized water instead of sample extract. Total phenolic content values were determined from a calibration curve prepared with a series of gallic acid equivalents (GAE), measured in milligrams per gram (dry weight).
The antioxidant capacity of garlic extract was measured using a method described by Sun et al. (51) using the free radical 2,2 diphyenyl-1-picrylhydrazyl (DPPH), with minor revisions. Garlic extract (0.1 ml) was added to 1 ml of DPPH solution, and the absorbance of the DPPH solution was determined at 515 nm after 30 min of incubation at room temperature (ca. 22°C). Methanolic solutions of Trolox were used for calibration to compare the antioxidant capacities of garlic concentrates. The antioxidant capacity of the sample was expressed as milligrams Trolox equivalents/gram (dry weight) sample.
Electron microscopy analysis.
Scanning electron microscopy (SEM) was performed to examine morphological changes of C. jejuni cells before and after treatment with garlic-derived organosulfur compounds (5 μM diallyl sulfide) in sterilized broth for 10 h at 22°C. Treated and untreated C. jejuni cells were harvested at 12,000 × g for 10 min. First, C. jejuni cells were fixed with 2% glutaraldehyde, 2% paraformaldehyde in 0.1 M phosphate buffer overnight. The samples were then rinsed with double-distilled water and freeze-dried in a VirTis lyophilizer (The VirTis Co., Inc., Gardiner, NY). The samples were mounted onto SEM stubs and sputter coated with a thin layer of gold. The coated samples were observed under an FEI Quanta 200F scanning electron microscope (Field Emission Instruments, Hillsboro, OR) using an accelerating voltage of 30 kV.
Transmission electron microscopy (TEM) was employed to study the bactericidal action of organosulfur compounds derived from garlic. C. jejuni cultures, untreated and treated with diallyl sulfide, were placed into the primary fixative overnight at 4°C. The samples were rinsed three times with 0.1 M phosphate buffer (10 min each) and postfixed by 2% osmium tetroxide for 2 h at room temperature. Next, the samples were quickly rinsed twice with 0.1 M buffer (10 min each), followed by dehydration with ethanol solutions (30%, 50%, 70%, 95%, and 100%), and then rinsed twice with 100% propylene oxide (10 min each). The bacterial samples were infiltrated by propylene oxide: 1:1 Spurr's resin overnight and then 100% Spurr's resin twice (overnight each time). Then, the samples were embedded in Spurr's resin. The stained samples were observed in a Philips electron microscope (Field Emission Instruments, Hillsboro, OR) operating at 200 kV.
FT-IR spectroscopy analysis.
At the end of each treatment, 100 ml of each sample was filtered through an aluminum oxide membrane filter (0.2-μm pore size, 25-mm optical density [OD]) (Anodisc; Whatman, Inc., Clifton, NJ) using a Whatman vacuum glass membrane filter holder (Whatman catalog no. 1960-032) to harvest bacterial cells. The Anodisc membrane filter does not contribute spectral features between the wave numbers of 4,000 and 1,000 cm−1 and provides a smooth and flat surface onto which the bacterial film can form. The Anodisc filters were then removed from the filtration apparatus and air dried under laminar flow at room temperature (ca. 22°C) for 60 min.
FT-IR spectra were collected using a Nicolet 380 FT-IR spectrometer (Thermo Electron, Inc., San Jose, CA). The aluminum oxide membrane filter, coated with a uniform thin layer of bacterial cells, was placed in direct contact with the diamond crystal cell (30,000 to 200 cm−1) of attenuated total reflectance (ATR). Infrared spectra were recorded from 4,002 to 399 cm−1 at a resolution of 4 cm−1. Each spectrum was acquired by adding together 32 interferograms. Six spectra were acquired at room temperature (ca. 22°C) for each sample to get a total of 18 spectra with each treatment. Triplicate experiments were conducted, and spectra from the first two experiments were used for establishment of chemometric models. Spectra from the third experiment were randomly selected for model validation.
Raman spectroscopy analysis.
A WITec alpha 300 Raman spectrometer system (WITec, Ulm, Germany) equipped with a WITec microscope was used in this study. This system is equipped with a 532-nm laser source and a 785-nm laser source. During the measurement, the 532-nm laser was focused onto the sample on the microscope stage through a 100× objective (Nikon, Melville, NY). Raman scattering signals were detected by a 1,600- by 200-pixel charge-coupled-device (CCD) array detector. The size of each pixel was 16 by 16 μm. Spectral data were collected with WITec project software v2.02 (WITec, Ulm, Germany). Spectra of each bacterial sample were collected using a 100× objective with a detection range from 3,700 to 200 cm−1 in the extended mode. The measurement was conducted with a 1-s integration time, with 50 spectral accumulations and approximately 2-mW incident laser powers. Spectral data for generating Raman maps were taken from the sample at specific wave numbers (300 to 1,800 cm−1).
Klarite (D3 Technologies Ltd., Glasgow, United Kingdom) SERS-active substrates were used in this study. These devices were fabricated on silicon wafers coated with gold. A 6- by 10-mm2 chip, including a 4- by 4-mm patterned SERS-active area and an unpatterned gold reference area, was adhered to a standard glass slide. Treated microbial cells (10 μl) were deposited onto the substrate, and Raman measurements were taken after 2 h of drying under a fume hood at room temperature (ca. 22°C).
Chemometric analysis and statistical analysis.
Vibrational (both infrared and Raman) spectra were initially preprocessed by EZ OMNIC 7.1a (Thermo Electron, Inc., Lafayette, CO). The raw spectra were subtracted from the relative background (control, aluminum membrane filter coated with residue after filtration). Then, automatic baseline correction was employed to flatten the baseline, followed by a smoothing of 5 (Gaussian function of 9.643 cm−1). The preprocessed spectra were read by Excel (Microsoft, Inc., Redmond, WA). The heights and areas of spectral bands were measured and calculated by OMNIC and Origin 8.1 (OriginLab Corp., Northampton, MA). Second-derivative transforms using a 9-point Savitzky-Golay filter and wavelet transforms (with a scale of 7) were performed for spectral processing in Matlab to enhance the resolution of superimposed bands and to minimize problems from unavoidable baseline shifts. The reproducibility of vibrational spectra from three independent experiments was investigated by calculating Dy1y2 according to the following equations. In the equations, y1i and y2i are signal intensities of two different spectra, while ȳ1 and ȳ2 are average values of signal intensities of two different spectra; n represents the data points in the selected wave number region. The Dy1y2 ranges from 0 to 2,000. The lower the value, the better the reproducibility of spectra, with 0 indicating spectral ranges that are identical, 1,000 indicating completely noncorrelated spectra, and 2,000 indicating completely negatively noncorrelated spectra (34).
The comparison of spectra was performed by calculating selectivity, which indicates the spectral variations between the reference samples (control garlic concentrate) and treated samples (bacteria inoculated with garlic concentrate). Factorization was performed on average spectra of the respective groups. Factor analysis extracts high-dimensional vibrational spectral data into factors or principal components and corresponding scores (12). These scores are used to calculate the spectral distance (SD).
Where Ti is the cluster radius score of the ith factor (1, 2… i factors used). Selectivity (S) is calculated as the ratio of spectral distance (SD) between average spectra and the sum of threshold values T1 and T2 (cluster radii) as follows: S = SD/(T1 + T2).
Chemometric models were established based on processed spectra, including cluster analysis (principal-component analysis [PCA]), dendrogram analysis (discriminant function analysis [DFA]), class analog analysis (soft independent modeling of class analogs [SIMCA]), loading plot analysis, and partial least-squares regression (PLSR). PCA is used to reduce the dimensionality of multivariate data while preserving most of the variances. The selected unrelated principal components (PCs) are plotted and visualized in cluster forms (18). DFA can construct branched dendrogram structures using prior knowledge of the composition of a biological sample (28). SIMCA is a supervised classification method. The test samples are compared to study their analogy to the training set of samples (3). The combination of different chemometric models may improve and finally validate the properties of test samples (i.e., injured C. jejuni) according to different treatments (i.e., garlic-derived organosulfur compounds). The PLSR was employed for quantitative analysis using Matlab. A total of 18 spectra from each sample were used to establish the calibration model. A leave-one-out cross validation was performed to evaluate the prediction power of the model by removing one standard from the data set at a time and applying a calibration to the remaining standards. The suitability of the developed models for predicting live-C. jejuni concentrations was assessed by determining the regression coefficient (R), latent variables, the root mean square error of estimation (RMSEE), and the root mean square error of cross validation (RMSECV), calculated using the following equations (12).
where SEE is the standard error of estimation, M is the total number of samples analyzed, R is the regression coefficient, Yimeas is a measured concentration value of sample i, and Yipred is a predicted concentration value of sample i. The overall suitability of the models in predicting the C. jejuni concentration was evaluated from the residual prediction deviation (RPD) values. RPD is the ratio of the standard variation to the standard error of prediction. Loading plots were derived from chemometric analyses and used for explaining segregation or linear regression of chemometric models based on molecular levels (3). The wave numbers between 1,800 cm−1 and 900 cm−1 were selected for infrared-based chemometric analyses, and the wave numbers between 1,800 cm−1 and 400 cm−1 were selected for Raman-based chemometric analyses in the current study.
The experiment was performed in three independent replicate trials. The results are expressed as the means of results of three independent replicates ± standard deviations. The significant difference (P < 0.05) between the band area of spectra and the regression coefficient of the loading plot (first three PCs) were determined by one-way analysis of variance (ANOVA) following the t test in Matlab.
RESULTS AND DISCUSSION
Inhibitory effects of garlic concentrate and organosulfur compounds on Campylobacter jejuni.
Organosulfur compounds and polyphenols have both antimicrobial and antioxidant capacity. The total phenolic content (TPC) of garlic extract was 3.57 ± 0.49 mg GAE/g (dry weight), and the total antioxidant capacity (TAC) measured as the DPPH radical was 0.83 ± 0.12 mg Trolox/g (dry weight). The purity and stability of diallyl constituents were monitored throughout the study by HPLC. Both diallyl sulfide and diallyl trisulfide were maintained at 97%, and the level of diallyl disulfide was higher than 95%. All the analytes used were stable during study, and the stability of analytes was monitored during the course of the experiment. It is worth noting that TAC, TPC, and organosulfur compounds were stable for up to a month when kept in the dark at 4°C and stored under conditions to reduce light exposure, which we did for 1 to 2 months. Therefore, purified organosulfur compounds were used within 2 weeks, and fresh garlic concentrate was prepared and used daily.
The bactericidal effect of garlic concentrate on a cocktail of C. jejuni is shown in Table 1. The bactericidal effect increased along with an increase in the concentration of the garlic concentrate. A 2- to 3-log-CFU/ml reduction was achieved when the garlic concentrate was higher than 25 μl/ml at 4°C for 24 h. At 22°C, the bactericidal effect was greater than that of the treatment at 4°C, with the effects in saline water being more pronounced than in nutrient broth at the treatment concentration because of the presence of buffering components. In the case of the 35°C treatment, C. jejuni cells were totally inactivated at concentrations tested after 24 h. The suppressive effect of garlic concentrate on a cocktail of C. jejuni cells is shown in Table 2. The suppressive effect increased along with an increase of the concentration of garlic concentrate in sterilized broth. Higher treatment temperatures (22 and 35°C) increased the effectiveness of the garlic treatment through increased diffusion of organosulfur components into the cell and also increased the effectiveness of the antimicrobial treatment; organosulfur components are known to have a greater antimicrobial effect on microbial cells during the log phase. The suppressive and bactericidal effects of organosulfur compounds derived from garlic are shown in Tables S1 and S2 in the supplemental material. As the number of sulfur atoms in organosulfur compounds increased, the antimicrobial effects also increased at 4, 22, and 35°C (diallyl sulfide < diallyl disulfide < diallyl trisulfide). These findings were in agreement with those of previous studies (41, 59). O'Gara et al. (41) demonstrated that the antimicrobial activities of the diallyl sulfides toward Helicobacter pylori increased with the number of sulfur atoms. Yin and Cheng (59) observed that diallyl disulfide was more efficient at eliminating major pathogenic microorganisms (i.e., Escherichia coli O157:H7 and Listeria monocytogenes) than diallyl sulfide in ground beef.
Table 1.
Effect of garlic concentrates on the growth and survival of Campylobacter jejuni in sterilized saline water at different temperatures and timesa
| Temp (°C) | Concn of garlic concentrate (μl/ml) | No. of viable cells (log CFU/ml) at: |
|||||
|---|---|---|---|---|---|---|---|
| 0 h | 1 h | 3 h | 5 h | 10 h | 24 h | ||
| 4 | 0 | 4.95 ± 0.29 | 4.93 ± 0.69 | 4.88 ± 0.52 | 4.89 ± 0.24 | 4.82 ± 0.71 | 4.25 ± 0.25 |
| 6.25 | 4.99 ± 0.05 | 4.94 ± 0.32 | 4.86 ± 0.35 | 4.62 ± 0.41 | 4.21 ± 0.33 | 3.95 ± 0.32 | |
| 12.5 | 5.01 ± 0.12 | 4.93 ± 0.19 | 4.58 ± 0.29 | 4.29 ± 0.08 | 4.07 ± 0.49 | 3.26 ± 0.19 | |
| 25 | 4.87 ± 0.09 | 4.83 ± 0.23 | 4.60 ± 0.46 | 4.11 ± 0.21 | 3.77 ± 0.15 | 2.68 ± 0.28 | |
| 37.5 | 4.95 ± 0.56 | 4.87 ± 0.27 | 4.52 ± 0.09 | 3.93 ± 0.24 | 3.67 ± 0.26 | 2.35 ± 0.31 | |
| 50 | 4.85 ± 0.25 | 4.72 ± 0.06 | 4.38 ± 0.08 | 3.69 ± 0.28 | 3.25 ± 0.34 | 2.07 ± 0.34 | |
| 22 | 0 | 5.02 ± 0.32 | 5.01 ± 0.59 | 4.93 ± 0.07 | 4.84 ± 0.24 | 4.85 ± 0.14 | 4.61 ± 0.25 |
| 6.25 | 4.92 ± 0.38 | 4.93 ± 0.30 | 4.91 ± 0.13 | 4.88 ± 0.08 | 4.81 ± 0.37 | 4.48 ± 0.31 | |
| 12.5 | 4.93 ± 0.09 | 4.87 ± 0.41 | 4.81 ± 0.23 | 4.72 ± 0.09 | 4.62 ± 0.28 | 3.54 ± 0.19 | |
| 25 | 5.03 ± 0.61 | 4.79 ± 0.14 | 4.52 ± 0.41 | 4.28 ± 0.17 | 3.36 ± 0.29 | 1.32 ± 0.08 | |
| 37.5 | 4.92 ± 0.48 | 4.61 ± 0.25 | 4.07 ± 0.26 | 3.78 ± 0.32 | 2.62 ± 0.52 | 0 | |
| 50 | 5.07 ± 0.22 | 4.61 ± 0.21 | 4.16 ± 0.53 | 3.57 ± 0.17 | 2.15 ± 0.17 | 0 | |
| 35 | 0 | 4.99 ± 0.49 | 4.92 ± 0.24 | 4.84 ± 0.09 | 4.53 ± 0.35 | 3.47 ± 0.14 | 0 |
| 6.25 | 4.89 ± 0.72 | 4.52 ± 0.31 | 4.16 ± 0.36 | 2.76 ± 0.45 | 1.38 ± 0.09 | 0 | |
| 12.5 | 4.91 ± 0.09 | 4.43 ± 0.47 | 3.96 ± 0.41 | 2.54 ± 0.12 | 0 | 0 | |
| 25 | 5.02 ± 0.21 | 4.17 ± 0.48 | 3.06 ± 0.28 | 0 | 0 | 0 | |
| 37.5 | 5.02 ± 0.38 | 4.19 ± 0.15 | 2.85 ± 0.33 | 0 | 0 | 0 | |
| 50 | 5.01 ± 0.56 | 4.07 ± 0.24 | 1.93 ± 0.23 | 0 | 0 | 0 | |
Garlic (Allium sativum) in sterilized saline (0.85%, wt/vol) affected the growth and survival of C. jejuni. Values are means ± standard deviations (n = 3).
Table 2.
Effect of garlic concentrate on the growth and survival of Campylobacter jejuni in sterilized campylobacter nonselective broth at different temperatures and times
| Temp (°C) | Concn of garlic concentrate (μl/ml) | No. of viable cells (log CFU/ml) ata: |
||
|---|---|---|---|---|
| 0 days | 1 day | 3 days | ||
| 4 | 0 | 4.98 ± 0.18 | 4.48 ± 0.13 | 3.43 ± 0.23 |
| 25 | 5.05 ± 0.32 | 3.75 ± 0.42 | 2.54 ± 0.26 | |
| 50 | 4.97 ± 0.29 | 2.89 ± 0.25 | 2.08 ± 0.09 | |
| 75 | 5.02 ± 0.56 | 2.64 ± 0.31 | 1.93 ± 0.06 | |
| 100 | 5.10 ± 0.45 | 2.71 ± 0.36 | 1.56 ± 0.12 | |
| 22 | 0 | 4.92 ± 0.37 | 3.94 ± 0.31 | 2.95 ± 0.15 |
| 25 | 5.12 ± 0.51 | 2.68 ± 0.38 | 0 | |
| 50 | 4.98 ± 0.32 | 0 | 0 | |
| 75 | 4.92 ± 0.35 | 0 | 0 | |
| 100 | 5.09 ± 0.72 | 0 | 0 | |
| 35 | 0 | 4.90 ± 0.44 | 7.59 ± 0.53 | 7.15 ± 0.79 |
| 25 | 4.89 ± 0.37 | 0 | 0 | |
| 50 | 4.99 ± 0.21 | 0 | 0 | |
| 75 | 5.01 ± 0.34 | 0 | 0 | |
| 100 | 5.12 ± 0.38 | 0 | 0 | |
Values are means ± standard deviations (n = 3).
Diallyl sulfide components are naturally formed in garlic, Chinese leek, and onion. The contents of diallyl sulfide and diallyl disulfide in garlic were 250 to 480 and 2,600 to 5,100 μg/kg garlic, respectively. In the current study, 10 ml of garlic concentrate (18 to 34 g fresh cloves) contains approximately 10 μM diallyl disulfide. Previous sensory studies were performed using diallyl disulfide, and a 10 μM concentration of this organosulfur compound did not produce a marked-off aroma in ground beef, while a strong garlic smell remained when 18 to 34 g garlic was used (59). Furthermore, it should be pointed out that organosulfur compounds provided a significant antioxidant capacity, and the uses of these agents at these concentrations in various food systems as an antioxidant and/or antimicrobial agent should be safe and acceptable, at least in savory and moderately to highly flavored meat- or vegetable-based foods. Furthermore, diallyl sulfide components (diallyl sulfide, diallyl disulfide, and diallyl trisulfide) compose approximately 80% of commercial garlic oil, and the significant antimicrobial effects of these organosulfur compounds may partially explain the antimicrobial effect of commercial garlic oil (41).
FT-IR and Raman spectral features of Campylobacter jejuni.
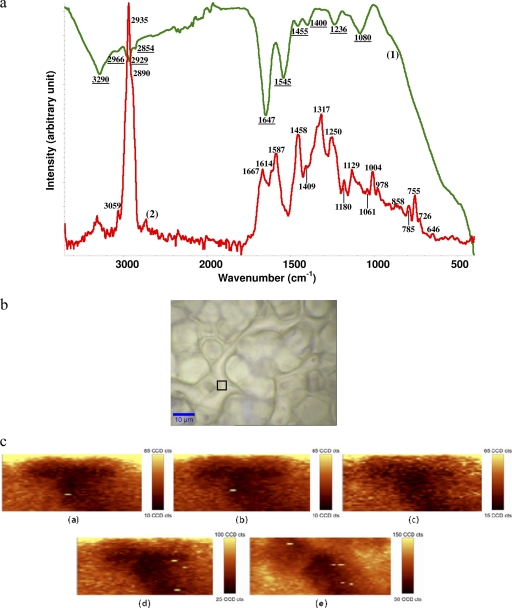
Infrared and Raman spectral features of C. jejuni intact cells are shown in Fig. 1. FT-IR spectral features of C. jejuni but not Raman spectral features of C. jejuni have been investigated (34, 35). The complementary (IR and Raman) spectral features provided more useful information about the biochemical components of the bacterial cell membrane than either alone. The assignment of bands frequently found in FT-IR and Raman spectra are summarized in Table S3 in the supplemental material. Both FT-IR and Raman spectroscopies provided a “fingerprint” region below the wave number of 1,800 cm−1, which reflects detailed information about the composition of C. jejuni cells. For Raman spectra, the bands at 646 and 1,614 cm−1 are assigned to tyrosine (37). The band at 726 cm−1 is assigned to C—S of the protein, CH2 rocking, and/or adenine (39). The band at 755 cm−1 is assigned to the symmetric breathing of tryptophan (37). The bands at 785 and 1,180 cm−1 are assigned to cytosine. The bands at 858 and 1,129 cm−1 are assigned to C—C stretching and the COC 1,4 glycosidic link (39). The bands at 1,004, 1,061, and 1,587 cm−1 are assigned to phenylalanine (30). The bands at 1,250 and 1,667 cm−1 are assigned to amide III and the α-helix structure of amide I, respectively (30). The band at 1,317 cm−1 is assigned to guanine (37). The band at 1,409 cm−1 is assigned to COO− stretching (37). The band at 1,458 cm−1 is assigned to nucleic acid (39). The band at 2,890 cm−1 is assigned to the CH2 asymmetric stretch of lipids and proteins (30). The distinctive band at 2,935 cm−1 is assigned to the CH3 and CH2 stretch (30). The band at 3,059 cm−1 is assigned to the (C C—H)(aromatic) stretch (37). For FT-IR spectra, the bands at 1,080 and 1,236 cm−1 are assigned to the symmetric and antisymmetric stretch of P
C—H)(aromatic) stretch (37). For FT-IR spectra, the bands at 1,080 and 1,236 cm−1 are assigned to the symmetric and antisymmetric stretch of P O of nucleic acids, respectively (37). The band at 1,400 cm−1 is assigned to the symmetric stretch of C—O of COO− groups (3). The band at 1,455 cm−1 is assigned to CH2 bending of lipids (34). The bands at 1,545 and 1,647 cm−1 are assigned, respectively, to amide II and amide I, the secondary structure of protein (36). The bands at 2,854, 2,929, and 2,966 cm−1 are related to methylene groups from lipids (35). The distinctive band at 3,290 cm−1 is assigned to the N—H stretch of proteins and O—H stretch of polysaccharides and water (39).
O of nucleic acids, respectively (37). The band at 1,400 cm−1 is assigned to the symmetric stretch of C—O of COO− groups (3). The band at 1,455 cm−1 is assigned to CH2 bending of lipids (34). The bands at 1,545 and 1,647 cm−1 are assigned, respectively, to amide II and amide I, the secondary structure of protein (36). The bands at 2,854, 2,929, and 2,966 cm−1 are related to methylene groups from lipids (35). The distinctive band at 3,290 cm−1 is assigned to the N—H stretch of proteins and O—H stretch of polysaccharides and water (39).
Fig. 1.
(a) Raw spectra of a C. jejuni IR transmission spectrum (line 1, green) and micro-Raman spectrum (line 2, red) with an excitation wavelength of 532 nm. (b) Micrograph of bacterial samples. The black frame indicates the mapping area for taking the Raman scans shown in panel a. (c) Raman mapping experiment of a single C. jejuni cell at selective wave numbers, namely, 713 to 780 cm−1 (image a), 1,084 to 1,200 cm−1 (image b), 1,272 to 1,388 cm−1 (image c), 1,518 to 1,661 cm−1 (image d), and 2,788 to 3,033 cm−1 (image e). cts, photon counts.
The Raman fingerprint region revealed features complementary to FT-IR spectroscopy for C. jejuni. These phenomena result from the different mechanisms of vibrational spectroscopy: Raman scattering relies on changes in the polarizability of functional groups as atoms vibrate, while IR absorption requires a change in the intrinsic dipole moment that occurs with molecular vibrations (39). Therefore, polar groups such as C O, N—H, and O—H have strong IR stretching vibrations in FT-IR spectroscopy, and nonpolar groups such as C—C and S—S have intense Raman bands, all of which add to the complementary natures of these two methods, justifying the use of both.
O, N—H, and O—H have strong IR stretching vibrations in FT-IR spectroscopy, and nonpolar groups such as C—C and S—S have intense Raman bands, all of which add to the complementary natures of these two methods, justifying the use of both.
Raman mapping (homogeneity) and spectral reproducibility studies.
The infrared spectrometer can determine properties only of bulk bacteria due to the laser width; however, the Raman spectrometer can detect features of a single bacterial cell, especially with the aid of confocal microscopy. A micrograph of C. jejuni is shown in Fig. 1b, and the frame indicates the mapping area of Raman images shown in the panel. The Raman mapping was performed on a single C. jejuni cell in five regions of different wave numbers (713 to 780 cm−1, 1,084 to 1,200 cm−1, 1,272 to 1,388 cm−1, 1,518 to 1,661 cm−1, and 2,788 to 3,033 cm−1) with a chosen step size of 3 by 3 μm2 (Fig. 1c), which provided major intensities of scattering bands in the Raman fingerprint region (Fig. 1a). The dark locations indicated lower contributions of band intensity, and light locations indicated higher contributions of band intensity. Therefore, it is important to understand that within the same bacteria, the band intensity is varied at different locations, and this reflects compositional differences at these different locations within the cell. Collectively, these maps are useful for quantifying biochemical changes occurring within individual cells as a result of an experimental treatment. For example, variation in the distributions of nucleic acids (Fig. 1c, image a), proteins (Fig. 1c, images b and c), and aliphatic lipids (Fig. 1c, image e) on the cell membrane can be observed. A combination of Raman mapping with Raman spectroscopy provides a method for localizing differences in chemical composition within the cell, making it possible to determine the specific site and type of cell injury.
The reproducibility of both FT-IR and Raman spectra from three independent experiments were calculated using the Pearson coefficient (expressed as the Dy1y2 value). Mean D values between 7 and 10 are considered normal when analyzing the first or second derivative of samples prepared from cultures grown in independent assays, and others have asserted that D values can be as high as 300 when microorganisms from different genera are compared (34). The D value of FT-IR spectra of C. jejuni is related to the following factors: wave number region (window) and culture age. First, five windows were selected to calculate D value: (i) the whole wave number region, i.e., 3,300 to 900 cm−1 (w1), (ii) 3,000 to 2,800 cm−1 (fatty acids; w2), (iii) 1,800 to 1,500 cm−1 (proteins and peptides; w3), (iv) 1,500 to 1,200 cm−1 (mixed region of proteins, fatty acids, and other phosphate-carrying compounds; w4), and (v) 1,200 to 900 cm−1 (carbohydrate; w5). The low D values were obtained from w1 (16.23 ± 0.39 to 24.13 ± 0.91) and w5 (19.91 ± 0.45 to 22.21 ± 1.14), and high D values were obtained from w2 (56.37 ± 7.31 to 81.29 ± 10.34), w3 (39.96 ± 4.59 to 50.21 ± 10.19), and w4 (66.46 ± 13.31 to 75.81 ± 10.29). Greater specificity may have been obtained for a single strain; a four-strain cocktail used in this study caused a higher D value. Second, the incubation time and its effect on spectral variation was also investigated and spectral reproducibility was consistent when the bacteria were cultivated within 48 h; however, as the cultivation time increased, the D value increased tremendously (higher than 400 at 72 h). Mouwen et al. indicated that reproducibility of infrared spectra of C. jejuni was strongly influenced by culture age (34). For C. jejuni, the shape of the cell (coccoid-spiral forms) and the chemical composition change as the culture ages, and both factors can significantly affect the reproducibility of vibrational spectral features in the current study. The D values for Raman spectra of C. jejuni were calculated from 1,800 to 400 cm−1 because spectral features were distinct. The D value was 3.32 ± 0.09 to 7.93 ± 1.15 after 48 h of cultivation, and it indicated that the reproducibility of Raman spectra is greater than that of FT-IR spectra. An additional increase in incubation time led to a higher D value and poorer reproducibility for both Raman and FT-IR spectroscopies, reducing the reproducibility of the Raman method to a range similar to that observed for FT-IR spectroscopy. For example, when cultivation times were greater than 48 h, the D value increased significantly (P < 0.05), from 7.93 at 48 h to 480 at 72 h. This finding emphasizes the necessity of following standardized procedures for vibrational spectral analysis, such as medium preparation, growth temperature, incubation time, and spectral measurement technique, if spectral reproducibility is to be obtained.
Second-derivative transformations to elucidate bacterial injury.
The variation of spectral features between garlic-treated (sulfide-treated) and non-garlic-treated (non-sulfide-treated) bacterial samples were not visually distinguishable, and second-derivative transformations needed to be performed to magnify minor differences in spectral features. The second-derivative transformations can reduce replicate variability, correct baseline shift, and resolve overlapping bands, therefore reducing the effect of band overlap (3). The selectivity values calculated at a 95% confidence interval for differentiating garlic concentrates and mixtures of bacteria and garlic concentrate were determined. Selectivity values greater than 1 were considered significant for detection of C. jejuni. Otherwise, overlapping clusters occurred (samples are not significantly different from the control). With the initial inoculation concentration of 105 CFU/ml, the selectivity value is higher than 1. This result was in agreement with a previous study (12) that found that 105 CFU/g was the detection limit for filtration by the FT-IR method to determine bacterial spectral features.
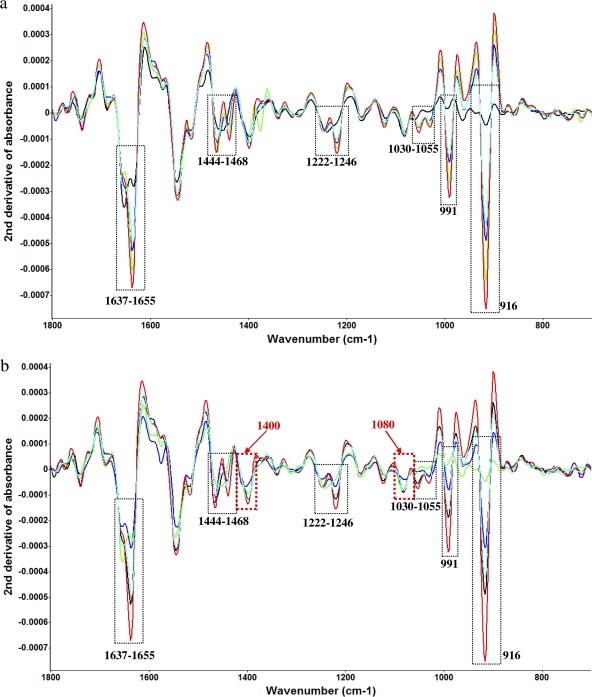
For the second-derivative transformation analyses of FT-IR spectra, the treatment using either garlic concentrate (25-μl/ml concentration) or diallyl sulfide (5 μM concentration) at 4°C in sterilized saline water (0.85% [wt/vol]) was performed to compare bacterial spectral variations. For both a garlic concentrate treatment (Fig. 2a) and a diallyl sulfide treatment (Fig. 2b), spectral variations were shown in several of the same band regions along with the time treatments (hour scale). The bands at 916 and 991 cm−1 are assigned to a phosphodiester (36). The band at 1,030 cm−1 is assigned to glycogen and CH2OH vibration (36). The band at 1,055 cm−1 is assigned to an oligosaccharide C—O bond in a hydroxyl group that might interact with some other membrane components, mainly from phospholipid phosphate and partly from oligosaccharide C—OH bonds (36). The band at 1,222 cm−1 is assigned to phosphate-stretching bands from phosphodiester groups of cellular nucleic acids (39). The band at 1,246 cm−1 is related to PO2− (asymmetric) (30). The band at 1,444 cm−1 is assigned to δCH2 lipids and fatty acids (36). The band at 1,468 cm−1 is assigned to δCH2 of lipids (36). The bands at 1,637 and 1,655 cm−1 are assigned to amide I of β-pleated sheet structures and amide I of α-helical structures, respectively (30).
Fig. 2.
(a) Second-derivative transformation of FT-IR spectral features of C. jejuni treated with 5 μM diallyl sulfide at 4°C for different time intervals (black, 0 h; indigo, 1 h; blue, 3 h; green, 5 h; red, 24 h). (b) Second-derivative transformation of FT-IR spectral features of C. jejuni treated with 25 μl/ml garlic concentrate at 4°C for different time intervals (green, 0 h; blue, 1 h; indigo, 3 h; black, 5 h; red, 24 h). The red columns show differences in spectral features treated with garlic concentrate and organosulfur compounds.
However, some differences were observed between garlic concentrate-treated samples and diallyl sulfide-treated samples when second-derivative transformations were employed. The band at 1,080 cm−1 is assigned to symmetric phosphate stretching (3). The band at 1,400 cm−1 is assigned to a symmetric stretch of methyl groups in (skeletal) proteins (30). The variations in these several bands may be due to the phenolic compounds in garlic concentrate which had additional antimicrobial effects on C. jejuni. Phenolics, which are somewhat hydrophobic, may act efficiently at the bacterial membrane-water interface, embedding into the membrane and thereby impairing the cell membrane and the transport processes (55). In the current study, the band intensities of only those bands that contributed significantly (P < 0.05) to the second-derivative transformations of absorption (y axis of Fig. 2) were calculated using Matlab and Origin. The spectral transformation and intensity calculations confirmed that organosulfur compounds derived from garlic provided the greatest contribution to the antimicrobial effects of garlic concentrate, with the phenolic compounds having a smaller effect.
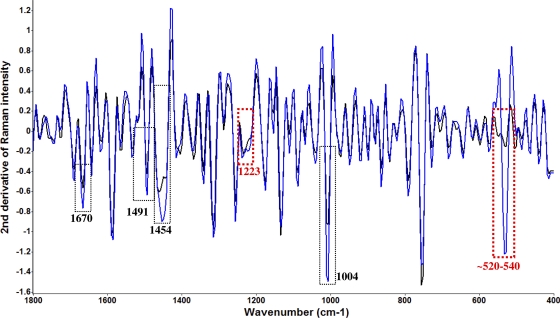
The second-derivative transformation of Raman spectra (Fig. 3) was performed to further study the bacterial cell variation under the treatment of organosulfur compounds derived from garlic. Raman spectra provided additional spectral information compared to FT-IR spectra. The bands at 520 to 540 cm−1 are assigned to S—S disulfide stretching in proteins (39). The band at 1,004 cm−1 is assigned to phenylalanine (39). The band at 1,223 cm−1 is assigned to cellular nucleic acid (37). The band at 1,454 cm−1 is assigned to CH2 stretching of phospholipids (37). The band at 1,491 cm−1 is assigned to C—N stretching vibration coupled with the in-plane C—H bending in amino radical cations (37). The band at 1,670 cm−1 is assigned to amide I (30). The significant (P < 0.05) variation in second-derivative transformations of spectral features around the wave numbers of the “sulfur” vibrational stretching area (520 to 540 cm−1) was a very important finding in this study. First, it validated the findings from previous studies (32) that sulfur compounds such as thiosulfinates are responsible for the antimicrobial activity of garlic. Inhibition of certain thiol-containing enzymes in microorganisms by the rapid reaction of thiosulfinates with thiol groups is assumed to be the main mechanism for antimicrobial activity, and allicin can freely penetrate the phospholipid bilayers of bacterial cell walls and interact with the thiol-containing enzymes (32). Here we have used vibrational spectroscopies, specifically Raman spectroscopy, to monitor the transmembrane transfer of sulfur-containing compounds into bacterial cells (Fig. 3), and we find that this is both a concentration and a time-dependent event. The concentration of intracellular sulfur compounds was inversely proportional to cell survival and to changes in the spectral features of bacterial proteins/enzymes indicative of denaturation (4, 40) that may be associated with binding of organosulfur compounds to thiol-containing proteins. Second, we demonstrated that using the combination of infrared and Raman spectroscopies provides complementary information and substantially more information than using a single technique for monitoring bacterial stress and injury. Some important markers (parameters), such as variation of sulfur compounds, were not available to be monitored by one technique (i.e., IR spectroscopy) but could be monitored by another one (i.e., Raman spectroscopy).
Fig. 3.
Second-derivative transformation of Raman spectral features of C. jejuni treated with 5 μM diallyl sulfide at 4°C for different time intervals (blue, treated for 4 h; black, control). The dotted red columns show differences of spectral features between FT-IR and Raman spectroscopies (especially sulfur compounds).
Electron microscope examination of cell injury.
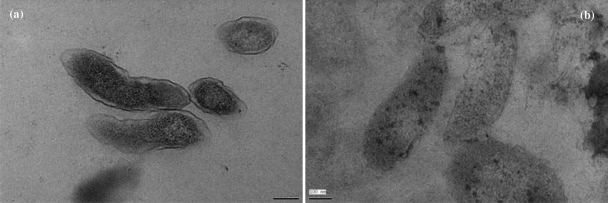
To correlate vibrational spectroscopic data with structure changes caused by garlic-derived organosulfur compounds, scanning electron micrograph data and transmission electron micrograph data were collected for nontreated and treated samples with 5 μM diallyl sulfide in sterilized broth for 10 h at 22°C. Figure S1 in the supplemental material and Fig. 4 show that untreated cells of C. jejuni had a uniform cellular structure with well-defined membranes and little debris in the cell's surrounding environment. Exposure to organosulfur compounds resulted in morphological damage, such as loss of the structural integrity of the cell wall, cell membrane, and intracellular matrix. Cell deformation, breakage of cell walls and membranes, condensation of cellular material, and the presence of significant amounts of cytoplasmic material and membrane fragments were observed in the damaged cells of C. jejuni.
Fig. 4.
Transmission electron microscope images of Campylobacter jejuni without (a) and with (b) treatment with organosulfur compounds (diallyl sulfide) derived from garlic (Allium sativum).
Cluster, dendrogram, and class analog analyses.
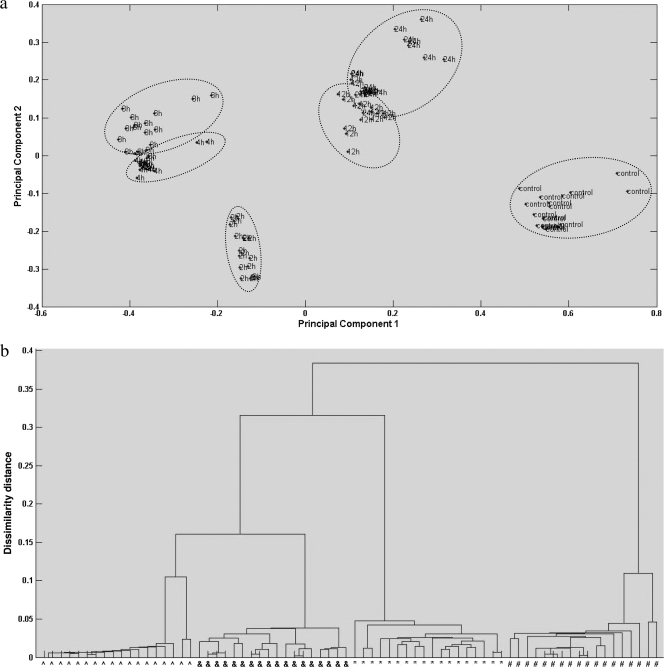
The injury levels of C. jejuni were investigated using three different segregation models, including cluster analysis (principal-component analysis [PCA]), dendrogram analysis (discriminant function analysis [DFA]), and class analog analysis (soft independent modeling of class analog [SIMCA]). All three analyses are based on principal-component selection; PCA is nonsupervised chemometrics, while DFA and SIMCA are supervised chemometrics (18). Due to the features of high-dimensional vectors for infrared and Raman spectra (wave number versus signal intensity), major PC extraction is important to chemometric-model analysis. Figure 5 shows representative clear segregation of C. jejuni samples treated with organosulfur compounds (diallyl sulfide) during various time intervals at 22°C. The spectra from the first two experiments were used to establish the segregation models, and the spectra from the third experiment were employed for model validation. Clear segregation (DFA) and tight clusters (PCA) indicated a significant (P < 0.05) difference in each group of samples, with interclass distance ranging from 3.15 to 29.86 based upon Mahalanobis distance measurements computed between the centroids of classes. Clusters with interclass distance values higher than 3 are believed to be significantly different from each other (12, 18). Class analog results were shown in Table 3, and an ∼90% correction rate for data classifying was achieved for both infrared and Raman spectra. Furthermore, in the current study, either sterilized saline water or campylobacter broth was used for analyses of all three segregation models at different treatment temperatures (4 or 22°C). The effect of temperature or extraneous material from either matrix (either broth or saline water) was negligible. Matrix components were removed by centrifugation and filtration through aluminum membrane filters to eliminate the effect of these components on spectral quality and reproducibility (see above) (3).
Fig. 5.
(a) Representative two-dimensional principal-component analysis of C. jejuni cocktail in sterilized campylobacter nonselective broth with 5 μM diallyl sulfide treatment for 0 (control), 2, 4, 8, 12, and 24 h at room temperature (22°C) using infrared spectra. (b) Representative dendrogram analysis of C. jejuni cocktail in sterilized campylobacter nonselective broth with 5 μM diallyl sulfide treatment for 0 (∧), 4 (&), 12 (*), and 24 h (#) at room temperature (22°C) using Raman spectra.
Table 3.
SIMCA classification results for each treatment compared to those for a controla
| Spectra | Treatment or treatment time (h) | No. of correctly classified spectra | % of correctly classified spectra |
|---|---|---|---|
| FT-IR | Control | 17 | 94 |
| 4 | 16 | 89 | |
| 8 | 16 | 89 | |
| 12 | 18 | 100 | |
| 24 | 17 | 94 | |
| Raman | Control | 9 | 100 |
| 4 | 8 | 89 | |
| 8 | 9 | 100 | |
| 12 | 8 | 89 | |
| 24 | 9 | 100 |
Five micromolar diallyl sulfide in sterilized broth at 22°C.
PLSR model analyses.
PLSR using a wave number below 1,800 cm−1 as x and an indicator variable (loading plot) for differentiation between the strains of injured levels as y was performed for both infrared and Raman spectroscopies. All the parameters related to the PLSR model are summarized in Table 4. Due to the limited number of samples, the leave-one-out method was performed as a cross validation step. A good PLSR model should have high values for the regression coefficient (R) (>0.95) and RPD (>5) and low values for RMSEE and RMSECV (<1) for calibration and cross validation (18). Furthermore, a reasonable number of latent variables (generally, <10) is desired for the PLSR model to avoid noise as useful bands in the model, vividly called “overfitting” (3). Both FT-IR and Raman spectrum-based PLSR models had promising results for predicting various concentrations of survival for C. jejuni in either sterilized broth or saline water treated with garlic-derived organosulfur compounds. Overall, both infrared- and Raman-based PLSR models provided similar models of behavior and prediction abilities on the basis of R, RPD, and RMSEE.
Table 4.
Partial least-squares regression models for quantification of live C. jejuni strains treated with garlic-derived organosulfur compoundsa
| Spectrab | Range | No. of samples | No. of latent variables | R calc | RMSEE cal | RPD cal | R vald | RMSE val | RPD val |
|---|---|---|---|---|---|---|---|---|---|
| FT-IR | 1.78–4.91 | 45 | 8 | ≥0.98 | ≤0.12 | ≥16.41 | ≥0.95 | ≤0.34 | ≥8.75 |
| Raman | 1.63–4.89 | 45 | 9 | ≥0.99 | ≤0.28 | ≥13.54 | ≥0.95 | ≤0.41 | ≥9.12 |
Five micromolar diallyl sulfide.
For FT-IR spectroscopy, the wave number from 1,800 to 700 cm−1 was used for model analyses; for Raman spectroscopy, the wave number from 1,800 to 400 cm−1 was used for model analyses.
cal, calibration.
val, validation.
Loading plot studies of the PLSR model.
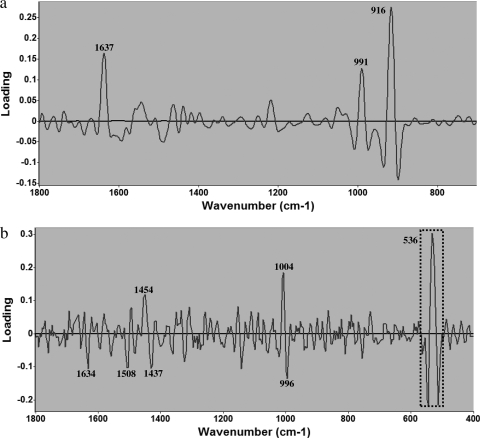
Analysis of FT-IR and Raman spectral loading plots derived from individual PLSR models showed that the chemical composition of bacterial cells was affected by the treatment of organosulfur compounds (Fig. 6). The major bands of the first principal component (PC) are most critical to elucidate the physiological changes of bacterial cells during various treatments, as determined by one-way ANOVA and the t test (P < 0.05). The first PC can explain the approximately 76% and 81% contributions to the regression coefficient (R) value for the FT-IR and Raman PLSR models, respectively. For an FT-IR loading plot (Fig. 6a), the distinctive band at 916 cm−1 is assigned to the phosphodiester region (30), and another band at 991 cm−1 is also assigned to phosphodiester stretching (39). The band at 1,637 cm−1 is assigned to amide I of β-pleated sheet structures (3). This indicates the importance of the integrity of phosphodiester and protein secondary structure to cell survivability under unfavorable treatments, such as exposure to organosulfur compounds. For the Raman loading plot (Fig. 6b), the distinctive band region around 520 to 540 cm−1 is assigned to S—S disulfide stretching in proteins (39). The band at 996 cm−1 is assigned to C—O ribose, C—C (36). The band at 1,004 cm−1 is assigned to phenylalanine (39). The band at 1,437 cm−1 is assigned to the CH2 deformation of lipids (36). The band at 1,454 cm−1 is assigned to CH2 stretching of phospholipids (30). The band at 1,508 cm−1 is assigned to cytosine (36), and the band at 1,634 cm−1 is assigned to amide I (39). This result demonstrates that the Raman loading plot confirms the complementary natures of Raman and FT-IR spectroscopies to determine type and degree of bacterial cell injury as indicated by variation in the chemical compositions of cells under unfavorable treatment. The distinction between cells exposed to different treatments around 520 to 540 cm−1 was impressive and reflected cellular uptake of sulfur compounds, providing evidence to support the hypothesis that the major bactericidal mechanism for cell inactivation by garlic is from damage caused by organosulfur compounds and is related to the combination of thiosulfinates with enzymes or functional proteins which possess sulfur atoms, thus altering the structures of those enzymes or proteins (5, 14, 32, 38).
Fig. 6.
Loading plots of first principal component obtained from PLSR for FT-IR spectroscopy (a) and Raman spectroscopy (b) for C. jejuni stress.
In conclusion, garlic concentrate was effective in inhibiting the growth of C. jejuni. This inhibition was proportional to the concentration of the organosulfur compounds, suggesting that the antimicrobial effect was dependent on the number of sulfur atoms in the diallyl sulfides. FT-IR spectroscopy verified that organosulfur compounds (diallyl sulfides and thiosulfinates) contributed most to the antimicrobial effect of garlic, substantially more than phenolic compounds. Raman spectroscopy conducted at a single-cell level provided information complementary to that provided by FT-IR spectroscopy and clearly showed that sulfur components were absorbed by C. jejuni cells (∼520 to 540 cm−1). This strongly supports the findings of previous studies showing that organosulfur compounds can freely penetrate cell membranes and combine with a thiol-containing enzyme and/or protein, altering their structures. Spectroscopy-based cluster analysis, dendrogram analysis, and class analog analysis can segregate C. jejuni based upon different injury levels, and this effect could be quantified using PLSR models to predict the actual survival of C. jejuni following diallyl sulfide treatment at different concentrations and at different temperatures. The specific biochemical components of the cell membrane most closely related to bacterial cell injury may be determined from spectroscopy-based loading plots. This is the first study to show how FT-IR and Raman spectroscopies can be used together to study bacterial stress and injury under unfavorable treatment conditions, and it shows the potential of this technique for monitoring pathogenic or spoilage microorganisms rapidly and precisely, potentially for field and online work.
Supplementary Material
ACKNOWLEDGMENTS
We deeply appreciate the assistance of Valerie Jean Lynch-Holm with electron microscopy at the Franceschi Microscopy and Imaging Center at Washington State University (WSU), Pullman, WA.
This work was supported from funds awarded to B.A.R. through a USDA special food security grant and from the School of Food Science at WSU. Research in the laboratory of M.E.K. is supported by the National Institutes of Health, Department of Health and Human Services, under contract number NO1-Al-30055 and, in part, from funds provided by the School of Molecular Biosciences at WSU. We also gratefully acknowledge the support of the National Science Foundation (award DMR-0619310) and the University of Idaho Biological Applications of Nanotechnology (BANTech) Center.
The chemometric models in the current study were developed with programming written by Xiaonan Lu using Matlab (version 2010a). The readers who are interested in Matlab programming codes used for PCA, DFA, SIMCA, PLSR, and other spectral preprocessing methods should send inquiries to xiaonan_lu@wsu.edu.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Ahn J., Grun I. U., Mustapha A. 2004. Antimicrobial and antioxidant activities of natural extracts in vitro and in ground beef. J. Food Prot. 67:148–155 [DOI] [PubMed] [Google Scholar]
- 2. Ahn J., Grun I. U., Mustapha A. 2007. Effects of plant extracts on microbial growth, color change, and lipid oxidation in cooked beef. Food Microbiol. 24:7–14 [DOI] [PubMed] [Google Scholar]
- 3. Al-Qadiri H. M., Lin M., Al-Holy M. A., Cavinato A. G., Rasco B. A. 2008. Detection of sublethal thermal injury in Salmonella enterica serotype Typhimurium and Listeria monocytogenes using Fourier transform infrared (FT-IR) spectroscopy (4,000 to 600 cm−1). J. Food Sci. 73:M54–M61 [DOI] [PubMed] [Google Scholar]
- 4. Alvarez-Ordóñez A., Prieto M. 2010. Changes in ultrastructure and Fourier transform infrared spectrum of Salmonella enterica serovar Typhimurium cells after exposure to stress conditions. Appl. Environ. Microbiol. 76:7598–7607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ankri S., Mirelman D. 1999. Antimicrobial properties of allicin from garlic. Microbes Infect. 2:125–129 [DOI] [PubMed] [Google Scholar]
- 6. Benkeblia N. 2004. Antimicrobial activity of essential oil extracts of various onions (Allium cepa) and garlic (Allium sativum). LWT Food Sci. Technol. 37:263–268 [Google Scholar]
- 7. Billing J., Sherman P. W. 1998. Antimicrobial functions of spices: why some like it hot. Q. Rev. Biol. 73:3–49 [DOI] [PubMed] [Google Scholar]
- 8. Castellano P., Vignolo G., Farías R. N., Arrondo J. L., Chehín R. 2007. Molecular view by Fourier transform infrared spectroscopy of the relationship between lactocin 705 and membranes: speculations on antimicrobial mechanism. Appl. Environ. Microbiol. 73:415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choo-Smith L.-P., et al. 2001. Investigating microbial (micro)colony heterogeneity by vibrational spectroscopy. Appl. Environ. Microbiol. 67:1461–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chung I., Kwon S. H., Shim S.-T., Kyung K. H. 2007. Synergistic antiyeast activity of garlic oil and allyl alcohol derived from alliin in garlic. J. Food Sci. 72:M437–M440 [DOI] [PubMed] [Google Scholar]
- 11. Cowan M. M. 1999. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 12:564–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davis R., Irudayaraj J., Reuhs B. L., Mauer L. J. 2010. Detection of E. coli O157:H7 from ground beef using Fourier transform infrared (FT-IR) spectroscopy and chemometrics. J. Food Sci. 75:M340–M346 [DOI] [PubMed] [Google Scholar]
- 13. Efrima S., Zeiri L. L. 2008. Understanding SERS of bacteria. J. Raman Spectrosc. 40:277–288 [Google Scholar]
- 14. Feldberg R. S., et al. 1988. In vitro mechanism of inhibition of bacterial cell growth by allicin. Antimicrob. Agents Chemother. 32:1763–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Friedman M., Henika P. R., Mandrell R. E. 2002. Bactericidal activities of plant essential oils and some of their isolated constituents against Campylobacter jejuni, Escherichia coli, Listeria monocytogenes, and Salmonella enterica. J. Food Prot. 65:1545–1560 [DOI] [PubMed] [Google Scholar]
- 16. Friedman M., Henika P. R., Mandrell R. E. 2003. Antibacterial activities of phenolic benzaldehydes and benzoic acids against Campylobacter jejuni, Escherichia coli, Listeria monocytogenes, and Salmonella enterica. J. Food Prot. 66:1811–1821 [DOI] [PubMed] [Google Scholar]
- 17. Ganan M., Martinez-Rodriguez A. J., Carrascosa A. V. 2009. Antimicrobial activity of phenolic compounds of wine against Campylobacter jejuni. Food Control 20:739–742 [Google Scholar]
- 18. Goodacre R. 2003. Explanatory analysis of spectroscopic data using machine learning of simple, interpretable rules. Vib. Spectrosc. 32:33–45 [Google Scholar]
- 19. Harris J. C., Cottrell S. L., Plummer S., Lloyd D. 2001. Antimicrobial properties of Allium sativum (garlic). Appl. Microbiol. Biotechnol. 57:282–286 [DOI] [PubMed] [Google Scholar]
- 20. Harz M., Rösch P., Popp J. 2009. Vibrational spectroscopy—a powerful tool for the rapid identification on microbial cells at the single-cell level. Cytometry A 75:104–113 [DOI] [PubMed] [Google Scholar]
- 21. Kim J. W., Kim Y. S., Kyung K. H. 2004. Inhibitory activity of essential oil of garlic and onion against bacteria and yeasts. J. Food Prot. 67:499–504 [DOI] [PubMed] [Google Scholar]
- 22. Kirschner C., et al. 2001. Classification and identification of enterococci: a comparative phenotypic, genotypic, and vibrational spectroscopic study. J. Clin. Microbiol. 39:1763–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumar M., Berwal J. S. 1998. Sensitivity of food pathogens to garlic (Allium sativum). J. Appl. Microbiol. 84:213–215 [DOI] [PubMed] [Google Scholar]
- 24. Kyung K. H., Lee Y. C. 2001. Antimicrobial activities of sulfur compounds derived from S-alk(en)yl-l-cysteine sulfoxides in Allium and Brassica. Food Rev. Int. 17:183–198 [Google Scholar]
- 25. Lacombe A., Wu V. C., Tyler S., Edwards K. 2010. Antimicrobial action of the American cranberry constituents; phenolics, anthocyanins, and organic acids, against Escherichia coli O157:H7. Int. J. Food Microbiol. 139:102–107 [DOI] [PubMed] [Google Scholar]
- 26. Lee C.-F., Han C.-K., Tsau J.-L. 2004. In vitro inhibitory activity of Chinese leek extract against Campylobacter species. Int. J. Food Microbiol. 94:169–174 [DOI] [PubMed] [Google Scholar]
- 27. Lin M., et al. 2004. Discrimination of intact and injured Listeria monocytogenes by Fourier transform infrared spectroscopy and principal component analysis. J. Agric. Food Chem. 52:5769–5772 [DOI] [PubMed] [Google Scholar]
- 28. López-Díez E. C., Winder C. L., Ashton L., Currie F., Goodacre R. 2005. Monitoring the mode of action of antibiotics using Raman spectroscopy: investigating subinhibitory effects on amikacin on Pseudomonas aeruginosa. Anal. Chem. 77:2901–2906 [DOI] [PubMed] [Google Scholar]
- 29. Maquelin K., et al. 2003. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J. Clin. Microbiol. 41:324–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maquelin K., et al. 2002. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods 51:255–271 [DOI] [PubMed] [Google Scholar]
- 31. Medina E., de Castro A., Romero C., Brenes M. 2006. Comparisons of the concentrations of phenolic compounds in olive oil and other plant oils: correlation with antimicrobial activity. J. Agric. Food Chem. 54:4954–4961 [DOI] [PubMed] [Google Scholar]
- 32. Miron T., Rabinkov A., Mirelman D., Wilchek M., Weiner L. 2000. The mode of action of allicin: its ready permeability through phospholipid membranes may contribute to its biological activity. Biochim. Biophys. Acta 1463:20–30 [DOI] [PubMed] [Google Scholar]
- 33. Moritz T. J., et al. 2010. Effect of cefazolin treatment on the nonresonant Raman signatures of the metabolic state of individual Escherichia coli cells. Anal. Chem. 82:2703–2710 [DOI] [PubMed] [Google Scholar]
- 34. Mouwen D. J., Weijtens M. J., Capita R., Alonso-Calleja C., Prieto M. 2005. Discrimination of enterobacterial repetitive intergenic consensus PCR types of Campylobacter coli and Campylobacter jejuni by Fourier transform infrared spectroscopy. Appl. Environ. Microbiol. 71:4318–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mouwen D. J., Capita R., Alonso-Calleja C., Prieto-Gómez J., Prieto M. 2006. Artificial neural network based identification of Campylobacter species by Fourier transform infrared spectroscopy. J. Microbiol. Methods 67:131–140 [DOI] [PubMed] [Google Scholar]
- 36. Movasaghi Z., Rehman S., Rehman I. U. 2008. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 43:134–179 [Google Scholar]
- 37. Movasaghi Z., Rehman S., Rehman I. U. 2007. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 42:493–541 [Google Scholar]
- 38. Naganawa R., et al. 1996. Inhibition of microbial growth by ajoene, a sulfur-containing compound derived from garlic. Appl. Environ. Microbiol. 62:4238–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Naumann D. 2001. FT-infrared and FT-Raman spectroscopy in biomedical research. Appl. Spectrosc. Rev. 36:239–298 [Google Scholar]
- 40. Neugebauer U., et al. 2007. The influence of fluoroquinolone drugs on the bacterial growth of S. epidermidis utilizing the unique potential of vibrational spectroscopy. J. Phys. Chem. A 111:2898–2906 [DOI] [PubMed] [Google Scholar]
- 41. O'Gara E. A., Hill D. J., Maslin D. J. 2000. Activities of garlic oil, garlic powder, and their diallyl constituents against Helicobacter pylori. Appl. Environ. Microbiol. 66:2269–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oust A., et al. 2006. Fourier transform infrared and Raman spectroscopy for characterization of Listeria monocytogenes strains. Appl. Environ. Microbiol. 72:228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Park S. F. 2002. The physiology of Campylobacter species and its relevance to their role as foodborne pathogens. Int. J. Food Microbiol. 74:177–188 [DOI] [PubMed] [Google Scholar]
- 44. Queiroz Y. S., Ishimoto E. Y., Bastos D. H., Sampaio G. R., Torres E. A. 2009. Garlic (Allium sativum L.) and ready-to-eat garlic products: in vitro antioxidant activity. Food Chem. 115:371–374 [Google Scholar]
- 45. Rees L. P., Minney S. F., Plummer N. T., Slater J. H., Skyrme D. A. 1993. A quantitative assessment of the anti-microbial activity of garlic (Allium sativum). World J. Microbiol. Biotechnol. 9:303–307 [DOI] [PubMed] [Google Scholar]
- 46. Rösch P., et al. 2005. Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations. Appl. Environ. Microbiol. 71:1626–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rose P., Whiteman M., Moore P. K., Zhu Y. Z. 2005. Bioactive S- alk(en)yl cysteine sulfoxide metabolites in the genus Allium: the chemistry of potential therapeutic agents. Nat. Prod. Rep. 22:351–368 [DOI] [PubMed] [Google Scholar]
- 48. Ross Z. M., O'Gara E. A., Hill D. J., Sleightholme H. V., Maslin D. J. 2001. Antimicrobial properties of garlic oil against human enteric bacteria: evaluation of methodologies and comparisons with garlic oil sulfides and garlic powder. Appl. Environ. Microbiol. 67:475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sivam G. P. 2001. Protection against Helicobacter pylori and other bacterial infections by garlic. J. Nutr. 131:1106S–1108S [DOI] [PubMed] [Google Scholar]
- 50. Stöckel S., et al. 2010. Raman spectroscopy-compatible inactivation method for pathogenic endospores. Appl. Environ. Microbiol. 76:2895–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sun T., Powers J. R., Tang J. 2007. Evaluation of the antioxidant activity of asparagus, broccoli and their juices. Food Chem. 105:101–106 [Google Scholar]
- 52. Unal R., Fleming H. P., McFeeters R. F., Thompson R. L., Breidt F., Jr., Giesbrecht F. G. 2001. Novel quantitative assays for estimating the antimicrobial activity of fresh garlic juice. J. Food Prot. 64:189–194 [DOI] [PubMed] [Google Scholar]
- 53. Valtierra-Rodríguez D., Heredia N. L., Garcia S., Sánchez E. 2010. Reduction of Campylobacter jejuni and Campylobacter coli in poultry skin by fruit extracts. J. Food Prot. 73:477–482 [DOI] [PubMed] [Google Scholar]
- 54. Willemse-Erix D. F. M., et al. 2009. Optical fingerprinting in bacterial epidemiology: Raman spectroscopy as a real-time typing method. J. Clin. Microbiol. 47:652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu V. C. H., Qiu X., Bushway A., Harper L. 2008. Antimicrobial effects of American cranberries (Vaccinium macrocarpon) on foodborne pathogens. LWT Food Sci. Technol. 41:1834–1841 [Google Scholar]
- 56. Wu V. C. H., Qiu X., de los Reyes B., Lin C.-S., Pan Y. J. 2009. Application of cranberry concentrate (Vaccinium macrocarpon) to control Escherichia coli O157:H7 in ground beef and its antimicrobial mechanism related to the downregulated slp, hdeA and cfa. Food Microbiol. 26:32–38 [DOI] [PubMed] [Google Scholar]
- 57. Wu V. C. H., Qiu X., Hsieh Y.-H. P. 2008. Evaluation of Escherichia coli O157:H7 in apple juice with Cornus fruit (Cornus officinalis Sieb. et Zucc.) extract by conventional media and thin agar layer method. Food Microbiol. 25:190–195 [DOI] [PubMed] [Google Scholar]
- 58. Wu V. C. H. 2008. A review of microbial injury and recovery methods in food. Food Microbiol. 25:735–744 [DOI] [PubMed] [Google Scholar]
- 59. Yin M.-C., Cheng W.-S. 2003. Antioxidant and antimicrobial effects of four garlic-derived organosulfur compounds in ground beef. Meat Sci. 63:23–28 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.