Abstract
Thiamine pyrophosphate (TPP), a biologically active form of thiamine (vitamin B1), is an essential cofactor in all living systems. Microorganisms either synthesize TPP via de novo biosynthesis pathways or uptake exogenous thiamine from the environment via specific transporters. The oral spirochete Treponema denticola is an important pathogen that is associated with human periodontal diseases. It lacks a de novo TPP biosynthesis pathway and needs exogenous TPP for growth, suggesting that it may obtain exogenous TPP via a thiamine transporter. In this study, we identified a gene cluster that encodes a TPP ABC transporter which consists of a TPP-binding protein (TDE0143), a transmembrane permease (TDE0144), and a cytosolic ATPase (TDE0145). Transcriptional and translational analyses showed that the genes encoding these three proteins are cotranscribed and form an operon (tbpABCTd) that is initiated by a σ70-like promoter. The expression level of this operon is negatively regulated by exogenous TPP and is mediated by a TPP-sensing riboswitch (Tdthi-box). Genetic and biochemical studies revealed that the TDE0143 deletion mutant (T. denticola ΔtbpA) had a decreased ability to transport exogenous TPP, and the mutant failed to grow when exogenous TPP was insufficient. These results taken together indicate that the tbpABCTd operon encodes an ABC transporter that is required for the uptake of exogenous TPP and that the expression of this operon is regulated by a TPP-binding riboswitch via a feedback inhibition mechanism.
INTRODUCTION
Human periodontal diseases are a group of infections that affect the structures surrounding teeth and occur in 80% of the adult population at some time in their lives (59, 72, 75). The diseases are caused primarily by polymicrobial infections followed by host factors (13). In the oral cavity, more than 700 different microorganisms have been identified (17, 41, 56), and they typically live in a synergetic manner (28, 72). In the symbiotic mode, the existence of one group of bacteria may create a nutritional and atmospheric environment that can facilitate the colonization and growth of other microorganisms (e.g., the metabolites from one group of bacteria can be used as an energy source by others). Thus, studying bacterial metabolism can help us to understand the ecology of these oral microorganisms and the etiology of periodontal diseases.
In the oral microflora there is a substantial amount of spirochetes, and more than 60 different species have been identified. 16S rRNA gene sequence analyses indicate that all of these oral spirochetes belong to one genus, Treponema (17, 56). However, due to their fastidious growth requirements and the lack of understanding of their nutritional metabolism, very few oral treponemes have been cultivated (8, 20). Treponema denticola is an oral spirochete that can be easily cultivated. Since its genome was sequenced and it can be genetically manipulated, T. denticola has been used as a model spirochete to study other oral spirochetes (20, 39, 44, 85). T. denticola is a member of the “red-complex” bacteria, which are a group of Gram-negative, anaerobic, and proteolytic oral bacteria that are highly associated with human periodontal diseases (13, 30). A group of studies have shown that T. denticola is associated with the incidence and severity of periodontal diseases, and several virulence-associated factors have been characterized (14, 20, 21, 46, 67).
T. denticola is a motile, fastidious, and obligate anaerobic bacterium that dwells in a complex and diverse microbial community within the oral cavity (29, 64). In order to survive in this highly specialized milieu and cause diseases, T. denticola has to acquire essential nutrients, overcome the competition with other bacterial species, and protect against the constant pressure of host defenses. Previous studies have focused on its genetics and pathogenesis, and very few studies have investigated the metabolism of T. denticola (4, 7, 12, 22, 29, 31, 35, 63). As an obligate bacterium, T. denticola needs exogenous fatty acids, amino acids, and vitamins to grow (4, 29). Among these nutrients, thiamine pyrophosphate (TPP), an active form of thiamine, is an essential cofactor of several important enzymes in carbohydrate and branched-chain amino acid metabolism, and it is needed by all living organisms (2, 18, 36, 62). In addition, thiamine biosynthesis (TBS) is often linked to other central metabolic pathways such as purine biosynthesis (2, 18, 36). Thus, studying the thiamine biosynthesis of a microorganism can help us to dissect its metabolic integration.
Bacteria either produce TPP via a de novo biosynthesis pathway or uptake exogenous TPP from the environment (2, 34). T. denticola needs exogenous TPP to support its growth (16), and yet neither de novo nor salvage TPP synthesis pathways have been identified, suggesting that T. denticola may uptake exogenous TPP via a specific transport system (70). In Salmonella enterica serovar Typhimurium, the thiBPQ operon encodes a TPP ABC transporter that consists of a thiamine-binding protein (ThiB), a transmembrane thiamine channel (ThiP), and an ATPase (ThiQ) (80). Genetic studies have shown that this transporter is required for the uptake of exogenous TPP, and it is essential for the growth of S. Typhimurium that has its de novo biosynthesis pathway disrupted (80). In this study, an operon that encodes a putative TPP ABC transporter was identified in T. denticola. Genetic and biochemical studies showed that this transporter is involved in the uptake of exogenous TPP, and it is essential for growth of T. denticola when the exogenous TPP is limited.
The riboswitch is a common regulatory mechanism that is often involved in the biosynthesis of amino acids, nucleotides, vitamins, and other molecules, and it has been identified in many organisms, including bacteria, archaea, fungi, and plants (5, 53, 60, 68, 83). Riboswitches are structured domains that usually reside in the noncoding regions of mRNAs (52, 68), where they bind metabolites and control gene expression. Each class of riboswitches identified so far forms a structured receptor, or “aptamer,” that directly binds to a specific metabolite (19, 69, 83). In Escherichia coli and S. Typhimurium, three thiamine biosynthetic operons (thiCEFSGH, thiMD, and thiBPQ) are negatively regulated by TPP and are mediated by the thiamine-sensing riboswitch (thi-box) (51, 69, 82). It has been experimentally demonstrated that the thi-box of thiM regulates the thiMD operon at the translational level by sequestering the Shine-Dalgarno (S/D) sequence. This prevents the ribosome from binding to the mRNA and translating the transcript (54, 82). The regulatory mechanism of thiC remains elusive. It has been postulated that the thi-box of thiC controls the thiCEFSGH operon by premature transcription termination (5, 82). In this study, it was found that the expression of the identified TPP transporter of T. denticola was regulated at the transcriptional level by a thi-box.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
T. denticola ATCC 35405 (wild type) (70) and the isogenic mutant described below were grown in OBGM medium with 10% heat-inactivated rabbit serum (55) at 37°C in an AS-580 anaerobic chamber (Anaerobe Systems, Morgan Hill, CA) with an atmosphere of 85% nitrogen, 5% carbon dioxide, and 10% hydrogen. For the semisolid medium, 0.7% low-melting-point SeaPlaque agarose (Lonza, Rockland, ME) was incorporated into the medium, and the plates were poured after inoculating a bacterial suspension into the medium at 37°C (44, 45). Escherichia coli strain TOP10 (Invitrogen, Carlsbad, CA) was used for DNA cloning and β-galactosidase assays, and the BL21 Codon Plus strain (Stratagene, La Jolla, CA) was used for preparing the recombinant protein. The E. coli strains were cultured in lysogeny broth (LB) supplemented with appropriate concentrations of antibiotics.
Preparation of TA medium.
To prepare a medium with a defined amount of TPP, the concentration of TPP in the OBGM medium was detected by high-performance liquid chromatography (HPLC) as described below. The concentrations of TPP in the OBGM medium, with and without additional TPP, were 14 μM and 1 μM, respectively. Analysis showed that brain heart infusion (BHI) and yeast extract are the main sources of TPP. To reduce the level of TPP in the medium, the amounts of BHI and yeast extract in the OBGM medium were decreased to 2.5 g liter−1 and 1.5 g liter−1, respectively, which reduced the concentration of TPP to approximately 100 nM. The rest of the components in the medium are the same as in the normal OBGM medium. The obtained medium was referred to as TPP assay (TA) medium. In this study, media containing three different concentrations of TPP (100 nM, 10 μM, and 50 μM) were prepared.
Detection of TPP by HPLC.
The TPP concentrations in the media and in the T. denticola cells were measured by high-performance liquid chromatography (HPLC) as previously described (37, 47) with some modifications. To detect intracellular TPP, 50 ml of stationary-phase T. denticola culture, which was cultivated in the TA media with three different concentrations of TPP, was harvested by centrifugation at 6,000 × g for 10 min. The resulting cell pellets were washed twice with phosphate-buffered saline (pH 7.4) (PBS) and then resuspended in PBS. The final cell densities were adjusted to approximately 1010 cells ml−1. The obtained samples were lysed with a French press and fractionated by centrifugation at 10,000 × g for 10 min. The resulting supernatants were first precipitated with a 3-fold volume of methanol and then centrifuged at 16,000 × g for 10 min. After the centrifugation, 100 μl of the supernatant was treated with a freshly prepared derivatization buffer (0.2 mM potassium ferricyanide and 5% sodium hydroxide) to oxidize TPP to the thiochrome derivative that can be fluorimetrically detected by HPLC. Five microliters of the obtained samples was processed for HPLC analysis.
Quantification of TPP was performed on an Agilent (Palo Alto, CA) 1100 series HPLC system consisting of an online vacuum degasser, quaternary pump, autosampler, thermostat-controlled column compartment, and fluorescence detector. Sample separation was achieved using a Phenomenex (Torrance, CA) Luna C18 column (100 mm by 2.0 mm by 3 μm). A gradient elution was employed to separate the samples. The elution parameters were as follows: an initial 3-min isocratic elution with 95% buffer A (0.02% trifluoroacetic acid in water) and 5% buffer B (0.02% trifluoroacetic acid in 90% acetonitrile), a linear increase of buffer B to 40% for 2 min and then elution with 40% buffer B for 7 min, followed by the initial condition from 13 min. TPP was detected at an excitation wavelength of 375 nm and an emission wavelength of 430 nm using a fluorescence detector. The calibration linear range of TPP was 0.15 to 15 pmol on the column. Three independent assays were conducted, and the intracellular TPP concentration was expressed as nanomolar.
Construction of tbpABCTd thi-box–lacZ fusions and TPP repression β-galactosidase assays.
The fragment spanning from nucleotide (nt) −366 to +69 (thi-box-L) of the tbpABCTd operon was PCR amplified from wild-type T. denticola chromosomal DNA using primer pair P15/P16 (Table 1), generating the amplicon with engineered EcoRI and BamHI cut sites at the 5′ and 3′ ends, respectively. The resultant amplicon was first cloned into the pGEM-T-Easy vector (Promega, Madison, WI) and then released by EcoRI and BamHI digestion. The obtained DNA fragment was fused to a promoterless lacZ gene in the pRS414 plasmid (a gift from R. Breaker, Yale University) (71, 82), in which the 23rd (thi-box-L) codon of TDE0143 resides in frame with the ninth usage codon of lacZ. Site-direct mutagenesis of thi-box-L was conducted using the QuikChange II site-directed mutagenesis kit (Stratagene) with primer pair P29/P30, according to the manufacturer's instructions. The mutation was confirmed by DNA sequencing (Roswell Park Cancer Institute DNA Sequencing Laboratory, Buffalo, NY), and the resultant construct was named thi-box-Lm.
Table 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′ → 3′)a | Commentb |
|---|---|---|
| P1 | GACAGTTTACTCGGCACG | Flanking region before TDE0143; F |
| P2 | ATCGATCAGCCCTTTGCGGGTATG | Flanking region before TDE0143; R |
| P3 | ATCGATCTCAAAGCCTCCTTCCCG | Flanking region after TDE0143; F |
| P4 | AGATATTTCGGTGAGGGC | Flanking region after TDE0143; R |
| P5 | ATCGATCCGATAGCTTCCGCTATTGC | erm cassette; F |
| P6 | ATCGATGGAAGCTGTCAGTAGTATACC | erm cassette; R |
| P7 | CGGAGAAGCTCCCATAGC | Co-RT-PCR, TDE0143-TDE0144; F |
| P8 | AAAGATGAGGCCGCCGAG | Co-RT-PCR, TDE0143-TDE0144; R |
| P9 | CCCATGTGCAAGAAGGGC | Co-RT-PCR, TDE0144-TDE0145; F |
| P10 | GACCATTCCGATTCCGCG | Co-RT-PCR, TDE0144-TDE0145; R |
| P11 | CATTACCCCTCTTGTCGAG | Co-RT-PCR, TDE0145-TDE0146; F |
| P12 | CGTTAATCTCGGCTATACC | Co-RT-PCR, TDE0145-TDE0146; R |
| P13 | CACCTGTGCCGCTCTTTTTGCA | rTbpATd; F |
| P14 | CTAAGCAAGTCCCTTGATAAC | rTbpATd; R |
| P15 | GAATTCGGACAACTTGTTATTATGCAC | 5′ of tbpATd thi-box; F |
| P16 | GGATCCTGCAAAAAGAGCGGCACAACC | 3′ of tbpATd thi-box-L; R |
| P17 | GGATCCCATATGCTCCCTCCGCCGGTA | 3′ of tbpATd thi-box-S; R |
| P18 | TAATTATCGGTTGTGCCGCT | TDE0143 5′-RACE sensor primer; F |
| P19 | CGGCAATACCCATACCTTCA | TDE0143 5′-RACE inner primer; R |
| P20 | TGCAGGTTTGGGAACATCCT | TDE0143 5′-RACE outer primer; R |
| P21 | CTGACGGAACAAAGGTTGTAG | qRT-PCR, TDE0143; F |
| P22 | CCTTACCTTCCAGTATGACTC | qRT-PCR, TDE0143; R |
| P23 | TAGGATTGCTGCCTTTACGG | qRT-PCR, TDE0144; F |
| P24 | ATTATTGCAGGCACGCAGAG | qRT-PCR, TDE0144; R |
| P25 | TCAGGCTGCGGCAAAACAAC | qRT-PCR, TDE0145; F |
| P26 | CATTGAGATGAGGGAAGAGG | qRT-PCR, TDE0145; R |
| P27 | CCGTCCATTGTAGGCTTTAC | qRT-PCR, dnaK; F |
| P28 | CCGTCAATGTCGATTCGTAC | qRT-PCR, dnaK; R |
| P29 | TGCTTTTACAAGCTAAAAACATACCCGCAA | TPP-binding site mutagenesis; F |
| P30 | TTGCGGGTATGTTTTTAGCTTGTAAAAGCA | TPP-binding site mutagenesis; R |
| P31 | GCAGCACATCCCCCTTTC | qRT-PCR, lacZ; F |
| P32 | CGTTGGTGTAGATGGGCG | qRT-PCR, lacZ; R |
| P33 | AACGTACTGGCAGATGCAG | qRT-PCR, groEL; F |
| P34 | GCGCTTTCAGTTCTTCAACTG | qRT-PCR, groEL; R |
The engineered restriction enzyme sites are underlined.
F, forward primer; R, reverse primer.
The thi-box-L--lacZ and thi-box-Lm–lacZ fusion constructs were transformed into E. coli TOP10 cells. A TPP repression β-galactosidase assay was conducted as previously described (82). Briefly, the E. coli TOP10 cells transformed with the lacZ fusion plasmids were grown in M9 glucose minimal medium plus 50 μg ml−1 vitamin assay Casamino Acids (Difco, Becton Dickinson and Company, Sparks, MD) to the mid-exponential phase either with or without TPP (100 μM). The β-galactosidase activity was measured with the β-galactosidase enzyme assay system (Promega) in 96-well plates. All experiments were repeated at least twice in duplicate.
Reverse transcription-PCR (RT-PCR), quantitative RT-PCR (qRT-PCR), and RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE).
RNA isolation was carried out as previously described (42, 65). For T. denticola, 100 ml of mid-log-phase cultures (∼ 5 × 107 cells ml−1) were harvested for RNA preparations; for E. coli, following the TPP repression β-galactosidase assays described above, 10 ml of the mid-exponential-phase cultures was harvested for RNA preparations. Total RNA was extracted using TRI reagent (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions. The resultant samples were treated with Turbo DNase I (Ambion, Austin, TX) at 37°C for 2 h to eliminate genomic DNA contamination. The resultant RNA samples were extracted using acid-phenol–chloroform (Ambion), precipitated in isopropanol, and washed with 70% ethanol. The RNA pellets were resuspended in RNase-free water. RNA (1 μg) was reverse transcribed using the avian myeloblastosis virus (AMV) reverse transcriptase (Promega) to generate cDNA.
For RT-PCR analysis, 1 μl of cDNA was PCR amplified using Taq DNA polymerase (Qiagen, Valencia, CA). The qRT-PCR analysis was conducted using iQ SYBR green Supermix and MyiQ thermal cycler (Bio-Rad, Hercules, CA), as previously described (74). The genes encoding heat shock proteins GroEL (E. coli) and DnaK (T. denticola TDE0628) were used as internal controls to normalize the qRT-PCR data. The primers used for the RT-PCR and qRT-PCR analyses are listed in Table 1. To determine the transcription start site upstream of TDE0143, 5′-RACE analysis was carried out using the FirstChoice RLM-RACE kit (Ambion) according to the manufacturer's protocol. Purified T. denticola RNA (10 μg) was reverse transcribed to cDNA with a 5′-RACE adapter, followed by PCR amplifications with primers P18, P19, and P20 (Table 1). The resultant PCR products were cloned into the pGEM-T-Easy vector and sequenced (Roswell Park Cancer Institute DNA Sequencing Laboratory).
Construction of a T. denticola tbpATd deletion mutant (T. denticola ΔtbpA).
To inactivate tbpATd (TDE0143), its flanking regions were PCR amplified from wild-type T. denticola genome DNA using the primer pairs P1/P2 and P3/P4 (Fig. 1 and Table 1). A previously described (45) ermF/AM erythromycin resistance cassette (Ermr) was PCR amplified using the primer pair P5/P6. The amplicons were individually cloned into the pGEM-T-Easy vector (Promega). The downstream fragment (flanking R2) was released by ClaI and SphI digestion and then ligated to the 3′ end of the upstream fragment (flanking R1) at the same cut sites. The Ermr cassette was then released by ClaI digestion and inserted into the obtained fragment described above, generating the tbpATd::erm plasmid, in which a 864 bp of TDE0143 fragment was deleted (Fig. 1). For the allelic replacement mutagenesis, the tbpATd::erm plasmid was linearized with NotI. Approximately 10 μg of the linearized plasmid was electroporated into wild-type T. denticola cells, and the transformed cells were selected on semisolid OBGM plates containing erythromycin (60 μg ml−1).
Fig. 1.
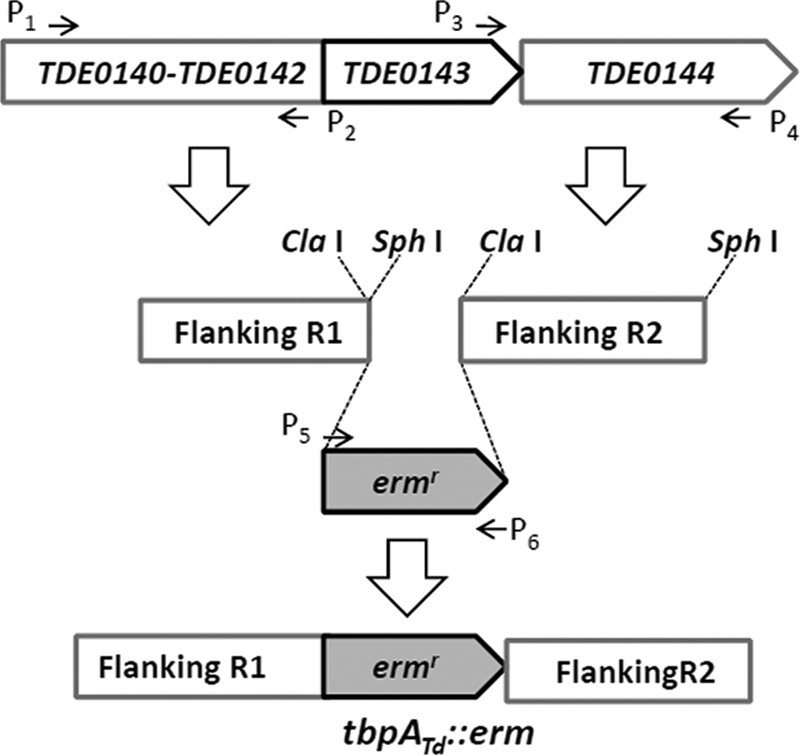
Diagram showing the targeted mutagenesis of the tbpATd gene. Arrows indicate the relative positions of PCR primers for constructing the targeted mutagenesis vector tbpATd::erm, in which an 864-bp TDE0143 fragment is deleted and replaced by the erythromycin resistance cassette (Ermr). The sequences of these primers are listed in Table 1.
Measurement of the growth rates of T. denticola.
To measure the growth curves of T. denticola strains, 50 μl of PBS-washed T. denticola cells (3 × 108 cells ml−1) were inoculated into 5 ml of TA medium (the initial cell density was 3 × 106 cells ml−1) containing different concentrations of TPP (100 nM, 10 μM, and 50 μM) and incubated at 37°C in an anaerobic chamber. The cultures were enumerated every 24 h using a Petroff-Hausser counting chamber (Hausser Scientific, Horsham, PA) for up to 7 days. Counts were repeated in triplicate with at least three independent samples. The results were expressed as mean cell numbers ± standard error of the mean (SEM).
Generation of TbpATd antiserum.
A DNA fragment that encodes the N-terminal 283 amino acids of TbpATd was PCR amplified using the primers P13/P14 (Table 1) and Platinum Pfx DNA polymerase (Invitrogen). The amplicon was cloned into the pET100/D-TOPO expression vector (Invitrogen), which generates a six-histidine tag at the N terminus of the recombinant protein. The resulting plasmid was then transformed into BL21 Codon Plus cells (Stratagene). The expression of recombinant TbpATd (rTbpATd) was induced using 0.1 M isopropyl-β-d-thiogalactoside (IPTG). The recombinant protein was purified at 4°C using HisTrap HP columns (GE Healthcare, Piscataway, NJ) under denaturing conditions. The final purified protein was dialyzed in buffer (0.01 M Tris base, pH 8.0) at 4°C overnight. To produce an antiserum against TbpATd, mice were immunized with 1 mg of rTbpATd in three inoculations during a 1-month period, and the obtained antiserum was tested with immunoblots as previously described (43).
Electrophoresis and Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed as previously described (42, 43). T. denticola cells were harvested at approximately 108 cells ml−1 in stationary phase. The same amount of whole-cell lysates (10 to 50 μg) was separated by SDS-PAGE and then transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). Immunoblots were probed with specific antibodies against TbpATd or FlaA (an internal control). Immunoblots were developed using horseradish peroxidase secondary antibody with the enhanced chemiluminescent detection system (ECL). Densitometry of immunoreactive proteins in the blots was used to determine the relative amounts of proteins as previously described (74). Densitometry was done using the Molecular Imager ChemiDoc XRS imaging system (Bio-Rad).
Construction of a homology model of TbpATd.
The crystal structure of E. coli TbpA protein (Protein Data Bank ID 2QRY) (73) was selected as the template for the homology modeling. Pairwise sequence alignment between TbpATd and the template was conducted using Clustal X. Modeler 9v7 (66) was used to construct the homology model. Chain A of 2QRY (73) together with TPP was used as the template. The automodel class with the conjugate gradient optimization method was adopted to produce the final model with the best objective function value among the five resultant candidates. Thiamine-binding residues were picked at relative positions comparable to those in 2QRY. All diagrams were produced in VMD 1.8.7.
RESULTS
Identification of a putative TPP ABC transporter.
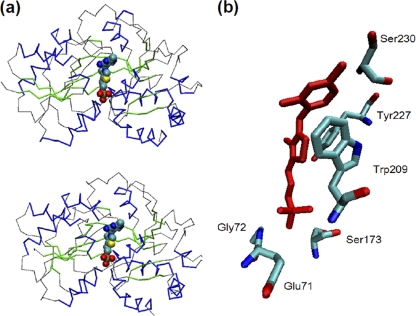
The TPP transporter of S. Typhimurium consists of a thiamine-binding protein (ThiB, also named TbpA), a transmembrane thiamine channel (ThiP), and an ATPase (ThiQ, also named FbpC) (15, 33). To determine if there is any similar transporter in T. denticola, the genes encoding these three proteins were used as queries to search the genome of T. denticola strain ATCC 35405 (70). The results revealed that a gene cluster encodes a putative TPP transporter that is a homolog of ThiBPQ, in which TDE0143 has 37% identity and 54% similarity to ThiB (ZP_03051779), TDE0144 has 29% identity and 49% similarity to ThiP (YP668009), and TDE0145 has 44% identity and 69% similarity to ThiQ (ZP_05623000). These three identified proteins were further used as queries to search the Pfam database and the Conserved Domain Database (CDD) (23, 48), and the results showed that they belong to the corresponding Pfam or CDD group. For example, TDE0143 (TbpATd) is a member of PRK11205, a family of thiamine transporter substrate-binding subunits (see Fig. S1 in the supplemental material). Structure modeling analysis further revealed that TbpATd has a structure similar to that of ThiB (Fig. 2a) and that it contains a well-conserved thiamine-binding motif (Fig. 2b), suggesting that TbpATd is a thiamine-binding protein. Collectively, these results suggest that the gene cluster of TDE0143 to -0145 encodes a putative TPP transporter, in which TDE0143 encodes a TPP-binding protein, TDE0144 (TbpBTd) encodes a transmembrane thiamine channel, and TDE0145 (TbpCTd) encodes an ATPase.
Fig. 2.
Structural characteristics of TbpATd. (a) Comparison of the overall structures of the E. coli TbpA protein (top) and TbpATd (bottom). The structural view of these two proteins is depicted by Cα trace with helices colored in blue, sheets in green, and the rest in gray. The TPP ligand is shown in a Van der Waals (VDW) representation. E. coli TbpA (Protein Data Bank ID 2QRY) (73) was selected for the structural modeling using the program Modeler 9v7 as previously described (66). (b) Predicted thiamine-binding site with the amino acids and the bound TPP (red).
Transcriptional analysis of the tbpABCTd operon.
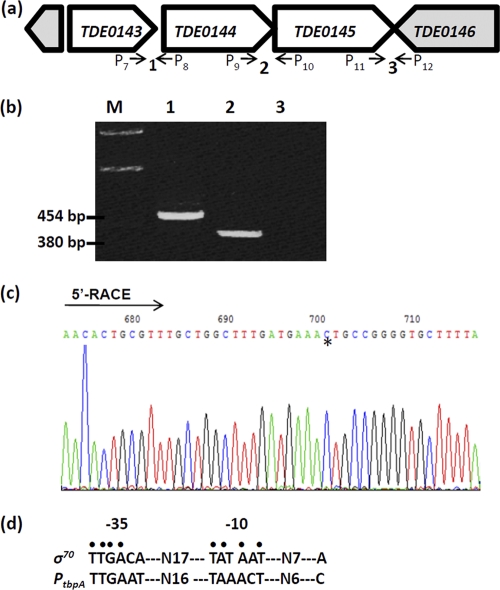
The open reading frames (ORFs) of TDE0143, TDE0144, and TDE0145 are in the same orientation and are adjacent to each other (the maximum intergenic space is 11 bp) (Fig. 3a), suggesting that these three genes are probably cotranscribed. To confirm this speculation, RT-PCR analysis described before (84) was conducted with three pairs of primers (P7 to P12 [Table 1]), spanning from TDE0143 to TDE0146 (Fig. 3a). As shown in Fig. 3b, the primers spanning from TDE0143 to TDE0145 yielded positive RT-PCR products of the predicted sizes. As expected, no product was detected between TDE0145 and TDE0146, since they are divergently transcribed (Fig. 3a and b). This analysis shows that these three genes form a polycistronic operon that is transcribed as a single transcript, and this gene cluster is referred to as the tbpABCTd operon.
Fig. 3.
Transcriptional analysis of the tbpABCTd operon. (a) Diagram showing the genes near TDE0143. The numbers and small arrows show the primers used for the RT-PCR analysis. (b) RT-PCR analysis. The lane numbers correspond to the primers labeled in panel a, and the sequences of these primers are described in Table 1. Lane M, 1.0-kb DNA ladder. (c) 5′-RLM-RACE analysis. The arrow shows the sequencing direction, and the asterisk indicates the identified transcriptional start site. (d) Comparison of the identified promoter sequence with the consensus sequence of the E. coli σ70 promoter. The conserved nucleotides are dotted.
To further delineate the regulation of tbpABCTd operon, RLM-RACE analysis was conducted to determine its transcriptional start site. The analysis mapped the start site to the nucleotide C that is 98 nt from the start codon of TDE0143 (Fig. 3c; see Fig. S2 in the supplemental material). A promoter-like consensus was identified at the −10 (TAAACT) and −35 (TTGAAT) regions (Fig. 3d), which is similar to those of the σ70 promoter of E. coli (26, 77), suggesting that tbpABCTd is probably regulated by a σ70-like promoter. The identified promoter was named PtbpA.
Identification of a thi-box at the 5′-UTR of tbpABCTd.
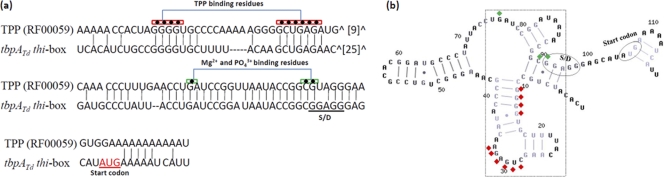
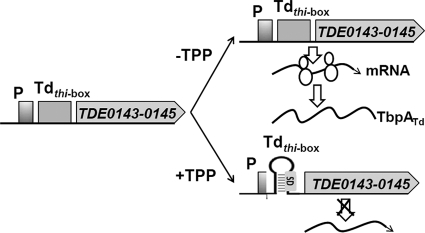
There is a 98-nt untranslated region (UTR) upstream of the tbpABCTd transcript, and a recent study projected that this region contains a putative thi-box (27). The predicted thi-box (designated Tdthi-box) is 117 nt and spans from nt −104 to +13 (counting from the start codon), which encompasses part of the PtbpA promoter and the whole S/D sequence (Fig. 4a). Sequence alignment analysis further confirmed that Tdthi-box belongs to the family of TPP riboswitches (RF00059) (27), and it contains the conserved residues involved in the binding of TPP and Mg2+ (19, 38, 82) (Fig. 4a). Secondary structure modeling predicted that Tdthi-box forms a conserved TPP aptamer that consists of five helices, three terminal loops, and two junction bulges (Fig. 4b). One of these junction bulges contains the UGAGA motif that is absolutely invariant in the family of TPP riboswitches (19, 38, 82).
Fig. 4.
Sequence and secondary structure of Tdthi-box. (a) Comparison between the 5′-untranslated region (UTR) of tbpATd (Tdthi-box) and the TPP riboswitch motif box (RF00059) (27). Diamonds represent the conserved residues required for the function and structure of TPP riboswitches. The underlined AUG is the translational start codon of tbpATd. (b) Diamonds represent the conserved residues identified in the riboswitches of thiM and thiC. The S/D sequence and the start codon are circled. The conserved TPP-sensing motif is boxed. The structure was predicted with the CONTRAfold program.
The expression level of the Tdthi-box–lacZ fusion reporter is negatively regulated by TPP.
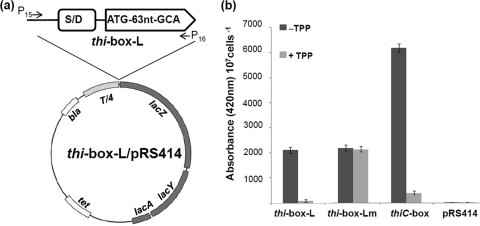
To further confirm the above proposition, TPP repression β-galactosidase assays were conducted using two Tdthi-box–lacZ fusion constructs (thi-box-L–lacZ and thi-box-Lm–lacZ) (Fig. 5a). The thiC-lacZ fusion acts as a TPP-sensing riboswitch positive control and the pRS414 plasmid as a negative control (71, 82). As expected, the β-galactosidase activity of thiC-lacZ was substantially reduced in the presence of TPP (100 μM), and no β-galactosidase activity was detected in the pRS414 plasmid (Fig. 5b). Similarly to that of thiC, the activity of thi-box-L--lacZ fusion was decreased approximately 20-fold by the addition of TPP (Fig. 5b). To determine if the reduction of β-galactosidase activity in thi-box-L-lacZ is mediated by TPP, the conserved TPP-binding site UGAGA (Fig. 4) was mutated to UAAAA in the thi-box-Lm–lacZ construct. As shown in Fig. 5b, the repression of Tdthi-box on the β-galactosidase activity was completely abolished in the mutated construct. Collectively, these results demonstrate that Tdthi-box is a TPP-sensing riboswitch.
Fig. 5.
Characterization of a TPP-sensing riboswitch in T. denticola (Tdthi-box). (a) Construction of Tdthi-box–lacZ fusions for the β-galactosidase assays. thi-box-L represents the fragment from nucleotide −366 to +69 of Tdthi-box. (b) TPP repression β-galactosidase assays. The β-galactosidase activity was expressed as Miller units/107 cells, and the levels represent means ± standard errors of the means from three independent experiments. Plasmid pRS414 containing the thiC-box was used as a positive control and plasmid pRS414 (82) as a negative control.
The action of Tdthi-box occurs at the transcriptional level.
The action of Tdthi-box can occur either at the translational level by preventing the ribosome from binding to the mRNA and translating the transcript or at the transcriptional level by premature transcription termination. To rule out one of these two possibilities, following the TPP repression β-galactosidase assays described above, the levels of lacZ mRNA were measured by qRT-PCR. As shown in Table 2, in the presence of TPP, the level of lacZ transcript was decreased about 16-fold (ranging from 13- to 20-fold) in the thi-box-L-–lacZ construct and 13-fold (ranging from 10- to 16-fold) in the thiC-lacZ construct. The level of lacZ transcript remained unchanged in the thi-box-Lm–lacZ construct. The decreased transcript level is proportional to the results from the TPP repression β-galactosidase assays (Fig. 5), indicating that the actions of Tdthi-box and the thi-box of thiC on the reporter gene expression occur primarily at the transcriptional level.
Table 2.
Effect of TPP on the lacZ transcript as evaluated by qRT-PCR analysis
| E. coli sample | TPP (μM)a | Mean ± SEMb |
Fold difference (2−ΔΔCT) | |||
|---|---|---|---|---|---|---|
|
CT value |
ΔCT | ΔΔCT | ||||
| lacZ | groEL | |||||
| thi-box-L–lacZ | 100 | 17.17 ± 0.07 | 19.44 ± 0.09 | −2.27 ± 0.09 | −4.03 ± 0.29 | 13.36–19.97 |
| 0 | 14.0 ± 0.19 | 20.30 ± 0.29 | −6.30 ± 0.29 | |||
| thi-box-Lm–lacZ | 100 | 16.61 ± 0.10 | 21.63 ± 0.53 | −5.03 ± 0.10 | −0.13 ± 0.13 | 1.0–1.20 |
| 0 | 16.48 ± 0.10 | 21.64 ± 0.04 | −5.16 ± 0.13 | |||
| thiC–lacZ | 100 | 16.66 ± 0.16 | 21.97 ± 0.31 | −5.31 ± 0.31 | −3.68 ± 0.31 | 10.34–15.89 |
| 0 | 12.83 ± 0.09 | 21.82 ± 0.31 | −8.99 ± 0.31 | |||
| Promoterless lacZ | 100 | 28.09 ± 1.58 | 19.28 ± 0.10 | 8.80 ± 1.58 | ||
| 0 | 26.77 ± 0.14 | 19.45 ± 0.13 | 7.33 ± 0.14 | |||
TPP added to the M9 medium for E. coli.
CT, threshold cycle. ΔΔCT = ΔCT without TPP − ΔCT with TPP.
Expression of the tbpABCTd operon is repressed by high concentrations of TPP.
To determine the effect of TPP on the expression of the tbpABCTd operon, T. denticola was cultured in the TPP assay (TA) medium with three different concentrations of TPP (100 nM, 10 μM, and 50 μM) as described in Materials and Methods. Among these three dosages, 10 μM TPP is similar to that in normal OBGM medium (55), 100 nM represents a reduced TPP level, and 50 μM represents an overabundance of TPP. The expression levels of tbpATd under these three different conditions were measured by qRT-PCR. Compared to that with the reduced TPP level, the amount of tbpATd transcript was decreased approximately 16-fold (ranging from 13- to 20-fold) in the presence of 10 μM TPP and 24-fold (ranging from 21- to 27-fold) in the presence of 50 μM TPP (Table 3). To further confirm the qRT-PCR results, Western blotting using a TbpATd antiserum was conducted. Compared to that with the reduced TPP level, the level of TbpATd was decreased 3.8-fold (10 μM) and 9.8-fold (50 μM), respectively (Fig. 6). The observed pattern is similar to that in the qRT-PCR analysis. Collectively, these results further confirm that Tdthi-box is a TPP-sensing riboswitch that negatively controls the expression level of the tbpABCTd operon at the transcriptional level.
Table 3.
Effect of TPP on the tbpATd transcript as evaluated by qRT-PCR analysis
| TPP (μM)a | Mean ± SEM |
Fold difference (2−ΔΔCT) | |||
|---|---|---|---|---|---|
|
CT value |
ΔCT | ΔΔCT | |||
| tbpATd | dnaK | ||||
| 50 | 25.00 ± 0.18 | 21.76 ± 0.08 | 3.20 ± 0.18 | −4.55 ± 0.19 | 20.53–26.72 |
| − | 21.58 ± 0.16 | 22.93 ± 0.19 | −1.35 ± 0.19 | ||
| 10 | 22.35 ± 0.17 | 19.69 ± 0.28 | 2.66 ± 0.28 | −4.01 ± 0.28 | 13.27–19.56 |
| − | 21.58 ± 0.16 | 22.93 ± 0.19 | −1.35 ± 0.19 | ||
−, reduced level of TPP (100 nM) in the OBGM medium for T. denticola.
Fig. 6.
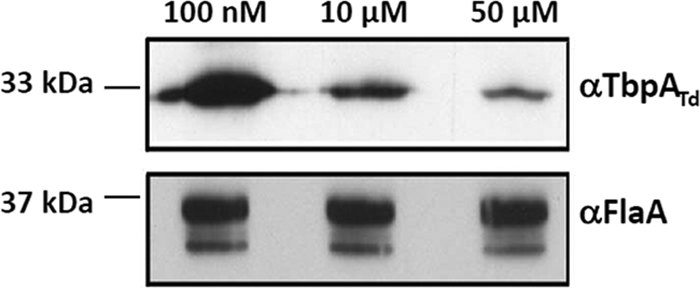
The TbpATd level is repressed by high concentrations of TPP. Wild-type T. denticola was cultivated in TA medium containing different concentrations of TPP (100 nM, 10 μM, and 50 μM), and the same amount of whole-cell lysates was analyzed by SDS-PAGE. Polyclonal antibodies against TbpATd or FlaA, a flagellin protein of T. denticola, were used as probes. FlaA was used as an internal control.
Isolation and characterization of the T. denticola ΔtbpA mutant.
To further study the function of the tbpABCTd operon, the TDE0143 gene was inactivated by targeted mutagenesis as illustrated in Fig. 1. Approximate 80 erythromycin-resistant (Ermr) colonies appeared 10 days after the plating, and these colonies were first screened by PCR with primers specific to Ermr. A total of 20 positive colonies were detected. One clone was further analyzed by PCR with different pairs of primers at the flanking region of TDE0143. The results showed that the target gene was deleted and replaced with the Ermr cassette as expected (data not shown). Western blotting with a specific antibody to TbpATd further showed that the cognate gene product was inactivated in the mutant (Fig. 7a). The obtained mutant was referred to as T. denticola ΔtbpA. Since the tbpATd operon is polycistronic, the insertion of the Ermr cassette in TDE0143 may alter the expression of downstream genes and thus complicate subsequent interpretations. However, the RT-PCR analysis showed that the insertion of Ermr did not have any impact on the expression of downstream genes TDE0144 and TDE0145 (Fig. 7b), which could rule out the existence of a polar effect in the T. denticola ΔtbpA mutant.
Fig. 7.
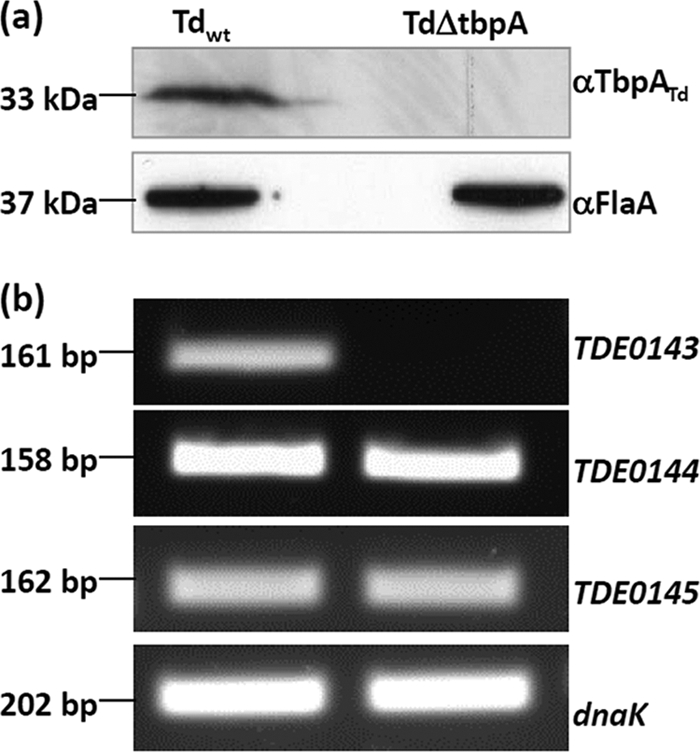
Characterization of the T. denticola ΔtbpA mutant. (a) Western blotting of the T. denticola ΔtbpA mutant. The same amounts of wild-type T. denticola and T. denticola ΔtbpA whole lysates were analyzed by SDS-PAGE. The immunoblotting was conducted as described for Fig. 5b. (b) RT-PCR analysis of the T. denticola ΔtbpA mutant. The transcripts of TDE0144 and TDE0145, the downstream genes of tbpATd, in wild-type T. denticola and T. denticola ΔtbpA were detected by RT-PCR.
TbpATd is required for the growth of T. denticola when exogenous TPP is limited.
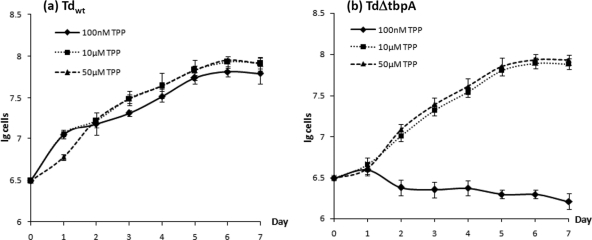
T. denticola needs exogenous TPP to grow (16), and TbpATd is a TPP-binding protein. Thus, we hypothesize that the inactivation of tbpATd may influence the transport of exogenous TPP and the growth of the mutant. To test this hypothesis, the wild-type strain and the T. denticola ΔtbpA mutant were cultivated in TA medium with three different concentrations of TPP (100 nM, 10 μM, and 50 μM), and their growth curves were measured. As shown in Fig. 8, at the concentrations of 10 μM and 50 μM, the growth patterns of wild-type T. denticola and the T. denticola ΔtbpA mutant were almost identical. However, at the low TPP concentration (100 nM), the mutant failed to grow. In contrast, the wild-type strain had only a slightly decreased growth rate compared to those under the other two culture conditions (10 μM and 50 μM). These results demonstrate that TbpATd is required for the growth of T. denticola when the exogenous TPP is limited.
Fig. 8.
Growth curves of wild-type T. denticola (a) and T. denticola ΔtbpA (b). The two strains were cultivated in TA medium containing three different concentrations of TPP (100 nM, 10 μM, and 50 μM). Cell counting was repeated in duplicate with at least three independent samples, and the results are expressed as mean ± SEM.
Inactivation of tbpATd represses the transport of exogenous TPP.
TbpA is a TPP-binding protein (73, 80). To determine whether TbpATd is involved in the transport of exogenous TPP, the intracellular level of TPP was measured by HPLC. As shown in Table 4, at the high TPP concentration (50 μM), the intracellular level of TPP in wild-type T. denticola (258.39 nM) was slightly higher than that in the mutant (230.34 nM). However, when the exogenous TPP was decreased (10 μM), the intracellular TPP level in the mutant (94.49 nM) was about 50% less than that in the wild-type strain (184.85 nM). These results show that the absence of TbpATd significantly reduces the transport of exogenous TPP, suggesting that TbpATd is required for the transport of TPP, in particular when the exogenous TPP is limited.
Table 4.
Comparison of intracellular TPP levels in wild-type T. denticola and the ΔtbpA mutanta
| TPP concn in TA medium | Mean intracellular TPP concn (nM) ± SEM in: |
|
|---|---|---|
| Wild-type T. denticola | ΔtbpA mutant | |
| 100 nM | 94.43 ± 8.96 | |
| 10 μM | 184.85 ± 4.52 | 94.49 ± 6.97 |
| 50 μM | 258.39 ± 7.21 | 230.34 ± 4.3 |
The two strains were cultivated in TA medium containing different concentrations of TPP (100 nM, 10 μM, and 50 μM). Three independent assays were conducted, and the intracellular concentrations of TPP were determined. The ΔtbpA mutant failed to grow at the low concentration of TPP (100 nM).
DISCUSSION
T. denticola lacks a de novo TPP biosynthesis pathway.
The de novo thiamine biosynthesis (TBS) pathway has been well studied in E. coli and S. Typhimurium, where it has been a model to understand TBS in other organisms (2, 34, 36). The synthesis of thiamine involves the separate formation of a thiazole moiety (THZ-P) and a pyrimidine moiety (HMP-P). HMP-P is typically synthesized by ThiC from an aminoimidazole ribotide that is derived from the purine biosynthesis pathway (10, 40). THZ-P is produced from pyruvate, tyrosine, and cysteine through a series of reactions that are catalyzed by several enzymes, such as ThiF, ThiS, ThiG, and ThiH (2, 34). The moieties of HMP-P and THZ-P are further condensed to TMP by ThiE (57, 78). However, all of these key enzymes are absent in the genome of T. denticola (70), suggesting that it lacks a de novo TBS pathway. Interestingly, a homolog of ThiD (TDE0693) is present in the genome of T. denticola. ThiD, a phosphomethyl pyrimidine kinase, converts HMP to HMP-P and then to HMP-PP (11). The function of this homolog is not clear. Given the absence of other key enzymes, it seems unlikely that TDE0693 is involved in de novo thiamine biosynthesis.
T. denticola lacks thiamine salvage biosynthesis pathways.
In addition to the de novo biosynthesis, a set of salvage pathways that utilize intermediate products or thiamine to synthesize TPP has been identified in different bacterial species (2, 34). In the enteric bacteria, thiazole alcohol can be converted to THZ-P by ThiM, a thiazole kinase. In Bacillus subtilis, HMP can be synthesized from formylaminopyrimidine, which is directly transported from the environment by the ThiXYZ transporter (32). However, genome mining analysis did not find any homologs of these salvage pathways in T. denticola. In addition, TPP can be directly derived from thiamine (49, 81). In the enteric bacteria, thiamine can be converted to TMP by ThiK (thiamine kinase) and then to TPP by ThiL. In B. subtilis, thiamine is directly converted to TPP by YloS, a thiamine pyrophosphokinase. However, none of these homologs were identified in the genome of T. denticola. To further determine whether T. denticola is able to directly use thiamine, the T. denticola ΔtbpA mutant was cultivated in TA medium containing different concentrations of thiamine (10 μM, 50 μM, and 100 μM). It was found that the mutant failed to grow in the presence of thiamine, suggesting that T. denticola is unable to produce TPP via the salvage pathway of metabolizing thiamine. Collectively, the above-described bioinformatic and genetic analyses have demonstrated that T. denticola lacks an endogenous TBS pathway, meaning that it has to utilize exogenous TPP.
Is TbpATd the sole TPP transporter in T. denticola?
Interestingly, when the exogenous TPP is sufficient, the T. denticola ΔtbpA mutant is still able to take up TPP. For example, in the presence of 50 μM TPP, the intracellular concentration of TPP in the mutant (230 nM) is only slightly lower than that in the wild type (258 nM), suggesting that the exogenous TPP is still able to enter into the mutant cells via certain mechanisms. A similar phenotype was observed in S. Typhimurium; a thiP thiH double mutant, in which both the de novo TBS pathway and the TPP transporter were blocked, failed to grow in a low concentration of TPP (100 nM) (80). The possible mechanism could be either that there is an additional unidentified TPP transporter that is independent of ThiBPQ or that the remaining part of the TPP ABC transporter (a permease and an ATPase) is still able to transport TPP. Several lines of evidence suggest that the first possibility is unlikely. First, the thiamine-binding proteins are well conserved. Besides ThiB, no additional homolog has yet been identified in the enteric bacteria or T. denticola. Second, T. denticola lacks a ThiBPQ-independent thiamine transporter (e.g., the ThiXYZ transporter of B. subtilis) (32, 62). Finally, both T. denticola ΔtbpA and the thiP thiH double mutant fail to grow when the exogenous TPP is limited (80), suggesting that either no additional TPP transporter exists or if it exists, the transport efficiency of exogenous TPP is quite limited. As such, the second mechanism proposed above is more compelling. As a thiamine-binding protein, TbpATd is probably able to concentrate TPP from the environment and facilitate the transport of TPP. In the absence of TbpATd, the remaining transporter (a permease and an ATPase) is still able to uptake exogenous TPP, but its transport efficiency is significantly decreased. Therefore, when the exogenous TPP is limited, the T. denticola ΔtbpA mutant is incapable of ensuring the minimal requirement of TPP, and thus the mutant fails to grow. This proposition was reinforced by our inability to inactivate either the TDE0144 (encoding a permease) or the TDE0145 (encoding an ATPase) gene.
Tdthi-box primarily regulates the tbpABCTd operon at the transcriptional level.
The TPP-sensing riboswitches thiC and thiM are composed of five helices, three junction bulges, and two terminal loops (69, 82). In the presence of TPP, two of the junction bulges bind to the pyrimidine ring and the pyrophosphate of TPP, forming a stable aptamer (19, 50, 69, 76). The thi-box of thiM incorporates the S/D sequence. It represses gene expression at the translational level by sequestering the S/D sequence and preventing translation (54, 82). Similar to the case for thiM, the predicted Tdthi-box incorporates the S/D sequence (Fig. 4). Therefore, it may regulate gene expression at the translational level, like its counterpart thiM. However, the qRT-PCR analysis showed that the addition of TPP significantly decreased the levels of lacZ and tbpATd transcripts (Tables 2 and 3). Such attenuations are proportional to the reduction of the β-galactosidase activity and TbpATd protein, suggesting that the action of Tdthi-box occurs primarily at the transcriptional level. Tdthi-box partially overlaps the identified PtbpA promoter (Fig. 4; see Fig. S2 in the supplemental material). It is most likely that in the presence of TPP, Tdthi-box form a stable aptamer that acts as a transcription terminator and blocks transcription. As the S/D sequence is sequestered by the aptamer of Tdthi-box, it is also possible that the translational regulation makes some contribution to the function of Tdthi-box. At this point, the existence of this possibility cannot be completely ruled out.
The regulatory mechanism of TPP transport in T. denticola.
Based on the results and the above discussions, a working model to illustrate the regulatory mechanism of TPP transport in T. denticola is proposed (Fig. 9). T. denticola lacks a de novo TBS pathway, and it depends on a TPP ABC transporter to take up exogenous TPP. The genes that encode this transport system form an operon (tbpABCTd) that is regulated by a TPP-sensing riboswitch (Tdthi-box). When the exogenous TPP is limited and the intracellular TPP is insufficient, Tdthi-box folds into a structure that allows the expression of the tbpABCTd operon. As such, more TPP transporter will be synthesized to transport exogenous TPP into the cells. When the intracellular TPP becomes sufficient, the free TPP binds to the Tdthi-box and forms a stable aptamer, which represses the transcription of the tbpABCTd operon and limits the uptake of exogenous TPP. This feedback inhibition mechanism can allow the spirochete to maintain a constant intracellular level of TPP to meet its growth requirements in response to the varied environments in the oral flora.
Fig. 9.
Schematic diagram showing the regulatory mechanism of TPP transport in T. denticola. T. denticola lacks a de novo TBS pathway, and it depends on the TPP transporter to take up exogenous TPP. The tbpABCTd operon encodes a TPP ABC transporter, which is regulated by a TPP-sensing riboswitch (Tdthi-box). When the intracellular TPP is insufficient, Tdthi-box folds into a structure that allows the transcription of the tbpABCTd operon, and more TPP transporter will be synthesized to take up exogenous TPP into the cells. When the intracellular TPP becomes sufficient, the free TPP binds to Tdthi-box and forms a stable aptamer, which in turn represses the transcription of the tbpABCTd operon and limits the uptake of exogenous TPP.
Thiamine metabolism in spirochetes.
TPP is an essential cofactor for several important enzymes of carbohydrate metabolism (2). For example, TPP is a coenzyme of pyruvate dehydrogenase that converts pyruvate to acetyl coenzyme A (acetyl-CoA). Acetyl-CoA can be further metabolized to produce ATP via the tricarboxylic acid (TCA) cycle. In addition, thiamine biosynthesis is linked to other central metabolism pathways (e.g., purine biosynthesis) (36). Thus, thiamine biosynthesis has been used in systems biology to dissect metabolic integration in microorganisms. Spirochetes are a group of medically important but poorly understood bacteria (1, 9). Most spirochetes have fastidious growth requirements. To begin to understand thiamine metabolism in spirochetes, we searched the genomes of sequenced spirochetes, including Borrelia spp., Brachyspira spp., Leptospira spp., and Treponema spp. It was found that Brachyspira spp. (B. hyodysenteriae and B. pilosicoli) and Leptospira spp. (L. biflexa, L. borgetersenii, and L. interrogans) contain a de novo TBS pathway (3, 6, 58, 61, 79). Along with T. denticola, T. pallidum and T. vincentii also lack a de novo TBS pathway (25). Similarly to T. denticola, they all contain a conserved TPP ABC transporter. For example, the homolog of TbpA in T. pallidum (TP0144) has 39% identity and 59% similarity to TbpATd. Interestingly, neither the de novo TBS pathway nor the TPP ABC transporter has been found in any Borrelia sp. (24).
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Limberger for providing T. denticola strains, R. Breaker for providing the pRS414 plasmid, and C. Fenno for providing the anti-FlaA serum.
This research was supported by Public Health Service grants DE019667 and AI078958 to C. Li.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 27 May 2011.
REFERENCES
- 1. Baranton G., Old I. G. 1995. The spirochetes: a different way of life. Bull. Inst. Pasteur 93:63–95 [Google Scholar]
- 2. Begley T. P., et al. 1999. Thiamin biosynthesis in prokaryotes. Arch. Microbiol. 171:293–300 [DOI] [PubMed] [Google Scholar]
- 3. Bellgard M. I., et al. 2009. Genome sequence of the pathogenic intestinal spirochete Brachyspira hyodysenteriae reveals adaptations to its lifestyle in the porcine large intestine. PLoS One 4:e4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blakemore R. P., Canale-Parola E. 1976. Arginine catabolism by Treponema denticola. J. Bacteriol. 128:616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breaker R. R. 2008. Complex riboswitches. Science 319:1795–1797 [DOI] [PubMed] [Google Scholar]
- 6. Bulach D. M., et al. 2006. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. U. S. A. 103:14560–14565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Capone R. F., Ning Y., Pakulis N., Alhazzazi T., Fenno J. C. 2007. Characterization of Treponema denticola pyrF encoding orotidine-5′-monophosphate decarboxylase. FEMS Microbiol. Lett. 268:261–267 [DOI] [PubMed] [Google Scholar]
- 8. Chan E. C., McLaughlin R. 2000. Taxonomy and virulence of oral spirochetes. Oral Microbiol. Immunol. 15:1–9 [DOI] [PubMed] [Google Scholar]
- 9. Charon N. W., Goldstein S. F. 2002. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu. Rev. Genet. 36:47–73 [DOI] [PubMed] [Google Scholar]
- 10. Chatterjee A., et al. 2008. Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nat. Chem. Biol. 4:758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng G., Bennett E. M., Begley T. P., Ealick S. E. 2002. Crystal structure of 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase from Salmonella typhimurium at 2.3 A resolution. Structure 10:225–235 [DOI] [PubMed] [Google Scholar]
- 12. Clark D. T., Soory M. 2006. The metabolism of cholesterol and certain hormonal steroids by Treponema denticola. Steroids 71:352–363 [DOI] [PubMed] [Google Scholar]
- 13. Darveau R. P. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8:481–490 [DOI] [PubMed] [Google Scholar]
- 14. Dashper S. G., Seers C. A., Tan K. H., Reynolds E. C. 2010. Virulence factors of the oral spirochete Treponema denticola. J. Dent. Res. doi: 0022034510385242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davidson A. L., Chen J. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73:241–268 [DOI] [PubMed] [Google Scholar]
- 16. De Ciccio A., McLaughlin R., Chan E. C. 1999. Factors affecting the formation of spherical bodies in the spirochete Treponema denticola. Oral Microbiol. Immunol. 14:384–386 [DOI] [PubMed] [Google Scholar]
- 17. Dewhirst F. E., et al. 2000. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol. Immunol. 15:196–202 [DOI] [PubMed] [Google Scholar]
- 18. Downs D. M. 2006. Understanding microbial metabolism. Annu. Rev. Microbiol. 60:533–559 [DOI] [PubMed] [Google Scholar]
- 19. Edwards T. E., Ferre-D'Amare A. R. 2006. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure 14:1459–1468 [DOI] [PubMed] [Google Scholar]
- 20. Ellen R. P., Galimanas V. B. 2005. Spirochetes at the forefront of periodontal infections. Periodontology 2000 38:13–32 [DOI] [PubMed] [Google Scholar]
- 21. Fenno J. C., McBride B. C. 1998. Virulence factors of oral treponemes. Anaerobe 4:1–17 [DOI] [PubMed] [Google Scholar]
- 22. Fenno J. C., et al. 2000. Identification of a Treponema denticola OppA homologue that binds host proteins present in the subgingival environment. Infect. Immun. 68:1884–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Finn R. D., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fraser C. M., et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586 [DOI] [PubMed] [Google Scholar]
- 25. Fraser C. M., et al. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375–388 [DOI] [PubMed] [Google Scholar]
- 26. Gaal T., et al. 2001. Promoter recognition and discrimination by EsigmaS RNA polymerase. Mol. Microbiol. 42:939–954 [DOI] [PubMed] [Google Scholar]
- 27. Gardner P. P., et al. 2009. Rfam: updates to the RNA families database. Nucleic Acids Res. 37:D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haffajee A. D., Socransky S. S. 2005. Microbiology of periodontal diseases: introduction. Periodontology 2000 38:9–12 [DOI] [PubMed] [Google Scholar]
- 29. Hespell R. B., Canale-Parola E. 1971. Amino acid and glucose fermentation by Treponema denticola. Arch. Mikrobiol. 78:234–251 [DOI] [PubMed] [Google Scholar]
- 30. Holt S. C., Ebersole J. L. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex,” a prototype polybacterial pathogenic consortium in periodontitis. Periodontology 2000 38:72–122 [DOI] [PubMed] [Google Scholar]
- 31. Jackson-Rosario S., Self W. T. 2009. Inhibition of selenium metabolism in the oral pathogen Treponema denticola. J. Bacteriol. 191:4035–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jenkins A. H., Schyns G., Potot S., Sun G., Begley T. P. 2007. A new thiamin salvage pathway. Nat. Chem. Biol. 3:492–497 [DOI] [PubMed] [Google Scholar]
- 33. Jones P. M., George A. M. 2004. The ABC transporter structure and mechanism: perspectives on recent research. Cell Mol. Life Sci. 61:682–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jurgenson C. T., Begley T. P., Ealick S. E. 2009. The structural and biochemical foundations of thiamin biosynthesis. Annu. Rev. Biochem. 78:569–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kent C., Gee P., Lee S. Y., Bian X., Fenno J. C. 2004. A CDP-choline pathway for phosphatidylcholine biosynthesis in Treponema denticola. Mol. Microbiol. 51:471–481 [DOI] [PubMed] [Google Scholar]
- 36. Koenigsknecht M. J., Downs D. M. 2010. Thiamine biosynthesis can be used to dissect metabolic integration. Trends Microbiol. 18:240–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Korner R. W., Vierzig A., Roth B., Muller C. 2009. Determination of thiamin diphosphate in whole blood samples by high-performance liquid chromatography—a method suitable for pediatric diagnostics. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 877:1882–1886 [DOI] [PubMed] [Google Scholar]
- 38. Kulshina N., Edwards T. E., Ferre-D'Amare A. R. 2010. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA 16:186–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuramitsu H. K. 2003. Molecular genetic analysis of the virulence of oral bacterial pathogens: an historical perspective. Crit. Rev. Oral Biol. Med. 14:331–344 [DOI] [PubMed] [Google Scholar]
- 40. Lawhorn B. G., Mehl R. A., Begley T. P. 2004. Biosynthesis of the thiamin pyrimidine: the reconstitution of a remarkable rearrangement reaction. Org. Biomol. Chem. 2:2538–2546 [DOI] [PubMed] [Google Scholar]
- 41. Lepp P. W., et al. 2004. Methanogenic archaea and human periodontal disease. Proc. Natl. Acad. Sci. U. S. A. 101:6176–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li C., Sal M., Marko M., Charon N. W. 2010. Differential regulation of the multiple flagellins in spirochetes. J. Bacteriol. 192:2596–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li C., Xu H., Zhang K., Liang F. T. 2010. Inactivation of a putative flagellar motor switch protein FliG1 prevents Borrelia burgdorferi from swimming in highly viscous media and blocks its infectivity. Mol. Microbiol. 75:1563–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li H., Ruby J., Charon N., Kuramitsu H. 1996. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J. Bacteriol. 178:3664–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Limberger R. J., Slivienski L. L., Izard J., Samsonoff W. A. 1999. Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. J. Bacteriol. 181:3743–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Loesche W. J. 1988. The role of spirochetes in periodontal disease. Adv. Dent. Res. 2:275–283 [DOI] [PubMed] [Google Scholar]
- 47. Lu J., Frank E. L. 2008. Rapid HPLC measurement of thiamine and its phosphate esters in whole blood. Clin. Chem. 54:901–906 [DOI] [PubMed] [Google Scholar]
- 48. Marchler-Bauer A., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Melnick J., et al. 2004. Identification of the two missing bacterial genes involved in thiamine salvage: thiamine pyrophosphokinase and thiamine kinase. J. Bacteriol. 186:3660–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miranda-Rios J. 2007. The THI-box riboswitch, or how RNA binds thiamin pyrophosphate. Structure 15:259–265 [DOI] [PubMed] [Google Scholar]
- 51. Miranda-Rios J., Navarro M., Soberon M. 2001. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc. Natl. Acad. Sci. U. S. A. 98:9736–9741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mironov A. S., et al. 2002. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111:747–756 [DOI] [PubMed] [Google Scholar]
- 53. Nudler E., Mironov A. S. 2004. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 29:11–17 [DOI] [PubMed] [Google Scholar]
- 54. Ontiveros-Palacios N., et al. 2008. Molecular basis of gene regulation by the THI-box riboswitch. Mol. Microbiol. 67:793–803 [DOI] [PubMed] [Google Scholar]
- 55. Orth R., O'Brien-Simpson N., Dashper S., Walsh K., Reynolds E. 2010. An efficient method for enumerating oral spirochetes using flow cytometry. J. Microbiol. Methods 80:123–128 [DOI] [PubMed] [Google Scholar]
- 56. Paster B. J., et al. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Peapus D. H., et al. 2001. Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase. Biochemistry 40:10103–10114 [DOI] [PubMed] [Google Scholar]
- 58. Picardeau M., et al. 2008. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One 3:e1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pihlstrom B. L., Michalowicz B. S., Johnson N. W. 2005. Periodontal diseases. Lancet 366:1809–1820 [DOI] [PubMed] [Google Scholar]
- 60. Regulski E. E., et al. 2008. A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Mol. Microbiol. 68:918–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ren S. X., et al. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888–893 [DOI] [PubMed] [Google Scholar]
- 62. Rodionov D. A., Vitreschak A. G., Mironov A. A., Gelfand M. S. 2002. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J. Biol. Chem. 277:48949–48959 [DOI] [PubMed] [Google Scholar]
- 63. Rother M., Bock A., Wyss C. 2001. Selenium-dependent growth of Treponema denticola: evidence for a clostridial-type glycine reductase. Arch. Microbiol. 177:113–116 [DOI] [PubMed] [Google Scholar]
- 64. Ruby J. D., et al. 1997. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J. Bacteriol. 179:1628–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sal M. S., et al. 2008. Borrelia burgdorferi uniquely regulates its motility genes and has an intricate flagellar hook-basal body structure. J. Bacteriol. 190:1912–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sali A., Blundell T. L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815 [DOI] [PubMed] [Google Scholar]
- 67. Sela M. N. 2001. Role of Treponema denticola in periodontal diseases. Crit. Rev. Oral Biol. Med. 12:399–413 [DOI] [PubMed] [Google Scholar]
- 68. Serganov A., Patel D. J. 2007. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 8:776–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Serganov A., Polonskaia A., Phan A. T., Breaker R. R., Patel D. J. 2006. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature 441:1167–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Seshadri R., et al. 2004. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc. Natl. Acad. Sci. U. S. A. 101:5646–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Simons R. W., Houman F., Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96 [DOI] [PubMed] [Google Scholar]
- 72. Socransky S. S., Haffajee A. D. 2005. Periodontal microbial ecology. Periodontology 2000 38:135–187 [DOI] [PubMed] [Google Scholar]
- 73. Soriano E. V., et al. 2008. Structural similarities between thiamin-binding protein and thiaminase-I suggest a common ancestor. Biochemistry 47:1346–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sze C. W., Li C. 2011. Inactivation of bb0184, which encodes carbon storage regulator A, represses the infectivity of Borrelia burgdorferi. Infect. Immun. 79:1270–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tatakis D. N., Kumar P. S. 2005. Etiology and pathogenesis of periodontal diseases. Dent. Clin. North Am. 49:491–516 [DOI] [PubMed] [Google Scholar]
- 76. Thore S., Leibundgut M., Ban N. 2006. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science 312:1208–1211 [DOI] [PubMed] [Google Scholar]
- 77. Typas A., Stella S., Johnson R. C., Hengge R. 2007. The −35 sequence location and the Fis-sigma factor interface determine sigmas selectivity of the proP (P2) promoter in Escherichia coli. Mol. Microbiol. 63:780–796 [DOI] [PubMed] [Google Scholar]
- 78. Vander Horn P. B., Backstrom A. D., Stewart V., Begley T. P. 1993. Structural genes for thiamine biosynthetic enzymes (thiCEFGH) in Escherichia coli K-12. J. Bacteriol. 175:982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wanchanthuek P., et al. 2010. The complete genome sequence of the pathogenic intestinal spirochete Brachyspira pilosicoli and comparison with other Brachyspira genomes. PLoS One 5:e11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Webb E., Claas K., Downs D. 1998. thiBPQ encodes an ABC transporter required for transport of thiamine and thiamine pyrophosphate in Salmonella typhimurium. J. Biol. Chem. 273:8946–8950 [DOI] [PubMed] [Google Scholar]
- 81. Webb E., Downs D. 1997. Characterization of thiL, encoding thiamin-monophosphate kinase, in Salmonella typhimurium. J. Biol. Chem. 272:15702–15707 [DOI] [PubMed] [Google Scholar]
- 82. Winkler W., Nahvi A., Breaker R. R. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419:952–956 [DOI] [PubMed] [Google Scholar]
- 83. Winkler W. C., Breaker R. R. 2005. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59:487–517 [DOI] [PubMed] [Google Scholar]
- 84. Yang Y., Li C. 2009. Transcription and genetic analyses of a putative N-acetylmuramyl-l-alanine amidase in Borrelia burgdorferi. FEMS Microbiol. Lett. 290:164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yang Y., Stewart P. E., Shi X., Li C. 2008. Development of a transposon mutagenesis system in the oral spirochete Treponema denticola. Appl. Environ. Microbiol. 74:6461–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.