Abstract
Free-living adult Amblyomma incisum ticks were collected in an Atlantic rainforest area at Intervales State Park, State of São Paulo, Brazil. From an A. incisum specimen, rickettsiae were successfully isolated in Vero cell culture by the shell vial technique. Rickettsial isolation was confirmed by optical microscopy, transmission electron microscopy, and PCRs targeting portions of the rickettsial genes gltA, htrA, rrs, and sca1 on infected cells. Fragments of 1,089, 457, 1,362, and 443 nucleotides of the gltA, htrA, rrs, and sca1 genes, respectively, were sequenced. By BLAST analysis, the partial sequence of rrs of the A. incisum rickettsial isolate was closest to the corresponding sequence of Rickettsia bellii (99.1% similarity). The gltA partial sequence was closest to the corresponding sequences of “Candidatus Rickettsia tarasevichiae” (96.1% similarity) and Rickettsia canadensis (95.8% similarity). The htrA partial sequence was closest to the corresponding sequence of R. canadensis (89.8% similarity). The sca1 partial sequence was closest to the corresponding sequence of R. canadensis (95.2% similarity). Since our rickettsial isolate was genetically distinct from other Rickettsia species, we propose a new species designated Rickettsia monteiroi sp. nov. Phylogenetic analyses indicated that R. monteiroi belongs to the canadensis group within the genus Rickettsia, together with the species R. canadensis and “Candidatus R. tarasevichiae”. Little or no antibody cross-reaction was observed between sera of R. monteiroi-inoculated guinea pigs and R. bellii-, Rickettsia rickettsii-, or R. canadensis-inoculated guinea pigs.
INTRODUCTION
The genus Rickettsia includes bacteria of the order Rickettsiales in the alpha subdivision of the class Proteobacteria. They are Gram-negative coccobacilli in obligate association with eukaryotic cells. A number of species have been identified in various terrestrial arthropods and more recently in leeches and amoebae (3, 22). Traditionally, pathogenic rickettsiae were classified into two groups: the typhus group (TG), composed of Rickettsia prowazekii and Rickettsia typhi and transmitted by lice and fleas, respectively, and the spotted fever group (SFG), composed of more than 20 species and transmitted mostly by ticks (24). Other rickettsiae have shown antigenic and genetic peculiarities that preclude their inclusion in either the TG or the SFG, such as Rickettsia bellii and Rickettsia canadensis, reported in ticks from the New World (15, 16). With the discovery of a variety of new rickettsiae in different orders of terrestrial arthropods, mostly free living, and also with genetic analysis of rickettsial plasmids, the genus Rickettsia has been reclassified into different groups, including the SFG, the TG, the transitional group (TRG), the bellii group (BG), the canadensis group (CG), and several other basal groups (5, 29). In Brazil, at least seven Rickettsia species have been reported, namely, the SFG species Rickettsia rickettsii, Rickettsia parkeri, “Candidatus Rickettsia amblyommii,” and Rickettsia rhipicephali, all associated with ticks; the TRG species Rickettsia felis and the TG species R. typhi, both associated with fleas; and the BG species R. bellii, associated with ticks (12, 23).
Many Rickettsia species cause diseases in humans and animals, to which they are transmitted by lice, fleas, ticks, or mites (20). Most of the recognized pathogenic Rickettsia species are classified into the SFG, which includes agents of spotted fever rickettsiosis that are transmitted by ticks to humans in different parts of the world (20). During the last decades, there has been an increasing number of new Rickettsia species of unknown pathogenicity, mostly isolated from ticks. Some of them, previously considered nonpathogenic, were recently shown to be pathogenic to humans, such as the SFG species Rickettsia slovaca, Rickettsia aeschlimannii, Rickettsia massiliae, and Rickettsia monacensis in Europe (9, 20). In addition, R. parkeri, an “old” SFG organism first reported in ticks in the 1939, was shown to be pathogenic 65 years later (19). These facts indicate that any novel described Rickettsia from invertebrate hosts, especially ticks, should be regarded as potentially pathogenic for humans.
In the present study, we describe a new Rickettsia species isolated from the tick Amblyomma incisum from southeastern Brazil. The organism was successfully isolated in cell culture; molecular characterization showed it to be distinct from any other previously described rickettsiae.
MATERIALS AND METHODS
Tick collection and hemolymph test.
During 2004 to 2006, free-living adult A. incisum ticks were collected from the vegetation in an Atlantic rainforest area at the Intervales State Park (24o18′S, 48o24′W), Ribeirão Grande Municipality, State of São Paulo, Brazil, as part of another study on the ecology of ticks in an Atlantic rainforest reserve (28). Ticks were brought alive to the laboratory, where they were held in an incubator at 32°C and 95% relative humidity for 5 days in order to stimulate rickettsial growth (2). Thereafter, each tick was individually submitted to the hemolymph test for detection of Rickettsia-like structures as previously described (2). Briefly, the distal portion of one leg of each tick was cut with scissors and a drop of hemolymph was air dried on a glass slide and stained by the Gimenez technique (6). Right after collection of hemolymph, live ticks were stored frozen at −80°C.
Isolation of rickettsiae.
Attempts to isolate rickettsiae in Vero cell culture were performed with ticks that were shown to contain Rickettsia-like structures by the hemolymph test. For this purpose, frozen ticks were thawed and subjected to the shell vial technique as previously described (10), with some modifications (13). Briefly, ticks were individually thawed in a water bath at 37°C and disinfected for 10 min in iodine-alcohol, followed by several washes in sterile water. Each tick was then triturated in 500 μl of brain heart infusion broth, and the resultant tick homogenate was inoculated into shell vials containing a monolayer of confluent Vero cells. After inoculation, the shell vials were centrifuged for 1 h at 700 × g at 22°C. Thereafter, the monolayer was washed once with minimal essential medium containing 5% bovine calf serum and then incubated at 28°C with medium containing antibiotics (1% penicillin and streptomycin). After 3 days, the medium was switched to antibiotic-free medium and the aspirated medium with some scraped cells was examined by Gimenez staining for the presence of Rickettsia-like organisms. If the result was positive, the monolayer of the shell vial was harvested and inoculated into a 25-cm2 flask containing a monolayer of confluent uninfected Vero cells. Cells in the 25-cm2 flask were evaluated by Gimenez staining until more than 90% of the cells were infected, when they were harvested and inoculated into 150-cm2 flasks of Vero cells. In all instances, inoculated Vero cells were incubated at 28°C. The level of infection of cells was monitored by Gimenez staining of scraped cells from each inoculated monolayer. A rickettsial isolate was considered established in the laboratory after at least three passages through 150-cm2 Vero cell flasks, each reaching more than 90% infected cells (13).
Molecular characterization.
Cell passages of isolates were genotypically identified by PCR amplification and DNA sequencing of the product of the resultant infected cells. For this purpose, DNA of infected cell passages was extracted by boiling (100°C for 10 min) as previously described (4) and subsequently tested by a battery of PCRs using all of the primer pairs listed in Table 1, which targeted fragments of five rickettsial genes, i.e., those for citrate synthase (gltA), the 17-kDa protein (htrA), the 16S rRNA (rrs), the 190-kDa outer membrane protein (ompA), and the Sca1 autotransporter protein (sca1). For each set of reactions, a negative control (5 μl of water) and a positive control (5 μl of DNA extracted from R. parkeri-infected cells) were included. PCR products of the expected size were purified using ExoSap (USB) and sequenced in an automated sequencer (ABI Prism 310 genetic analyzer; Applied Biosystems/Perkin-Elmer) according to the manufacturer's protocol. Partial sequences obtained were submitted to BLAST analysis (1) to determine similarities to other Rickettsia species.
Table 1.
Primer pairs used for amplification of rickettsial genes
| Target gene, primer pair no., and primer | Nucleotide sequence (5′-3′) | Reference |
|---|---|---|
| gltA | ||
| 1 | 13 | |
| CS-78 | GCAAGTATCGGTGAGGATGTAAT | |
| CS-323 | GCTTCCTTAAAATTCAATAAATCAGGAT | |
| 2 | 14 | |
| CS-239 | GCTCTTCTCATCCTATGGCTATTAT | |
| CS-1069 | CAGGGTCTTCGTGCATTTCTT | |
| htrA | ||
| 3 | 13 | |
| 17k-5 | GCTTTACAAAATTCTAAAAACCATATA | |
| 17k-3 | TGTCTATCAATTCACAACTTGCC | |
| rrs | ||
| 4 | 30 | |
| fD1 | AGAGTTTGATCCTGGCTCAG | |
| 800r | CTACCAGGGTATCTAAT | |
| rP2 | ACGGCTACCTTGTTACGACTT | |
| ompA | ||
| 5 | 25 | |
| Rr190.70p | ATGGCGAATATTTCTCCAAAA | |
| Rr190.602n | AGTGCAGCATTCGCTCCCCCT | |
| sca1 | ||
| 6 | 17 | |
| F1MAX | AAGAGGTYTRTGGATGCGT | |
| RMAX | GAYAATATATTATTYTCTTTC | |
Phylogenetic analyses were performed using the program MEGA version 3.1 (11). Partial DNA sequences obtained from the amplified PCR products (gltA, htrA, rrs, and sca1) were aligned with the corresponding sequences of other Rickettsia species available in GenBank using the CLUSTAL algorithm. Phylogenetic distances between homologous sequences were calculated by using the Kimura two-parameter model. First, for each gene analyzed, a phylogram was constructed by the neighbor-joining method. The sequences of the four genes were then concatenated (rrs-gltA-htrA-sca1) and manually aligned using GeneDoc software. Phylogenetic trees were inferred by the neighbor-joining and maximum-parsimony methods (11) with 1,000 replicates of random-addition taxa and tree bisection and reconnection branch swapping; all positions were equally weighted.
Serologic tests.
Three male guinea pigs were each injected intraperitoneally with ∼1 × 106 Vero cells infected with an A. incisum rickettsial isolate derived from a fresh monolayer containing >90% infected cells. Another group of three male guinea pigs were injected intraperitoneally with ∼1 × 106 Vero cells infected with R. rickettsii strain Taiaçu, another three guinea pigs were injected with ∼1 × 106 Vero cells infected with R. bellii strain Mogi, and another group of three guinea pigs were injected with ∼1 × 106 Vero cells infected with R. canadensis strain McKiel, which had been grown in Vero cells in our laboratories. Except for the guinea pigs injected with R. canadensis, the remaining guinea pigs were examined daily for fever (if the rectal temperature was >39.5°C) and scrotal reactions. After 3 weeks, all 12 guinea pigs were bled and their sera were individually tested by immunofluorescence assay (IFA) as previously described (7), employing crude antigens of either the A. incisum rickettsial isolate, R. rickettsii strain Taiaçu, R. bellii strain Mogi, or R. canadensis strain McKiel. Antigens were prepared using whole infected Vero cells as previously described (7). Sera were diluted in 2-fold increments, starting from a 1:64 dilution, and tested with fluorescein isothiocyanate-labeled rabbit anti-guinea pig IgG (Sigma, St. Louis, MO). Endpoint titers for both homologous and heterologous reactions were determined.
Transmission electron microscopy.
Infected Vero cell monolayers were immersed in Ito's fixative, a mixture of 1.25% formaldehyde, 2.5% glutaraldehyde, 0.03% trinitrophenol, 0.03% CaCl2, and 0.05 M cacodylate buffer at pH 7.3 (8); postfixed in 1% osmium tetroxide for 1 h; and stained en bloc in 1% uranyl acetate-0.1 M maleate buffer (pH 5.2). Pellets were dehydrated in cetonic series, embedded in Spurr resin, and polymerized at 60°C overnight. Ultrathin sections (thickness, 70 nm) were prepared by using an LKB Ultratome, placed on copper grids, stained with uranyl acetate and lead citrate, and examined in a Philips EM 208 electron microscope at 80 kV.
Nucleotide sequence accession numbers.
The GenBank nucleotide sequence accession numbers of the partial sequences of the rickettsial isolate generated in this study are FJ269035 for the gltA gene, FJ269036 for the htrA gene, FJ269037 for the rrs gene, and JF734727 for the sca1 gene.
RESULTS
A total of 388 A. incisum ticks were collected (180 males, 208 females). By the hemolymph test, only two male ticks were shown to contain Rickettsia-like organisms within their hemocytes. One of them was processed in another study and was shown to be infected by R. bellii (18). The second hemolymph-positive tick was processed by the shell vial technique in the present study, and rickettsiae were successfully isolated and established in Vero cell culture (see Fig. S1 in the supplemental material). DNA of infected cells at the fourth rickettsial passage was subjected to PCRs targeting the gltA, ompA, htrA, rrs, and sca1 genes. PCR products of the expected size were obtained with the gltA, htrA, rrs, and sca1 primers, but no product was obtained with the ompA primers (these primers are specific for SFG rickettsiae). We sequenced 1,089, 457, 1,362, and 443 nucleotides (nt) of the gltA, htrA, rrs, and sca1 genes, respectively. By BLAST analysis, the rrs partial sequence of the A. incisum rickettsial isolate was most similar (99.1% identity; 1,350/1,362 nt) to the corresponding sequence of two strains of R. bellii (CP000087 and L36103). The gltA partial sequence was most similar (96.1% identity; 1,041/1,083 nt, excluding indels) to the corresponding sequence of four strains of “Candidatus R. tarasevichiae” (AF503167, DQ168983, DQ168982, and DQ168981) detected in Ixodes persulcatus ticks from Russia (26) and 95.8% (1,040/1,085 nt, excluding indels) similar to the corresponding sequence of R. canadensis (U59713). The htrA partial sequence was most similar (89.8%; 396/441 nt, excluding indels) to the corresponding sequence of R. canadensis (AF445381) and 86.1% (383/445 nt) similar to the corresponding sequence of Rickettsia sibirica (AF445384). The sca1 partial sequence was most similar (95.2%; 419/440 nt, excluding indels) to corresponding sequence of R. canadensis (DQ306905 and AY355367). Since our rickettsial isolate was shown to be genetically distinct from other Rickettsia species, we propose a new species designated Rickettsia monteiroi sp. nov. The name of the species is for José Lemos Monteiro, an outstanding pioneer of Brazilian rickettsiology. Monteiro died of Rocky Mountain spotted fever in 1935 while working with ticks and R. rickettsii in his laboratory at the Instituto Butantan, São Paulo, Brazil.
None of the guinea pigs inoculated with R. monteiroi or R. bellii developed a fever or a scrotal reaction during the 21 days postinoculation. Conversely, all three guinea pigs inoculated with R. rickettsii developed a high fever (rectal temperature, >40°C) and a scrotal reaction starting 3 or 4 days after inoculation. On the 6th day, these three guinea pigs were treated with oxytetracycline HCl (10 mg/kg given intramuscularly) in order to prevent death. All of the guinea pigs, including those inoculated with R. canadensis, were bled 21 days after inoculation to obtain sera to be tested by IFA. The IgG endpoint titers of homologous and heterologous reactions for the four rickettsial antigens are shown in Table 2. For each rickettsial antigen, the highest titers were obtained for homologous sera, whereas lower titers or no detectable reactivity (titer of <64) was observed for heterologous reactions (see Fig. S1 in the supplemental material).
Table 2.
Homologous and heterologous endpoint titers of IgG to three Rickettsia species in sera of guinea pigs inoculated with the agents
| Inoculum and guinea pig no. | Endpoint titer for following antigen: |
|||
|---|---|---|---|---|
| R. monteiroi | R. bellii | R. rickettsii | R. canadensis | |
| R. monteiroi | ||||
| 1 | 2,048 | <64 | <64 | 512 |
| 2 | 8,192 | <64 | <64 | 512 |
| 3 | 2,048 | <64 | 1,024 | 512 |
| R. bellii | ||||
| 4 | <64 | 512 | 256 | <64 |
| 5 | <64 | 1,024 | 256 | <64 |
| 6 | <64 | 256 | 128 | <64 |
| R. rickettsii | ||||
| 7 | <64 | 256 | 32,768 | <64 |
| 8 | <64 | 4,096 | 32,768 | <64 |
| 9 | <64 | 512 | 16,384 | <64 |
| R. canadensis | ||||
| 10 | 256 | <64 | <64 | 32,768 |
| 11 | 512 | <64 | <64 | 16,384 |
| 12 | 512 | <64 | <64 | 32,768 |
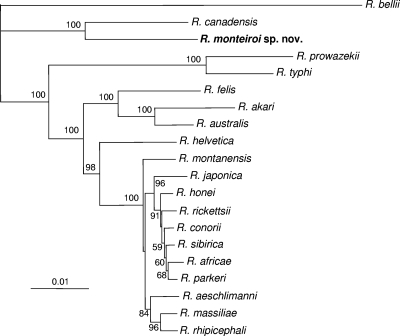
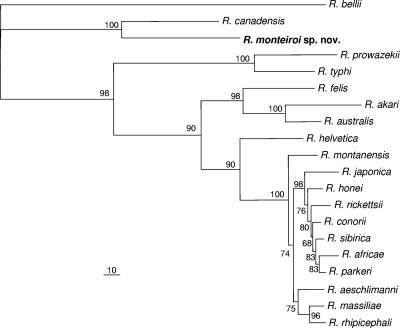
Due to the limited partial sequence fragments of many rickettsiae available in GenBank, phylogenetic analyses were performed using sequences of 1,321, 829, 386, and 443 nt of the rrs, gltA, htrA, and sca1 genes, respectively, with each gene analyzed separately. Analysis of the rrs, htrA, and sca1 partial sequences showed that R. monteiroi segregated closest to R. canadensis, supported by high bootstrap values (94, 92, and 100%, respectively). In the analysis of the gltA gene, we included a larger number of representative partial sequences from different rickettsial groups, such as the BG, the CG, and the adalia group (there was no sequence of sufficient length for these additional organisms to be included in the other gene analyses). R. monteiroi formed a distinct clade with R. canadensis and “Candidatus R. tarasevichiae,” supported by a 100% bootstrap value, indicating that R. monteiroi is a CG rickettsia (see Fig. S2 to S5 in the supplemental material). In the concatenated analyses, which included a total of 3,293 nt (1,362, 1,089, 398, and 443 nt for the rrs, gltA, htrA, and sca1 genes, respectively), R. monteiroi formed a distinct clade with R. canadensis with 100% bootstrap support by either neighbor-joining or maximum-parsimony analysis (Fig. 1 and 2).
Fig. 1.
Molecular phylogenetic analysis of R. monteiroi isolated from the tick A. incisum from Brazil. A total of 3,293 unambiguously aligned nucleotide sites of the rickettsial genes rrs, gltA, htrA, and sca1 were concatenated and subjected to analysis by the neighbor-joining method. Bootstrap values are shown at the nodes. Bar, 0.01 substitution. The GenBank accession numbers of the sequences included in this analysis are shown in Fig. S2 to S5 in the supplemental material.
Fig. 2.
Molecular phylogenetic analysis of R. monteiroi isolated from the tick A. incisum from Brazil. A total of 3,293 unambiguously aligned nucleotide sites of the rickettsial genes rrs, gltA, htrA, and sca1 were concatenated and subjected to analysis for maximum-parsimony inferences. Bootstrap values are shown at the nodes. Bar, 10 substitutions. The GenBank accession numbers of the sequences included in this analysis are shown in Fig. S2 to S5 in the supplemental material.
Ultrastructurally, R. monteiroi was morphologically identified within heavily infected Vero cells. The rickettsiae possessed typical bacillary morphology and were observed free in the cytosol. Most of the rickettsiae ranged from 0.41 to 0.53 μm in width and from 1.0 to 1.5 μm in length. A higher magnification of the cell wall revealed typical Gram-negative morphology consistent with rickettsial species, including a cytoplasmic membrane, a periplasmic space, and an outer membrane with an inner leaflet slightly thicker than the outer leaflet (see Fig. S6 in the supplemental material).
DISCUSSION
In the present study, we describe a new Rickettsia species, R. monteiroi, isolated from the tick A. incisum collected in an Atlantic rainforest reserve in southeastern Brazil. Through the phylogenetic analyses of four rickettsial genes, R. monteiroi was demonstrated to be always closest to R. canadensis, with high bootstrap support values. However, the genetic distance between these two rickettsiae (as shown by branch lengths in Fig. 1) is greater than the distance between any two species of the SFG and greater than the distance between the only two TG species, R. prowazekii and R. typhi. In addition, serological analyses showed that despite some cross-reactivity between R. monteiroi and R. canadensis guinea pig sera, homologous endpoint titers were always ≥4-fold higher than heterologous titers (Table 2). All of these facts support the status of R. monteiroi as a new species.
Molecular characterization indicates that R. monteiroi belongs to the CG of rickettsiae, which is currently composed of R. canadensis, “Candidatus R. tarasevichiae,” and an unnamed Rickettsia recently detected in the beetle Coccotrypes dactyliperda (Coleoptera: Curculionidae) (29). None of the CG rickettsiae has been isolated from humans or animals, but there has been serological evidence of human illness caused by R. canadensis in the United States (21). Until something is known about the pathogenicity of R. monteiroi for humans, this organism should be considered a potential human pathogen because its invertebrate host (A. incisum) is the most important human-biting tick in the primary Atlantic rainforest in the state of São Paulo, southeastern Brazil (27).
Two out of three guinea pigs inoculated with R. monteiroi in the present study showed no serological reaction against R. rickettsii, the most important rickettsial pathogen in the western hemisphere. In addition, none of the R. rickettsii- or R. bellii-infected guinea pigs showed a serological cross-reaction with R. monteiroi. In Brazil, serologic diagnosis of rickettsial infection in humans has employed solely R. rickettsii antigens (12). Based on our serologic results with guinea pigs, we can infer that a potential human or animal natural infection due to R. monteiroi would hardly be detected by serology if employing solely R. rickettsii antigens; i.e., if R. monteiroi is pathogenic for humans, routine laboratory serologic tests currently used in Brazil would possibly give false-negative results.
In the present study, 21 tick specimens that showed inconclusive results by the hemolymph test (due to a lack of hemocytes or excessive staining) and 28 hemolymph-negative specimens were individually tested by PCR targeting the rickettsial gltA gene using primers CS-78 and CS-323 (Table 1), resulting in no visible amplification (data not shown). These results indicate that the rate of infection of A. incisum ticks by R. monteiroi is very low, possibly lower than or around 1 to 2%, as shown by the hemolymph test and PCR.
R. monteiroi is the eighth Rickettsia species described in Brazil and, at the same time, the sixth Rickettsia species isolated from Brazilian ticks. R. monteiroi joins a growing list of tick-associated rickettsiae whose infectivity and pathogenicity for vertebrates are unknown. Further serosurvey studies employing R. monteiroi antigens, in comparison with other rickettsial antigens, should determine the potential of this novel Rickettsia to infect humans and animals in Brazil.
Description of Rickettsia monteiroi sp. nov.
Rickettsia monteiroi (mon.tei′roi. N.L. gen. masc. n. monteiroi of Monteiro, named in honor of the Brazilian rickettsiologist José Lemos Monteiro, who contributed to our knowledge of rickettsiae and rickettsioses in Brazil). Gram-negative, obligately intracellular bacterium. Grows on Vero cells at 28°C in minimal essential medium supplemented with 5% bovine calf serum. Nonmotile. rrs (16S rRNA), gltA, htrA, and sca1 gene sequencing indicates that this rickettsia is clearly different from all other recognized rickettsial species, the most closely related organisms being R. canadensis, R. bellii, and “Candidatus R. tarasevichiae.” No information is available about the possible pathogenicity of this organism for vertebrate hosts. The known geographical distribution of this bacterium is restricted to Brazil. The type strain is IntervalesT, which was isolated from an A. incisum tick collected in the Intervales State Park, State of São Paulo, Brazil. The type strain has been deposited in the Rickettsial Collection of the Laboratory of Parasitic Diseases of the Faculty of Veterinary Medicine, University of São Paulo, São Paulo, Brazil, and in the Rickettsial and Ehrlichial Diseases Research Laboratory at the University of Texas Medical Branch, Galveston, TX.
Supplementary Material
ACKNOWLEDGMENTS
We thank David H. Walker (University of Texas Medical Branch) for his technical support during this study.
This work was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
This work was performed in the Faculty of Veterinary Medicine of the University of São Paulo, São Paulo, Brazil.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 17 June 2011.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Burgdorfer W. 1970. The hemolymph test. Am. J. Trop. Med. Hyg. 19:1010–1014 [PubMed] [Google Scholar]
- 3. Dyková I., Veverkova M., Fiala I., Machackova B., Peckova H. 2003. Nuclearia pattersoni sp. n. (Filosea), a new species of amphizoic amoeba isolated from gills of roach (Rutilus rutilus), and its rickettsial endosymbiont. Folia Parasitol. 50:161–170 [PubMed] [Google Scholar]
- 4. Eremeeva M. E., Balayeva N. M., Ignatovich V. F., Raoult D. 1993. Proteinic and genomic identification of spotted fever group rickettsiae isolated in the former USSR. J. Clin. Microbiol. 31:2625–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gillespie J. J., et al. 2007. Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS One 2:e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gimenez D. F. 1964. Staining rickettsiae in yolk-sac cultures. Stain Technol. 39:135–140 [DOI] [PubMed] [Google Scholar]
- 7. Horta M. C., et al. 2004. Prevalence of antibodies to spotted fever group rickettsiae in humans and domestic animals in a Brazilian spotted fever-endemic area in the State of São Paulo, Brazil: serologic evidence for infection by Rickettsia rickettsii and another spotted fever group Rickettsia. Am. J. Trop. Med. Hyg. 71:93–97 [PubMed] [Google Scholar]
- 8. Ito S., Rikihisa Y. 1981. Techniques for electron microscopy of rickettsiae, p. 213–227 In Burgdorfer W., Anacker R. L. (ed.), Rickettsiae and rickettsial diseases. Academic Press, New York, NY [Google Scholar]
- 9. Jado I., et al. 2007. Rickettsia monacensis and human disease, Spain. Emerg. Infect. Dis. 13:1405–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kelly P. J., Raoult D., Mason P. R. 1991. Isolation of spotted fever group rickettsias from triturated ticks using a modification of the centrifugation-shell vial technique. Trans. R. Soc. Trop. Med. Hyg. 85:397–398 [DOI] [PubMed] [Google Scholar]
- 11. Kumar S., Tamura K., Nei M. 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150–163 [DOI] [PubMed] [Google Scholar]
- 12. Labruna M. B. 2009. Ecology of Rickettsia in South America. Ann. N. Y. Acad. Sci. 1166:156–166 [DOI] [PubMed] [Google Scholar]
- 13. Labruna M. B., et al. 2004. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of Sao Paulo, Brazil, where Brazilian spotted fever is endemic. J. Clin. Microbiol. 42:90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Labruna M. B., et al. 2004. Molecular evidence for a spotted fever group Rickettsia species in the tick Amblyomma longirostre in Brazil. J. Med. Entomol. 41:533–537 [DOI] [PubMed] [Google Scholar]
- 15. Labruna M. B., et al. 2007. Infection by Rickettsia bellii and Candidatus “Rickettsia amblyommii” in Amblyomma neumanni ticks from Argentina. Microb. Ecol. 54:126–133 [DOI] [PubMed] [Google Scholar]
- 16. McKiel J. A., Bell E. J., Lackman D. B. 1967. Rickettsia canada: a new member of the typhus group of rickettsiae isolated from Haemaphysalis leporispalustris ticks in Canada. Can. J. Microbiol. 13:503–510 [DOI] [PubMed] [Google Scholar]
- 17. Ngwamidiba M., Blanc G., Raoult D. D., Fournier P. E. 2006. Sca1, a previously undescribed paralog from autotransporter protein-encoding genes in Rickettsia species. BMC Microbiol. 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pacheco R., Rosa S., Richtzenhain L., Szabó M. P. J., Labruna M. B. 2008. Isolation of Rickettsia bellii from Amblyomma ovale and Amblyomma incisum ticks from southern Brazil. Rev. MVZ Córdoba 13:1273–1279 [Google Scholar]
- 19. Paddock C. D., et al. 2004. Rickettsia parkeri: A newly recognized cause of spotted fever rickettsiosis in the United States. Clin. Infect. Dis. 38:805–811 [DOI] [PubMed] [Google Scholar]
- 20. Parola P., Davoust B., Raoult D. 2005. Tick- and flea-borne rickettsial emerging zoonoses. Vet. Res. 36:469–492 [DOI] [PubMed] [Google Scholar]
- 21. Parola P., Labruna M. B., Raoult D. 2009. Tick-borne rickettsioses in America: unanswered questions and emerging diseases. Curr. Infect. Dis. Rep. 11:40–50 [DOI] [PubMed] [Google Scholar]
- 22. Perlman S. J., Hunter M. S., Zchori-Fein E. 2006. The emerging diversity of Rickettsia. Proc. Biol. Sci. 273:2097–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pinter A., Labruna M. B. 2006. Isolation of Rickettsia rickettsii and Rickettsia bellii in cell culture from the tick Amblyomma aureolatum in Brazil. Ann. N. Y. Acad. Sci. 1078:523–529 [DOI] [PubMed] [Google Scholar]
- 24. Raoult D., Roux V. 1997. Rickettsioses as paradigms of new or emerging infectious diseases. Clin. Microbiol. Rev. 10:694–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Regnery R. L., Spruill C. L., Plikaytis B. D. 1991. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J. Bacteriol. 173:1576–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shpynov S., Fournier P. E., Rudakov N., Tarasevich I., Raoult D. 2006. Detection of members of the genera Rickettsia, Anaplasma, and Ehrlichia in ticks collected in the Asiatic part of Russia. Ann. N. Y. Acad. Sci. 1078:378–383 [DOI] [PubMed] [Google Scholar]
- 27. Szabó M. P., et al. 2006. Ticks (Acari: Ixodidae) parasitizing humans in an Atlantic rainforest reserve of southeastern Brazil with notes on host suitability. Exp. Appl. Acarol. 39:339–346 [DOI] [PubMed] [Google Scholar]
- 28. Szabó M. P., et al. 2009. Ecological aspects of the free-living ticks (Acari: Ixodidae) on animal trails within Atlantic rainforest in south-eastern Brazil. Ann. Trop. Med. Parasitol. 103:57–72 [DOI] [PubMed] [Google Scholar]
- 29. Weinert L. A., Werren J. H., Aebi A., Stone G. N., Jiggins F. M. 2009. Evolution and diversity of Rickettsia bacteria. BMC Biol. 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.