Abstract
The stator proteins PomA and PomB form a complex that couples Na+ influx to torque generation in the polar flagellar motor of Vibrio alginolyticus. This stator complex is anchored to an appropriate place around the rotor through a putative peptidoglycan-binding (PGB) domain in the periplasmic region of PomB (PomBC). To investigate the function of PomBC, a series of N-terminally-truncated and in-frame mutants with deletions between the transmembrane (TM) segment and the PGB domain of PomB was constructed. A PomBC fragment consisting of residues 135 to 315 (PomBC5) formed a stable homodimer and significantly inhibited the motility of wild-type cells when overexpressed in the periplasm. A fragment with an in-frame deletion (PomBΔL) of up to 80 residues retained function, and its overexpression with PomA impaired cell growth. This inhibitory effect was suppressed by a mutation at the functionally critical Asp (D24N) in the TM segment of PomB, suggesting that a high level of Na+ influx through the mutant stator causes the growth impairment. The overproduction of functional PomA/PomBΔL stators also reduced the motile fractions of the cells. That effect could be slightly relieved by a mutation (L168P) in the putative N-terminal α-helix that connects to the PGB domain without affecting the growth inhibition, suggesting that a conformational change of the region including the PGB domain affects stator assembly. Our results reveal common features of the periplasmic region of PomB/MotB and demonstrate that a flexible linker that contains a “plug” segment is important for the control of Na+ influx through the stator complex as well as for stator assembly.
INTRODUCTION
The flagellum is used by most bacteria to swim in a liquid environment and to swarm on a surface. It consists of a flagellar filament, a hook, and a basal body surrounded by multiple stator complexes (3, 22). The filament is a helical structure that serves as a screw propeller to change the rotary motion of the motor into thrust. The hook is a short tubular structure that serves as a universal joint to smoothly transmit the torque produced by the motor to the filament. The basal body of Gram-negative bacteria is a supramolecular complex composed of the rod and the MS, P, L, and C rings (23). The fuel for motor rotation is the transmembrane (TM) gradient of ions, H+ or Na+, which go through the stator complex and are coupled to the torque generation to rotate the flagellum (37). However, the mechanism by which the potential electrochemical energy is converted into mechanical work is still not well understood.
Genetic, biochemical, and biophysical studies to characterize the mechanism of rotation of the flagellar motor in Escherichia coli and Salmonella enterica serovar Typhimurium have been actively carried out. In those motors, only 5 proteins, MotA, MotB, FliG, FliM, and FliN, are in charge of the torque generation of the motor (4). The soluble rotor proteins FliG, FliM, and FliN, which form the C-ring structure in the rotor (9, 39), are also responsible for switching the direction of motor rotation (43, 44). The two membrane proteins, MotA and MotB, which form the stator complex in an A4B2 stoichiometry, serve as a proton channel, and they couple proton flux to torque generation (6, 7, 17, 18, 34). In the fully functioning motor, about a dozen stator complexes are assembled around the rotor to generate torque (31).
The marine bacterium Vibrio alginolyticus has a Na+-driven single-polar flagellum, and its motor requires PomA, PomB, MotX, and MotY for torque generation (1, 27, 30). The stator proteins PomA and PomB are homologs of MotA and MotB, respectively, and form the PomA4PomB2 complex that serves as a Na+ channel (32, 33, 45). PomB contains 315 amino acids and has a predicted single TM segment (residues 15 to 39) at its N terminus and a large C-terminal periplasmic region (Fig. 1). Asp24 in the TM segment (Asp32 in MotB of E. coli) is absolutely conserved in all PomB/MotB orthologs and is thought to be the Na+ binding site that is critical for motor function (38). Recently, Na+ binding to the Asp24 residue was directly shown by attenuated total reflectance-Fourier transform infrared (ATR-FTIR) spectroscopy, providing the first evidence for the interaction between the stator and its coupling ion (35). The point mutations D24N and D24C do not confer any motility and reduce the polar localization of the PomA/PomB stator complex, suggesting that Na+ binding at this critical aspartate residue is required for incorporation of the stator into the motor (10). Our further analyses revealed that PomA/PomB stator assembly is a dynamic and reversible process that depends on the Na+ flux through the motor (10) and also requires the T ring structure of the basal body, which is composed of MotX and MotY (36). Thus, the assembly mechanism of the stator in Vibrio should be considered in these contexts, but our knowledge is still limited. The membrane localizations of the PomA/PomB complex and of PomB alone were changed from the inner membrane to the intermediate fraction between the inner and outer membrane when coexpressed with MotX, suggesting an interaction between MotX and PomB (28).
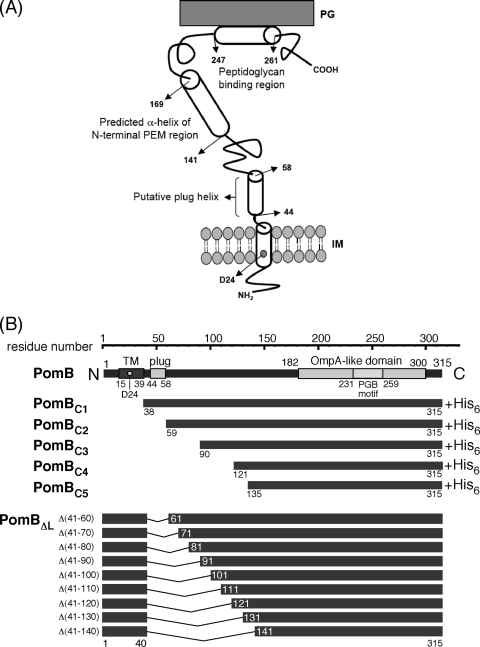
Fig. 1.
The PomB topology model and constructs used in this study. (A) The putative topology of PomB. PomB contains 315 amino acids and has a single TM segment (residues 15 to 39) and a plug segment (residues 44 to 58, speculated to prevent premature Na+ translocation across the cell membrane) at its N terminus. D24 is regarded as the critical Na+ binding site. Amino acids 143 to 169 are predicted to form a putative N-terminal helix of the PEM region. The large periplasmic region includes an OmpA-like domain (residues 182 to 300) with a PGB motif (residues 231 to 259). The putative helix (residues 247 to 261) that may form the dimer interface in the PGB domain is also indicated. (B) Primary structure of PomB and schematic representation of N-terminally-truncated PomB fragments (PomBC) and in-frame deletions (PomBΔL). The in-frame-deletion derivatives constructed in this study lack the putative plug segment and part of the periplasmic linker region. IM, inner membrane.
When assembled, the stator is anchored at the peptidoglycan (PG) layer near the rotor in the periplasmic region of the B subunit (PomB or MotB) (42). Residues 182 to 300 in the periplasmic region of PomB show sequence similarity to the OmpA-like proteins and include a putative peptidoglycan-binding (PGB) motif, which is well conserved among proteins like OmpA and Pal (Fig. 1) (1). These kinds of proteins are outer membrane proteins that interact with the PG layer noncovalently (8, 15). The PGB motif of PomB (MotB) is believed to associate with the PG layer to anchor the PomA/PomB (MotA/MotB) complex around the rotor. Many mutations in this region of E. coli or Salmonella MotB completely abolish motility (5, 40). Recently, the crystal structure of the entire periplasmic region essential for motility (called the PEM) of Salmonella MotB was reported, and it appears to be a single-domain structure with a long N-terminal α-helix that protrudes from the domain (20). The core of the domain has a typical OmpA-like structure and shows considerable structural similarity to other PGB domains. Functional analyses based on the structural information revealed that the PGB domains must dimerize in order to form a proton-conducting channel, and drastic conformational changes in the N-terminal portion of the PEM region are required for both PG binding and proton channel activation.
Given the secondary-structure prediction and the multiple-sequence alignment of PomB orthologs, it is plausible that the periplasmic region between residues 41 to 140 of PomB can serve as a tolerant flexible linker that connects the TM segment and the putative PEM region containing the PGB domain. Several in-frame deletions in the corresponding linker region (residues 51 to 110) of E. coli MotB showed that deletion of residues 51 to 60, 51 to 60, 51 to 70, 51 to 80, and 51 to 90 caused growth inhibition but did not impair motility when the deletion mutants were overproduced. This finding indicated that deletion of the “plug” segment (Fig. 1, residues P53 to P66 in Salmonella MotB, P52 to P65 in E. coli MotB), which consists of an amphipathic α-helix, induces a massive H+ influx through the stator channel that inhibits cell growth without affecting torque generation (13, 24). The Salmonella MotB mutant with residues 51 to 100 deleted [MotB(Δ51-100)] did not cause growth impairment and produced a functional and stable stator complex (20, 25). Interestingly, when mutation L119P or L119E in the N-terminal helix of the PEM region was introduced into the MotB(Δ51-100) mutant, growth impairment that did not affect motility was again observed, suggesting that the N-terminal region of PEM regulates the H+ translocation of the MotA/MotB stator complex (20). The function and characteristics of the linker and PEM regions of PomB are still unclear.
In this study, based on the sequence alignment of PomB with its orthologs and the crystal structure of the PEM region of Salmonella MotB, we carried out mutational and biochemical studies on a series of N-terminally-truncated and in-frame-deletion mutants of PomB in V. alginolyticus. We found that the periplasmic PomB fragments form a homodimer and that deletions in the PomB linker region cause growth impairment of the cells. We also investigated several mutations in the PEM region, measuring their effects on motility and growth.
MATERIALS AND METHODS
Bacterial strains, plasmids, and mutagenesis.
Bacterial strains and plasmids used in this study are listed in Table 1. The vector plasmid pTY57 was a kind gift from T. Yorimitsu, and the pomB fliG double-deletion strain NMB200 was a kind gift from N. Nonoyama (S. Kojima, N. Nonoyama, N. takekawa, H. Fukuoka, and M. Homma, submitted for publication). The design of the N-terminally-truncated fragments of V. alginolyticus PomB and site-directed mutagenesis were carried out according to a web-based secondary structure prediction program, PSIPRED (14), and the multiple-sequence alignment of PomB orthologs from various bacteria. Point mutations were generated using the QuikChange site-directed mutagenesis protocol (Stratagene). In-frame deletions of PomB were constructed according to the two-step-PCR method reported previously (41) or the one-step-PCR-based method. In the latter case, we designed a sense primer that anneals to the sequence encoding the ∼7 downstream C-terminal residues at the junction of the deletion and an antisense primer that anneals to the sequence encoding the ∼7 upstream N-terminal residues at the junction. The nicked circular strands with the desired deletions were generated by PCR using these two primers, and the 5′ end was phosphorylated by polynucleotide kinase (Takara) and then ligated to produce the designed in-frame-deletion constructs. The addition of the MotY leader sequence (residues 1 to 22) to the N terminus of each PomB fragment was carried out by a two-step-PCR method using the plasmid pAS101 (21) as a template of motY as described previously (16). Each mutation and plasmid construction was confirmed by DNA sequencing with an ABI Prism 3130 genetic analyzer instrument (Applied Biosystems).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| V. alginolyticus | ||
| VIO5 | Rifr Pof+ Laf− | 30 |
| NMB191 | Rifr Pof+ Laf− ΔpomAB | 46 |
| NMB200 | RIfr Pof+ Laf− ΔpomB ΔfliG | N. Nonoyama |
| E. coli | ||
| BL21(DE3) | Host for overexpression from the T7 promoter | |
| DH5α | Recipient for cloning experiments | |
| Plasmids | ||
| pET19b | T7 expression vector | Novagen |
| pBAD33 | Cmr PBAD | 12 |
| pTY57 | Cmr PBAD with a multicloning site of pBAD24 | T. Yorimitsu |
| pSU41 | Kmr PlaclacZα | 2 |
| pAS101 | pSU41/MotY | 21 |
| pHFAB | pBAD33/PomA+PomB | 11 |
| pTSK32 | pET19b/PomBC1-His6 | This study |
| pTSK33 | pET19b/PomBC2-His6 | This study |
| pTSK34 | pET19b/PomBC3-His6 | This study |
| pTSK35 | pET19b/PomBC4-His6 | This study |
| pTSK36 | pET19b/PomBC5-His6 | This study |
| pLSK5 | pTY57/MotYL::PomBC4-His6a | This study |
| pLSK6 | pTY57/PomBC4-His6 | This study |
| pLSK7 | pTY57/MotYL::PomBC5-His6a | This study |
| pLSK8 | pTY57/PomBC5- His6 | This study |
| pLSK9 | pBAD33/PomA+PomBΔL(Δ41-60) | This study |
| pLSK10 | pBAD33/PomA+PomBΔL(Δ41-70) | This study |
| pLSK11 | pBAD33/PomA+PomBΔL(Δ41-80) | This study |
| pLSK12 | pBAD33/PomA+PomBΔL(Δ41-90) | This study |
| pLSK13 | pBAD33/PomA+PomBΔL(Δ41-100) | This study |
| pLSK14 | pBAD33/PomA+PomBΔL(Δ41-110) | This study |
| pTSK37 | pBAD33/PomA+PomBΔL(Δ41-120) | This study |
| pTSK38 | pBAD33/PomA+PomBΔL(Δ41-130) | This study |
| pTSK39 | pBAD33/PomA+PomBΔL(Δ41-140) | This study |
| pTSK37-D24N | pBAD33/PomA+PomBΔL(Δ41-120, D24N) | This study |
| pTSK37-M153P | pBAD33/PomA+PomBΔL(Δ41-120, M153P) | This study |
| pTSK37-M157P | pBAD33/PomA+PomBΔL(Δ41-120, M157P) | This study |
| pTSK37-L160P | pBAD33/PomA+PomBΔL(Δ41-120, L160P) | This study |
| pTSK37-I164P | pBAD33/PomA+PomBΔL(Δ41-120, I164P) | This study |
| pTSK37-L168P | pBAD33/PomA+PomBΔL(Δ41-120, L168P) | This study |
Each PomB fragment was fused to a MotY leader sequence (MotYL). Rifr, rifampin resistant; Cmr, chloramphenicol resistant; Pof+, normal polar flagellar formation, Laf−, defective in lateral flagellar formation; PBAD, arabinose promoter.
Culture of cells and motility assay.
V. alginolyticus cells were cultured at 30°C in VC medium (0.5% [wt/vol] polypeptone, 0.5% [wt/vol] yeast extract, 0.4% [wt/vol] K2HPO4, 3% [wt/vol] NaCl, 0.2% [wt/vol] glucose) or in VPG500 medium (1% [wt/vol] polypeptone, 0.4% [wt/vol] K2HPO4, 500 mM NaCl, 0.5% [wt/vol] glycerol). E. coli cells were cultured at 37°C in LB broth (1% [wt/vol] Bacto tryptone, 0.5% [wt/vol] NaCl, 0.5% [wt/vol] yeast extract) for DNA manipulations or in SB broth (1.2% [wt/vol] Bacto tryptone, 2.4% [wt/vol] yeast extract, 0.5% [wt/vol] glycerol, 1.25% [wt/vol] K2HPO4, 0.38% [wt/vol] KH2PO4) for protein purification. For the motility assay of Vibrio cells on soft-agar plates, 2 μl of concentrated overnight cultures in VC medium with an optical density at 660 nm (OD660) of 5 were spotted on VPG500 (or VPG for experiment shown in Fig. 4B only) soft-agar plates (0.25% agar plates containing 2.5 μg/ml of chloramphenicol and 0.2% or 0.02% arabinose). Measurement of the motile fraction of Vibrio cells was carried out as follows. Cells were cultured for 4 h at 30°C in VPG500 broth containing appropriate concentrations of arabinose and chloramphenicol. The cultures were then diluted 100-fold with VPG500 broth, and motility was immediately observed and recorded using dark-field microscopy. Using this recorded data, we counted the numbers of motile cells in the observed fields, and the percentage of motile cells among the cells observed is shown as the motile fraction. The recorded data were also used for the swimming-speed measurement analyzed by software for motion analysis (Move-tr/2D; Library Co., Tokyo, Japan).
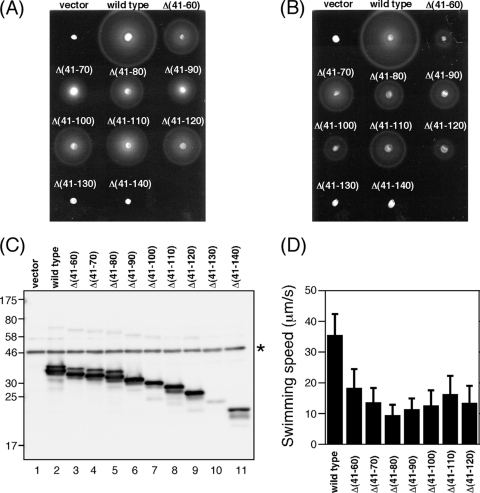
Fig. 4.
Effect of in-frame deletions in PomB on motility. (A and B) Motility of NMB191 (ΔpomAB) cells coexpressing PomA and PomBΔL proteins. NMB191 cells harboring plasmid pHFAB, pLSK9, pLSK10, pLSK11, pLSK12, pLSK13, pLSK14, pTSK37, pTSK38, or pTSK39 were inoculated into VPG semisolid agar containing 0.02% (A) or 0.2% (B) arabinose and were incubated at 30°C for 6 h. For the vector control, we used pBAD33. (C) Immunoblot detection of PomBΔL proteins. NMB191 cells harboring the same plasmids as for panel A or B were grown in VPG broth containing 0.02% arabinose at 30°C for 4 h, after which whole-cell proteins were prepared and then analyzed by immunoblotting with the anti-PomB antibody. The asterisk indicates a nonspecific band detected by the anti-PomB antibody. (D) Swimming speeds of cells expressing PomA/PomBΔL stators. NMB191 cells harboring the plasmids listed above were grown in VPG broth containing 0.02% arabinose at 30°C for 4 h, and then their swimming motilities were observed, using dark-field microscopy. Swimming speeds were analyzed by motion analysis software.
Growth curves.
Growth of Vibrio cells was monitored as previously described (20). Overnight cultures of Vibrio cells in VC medium were diluted 1:100 in 3 ml of VPG500 medium with (0.2% at final concentration) or without arabinose and were shaken at 30°C. Every h the culture was diluted 1:10 in VPG500 broth to measure the value at OD660.
Preparation of whole-cell extracts and the periplasmic fraction.
The periplasmic fraction of Vibrio cells was prepared using the method described by Nakamaru et al. (26). Vibrio cells were cultured at 30°C for 4 h in VPG500 or VPG broth containing an appropriate concentration of arabinose. Cells were harvested and suspended in buffer A (1 M NaCl, 50 mM Tris-HCl [pH 7.5]) at a cell concentration equivalent to an optical density at 660 nm of 5. This cell suspension was diluted 4 times with water to prepare the whole-cell samples. The cells were collected again and resuspended in buffer B (50 mM Tris-HCl [pH 8.5], 2 mM EDTA, 0.95 M sucrose, and 1 mg/ml of lysozyme) and then incubated at 30°C for 20 min. After centrifugation (17,000 × g, 10 min), the supernatants that contained the periplasmic proteins were collected as the periplasmic samples and the pellets were resuspended in water with the same volume of periplasmic samples to prepare the spheroplast samples.
Detection of proteins.
Protein samples were suspended in sodium dodecyl sulfate (SDS) loading buffer containing 5% (vol/vol) β-mercaptoethanol and were boiled for 5 min. Proteins were separated by SDS-PAGE, and immunoblotting was performed using an anti-PomB antibody (PomBC2B0455) (38a).
Purification of PomB fragments.
Fragments of several N-terminally-truncated variants of PomB (PomBC) were overproduced from the plasmids pTSK32 (PomBC1), pTSK33 (PomBC2), pTSK34 (PomBC3), pTSK35 (PomBC4), and pTSK36 (PomBC5) in E. coli BL21(DE3). Fresh colonies were inoculated into 1 liter of SB medium containing 100 μg/ml of ampicillin, and cells were grown at 37°C to an OD660 of ∼0.2 and then cooled to room temperature and induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cultures were continued for about 4 more h at 24°C. Cells were collected by centrifugation (7,000 × g) and then were resuspended in 50 ml of buffer C (20 mM Tris-HCl [pH 8.0], 150 mM NaCl) containing one tablet of Complete protease inhibitor cocktail (Roche Diagnostics). Cells were then disrupted using a French press (Ohtake Works) and centrifuged at 100,000 × g for 30 min, and the supernatant (soluble fraction) was loaded onto a HisTrap (GE Healthcare) column. The bound proteins were eluted with a linear 0 to 500 mM gradient of imidazole in buffer C, and the peak fractions were collected.
To purify the PomBC5 proteins from the periplasms of Vibrio cells, overnight cultures of VIO5 cells harboring the plasmid pLSK7 were diluted 1:100 in 1 liter of VPG500 broth containing 0.2% arabinose and were grown at 30°C for 4 h. Cells were collected by centrifugation (7,000 × g), washed once with buffer A, and then resuspended in buffer B. After incubation at 30°C for 20 min, the supernatants were collected by centrifugation (7,000 × g), after which imidazole and MgCl2 were added at final concentrations of 5 mM and 2 mM, respectively. After ultracentrifugation (186,000 × g, 30 min), the supernatants were loaded onto a HisTrap column and proteins were purified as described above.
Analytical size exclusion column chromatography.
Analytical size exclusion column chromatography of purified PomBC fragments was performed with a Superdex 75 10/300 GL column (GE Healthcare) using an Äkta system (GE Healthcare). The column was equilibrated with buffer C and eluted at a flow rate of 0.6 ml/min. Conalbumin (75 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), RNase A (13.7 kDa), and aprotinin (6.5 kDa) were used as size markers (GE Healthcare).
RESULTS
Dimerization of the periplasmic fragments of PomB.
It is presumed that the periplasmic region of PomB has functional importance with respect to stator assembly and activation and that its dimer formation is crucial to the formation of the Na+-conducting channel. To characterize the periplasmic region of V. alginolyticus PomB, we constructed several N-terminally-truncated variants of PomB (PomBC) that lack the TM and plug segments based on the multiple-sequence alignment and secondary-structure prediction of the PomB orthologs. (Fig. 1B). We purified the His-tagged fragments and analyzed them by analytical size exclusion chromatography to evaluate their oligomerization. PomBC1 did not behave well and degraded during the purification, so we did not further study that fragment. All other fragments were stable and well behaved, having single discrete peaks in their chromatograms (Fig. 2). The estimated molecular masses from the elution profiles were 68 kDa, 58 kDa, 49 kDa, and 41 kDa, for PomBC2, PomBC3, PomBC4, and PomBC5, respectively, indicating that the sizes of the fragments are nearly twice the deduced molecular masses of the PomBC monomers (29 kDa, 27 kDa, 23 kDa, and 22 kDa, respectively), suggesting that the PomBC fragments also formed dimers in solution.
Fig. 2.
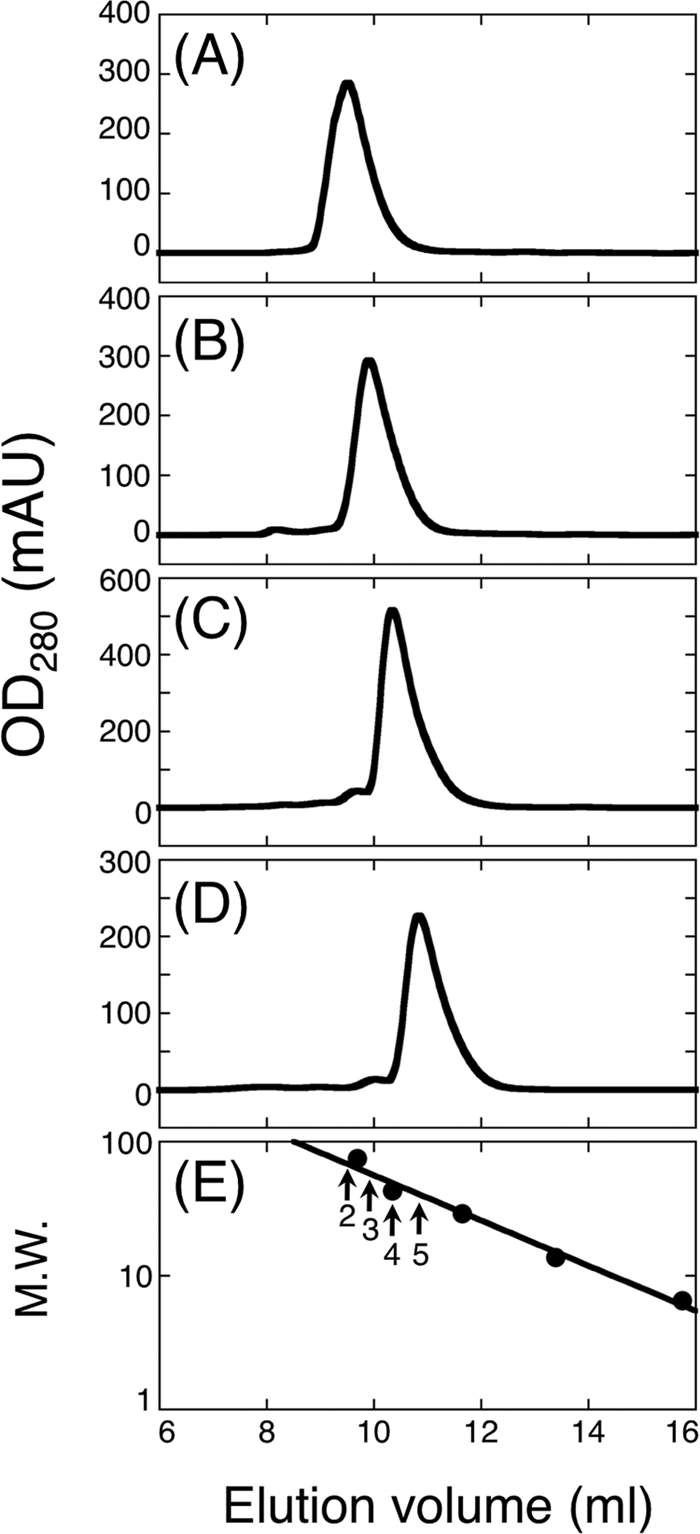
Elution profiles of purified PomB fragments by analytical size exclusion chromatography using a Superdex 75 10/300 GL column. PomBC2 (A), PomBC3 (B), PomBC4 (C), PomBC5 (D), and standard marker proteins (E) were analyzed. Arrows in panel E indicate the elution peaks of the PomB fragments with their numbers (e.g., “2” means PomBC2). Closed symbols in panel E indicate the elution peaks of the following marker proteins: conalbumin (9.69 ml; molecular weight, 75), ovalbumin (10.35 ml; molecular weight, 43), carbonic anhydrase (11.65 ml; molecular weight, 29), RNase A (13.39 ml; molecular weight, 13.7), and aprotinin (15.75 ml; molecular weight, 6.5). Molecular weights are given in thousands. mAU, milli absorbance units; M.W., molecular weight.
Multicopy effects of PomBC fragments on motility.
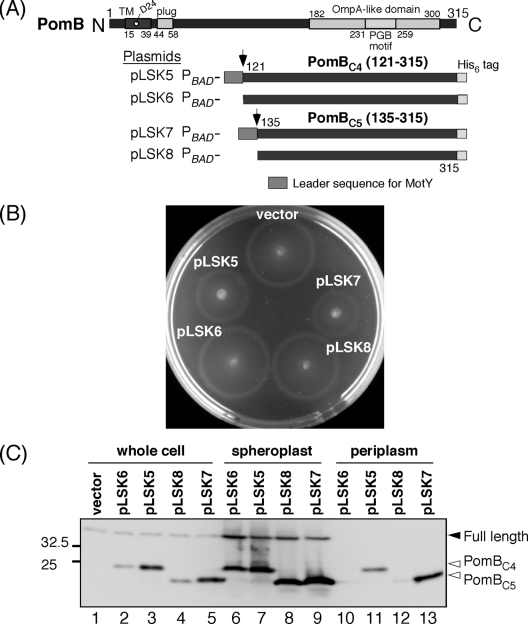
A previous study had shown that a Salmonella MotBC fragment significantly inhibits the motility of wild-type cells when overproduced in the periplasm (19). To test whether PomBC fragments exhibit a similar effect on motility, we fused two smaller PomBC fragments (PomBC4 and PomBC5), which included the corresponding region of MotBC6, to a leader sequence of MotY at their N termini to direct their location to the periplasmic space (Fig. 3 A). We introduced the plasmids pLSK5 (MotY leader::PomBC4-His6) and pLSK7 (MotY leader::PomBC5-His6) into VIO5, the wild-type strain for Na+-driven polar flagellar motility, and their motilities were examined on soft-agar plates that included 0.2% arabinose. VIO5 cells harboring pLSK5 or pLSK7 showed significantly smaller motility rings than VIO5 cells harboring the vector, the plasmid pLSK6 or pLSK8, which expressed leaderless PomBC fragments (Fig. 3B). The effect of PomBC5 on motility was slightly greater than that of PomBC4. Motility inhibition was not observed on plates without arabinose (data not shown). We detected the PomBC fragments in whole-cell lysates, as well as in spheroplast and periplasmic fractions from these strains (Fig. 3C). In whole-cell lysates, PomBC4 and PomBC5 fragments fused to the MotY leader sequence were more abundant than those without the MotY leader sequence. In the spheroplast fraction, PomBC fragments were detected in similar numbers regardless of the presence or absence of the leader sequence. In the periplasmic fraction, only PomBC fragments with the leader sequence were detected, with a higher number of PomBC5 than PomBC4, which is consistent with the slightly stronger motility inhibition of PomBC5 than of PomBC4. To further confirm the periplasmic location of PomBC, we purified PomBC5 protein from the periplasmic fraction and analyzed it using mass spectroscopy. The mass of purified PomBC5 was almost the same as that of the leaderless fragment, confirming the cleavage of the MotY signal peptide during the export into the periplasm. These results indicate that although the effects of PomBC are weaker than those of MotBC, the periplasmic overproduction of PomBC still significantly interferes with the motility of wild-type cells. We speculate that PomBC fragments in the periplasm are folded and can associate with endogenous motor proteins to interfere with motor rotation.
Fig. 3.
Overexpression of PomBC fragments in the periplasm interferes with motility. (A) Schematic representation of PomBC fragments designed for periplasmic localization. Plasmids pLSK5 and pLSK7 encode the PomBC4 and PomBC5 fragments (residues 121 to 315 and 131 to 315), respectively. Their N termini were fused to a MotY signal peptide (residues 1 to 22 of MotY), and their C termini were fused to a hexahistidine tag. The plasmids pLSK6 and pLSK8 do not contain the MotY signal peptide at their N termini. All constructs were placed under the control of the arabinose promoter. (B) Motility assay of VIO5 harboring pTY57 (vector), pLSK5, pLSK6, pLSK7, or pLSK8 on VPG500 soft-agar plates containing 0.2% arabinose. Plates were incubated at 30°C for 4 h. (C) Detection of PomBC fragments. Whole-cell proteins and spheroplast and periplasmic fractions were prepared from VIO5 cells that harbor pTY57 (vector control), pLSK5, pLSK6, pLSK7, or pLSK8 grown in VPG500 broth at 30°C for 4 h in the presence of 0.2% arabinose; they were analyzed by SDS-PAGE followed by immunoblotting with the anti-PomB antibody.
Function of in-frame-deletion constructs of PomB.
To test whether the linker region of V. alginolyticus PomB (residues 41 to 140) contains amino acids that are important for function, we also constructed a series of in-frame-deletion constructs of PomB (hereafter called PomBΔL) that lack residues 41 to 60 (Δ41-60), 41 to 70 (Δ41-70), 41 to 80 (Δ41-80), 41 to 90 (Δ41-90), 41 to 100 (Δ41-100), 41 to 110 (Δ41-110), 41 to 120 (Δ41-120), 41 to 130 (Δ41-130), or 41 to 140 (Δ41-140) (Fig. 1B). These constructs were coexpressed with PomA in the pomAB null strain NMB191 from the arabinose-inducible plasmids, and the motilities of the cells were assayed on soft-agar plates containing 0.02% arabinose (Fig. 4 A). The constructs with the two largest deletions (residues 41 to 130 and 41 to 140) completely impaired motility, but the mutants with other deletions retained function, forming motility rings with slightly reduced sizes compared to that of the wild type. Therefore, the PEM for PomB is defined for the region containing residues 121 to 315. When PomBΔL proteins were coexpressed with PomA at a higher level (induced by 0.2% arabinose; Fig. 4B), the ring sizes of the Δ41-60, Δ41-100, and Δ41-120 mutants were significantly smaller than those of the Δ41-70, Δ41-80, Δ41-90, and Δ41-110 mutants, whose ring sizes were slightly smaller than that of the wild type. Therefore, some of the deletion constructs show a multicopy effect on motility. The expression and stability of the deletion constructs were analyzed by immunoblotting of whole-cell extracts (Fig. 4C). Only faint or weak bands were detected for the nonfunctional derivatives (the Δ41-130 and Δ41-140 mutants). These deletions seemed to affect protein stability or expression, so mutant stators may not support motility. All other proteins were detected at a level comparable to that of the wild type. We next investigated the free-swimming motility of cells expressing each PomA/PomBΔL stator, using dark-field microscopy. When stators were induced by 0.02% arabinose, all in-frame-deletion strains swam at reduced and somewhat similar speeds, less than one-half that of the wild type (Fig. 4D). There seems to be no significant correlation between the swimming speed and the length of the deletion, and that trend was also observed for the motile fraction of the swimming cells (Table 2, left column). Motile fractions of the deletion constructs were within 23% to ∼34%, about one-half that of the wild-type (47%). When stator expression was induced at a high level (by 0.2% arabinose), the motile fractions of all deletion strains were reduced (Table 2, right column), especially strains that form smaller motility rings with 0.2% arabinose (only 2% to ∼6% for the Δ41-60, Δ41-100, and Δ41-120 mutants; Fig. 4B), further showing that these in-frame deletions in PomB cause a multicopy effect on motility.
Table 2.
Motile fraction of cells expressing PomB deletion variants
| Strain | Percentage of motile cells cultured with indicated amt of arabinosea (no. of cells analyzed) |
|
|---|---|---|
| 0.02% | 0.2% | |
| pBAD33 (vector) | 0 (213) | 0 (241) |
| pHFAB (full length) | 47 (275) | 38 (294) |
| pLSK9 (Δ41-60) | 24 (207) | 2 (370) |
| pLSK10 (Δ41-70) | 34 (340) | 20 (333) |
| pLSK11 (Δ41-80) | 29 (207) | 13 (245) |
| pLSK12 (Δ41-90) | 31 (270) | 14 (220) |
| pLSK13 (Δ41-100) | 30 (227) | 6 (311) |
| pLSK14 (Δ41-110) | 34 (311) | 31 (279) |
| pTSK37 (Δ41-120) | 23 (250) | 2 (415) |
| pTSK38 (Δ41-130) | 0 (202) | 0 (210) |
| pTSK39 (Δ41-140) | 0 (217) | 0 (245) |
The NMB191 (ΔpomAB) strain containing each plasmid was cultured at 30°C in VPG500 broth for 4 h with 0.02% or 0.2% arabinose, and swimming motility was observed using dark-field microscopy. Motile fractions were measured as described in Materials and Methods.
The effect of PomBΔL overproduction on cell growth.
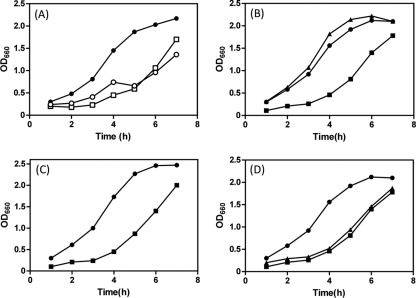
In V. alginolyticus PomB, residues V44 to F58 (corresponding to P52 to P65 in E. coli MotB) have been predicted to be a plug segment for the Na+ channel in the PomA/PomB stator complex. We speculated that the multicopy effect on motility observed for some in-frame-deletion constructs (Fig. 4B and Table 2) might be caused by the growth impairment. To test this possibility, we monitored cell growth in VPG500 broth containing 0.2% arabinose. When PomBΔL was cooverexpressed with PomA in NMB191 from the plasmid, a dramatic decrease in cell growth was observed in all deletion constructs. Figure 5 A shows typical growth curves observed for cells expressing the Δ41-60 mutant (strong multicopy effect), the Δ41-110 mutant (did not show a multicopy effect), and the wild-type stator, which demonstrates the greatly reduced cell growth for deletion constructs compared to that for the wild type, especially as seen at the log phase. Similar growth impairment was observed for all other deletion constructs (data not shown). These results indicate that the predicted segment 44 to 58 can function as the plug in PomB, and if the deleted region(s) contains this segment, overproduction of the PomA/PomBΔL mutant stator will impair cell growth. Since all deletion constructs caused similar growth impairment, hereafter we focused on the shortest functional construct, the Δ41-120 mutant, in this study.
Fig. 5.
Overproduction of PomBDΔL impairs cell growth. (A) NMB191 cells harboring the plasmid pHFAB (•), pLSK9 (□), or pLSK14 (○) were cultured in VPG500 broth at 30°C for 7 h with 0.2% arabinose added at the beginning (at 0 h). Cell growth was monitored at an absorbance of 660 nm every h. (B) NMB191 cells harboring the plasmid pHFAB (•), pTSK37 (▪), or pTSK37 D24N (▴) were cultured in VPG500 broth at 30°C for 7 h with 0.2% arabinose added at the beginning (at 0 h). Cell growth was monitored at an absorbance of 660 nm every h. (C) NMB200 cells harboring the plasmid pTSK37 (▪) or pTSK37 D24N (•) were cultured at 30°C for 7 h with 0.2% arabinose added at the beginning. (D) Effect of mutations at the putative helix of the N-terminal PEM region on growth impairment. Strains with pHFAB (•), pTSK37 (▪), or pTSK37 L168P (▴) were cultured in VPG500 broth at 30°C for 7 h with 0.2% arabinose added at the beginning.
If this growth impairment is caused by the massive Na+ influx through the PomA/PomBΔL stator complex, as seen in the MotA/MotB H+-driven stator (13, 24), a mutation that disrupts the Na+ binding site in the stator (PomB D24N) should eliminate such a deleterious effect. Therefore, we introduced the mutation D24N into PomBΔL(Δ41-120) and monitored cell growth (Fig. 5B). As expected, the D24N mutation eliminated the growth inhibition but did not affect protein stability or expression (data not shown). Moreover, as the Na+ concentration was decreased in the growth medium, the inhibitory effect became weaker (data not shown). This suggests that the growth impairment can be mainly attributed to the Na+ influx through the PomA/PomBΔL(Δ41-120) complex rather than being a general effect due to the overproduction of membrane protein. Thus, this region, especially the plug segment, appears to be important for the regulation of Na+ influx. A similar growth inhibition was observed when the PomA/PomBΔL(Δ41-120) complex was overexpressed in NMB200 (ΔpomB ΔfliG), which does not have the hook-basal body structure (Fig. 5C), and in the wild-type strain VIO5 (data not shown), showing that this inhibitory effect does not require the flagellar basal body.
Effects of mutations in the putative helix of the N-terminal PEM region in PomBΔL(Δ41-120) on motility.
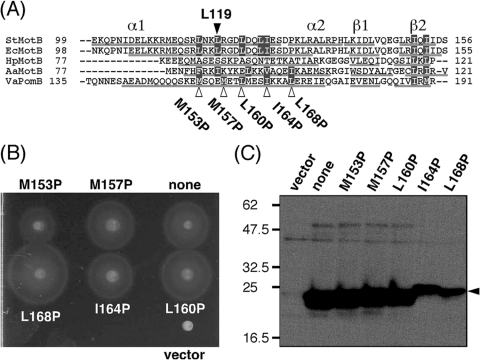
The crystal structure of Salmonella MotBC and subsequent functional analyses suggested that large conformational changes in the PEM region are required to anchor the stator to the PG layer and to activate its H+-conducting activity (20). To investigate the function of the PEM region in PomB, we again aligned PomB orthologs and predicted the secondary structure at the N-terminal PEM region (Fig. 6 A). Since mutation L119P or L119E in the amphipathic helix 1 of Salmonella MotBC would be predicted to disrupt hydrophobic interactions between this helix and the PGB core domain, we mutated the semiconserved hydrophobic residues predicted to be on helix 1 of PomBC. Point mutations M153P, M157P, L160P, I164P, and L168P were introduced into PomBΔL(Δ41-120) (Fig. 6A). When mutant PomBΔL(Δ41-120) was coexpressed with PomA in NMB191 in the presence of 0.02% arabinose, all mutants formed the same size motility ring as wild-type PomBΔL(Δ41-120) (data not shown). In the presence of 0.2% arabinose, a slightly larger motility ring was formed by the L168P mutant after 7 h of incubation at 30°C (Fig. 6B). We then investigated the motile fraction of the cells under these conditions (Table 3). The motile fraction of cells that overproduced the L168P mutant was larger than for any other mutant or the wild-type PomBΔL(Δ41-120) (Table 3). This indicates that the larger motile fraction of the L168P mutant contributes to the slightly larger motility ring formation, as seen in some of the deletion constructs (Fig. 4B and Table 2). Immunoblot analysis of the whole-cell extracts of NMB191 cells expressing the mutant PomA/PomBΔL(Δ41-120) complex induced by 0.2% arabinose showed that the amount of L168P protein decreased greatly (Fig. 6C), and growth inhibition by wild-type PomBΔL(Δ41-120) was not rescued by this mutation (Fig. 5D). These results suggest that the conformation of the PomA/PomBΔL(Δ41-120) complex might be altered into a more favorable state for stator assembly and anchoring to the PG layer by the L168P mutation.
Fig. 6.
Effects of mutations in the putative helix of the N-terminal PEM region in PomBΔL(Δ41-120) on motility. (A) Multiple-sequence alignment of PomB orthologs at the putative helix of the N-terminal PEM region. The aligned sequences are Salmonella enterica serovar Typhimurium MotB (StMotB); Escherichia coli MotB (EcMotB); Helicobacter pylori MotB (HpMotB); Aquifex aeolicus MotB (AaMotB); and Vibrio alginolyticus PomB (VaPomB). Alignment was carried out using ClustalW software. Secondary structures were predicted by the web-based program PSIPRED (14), and the predicted α-helix and β-sheet are indicated as a line below each sequence except for Salmonella MotB, whose crystal structure (MotBC2) has been determined. Its structural elements are indicated above the sequence. Residues emphasized by the gray boxes are the semiconserved hydrophobic ones. L119 of Salmonella MotB and the mutations generated are indicated by triangles. (B) Motility of NMB191 cells expressing mutant PomBΔL(41-120) proteins. Overnight cultures of NMB191 cells harboring the plasmid pTSK37 with each mutation were spotted on VPG500 soft-agar plates containing 0.2% arabinose at 30°C for 7 h. For the vector control, we used pBAD33. (C) Detection of mutant PomBΔL(41-120) proteins. Cells were cultured in VPG500 broth at 30°C for 4 h with 0.2% arabinose. Whole-cell proteins were prepared and then analyzed by immunoblotting. Vector, pBAD33 in NMB191.
Table 3.
Motile fraction of cells expressing PomBΔL(Δ41-120) with mutations in the putative PEM region
| Strain | Percentage of motile cellsa (no. of cells analyzed) |
|---|---|
| pHFAB | 26 (358) |
| pTSK37 | 1 (265) |
| pTSK37 M153P | 2 (180) |
| pTSK37 M157P | 0 (316) |
| pTSK37 L160P | 4 (224) |
| pTSK37 I164P | 4 (283) |
| pTSK37 L168P | 10 (225) |
The NMB191 strain containing each plasmid was cultured at 30°C in VPG500 broth for 4 h with 0.2% arabinose, and swimming motility was observed using dark-field microscopy. Motile fractions were measured as described in Materials and Methods.
DISCUSSION
Although the basic mechanism of torque generation is shared with the H+-driven motor, the Na+-driven polar flagellar motor of V. alginolyticus has some distinct characteristics. The PomA/PomB stator assembly depends on Na+ (10) and also requires the T ring structure that consists of MotX and MotY (36). Such distinct characteristics might be assigned to the specific functional roles of the periplasmic region of PomB, since the structure-based functional analysis of the corresponding region of Salmonella MotB revealed its functional importance, especially for stator assembly and activation (20). In this study, we investigated the role of the periplasmic region of PomB, using mutational and biochemical approaches.
We constructed 5 PomB fragments (PomBC) with N-terminal truncations of 37, 58, 89, 120, or 134 residues. Analytical size exclusion chromatography showed that these soluble PomBC fragments form dimers, and the two smallest constructs, PomBC4 and PomBC5, impaired the motility of wild-type cells when overproduced in the periplasm. This finding suggests that these PomBC fragments are properly folded so that they can associate with motor proteins to interfere with rotation, as has been reported for Salmonella MotB (19). It has been shown that MotX is likely to interact with PomB (28). Thus, it is also possible that the inhibition of motility might be caused by the titration of MotX from the motor by the overproduction of PomBC in the periplasm. However, we could not detect MotX in the periplasmic fraction when PomBC5 was overproduced in the periplasm, and it was not copurified with PomBC5 by affinity chromatography (data not shown). This indicates that MotX, which is not a membrane protein but has a strong insoluble nature (28, 29), is not solubilized by the periplasmic PomBC5. Therefore, the MotX-PomB interaction may be weak outside of the motor and may require conformational changes in PomB or MotX that are induced to strengthen the interactions only when the stator is located around the rotor. For further investigation of the MotX-PomB interaction, more mutational analyses will be required for both proteins. For example, the mutations in PomB or MotX that make the stator constitutively localize around the rotor despite the Na+ concentration in the medium could cause stronger interactions between them.
The region that connects the TM segment (residues 15 to 39) and the putative PEM region (residue 121 to the C terminus) is slightly longer than the Salmonella or E. coli MotB and is predicted to be a tolerant flexible linker, with the putative plug segment at its N terminus. We constructed 9 in-frame-deletion mutants of PomB at this region (PomBΔL proteins), and found that most PomBΔL proteins can form a functional stator, the exceptions being the two largest deletion constructs (Δ41-130 and Δ41-140). These two proteins were detected in small amounts in whole-cell extracts, indicating that reduced protein stability or expression might be the reason for the loss of function. However, our attempts to isolate motile revertants from the strains expressing these mutant stators have not been successful so far, so it is also possible that the critical length or region of PomB exists between residues 121 to 131. A comparative study of Salmonella MotB showed that deletion of up to 40 residues from the corresponding linker region did not affect the function of the constructs (25). On the other hand, 7 functional deletion constructs of PomB (deletions of up to 80 residues) retained reduced but similar levels of function, showing swimming speeds and motile fractions of about one-half those of the wild-type when induced by 0.02% arabinose (Fig. 4 and Table 2). All these constructs lack the plug segment, so somehow deletion of the plug caused the reduction in PomB function to a certain extent. Smaller motile fractions may be attributed to inefficient stator assembly around the rotor that could cause a decreased swimming speed, so we speculate that deletion of the plug may affect the stator assembly.
Overproduction of all functional PomA/PomBΔL stators impaired cell growth in the presence of 0.2% arabinose (Fig. 5). A similar inhibition was observed when the stator complex with the shortest functional PomB, PomBΔL(Δ41-120), was overexpressed in the pomB fliG double-deletion mutant or in wild-type cells, indicating that this growth impairment occurs without the flagellar basal body structure. This inhibitory effect was eliminated by the introduction of the D24N mutation. Moreover, when the concentration of Na+ was reduced in the medium, the growth inhibition was weakened (data not shown), which suggests that the growth impairment is related to the Na+ influx through the PomA/PomBΔL stator. This implies that the growth inhibition may be caused by the lack of the plug, which regulates the ion flow of the stator complex composed of PomA and PomB and prevents the premature ion translocation across the cell membrane when the stator is not incorporated into the motor and freely diffuses in the membrane. Our results suggest that the deletion of 80 residues in PomB is sufficient to alter the conformation of the PomA/PomB complex to allow active Na+ translocation. The plug function may be commonly conferred in the stator complex of either the Na+-driven type or the H+-driven type.
The motile fractions of cells overexpressing PomA/PomBΔL stators (induced by 0.2% arabinose) correlated with the levels of motility reduction, showing that all mutant strains exhibit smaller motile fractions than the wild-type, especially the 3 mutants that formed smaller motility rings (Fig. 4B and Table 2). Since overproduction of all functional PomA/PomBΔL stators caused growth inhibition, this multicopy effect on motility may be caused by both growth impairment and smaller motile fractions. The reason the higher level of expression caused a reduction in the motile fraction is not clear. It is possible that these mutant stators may conduct larger amounts of Na+ than others, whose activity could not be distinguished by the simple growth inhibition assay performed here. If so, the higher level of Na+ influx may cause a slight reduction of the Na+ motive force that may affect Na+-dependent stator assembly and thereby cause a reduction in the motile fraction. To test this possibility, we need more direct measurements of the Na+-conducting activity of mutant stators. Some of the PomBΔL proteins did not show a multicopy effect on motility, so we speculated that proper anchoring to the PG layer and assembly around the rotor depend on the conformation of the PEM region of PomB, which can be varied by the deletions. Therefore, we investigated mutations at the putative helix in the N-terminal PEM region. In Salmonella MotB, a large conformational change in this region has been proposed to affect the assembly and activation of the MotA/BΔL complex, and the mutation L119P or L119E in this region affects cell growth without impairing motility (20). We mutated the corresponding hydrophobic residues of PomBΔL(Δ41-120) to proline and expected to observe effects similar to those seen in MotBΔL. However, a significant effect on motility was seen only in the highly overproduced L168P mutant (induced by 0.2% arabinose) (Fig. 6B). That mutation did not affect growth (Fig. 5D) but showed a significantly larger motile fraction of cells than the other mutations in this region (Table 3). That could be the main reason for the slightly improved motility on soft-agar plates (Fig. 6B), which suggests that the stator assembly was improved. A phenotype similar to that of the Δ41-120/L168P mutant was observed for the deletion constructs (Δ41-70, Δ41-80, Δ41-90, and Δ41-110) and formed significantly larger motility rings and showed higher motile fractions than the Δ41-120 construct with 0.2% arabinose (Fig. 4B). We speculate that the L168P mutation alters the conformation of the PEM region to allow more efficient PG anchoring, so this mutant stator complex could be more efficiently incorporated into the motor. Similarly, the deletion constructs described above may have conformations that allow them to be anchored around the rotor. Therefore, the putative helix of the N-terminal PEM region of PomB and its connected linker region play key roles in stator assembly and activation.
In summary, our results show that the plug is a common feature for PomB/MotB proteins that is important for stator function and that the flexible linker that connects the TM segment and the PEM region is dispensable, but its appropriate length or conformation is required for proper function. The basic properties and roles of the PEM region of PomB are similar to those of MotB, but there are distinct features, such as the interaction with the T ring (presumably with MotX). The region in PomB responsible for interactions was not identified in this study and should be unveiled in the future. We emphasize that the in-frame-deletion construct PomBΔL(Δ41-120), which is the shortest functional PomB protein that lacks the periplasmic linker region (including the plug segment), shows an inhibitory effect on growth when overproduced. That property is quite useful for evaluating the Na+-conducting activity of the stator in vivo, as shown in this study, as well as the in vitro reconstitution system using the purified stator complex.
ACKNOWLEDGMENTS
We thank David Blair for careful reading of the manuscript and comments; Tomohiro Yorimitsu and Natsumi Nonoyama for kindly providing plasmid pTY57 and strain NMB200, respectively; and Sachi Tatematsu and Mayumi Taniguchi for technical assistance.
This work was supported by Grants-in-Aid for Scientific Research (20051009 to S.K. and 18074003 to M.H.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Asai Y., et al. 1997. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J. Bacteriol. 179:5104–5110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartolomé B., Jubete Y., Martinez E., Cruz F. D. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75–78 [DOI] [PubMed] [Google Scholar]
- 3. Berg H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19–54 [DOI] [PubMed] [Google Scholar]
- 4. Blair D. F. 2003. Flagellar movement driven by proton translocation. FEBS Lett. 545:86–95 [DOI] [PubMed] [Google Scholar]
- 5. Blair D. F., Kim D. Y., Berg H. C. 1991. Mutant MotB proteins in Escherichia coli. J. Bacteriol. 173:4049–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Braun T. F., Al-Mawsawi L. Q., Kojima S., Blair D. F. 2004. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 43:35–45 [DOI] [PubMed] [Google Scholar]
- 7. Dean G. D., Macnab R. M., Stader J., Matsumura P., Burks C. 1984. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J. Bacteriol. 159:991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Mot R., Vanderleyden J. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333–334 [DOI] [PubMed] [Google Scholar]
- 9. Francis N. R., Sosinsky G. E., Thomas D., Derosier D. J. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261–1270 [DOI] [PubMed] [Google Scholar]
- 10. Fukuoka H., Wada T., Kojima S., Ishijima A., Homma M. 2009. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol. Microbiol. 71:825–835 [DOI] [PubMed] [Google Scholar]
- 11. Fukuoka H., Yakushi T., Kusumoto A., Homma M. 2005. Assembly of motor proteins, PomA and PomB, in the Na+-driven stator of the flagellar motor. J. Mol. Biol. 351:707–717 [DOI] [PubMed] [Google Scholar]
- 12. Guzman L. M., Belin D., Carson M. J., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hosking E. R., Vogt C., Bakker E. P., Manson M. D. 2006. The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 364:921–937 [DOI] [PubMed] [Google Scholar]
- 14. Jones D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195–202 [DOI] [PubMed] [Google Scholar]
- 15. Koebnik R. 1995. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol. Microbiol. 16:1269–1270 [DOI] [PubMed] [Google Scholar]
- 16. Kojima S., Asai Y., Atsumi T., Kawagishi I., Homma M. 1999. Na+-driven flagellar motor resistant to phenamil, an amiloride analog, caused by mutations in putative channel components. J. Mol. Biol. 285:1537–1547 [DOI] [PubMed] [Google Scholar]
- 17. Kojima S., Blair D. F. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041–13050 [DOI] [PubMed] [Google Scholar]
- 18. Kojima S., Blair D. F. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43:26–34 [DOI] [PubMed] [Google Scholar]
- 19. Kojima S., Furukawa Y., Matsunami H., Minamino T., Namba K. 2008. Characterization of the periplasmic domain of MotB and implications for its role in the stator assembly of the bacterial flagellar motor. J. Bacteriol. 190:3314–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kojima S., et al. 2009. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol. Microbiol. 73:710–718 [DOI] [PubMed] [Google Scholar]
- 21. Kojima S., et al. 2008. Insights into the stator assembly of the Vibrio flagellar motor from the crystal structure of MotY. Proc. Natl. Acad. Sci. U. S. A. 105:7696–7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Macnab R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77–100 [DOI] [PubMed] [Google Scholar]
- 23. Minamino T., Namba K. 2004. Self-assembly and type III protein export of the bacterial flagellum. J. Mol. Microbiol. Biotechnol. 7:5–17 [DOI] [PubMed] [Google Scholar]
- 24. Morimoto Y. V., Che Y. S., Minamino T., Namba K. 2010. Proton-conductivity assay of plugged and unplugged MotA/B proton channel by cytoplasmic pHluorin expressed in Salmonella. FEBS Lett. 584:1268–1272 [DOI] [PubMed] [Google Scholar]
- 25. Muramoto K., Macnab R. M. 1998. Deletion analysis of MotA and MotB, components of the force-generating unit in the flagellar motor of Salmonella. Mol. Microbiol. 29:1191–1202 [DOI] [PubMed] [Google Scholar]
- 26. Nakamaru Y., Takahashi Y., Unemoto T., Nakamura T. 1999. Mechanosensitive channel functions to alleviate the cell lysis of marine bacterium, Vibrio alginolyticus, by osmotic downshock. FEBS Lett. 444:170–172 [DOI] [PubMed] [Google Scholar]
- 27. Okabe M., Yakushi T., Asai Y., Homma M. 2001. Cloning and characterization of motX, a Vibrio alginolyticus sodium-driven flagellar motor gene. J. Biochem. 130:879–884 [DOI] [PubMed] [Google Scholar]
- 28. Okabe M., Yakushi T., Homma M. 2005. Interactions of MotX with MotY and with the PomA/PomB sodium ion channel complex of the Vibrio alginolyticus polar flagellum. J. Biol. Chem. 280:25659–25664 [DOI] [PubMed] [Google Scholar]
- 29. Okabe M., Yakushi T., Kojima M., Homma M. 2002. MotX and MotY, specific components of the sodium-driven flagellar motor, colocalize to the outer membrane in Vibrio alginolyticus. Mol. Microbiol. 46:125–134 [DOI] [PubMed] [Google Scholar]
- 30. Okunishi I., Kawagishi I., Homma M. 1996. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol. 178:2409–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reid S. W., et al. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. U. S. A. 103:8066–8071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sato K., Homma M. 2000. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:5718–5722 [DOI] [PubMed] [Google Scholar]
- 33. Sato K., Homma M. 2000. Multimeric structure of PomA, a component of the Na+-driven polar flagellar motor of Vibrio alginolyticus. J. Biol. Chem. 275:20223–20228 [DOI] [PubMed] [Google Scholar]
- 34. Stader J., Matsumura P., Vacante D., Dean G. E., Macnab R. M. 1986. Nucleotide sequence of the Escherichia coli MotB gene and site-limited incorporation of its product into the cytoplasmic membrane. J. Bacteriol. 166:244–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sudo Y., et al. 2009. Interaction between Na+ ion and carboxylates of the PomA-PomB stator unit studied by ATR-FTIR spectroscopy. Biochemistry 48:11699–11705 [DOI] [PubMed] [Google Scholar]
- 36. Terashima H., Fukuoka H., Yakushi T., Kojima S., Homma M. 2006. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na-driven flagella and required for stator formation. Mol. Microbiol. 62:1170–1180 [DOI] [PubMed] [Google Scholar]
- 37. Terashima H., Kojima S., Homma M. 2008. Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270:39–85 [DOI] [PubMed] [Google Scholar]
- 38. Terashima H., Kojima S., Homma M. 2010. Functional transfer of an essential aspartate for the ion-binding site in the stator proteins of the bacterial flagellar motor. J. Mol. Biol. 397:689–696 [DOI] [PubMed] [Google Scholar]
- 38a. Terauchi T., Terashima H., Kojima S., Homma M. The critical role of a conserved residue, PomB-F22, in the transmembrane segment of the flagellar stator complex for conducting ions and generating torque. Microbiology, in press [DOI] [PubMed] [Google Scholar]
- 39. Thomas D. R., Francis N. R., Xu C., DeRosier D. J. 2006. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:7039–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Togashi F., Yamaguchi S., Kihara M., Aizawa S. I., Macnab R. M. 1997. An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB. J. Bacteriol. 179:2994–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Toker A. S., Kihara M., Macnab R. M. 1996. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J. Bacteriol. 178:7069–7079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yakushi T., Hattori N., Homma M. 2005. Deletion analysis of the carboxyl-terminal region of the PomB component of the Vibrio alginolyticus polar flagellar motor. J. Bacteriol. 187:778–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamaguchi S., et al. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamaguchi S., Fujita H., Ishihara H., Aizawa S., Macnab R. M. 1986. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J. Bacteriol. 166:187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yorimitsu T., Kojima M., Yakushi T., Homma M. 2004. Multimeric structure of the PomA/PomB channel complex in the Na+-driven flagellar motor of Vibrio alginolyticus. J. Biochem. 135:43–51 [DOI] [PubMed] [Google Scholar]
- 46. Yorimitsu T., Sato K., Asai Y., Kawagishi I., Homma M. 1999. Functional interaction between PomA and PomB, the Na+-driven flagellar motor components of Vibrio alginolyticus. J. Bacteriol. 181:5103–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]