Abstract
Celiac disease (CD) is an immune-mediated enteropathy involving genetic and environmental factors whose interaction might influence disease risk. The aim of this study was to determine the effects of milk-feeding practices and the HLA-DQ genotype on intestinal colonization of Bacteroides species in infants at risk of CD development. This study included 75 full-term newborns with at least one first-degree relative suffering from CD. Infants were classified according to milk-feeding practice (breast-feeding or formula feeding) and HLA-DQ genotype (high or low genetic risk). Stools were analyzed at 7 days, 1 month, and 4 months by PCR and denaturing gradient gel electrophoresis (DGGE). The Bacteroides species diversity index was higher in formula-fed infants than in breast-fed infants. Breast-fed infants showed a higher prevalence of Bacteroides uniformis at 1 and 4 months of age, while formula-fed infants had a higher prevalence of B. intestinalis at all sampling times, of B. caccae at 7 days and 4 months, and of B. plebeius at 4 months. Infants with high genetic risk showed a higher prevalence of B. vulgatus, while those with low genetic risk showed a higher prevalence of B. ovatus, B. plebeius, and B. uniformis. Among breast-fed infants, the prevalence of B. uniformis was higher in those with low genetic risk than in those with high genetic risk. Among formula-fed infants, the prevalence of B. ovatus and B. plebeius was increased in those with low genetic risk, while the prevalence of B. vulgatus was higher in those with high genetic risk. The results indicate that both the type of milk feeding and the HLA-DQ genotype influence the colonization process of Bacteroides species, and possibly the disease risk.
INTRODUCTION
The newborn intestine is colonized immediately after birth by microorganisms from the mother and the environment (12, 21). At birth, the intestinal milieu of neonates shows a positive redox potential, and early bacterial colonization begins with facultative anaerobes (Enterobacteriaceae, Lactobacillus, etc.) that gradually consume the oxygen, permitting the growth of strict anaerobes (Bifidobacterium, Bacteroides, Clostridium, etc.) (18, 22). Subsequently, milk-feeding practices play an important role in the microbiota composition of the infant gut (2, 18, 22). In breast-fed infants, the microbiota is less diverse and is dominated by Bifidobacterium species, while a more diverse microbiota develops only after complementary feeding commences. In contrast, the bacterial composition of formula-fed infants is dominated by members of diverse genera (Enterobacteriaceae, Streptococcus, Bacteroides, Clostridium, and Bifidobacterium) (1, 9). Intestinal colonization influences diverse physiological functions, which may have an impact on the host's health and disease risk (16, 18, 25). Nevertheless, there is still limited information on the initial establishment of Bacteroides species and its possible influence on health (11, 19, 30).
Celiac disease (CD) is a multifactorial disorder involving both genetic and environmental factors. This disease is associated with human leukocyte antigen (HLA) genes of the major histocompatibility complex (MHC), and approximately 95% of patients are positive for HLA-DQ2 or -DQ8 (28). Studies of twins also showed that in 25% of cases, one twin of the pair did not develop CD, supporting the role of environmental factors in disease development (10, 20). Breast-feeding seems to exert a protective effect against CD development (4, 15), but its possible connection with modulation of the intestinal microbiota is unknown. A preliminary study suggested an association between increased Bacteroides-Prevotella group proportions and the HLA-DQ genotype, but the sample size was limited (8).
The aim of this study was to determine the influence of milk-feeding practices (breast-feeding versus formula feeding) and HLA-DQ genotype on the intestinal colonization process of Bacteroides species in a representative group of infants with a familial risk of developing CD. To do so, PCR and denaturing gradient gel electrophoresis (DGGE) analyses were performed. The final purpose of the study was to shed light on the interactions between the intestinal colonization process, diet, and genotype and on their overall influence on CD risk.
MATERIALS AND METHODS
Subjects.
This study included 75 full-term newborns with at least one first-degree relative suffering from CD, selected from an ongoing prospective observational 3-year study. Seventy-four percent of the infants had a sibling suffering from CD, 17% of the infants had a parent with CD, and 9% of the infants had both one sibling and one parent with CD. The distributions of the type and number of relatives suffering from CD were similar for the subgroups of infants considered for statistical comparisons (Table 1). Fifty-one percent of the samples were from infants born in Madrid (center of Spain), and the rest (49%) were from infants born in Eastern Spain. Exclusion criteria included prematurity, maternal infections or clinical illness during pregnancy, maternal antibiotic or probiotic administration during the last 2 weeks of gestation and intrapartum, and babies given antibiotic prophylaxis or therapy. Infants were divided according to feeding practice, into formula-fed infants (n = 35), eligible if they were fed exclusively with formula from birth, and breast-fed infants (n = 40), eligible if they were breast-fed exclusively during the 4-month period under study. Infants were also classified into two main CD genetic risk groups, a high-risk group (n = 39) and a low-risk group (n = 36), on the basis of their HLA-DQ genotype. The classification was based on the criteria of Bourgey et al. (3) and considering the HLA distribution of the Eastern Spanish population (5). The demographic data and dietary history of every subject were recorded (Table 1). The study was conducted in accordance with the ethical rules of the Helsinki Declaration (Hong Kong revision, September 1989), following EEC Good Clinical Practice guidelines (document 111/3976/88, July 1990) and current Spanish law, which regulates clinical research in humans (Royal Decree 561/1993 regarding clinical trials). The study was approved by the local ethics committees of the CSIC and the hospitals involved (Hospital Universitario Sant Joan de Reus, Hospital Universitario Sant Joan de Deu, Hospital Clínico Universitario of Valladolid, Hospital Universitario La Paz, Hospital Infantil Universitario La Fe, Hospital Universitario Infantil Niño Jesús, and Hospital Universitario Nuestra Señora de Candelaria). Written informed consent was obtained from the parents of children included in the study.
Table 1.
Demographic characteristics of the study cohort of infantsa
| Parameter | Value for low-risk infants (<1% probability of CD) (n = 36) |
Value for high-risk infants (7-28% probability of CD) (n = 39) |
P value | ||
|---|---|---|---|---|---|
| Breast-fed (n = 20) | Formula-fed (n = 16) | Breast-fed (n = 20) | Formula-fed (n = 19) | ||
| No. of infants with first-degree relative(s) with CD | 0.923 | ||||
| Parent | 4 | 2 | 5 | 3 | |
| Sibling | 14 | 12 | 12 | 14 | |
| Parent and sibling | 2 | 2 | 3 | 4 | |
| No. of infants with delivery type | 0.900 | ||||
| Vaginal | 17 | 13 | 18 | 16 | |
| Caesarean | 3 | 3 | 2 | 3 | |
| Length (cm) | 49.2 ± 1.4 | 50.0 ± 1.9 | 49.6 ± 2.2 | 50.5 ± 1.7 | 0.309 |
| Weight (g) | 3,253.0 ± 307.7 | 3,269.0 ± 519.3 | 3,364.0 ± 514.7 | 3,459.0 ± 795.3 | 0.640 |
| Length of gestation (wk) | 38.3 ± 3.7 | 38.3 ± 1.3 | 39.3 ± 1.1 | 39.5 ± 1.4 | 0.332 |
Bacterial strains.
The reference strains used as ladders for Bacteroides species identification by DGGE were Bacteroides distasonis DSM 20701, B. fragilis DSM 2451, B. ovatus DSM 1896, B. thetaiotaomicron DSM 2079, and B. vulgatus DSM 1447. Another six strains used as ladders were isolated from human stools and identified by 16S rRNA gene sequencing with primers 27d and 1401r as described elsewhere (13), using an ABI Prism 3130XL genetic analyzer (Applied Biosystems, CA). Search analyses were conducted in GenBank, using the Basic Local Alignment Search Tool (BLAST) algorithm. Sequences of our isolates showed >97% similarity to sequences of the species B. dorei, B. massiliensis, B. caccae, B. coprocola, B. intestinalis, and B. uniformis (GenBank accession numbers EU722737.1, AB510703.1, AB510697.1, AB200223.1, AB437413.1, and AB247141.1, respectively) and were assigned to these species. Bacteroides strains were grown in Schaedler agar medium (Scharlau, Barcelona, Spain) supplemented with kanamycin (100 mg/liter), vancomycin (7.5 mg/liter), and vitamin K (0.5 mg/liter) and incubated under anaerobic conditions (AnaeroGen; Oxoid, Hampshire, United Kingdom) at 37°C.
DNA extraction, PCR amplification, and DGGE analysis.
Stool samples were collected from every infant at 7 days, 1 month, and 4 months of age and kept at −20°C until analysis. Samples were diluted and homogenized. DNAs from diluted stool samples and from bacterial strains used as ladders were extracted by using a QIAamp DNA stool minikit (Qiagen, Hilden, Germany) following the manufacturer's instructions.
Bacteroides genus-specific PCR was performed with 16S rRNA gene-targeted primers Bfra531-f (5′-ATACGGAGGATCCGAGCGTTA-3′) and Bfra766-GC-r (5′-CTGTTTGATACCCACACTGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG-3′). Each reaction mixture of 30 μl was composed of 3 μl 10× buffer stock (containing 1.5 mM MgCl2), 1.5 μl bovine serum albumin (10 mg/ml), a 0.5 μM concentration of each deoxynucleoside triphosphate, 1 μM (each) primers, 2.5 U Taq polymerase (Ecotaq; Ecogen, Barcelona, Spain), and 30 ng DNA. DGGE analysis was carried out on a Dcode universal mutation detection system (Bio-Rad, Richmond, CA) (23). Unknown DGGE bands were excised from the gels, reamplified, and purity checked by DGGE. The PCR products were purified using a GFX PCR DNA and gel band DNA purification kit (GE Healthcare, Buckinghamshire, United Kingdom) and were identified by DNA sequencing using an ABI Prism 3130XL genetic analyzer (Applied Biosystems, CA). Search analyses were conducted as described above, and sequences with >97% similarity were considered to be of the same species.
Statistical and cluster analyses.
Gels were aligned using ladders, and bands were estimated visually and coded as present or absent. The relatedness of microbial communities was expressed as similarity clusters, using the Dice coefficient and the unweighted-pair group method using average linkages (UPGMA), and the diversity of taxa present in fecal samples was calculated with the Shannon-Wiener index (H′), using PAST (Palaeontological Statistics) software. The Levene test was used to establish variance homogeneity and diversity data distributions, and the effects of feeding practices and genetic risk on Bacteroides diversity were determined by using a linear mixed-model analysis for repeated measures in which sampling time was the repeated variable, using SPSS 17.0 software (SPSS Inc., Chicago, IL). P values of <0.05 were considered statistically significant. The interaction between feeding practices and genetic risk of CD in the studied cohort of infants was not statistically significant by applying the linear mixed-model analysis. Differences in Bacteroides species prevalence were established by applying χ2 tests and, where appropriate, by two-tailed Fisher's exact test. Analyses were carried out with Statgraphics software (Manugistics, Rockville, MD), and statistical differences were established in cases with P values of <0.05.
RESULTS
Infant gut colonization by Bacteroides species during the first months of life.
In considering the whole cohort of infants, significant differences in Bacteroides species diversity indexes (H′) were not detected based on the infants' ages, with values of 1.45 at 7 days, 1.42 at 1 month, and 1.36 at 4 months.
Some profiles clustered according to infant age, but others were subject specific, so cluster analysis did not classify Bacteroides profiles according to infant age (data not shown), nor did the prevalence of Bacteroides species differ significantly in the cohort of infants at different ages (data not shown).
Influence of milk-feeding method on Bacteroides species colonization.
The Bacteroides diversity index (H′) was significantly higher for formula-fed infants than for breast-fed infants during the study period, as determined by applying a linear mixed-model analysis for repeated measures (P = 0.01).
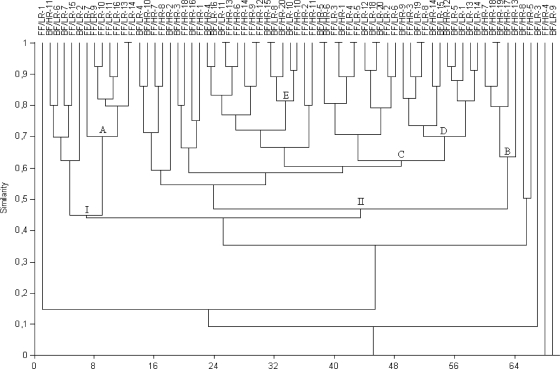
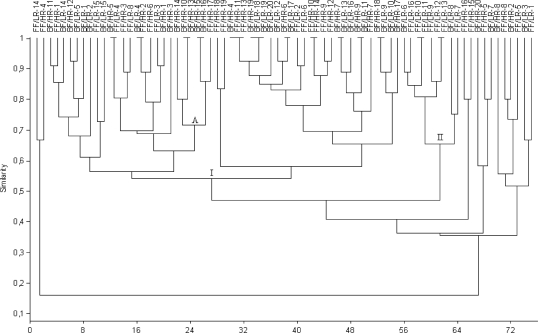
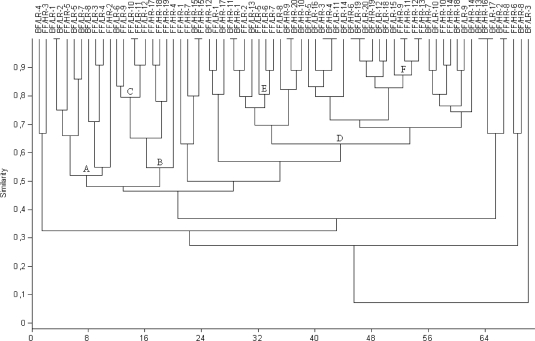
Clustering analysis of DGGE profiles obtained for infant stools at 7 days, 1 month, and 4 months of age revealed some associations between the types of milk-feeding practices. At 7 days of age, some breast-fed and formula-fed infant profiles were grouped together. Cluster A contained 7 formula-fed infant profiles (70% similarity). Moreover, another three clusters containing breast-fed infant profiles were found: cluster B contained 5 profiles, cluster D contained 9 profiles, and cluster E contained 4 profiles, with 63%, 70%, and 82% similarity, respectively (Fig. 1). At 1 month of age, cluster I included most of the breast-fed infant profiles (33), with 55% similarity, whereas cluster II contained 7 formula-fed infant profiles (65% similarity) (Fig. 2). At 4 months of age, 24 breast-fed infant profiles were grouped in cluster D (63% similarity). However, this cluster also contained another 2 subclusters (E and F) including 4 formula-fed infant profiles each, with similarities of >80%. This analysis also revealed another cluster (B), grouping 8 formula-fed infant profiles at 54% similarity (Fig. 3).
Fig. 1.
Dendrogram derived from DGGE analysis of Bacteroides species in fecal samples of the study cohort of infants at 7 days of age, based on Dice's similarity index and the UPGMA clustering algorithm. Infants were divided according to feeding practice (breast-fed [BF] and formula-fed [FF]) and genetic risk of CD development (low risk [LR] and high risk [HR]). Letters correspond to clusters of samples.
Fig. 2.
Dendrogram derived from DGGE analysis of Bacteroides species in fecal samples of the study cohort of infants at 1 month of age, based on Dice's similarity index and the UPGMA clustering algorithm. Infants were divided according to feeding practice (breast-fed [BF] and formula-fed [FF]) and genetic risk of CD development (low risk [LR] and high risk [HR]). Letters correspond to clusters of samples.
Fig. 3.
Dendrogram derived from DGGE analysis of Bacteroides species in fecal samples of the study cohort of infants at 4 months of age, based on Dice's similarity index and the UPGMA clustering algorithm. Infants were divided according to feeding practice (breast-fed [BF] and formula-fed [FF]) and genetic risk of CD development (low risk [LR] and high risk [HR]). Letters correspond to clusters of samples.
The prevalences of Bacteroides species in the cohort of infants, given according to feeding practices, are shown in Table 2. At 7 days of age, the prevalence of B. vulgatus was higher in breast-fed infants than in formula-fed infants (P = 0.03). In contrast, B. caccae and B. intestinalis were detected more frequently in formula-fed infants than in breast-fed infants (P < 0.01). At 1 month of age, the prevalence rates of B. vulgatus and B. caccae were equal in both the breast-fed and formula-fed groups; nevertheless, the prevalence of B. intestinalis continued to be increased in formula-fed infants compared with breast-fed infants (P < 0.01). In contrast, B. uniformis was more prevalent in breast-fed infants than in formula-fed infants (P = 0.04). Interestingly, at 4 months of age, the prevalence of B. uniformis was also increased in breast-fed compared with formula-fed infants (P = 0.04), while that of B. intestinalis was higher in formula-fed infants than in breast-fed infants (P < 0.01). Furthermore, the prevalences of B. caccae and B. plebeius were higher in formula-fed infants than in breast-fed infants (P = 0.01 and P = 0.03, respectively).
Table 2.
Prevalence of Bacteroides species detected by PCR-DGGE analysis, using Bfra531-f and Bfra766-GC-r primers and fecal DNAs from the study infants at 7 days, 1 month, and 4 months of age, stratified according to feeding practices
| Species | No. (%) of infants carrying speciesa |
|||||
|---|---|---|---|---|---|---|
| 7 days |
1 mo |
4 mo |
||||
| Breast-fed (n = 39) | Formula-fed (n = 31) | Breast-fed (n = 40) | Formula-fed (n = 35) | Breast-fed (n = 40) | Formula-fed (n = 30) | |
| B. caccae | 4 (10) | 12 (39)* | 9 (23) | 12 (34) | 5 (13) | 12 (40)* |
| B. coprocola | 16 (41) | 14 (45) | 19 (48) | 17 (49) | 23 (58) | 16 (53) |
| B. distasonis | 18 (46) | 15 (48) | 15 (38) | 20 (57) | 16 (40) | 14 (47) |
| B. dorei | 12 (31) | 6 (19) | 9 (23) | 8 (23) | 7 (18) | 5 (17) |
| B. fragilis/B. thetaiotaomicron | 28 (72) | 27 (87) | 31 (78) | 30 (86) | 31 (78) | 27 (90) |
| B. intestinalis | 0 | 9 (29)* | 0 | 4 (11)* | 0 | 6 (20)* |
| B. massiliensis | 14 (36) | 5 (16) | 11 (28) | 7 (20) | 10 (25) | 5 (17) |
| B. ovatus | 13 (33) | 17 (55) | 18 (45) | 20 (57) | 17 (43) | 17 (57) |
| B. plebeiusb | 4 (10) | 8 (26) | 4 (10) | 9 (26) | 2 (5) | 7 (23)* |
| B. uniformis | 24 (62) | 17 (55) | 27 (68) | 15 (43)* | 29 (73) | 14 (47)* |
| B. vulgatus | 33 (85) | 19 (61)* | 32 (80) | 23 (66) | 25 (63) | 17 (57) |
*, significant differences in Bacteroides species between breast-fed and formula-fed infants (P < 0.05; Fisher's exact test).
Species identification was done by sequencing of PCR-DGGE bands.
Influence of HLA-DQ genotype on Bacteroides species colonization.
The diversity index of Bacteroides species colonizing the infant gut, calculated by applying a linear mixed-model analysis for repeated measures, proved to be significantly higher in infants with low genetic risk than in those with high genetic risk of CD development (P = 0.04).
Clustering analysis of DGGE profiles obtained for infant fecal samples at 7 days and 1 and 4 months of age revealed some associations between the Bacteroides species profiles and the genetic risk of CD development. At 7 days of age, DGGE profiles were divided into two main groups: cluster I contained 11 profiles of infants with low genetic risk (45% similarity), and cluster II contained almost all profiles (31) for infants with high genetic risk (48% similarity) but also included one subgroup (C) containing 15 profiles of infants with low genetic risk (62% similarity) (Fig. 1). At 1 month of age, only two small clusters were detected: cluster II included 9 profiles for infants with low genetic risk (65% similarity), and cluster A included 6 profiles for infants with high genetic risk (70% similarity) (Fig. 2). At 4 months of age, 12 profiles for infants with low genetic risk were grouped into two different clusters (A and C), with 52% and 80% similarity, respectively, while others could not be differentiated clearly from those for infants with high genetic risk (Fig. 3).
The prevalences of Bacteroides species in the cohort of infants, stratified according to their genetic risk of CD development, are shown in Table 3. At 7 days of age, the prevalence of B. vulgatus was higher in high-risk than in low-risk infants (P < 0.01); in contrast, B. ovatus, B. plebeius, and B. uniformis were more prevalent in low-risk than in high-risk infants (P < 0.01). The same differences were detected for these bacterial species at 1 month of age (P = 0.03, P = 0.04, P < 0.01, and P < 0.01, respectively) and 4 months of age (P = 0.02, P = 0.04, P = 0.14, and P = 0.02, respectively).
Table 3.
Prevalence of Bacteroides species detected by PCR-DGGE analysis, using Bfra531-f and Bfra766-GC-r primers and fecal DNAs from the study infants with family risk of CD development at 7 days, 1 month, and 4 months of age, stratified according to genetic risk
| Species | No. (%) of infants carrying speciesa |
|||||
|---|---|---|---|---|---|---|
| 7 days |
1 mo |
4 mo |
||||
| Low risk | High risk | Low risk | High risk | Low risk | High risk | |
| B. caccae | 8 (24) | 8 (22) | 10 (28) | 11 (28) | 8 (25) | 9 (24) |
| B. coprocola | 13 (38) | 17 (47) | 18 (50) | 18 (46) | 17 (53) | 22 (58) |
| B. distasonis | 16 (47) | 17 (47) | 19 (53) | 16 (41) | 14 (44) | 16 (42) |
| B. dorei | 7 (21) | 11 (31) | 7 (19) | 10 (26) | 4 (13) | 8 (21) |
| B. fragilis/B. thetaiotaomicron | 26 (76) | 29 (81) | 27 (75) | 34 (87) | 26 (81) | 32 (84) |
| B. intestinalis | 2 (5.9) | 7 (19) | 1 (2.8) | 3 (7.7) | 1 (3.1) | 5 (13) |
| B. massiliensis | 12 (35) | 7 (19) | 10 (28) | 8 (21) | 10 (31) | 5 (13) |
| B. ovatus | 22 (65) | 8 (22)* | 26 (72) | 12 (31)* | 21 (66) | 13 (34)* |
| B. plebeiusb | 10 (29) | 0* | 13 (36) | 0* | 9 (28) | 0* |
| B. uniformis | 24 (71) | 17 (47)* | 25 (69) | 17 (44)* | 23 (72) | 20 (53) |
| B. vulgatus | 20 (59) | 32 (89)* | 22 (61) | 33 (85)* | 14 (44) | 28 (74)* |
*, significant differences in Bacteroides species between infants with low genetic risk (<1% probability) and high genetic risk (7 to 28% probability) of developing CD (P < 0.05; Fisher's exact test).
Species identification was done by sequencing PCR-DGGE bands.
Influence of genotype on Bacteroides species colonizing the guts of breast-fed and formula-fed infants.
The prevalences of Bacteroides species are shown in Table 4, presented according to the genotypes of breast-fed and formula-fed infants. Among breast-fed infants, the prevalence of B. uniformis was significantly higher in infants with low genetic risk than in those with high genetic risk during the study period, but significant differences were detected only at 7 days and 1 month of age (P < 0.01 and P = 0.02, respectively). Among formula-fed infants, the prevalence rates of B. ovatus and B. plebeius were higher (P = <0.001 to 0.002) in infants with low genetic risk than in those with high genetic risk of CD development, while the prevalence of B. vulgatus was higher in infants with high genetic risk (P = 0.003 to 0.030).
Table 4.
Prevalence of Bacteroides species detected by DGGE analysis, using the primer pair Bfra531-f and Bfra766-GC-r and fecal DNAs from either breast-fed or formula-fed infants, as a function of the genetic risk of CD development (low risk, <1% probability; and high risk, 7 to 28% probability) at 7 days, 1 month, and 4 months of age
| Species | Infant age | No. of breast-fed infants carrying species/total no. of infants (%) |
P valuea | No. of formula-fed infants carrying species/total no. of infants (%) |
P valuea | ||
|---|---|---|---|---|---|---|---|
| Low risk | High risk | Low risk | High risk | ||||
| B. caccae | 7 days | 3/19 (15.8) | 1/20 (5) | 0.342 | 5/15 (33.3) | 7/16 (43.8) | 0.716 |
| 1 mo | 5/20 (25) | 4/20 (20) | 1 | 5/16 (31.3) | 7/19 (36.8) | 1 | |
| 4 mo | 4/20 (20) | 1/20 (5) | 0.342 | 4/12 (33.3) | 8/18 (44.4) | 0.709 | |
| B. coprocola | 7 days | 6/19 (31.6) | 10/20 (50) | 0.333 | 7/15 (46.7) | 7/16 (43.8) | 1 |
| 1 mo | 10/20 (50) | 9/20 (45) | 1 | 8/16 (50) | 9/19 (47.4) | 1 | |
| 4 mo | 10/20 (50) | 13/20 (65) | 0.523 | 7/12 (58.3) | 9/18 (50) | 0.722 | |
| B. distasonis | 7 days | 8/19 (42.1) | 10/20 (50) | 0.751 | 8/15 (53.3) | 7/16 (43.8) | 0.724 |
| 1 mo | 9/20 (45) | 6/20 (30) | 0.353 | 10/16 (62.5) | 10/19 (52.6) | 0.734 | |
| 4 mo | 9/20 (45) | 7/20 (35) | 0.748 | 5/12 (41.7) | 9/18 (50) | 0.722 | |
| B. dorei | 7 days | 6/19 (31.6) | 6/20 (30) | 1 | 1/15 (6.7) | 5/16 (31.3) | 0.172 |
| 1 mo | 6/20 (30) | 3/20 (15) | 0.289 | 1/16 (6.3) | 7/19 (36.8) | 0.047* | |
| 4 mo | 4/20 (20) | 3/20 (15) | 1 | 0/12 (0) | 5/18 (27.8) | 0.066 | |
| B. fragilis/B. thetaiotaomicron | 7 days | 13/19 (68.4) | 15/20 (75) | 0.731 | 13/15 (86.7) | 14/16 (87.5) | 1 |
| 1 mo | 14/20 (70) | 17/20 (85) | 0.289 | 13/16 (81.3) | 17/19 (89.5) | 0.642 | |
| 4 mo | 15/20 (75) | 16/20 (80) | 1 | 11/12 (91.7) | 16/18 (88.9) | 1 | |
| B. intestinalis | 7 days | 0/19 (0) | 0/20 (0) | 2/15 (13.3) | 7/16 (43.8) | 0.113 | |
| 1 mo | 0/20 (0) | 0/20 (0) | 1/16 (6.3) | 3/19 (15.8) | 0.608 | ||
| 4 mo | 0/20 (0) | 0/20 (0) | 1/12 (8.3) | 5/18 (27.8) | 0.358 | ||
| B. massiliensis | 7 days | 10/19 (52.6) | 4/20 (20) | 0.048* | 2/15 (13.3) | 3/16 (18.8) | 1 |
| 1 mo | 8/20 (40) | 3/20 (15) | 0.155 | 2/16 (12.5) | 5/19 (26.3) | 0.415 | |
| 4 mo | 8/20 (40) | 2/20 (10) | 0.065 | 2/12 (16.7) | 3/18 (16.7) | 1 | |
| B. ovatus | 7 days | 8/19 (42.1) | 5/20 (25) | 0.320 | 14/15 (93.3) | 3/116 (18.8) | <0.001* |
| 1 mo | 12/20 (60) | 6/20 (30) | 0.111 | 14/16 (87.5) | 6/19 (31.6) | 0.002* | |
| 4 mo | 9/20 (45) | 8/20 (40) | 0.762 | 12/12 (100) | 5/18 (27.8) | <0.001* | |
| B. plebeius | 7 days | 2/19 (10.5) | 2/20 (10) | 1 | 8/15 (53.3) | 0/16 (0) | 0.001* |
| 1 mo | 4/20 (20) | 0/20 (0) | 0.106 | 9/16 (56.3) | 0/19 (0) | <0.001* | |
| 4 mo | 2/20 (10) | 0/20 (0) | 0.244 | 7/12 (58.3) | 0/18 (0) | 0.002* | |
| B. uniformis | 7 days | 16/19 (84.2) | 8/20 (40) | 0.008* | 8/15 (53.3) | 9/16 (56.3) | 1 |
| 1 mo | 18/20 (90) | 9/20 (45) | 0.019* | 7/16 (43.8) | 8/19 (42.1) | 1 | |
| 4 mo | 17/20 (85) | 12/20 (60) | 0.155 | 6/12 (50) | 8/18 (44.4) | 1 | |
| B. vulgatus | 7 days | 15/19 (78.9) | 18/20 (90) | 0.695 | 5/15 (33.3) | 14/16 (87.5) | 0.003* |
| 1 mo | 15/20 (75) | 17/20 (85) | 0.695 | 7/16 (43.8) | 16/19 (84.2) | 0.030* | |
| 4 mo | 11/20 (55) | 14/20 (70) | 0.353 | 3/12 (25) | 14/18 (77.8) | 0.008* | |
*, statistical differences between the genetic risk groups. Statistical differences were calculated by using Fisher's exact test and considering differences significant if the P value was <0.05.
DISCUSSION
The present study provides the first evidence of the effects exerted by both milk-feeding practices and CD genetic risk factors on the intestinal colonization process of Bacteroides species in infants at early life.
Our study indicates that milk-feeding practices influence both Bacteroides diversity and species prevalence. This study reports an increased prevalence of B. vulgatus in breast-fed infants at 7 days of age, which is not in agreement with the results of the only published related study, which reported that B. vulgatus and B. thetaiotaomicron were predominant in fecal samples from 5- to 6-day-old formula-supplemented newborns (23).
Genetic predisposition to CD has been linked to the MHC region on chromosome 6p21, with over 95% of CD patients expressing HLA-DQ2 or -DQ8 (26). In our study, infants were classified into two groups according to HLA-DQ genotype, with one including those with a high probability of developing CD (7 to 28%) and the other including those with a low probability (<1%) of developing the disease (5). Infants with high genetic risk had an increased prevalence of B. vulgatus and a reduced prevalence of B. ovatus, B. plebeius, and B. uniformis compared with infants with low genetic risk. Enterocytes that are in close proximity with the intestinal contents and bacteria can express HLA-II molecules of the MHC to a certain extent and are able to act as antigen-presenting cells (16). Moreover, HLA-II molecules are expressed primarily by dendritic cells present in the lamina propria, which can extend dendrites across this epithelium, capture bacteria and other antigens at the mucosal surface, and present the antigens to B or T lymphocytes (6). This process is a critical step in the initiation of a mucosal immune response, and its possible role in restricting bacterial colonization cannot be disregarded (14). In fact, the polysaccharide of B. fragilis has been proven to associate with MHC-II molecules with a high affinity, through a mechanism mirroring peptide presentation, which could mediate MHC-II antigen presentation and, ultimately, T cell recognition during bacterial infec- tion (7).
In a preliminary study of exclusively breast-fed infants, larger proportions of the Bacteroides-Prevotella group were detected in infants with high genetic risk than in those with low genetic risk of CD development (8); however, the effect of milk-feeding type was not considered. The present study provides a wider characterization of the Bacteroides population at the species level and also takes into account the variable of milk-feeding type. In this study, we demonstrated that the prevalence of B. vulgatus was increased in infants with high genetic risk, considering both the total cohort and formula-fed infants only. Furthermore, in the whole cohort of infants, prevalence rates for B. uniformis, B. ovatus, and B. plebeius were also increased in the group with low genetic risk, and this trend was confirmed by considering the subgroup of either breast-fed or formula-fed infants. These results indicate that the HLA-DQ genotype has an independent effect on the colonization of these species, in addition to that exerted by the type of milk feeding, which could also influence the disease risk.
The colonization process of the intestine shortly after birth seems to be important for adequate immune response maturation and to the risk of suffering immune-mediated diseases. A few studies suggest that B. fragilis could be involved in this process, influencing Toll-like receptor 4 (TLR4) mRNA in peripheral blood monocytes (27) or the number of circulating IgA and IgM antibody-producing cells (11, 19). In vitro studies also indicate that exposure to polysaccharide A of B. fragilis activates CD4+ T cells, resulting in a Th1 response, which could modify the risk of suffering CD (29). Nevertheless, neither in vitro nor in vivo information is available regarding the potential immunological role of other Bacteroides species.
In summary, the results indicate that both the milk-feeding type and the HLA-DQ genotype influence the colonization process of Bacteroides species early in life. An increased B. vulgatus prevalence was associated with the genotype of infants at high risk of CD development, while increased B. uniformis prevalence was associated with the genotype of infants at low risk of CD development and with breast-feeding, which could partly explain the protective effect of breast-feeding. Further research is under way to disclose a possible relationship between these findings and subsequent development of CD in a follow-up study of this cohort of infants.
ACKNOWLEDGMENTS
This work was supported by public grants AGL2007-66126-C03-01-03/ALI and Consolider Fun-C-Food CSD2007-00063 from the Spanish Ministry of Science and Education (MEC). Scholarships to E. Sánchez from the Institute Danone and to G. De Palma and T. Pozo from CSIC are fully acknowledged.
The assistance of Laura Barrios in statistical analysis is also fully acknowledged.
Footnotes
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Bezirtzoglou E. 1997. The intestinal microflora during the first weeks of life. Anaerobe 3:173–177 [DOI] [PubMed] [Google Scholar]
- 2. Biasucci G., Benenati B., Morelli L., Bessi F., Boehm G. 2008. Cesarean delivery may affect the early biodiversity of intestinal bacteria. J. Nutr. 138:1796S–1800S [DOI] [PubMed] [Google Scholar]
- 3. Bourgey M., et al. 2007. HLA related genetic risk for coeliac disease. Gut 56:1054–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Branski D., Fasano A., Troncone R. 2006. Latest developments in the pathogenesis and treatment of celiac disease. J. Pediatr. 149:295–300 [DOI] [PubMed] [Google Scholar]
- 5. Capilla A., et al. 2007. Genetic analyses of celiac disease in a Spanish population confirm association with CELIAC3 but not with CELIAC4. Tissue Antigens 70:324–329 [DOI] [PubMed] [Google Scholar]
- 6. Chieppa M., Rescigno M., Huang A. Y., Germain R. N. 2006. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J. Exp. Med. 203:2841–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cobb B. A., Kasper D. L. 2008. Characteristics of carbohydrate antigen binding to the presentation protein HLA-DR. Glycobiology 18:707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Palma G., et al. 2009. Interplay between human leukocyte antigen genes and the microbial colonization process of the newborn intestine. Curr. Issues Mol. Biol. 12:1–10 [PubMed] [Google Scholar]
- 9. Favier C. F., Vaughan E. E., de Vos W. M., Akkermans A. D. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Greco L., et al. 2002. The first large population based twin study of coeliac disease. Gut 50:624–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gronlund M. M., Arvilommi H., Kero P., Lehtonen O. P., Isolauri E. 2000. Importance of intestinal colonisation in the maturation of humoral immunity in early infancy: a prospective follow up study of healthy infants aged 0-6 months. Arch. Dis. Child. Fetal Neonatal Ed. 83:F186–F192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gronlund M. M., Lehtonen O. P., Eerola E., Kero P. 1999. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J. Pediatr. Gastroenterol. Nutr. 28:19–25 [DOI] [PubMed] [Google Scholar]
- 13. Gueimonde M., et al. 2004. Viability and diversity of probiotic Lactobacillus and Bifidobacterium populations included in commercial fermented milks. Food Res. Int. 37:839–850 [Google Scholar]
- 14. Hapfelmeier S., et al. 2008. Microbe sampling by mucosal dendritic cells is a discrete, MyD88-independent step in ΔinvG S. Typhimurium colitis. J. Exp. Med. 205:437–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ivarsson A., Hernell O., Stenlund H., Persson L. A. 2002. Breast-feeding protects against celiac disease. Am. J. Clin. Nutr. 75:914–921 [DOI] [PubMed] [Google Scholar]
- 16. Kaiserlian D., Vidal K. 1993. Antigen presentation by intestinal epithelial cells. Immunol. Today 14:144. [DOI] [PubMed] [Google Scholar]
- 17. Reference deleted.
- 18. Mackie R. I., Sghir A., Gaskins H. R. 1999. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 69:1035S–1045S [DOI] [PubMed] [Google Scholar]
- 19. Moreau M. C., Ducluzeau R., Guy-Grand D., Muller M. C. 1978. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect. Immun. 21:532–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nistico L., et al. 2006. Concordance, disease progression, and heritability of coeliac disease in Italian twins. Gut 55:803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Orrhage K., Nord C. E. 1999. Factors controlling the bacterial colonization of the intestine in breastfed infants. Acta Paediatr. Suppl. 88:47–57 [DOI] [PubMed] [Google Scholar]
- 22. Penders J., et al. 2006. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511–521 [DOI] [PubMed] [Google Scholar]
- 23. Rotimi V. O., Duerden B. I. 1981. Bacteroides species in the normal neonatal faecal flora. J. Hyg. (London) 87:299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reference deleted.
- 25. Sanz Y., De Palma G. 2009. Gut microbiota and probiotics in modulation of epithelium and gut-associated lymphoid tissue function. Int. Rev. Immunol. 28:397–413 [DOI] [PubMed] [Google Scholar]
- 26. Silano M., Agostoni C., Guandalini S. 2010. Effect of the timing of gluten introduction on the development of celiac disease. World J. Gastroenterol. 16:1939–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sjogren Y. M., et al. 2009. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin. Exp. Allergy 39:1842–1851 [DOI] [PubMed] [Google Scholar]
- 28. van Heel D. A., Hunt K., Greco L., Wijmenga C. 2005. Genetics in coeliac disease. Best Pract. Res. Clin. Gastroenterol. 19:323–339 [DOI] [PubMed] [Google Scholar]
- 29. Wang Q., et al. 2006. A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. J. Exp. Med. 203:2853–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zocco M. A., Ainora M. E., Gasbarrini G., Gasbarrini A. 2007. Bacteroides thetaiotaomicron in the gut: molecular aspects of their interaction. Dig. Liver Dis. 39:707–712 [DOI] [PubMed] [Google Scholar]