Abstract
Members of the Burkholderia cepacia complex (BCC) are serious respiratory pathogens in immunocompromised individuals and in patients with cystic fibrosis (CF). They are exceptionally resistant to many antimicrobial agents and have the capacity to spread between patients, leading to a decline in lung function and necrotizing pneumonia. BCC members often express a mucoid phenotype associated with the secretion of the exopolysaccharide (EPS) cepacian. There is much evidence supporting the fact that cepacian is a major virulence factor of BCC. UDP-glucose dehydrogenase (UGD) is responsible for the NAD-dependent 2-fold oxidation of UDP-glucose (UDP-Glc) to UDP-glucuronic acid (UDP-GlcA), which is a key step in cepacian biosynthesis. Here, we report the structure of BceC, determined at 1.75-Å resolution. Mutagenic studies were performed on the active sites of UGDs, and together with the crystallographic structures, they elucidate the molecular mechanism of this family of sugar nucleotide-modifying enzymes. Superposition with the structures of human and other bacterial UGDs showed an active site with high structural homology. This family contains a strictly conserved tyrosine residue (Y10 in BceC; shown in italics) within the glycine-rich motif (GXGYXG) of its N-terminal Rossmann-like domain. We constructed several BceC Y10 mutants, revealing only residual dehydrogenase activity and thus highlighting the importance of this conserved residue in the catalytic activity of BceC. Based on the literature of the UGD/GMD nucleotide sugar 6-dehydrogenase family and the kinetic and structural data we obtained for BceC, we determined Y10 as a key catalytic residue in a UGD rate-determining step, the final hydrolysis of the enzymatic thioester intermediate.
INTRODUCTION
The Burkholderia cepacia complex (BCC) consists of at least 17 species that have been isolated from sputum of cystic fibrosis (CF) patients. Members of the BCC have emerged as highly problematic opportunistic human pathogens in immunocompromised individuals and in patients with CF (31, 33). CF patients are likely to see their disease accelerating, as BCC pulmonary colonization leads to a rapid decline of the lung function and in some cases the development of the “cepacian syndrome,” a fatal condition that is characterized by necrotizing pneumonia and bacteremia (31, 33). Treatment of BCC-infected patients has proved to be of great difficulty as these bacteria are intrinsically resistant to multiple antibiotics.
The distribution of BCC species recovered from CF patients varies worldwide and over the period of time under analysis. In general, B. cenocepacia predominates, followed by Burkholderia multivorans (2, 4, 5, 31, 33, 44). However, the species B. cepacia, found infrequently in CF patients worldwide, has an exceptionally high representation among the BCC isolates in Portugal, accounting for one-half of the number of the isolates retrieved from 1995 to 2006 (12). Intrinsically contaminated saline solutions for nasal application and the environment are likely sources for B. cepacia infection (12). During the epidemiological survey carried out in Portugal, it was found that B. cepacia isolates may also be involved in the progressive deterioration of the CF lung during persistent infection (11).
BCC possesses many of the virulence factors that are known to play a role in Pseudomonas aeruginosa CF infection (31). Despite all the studies performed on BCC, little is known about the pathogenic mechanisms of these bacteria. In the lungs of CF patients, P. aeruginosa secretes abundant amounts of the exopolysaccharide (EPS) alginate, which is believed to give rise to protection from the host immune system response and antibiotic therapy and promotes persistence of the bacteria in the respiratory tract (51). A vast range of BCC isolates from CF patients was shown to express a mucoid phenotype associated with the production of EPS. Moreover, BCC mutants defective in EPS synthesis were found to be less virulent in a lung infection model in mice with a null allele of the gene involved in X-linked chronic granulomatous disease (CGD) (53).
Cepacian is the main EPS produced by BCC isolates, in particular, the B. cepacia IST408 isolate retrieved from a Portuguese CF patient (11). It is composed of a branched acetylated heptasaccharide repeat unit with d-glucose, d-rhamnose, d-mannose, d-galactose, and d-glucuronic acid in the ratio of 1:1:1:3:1, respectively (50). Cepacian biosynthesis is a multistep process directed by genes belonging to the bce-I and bce-II cepacian gene clusters (21, 38).
UDP-glucose dehydrogenase (UGD) (EC 1.1.1.22) catalyzes two successive NAD-dependent oxidations of UDP-glucose (UDP-Glc) (6, 15). Due to the striking similarity in the primary amino acid sequences of the active sites in UGDs from different species, a common reaction mechanism has been proposed among the family members, from bacteria to animals and plants (23, 26, 48). In fact, numerous studies on the reaction mechanism have been carried out on both bacterial and mammalian enzymes (6, 15, 23, 52). Both hydride transfer steps between the substrate C-6′ center and NAD+ have previously been studied at the atomic level (15, 23), but the final reaction step has remained an open issue with respect to how the covalently bound reaction intermediate is hydrolyzed and what are the critical residues in this process (23).
In this work, the structure of B. cepacia UGD (BceC) was determined, and its biochemical characterization was performed. Given the lethal impact of B. cepacia infection in human CF patients, the BceC catalytic site was analyzed, taking into account the previously described Streptococcus pyogenes UGD (6) and available human structures. Mutagenic studies were performed on a strictly conserved residue among all UGDs, Tyr10 in the first Rossmann fold motif, GXGYXG, of BceC. Several Tyr10 mutants were constructed, producing essentially inactive proteins, indicating that this residue plays an important role in the enzyme's activity.
MATERIALS AND METHODS
DNA manipulation and generation of Y10 mutants.
Point mutants of the BceC protein were generated from the wild-type pBceC construct using a protocol based on a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) (47). Based on the nucleotide sequence of the bceC gene (GenBank accession number GQ451909), the oligonucleotide pairs Y10S_up (5′-ATC ATC GGC AGC GGT TCC GTA GGT CTT GTC ACC-3′) and Y10S_rev (5′-GGT GAC AAG ACC TAC GGA ACC GCT GCC GAT GAT-3′), Y10K_up (5′-CT ATC ATC GGC AGC GGT AAG GTA GGT CTT GTC ACC GG-3′) and Y10K_rev (5′-CC GGT GAC AAG ACC TAC CTT ACC GCT GCC GAT GAT AG-3′), and Y10F_up (5′-CT ATC ATC GGC AGC GGT TTC GTA GGT CTT GTC AC-3′) and Y10F_rev (5′-GT GAC AAG ACC TAC GAA ACC GCT GCC GAT GAT AG-3′) were designed for mutants Y10S, Y10K, and Y10F, respectively. Each amino acid substitution was confirmed by N-terminal sequencing, and the plasmids containing the altered bceC genes were transformed into Escherichia coli SURE strain (Stratagene, La Jolla, CA). Oligonucleotides were purchased from MWG Biotech, restriction enzymes were from Gibco BRL, and DNA polymerases were from Roche Diagnostics (Mannheim, Germany).
Protein purification and crystallization.
BceC wild-type and Y10 mutants were expressed and purified as previously described (47). Briefly, the cells harboring the recombinant constructs of pBceC, pBceC_y10f, pBceC_y10s, and pBceC_y10k were grown at 37°C to an A600 of approximately 0.6 in LB medium supplemented with 100 mg ml−1 ampicillin and induced at 28°C for 6 h by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.3 mM. Cells were harvested by centrifugation (7,000 × g for 40 min at 4°C), resuspended in 20 mM phosphate buffer, pH 7.4, containing 1 M NaCl, 20 mM imidazole, and a protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany), and disrupted on a French press. Nucleic acids and cell debris were removed by centrifugation at 17,000 × g for 40 min at 4°C, and the supernatant was applied onto a 5-ml Ni-chelating column (HisTrap FF; GE Healthcare, United Kingdom), previously equilibrated in the same phosphate buffer. Proteins were eluted using a linear imidazole gradient up to 500 mM, pure fractions were pooled, and the buffer was exchanged to 25 mM Tris-HCl, pH 8.3, 50 mM NaCl, and 2.5 mM dithiothreitol (DTT). Protein batches for crystallization purposes were complemented with 0.25 mM UDP-GlcA and 0.5 mM NAD (NAD+). All proteins were stored at −80°C, with a final concentration of approximately 10 mg·ml−1. UDP-GlcA and NAD+ were obtained from Sigma-Aldrich.
Standard enzymatic activity assays.
The catalytic oxidation of UDP-glucose to UDP-glucuronic acid by BceC is concomitant with the reduction of two NAD+ to NADH molecules. The determination of UGD activity in native BceC or its mutants was monitored by measuring the absorbance at 340 nm, which accompanies the reduction of NAD+ to NADH. Assays were performed during 60 min at 30°C in a reaction volume of 1 ml containing 100 mM Tris-HCl, pH 8.7, 10 mM MgCl2, 2 mM DTT, 2 mM UDP-Glc, and 1.5 mM NAD+. The reaction was initiated with the addition of 3 μg of pure protein to the cuvette. Control assays were performed with enzyme reaction mixtures lacking the enzyme or the substrate. Assays were performed in duplicate or triplicate, and specific activities were calculated from absorbance values using an NADH molar extinction coefficient of 6,220 M−1 cm−1. UDP-Glc and NAD+ were acquired from Sigma-Aldrich. Apparent kinetic parameters (Km and Vmax) for substrate (UDP-Glc) and cofactor (NAD+) were obtained by measuring the level of NAD+ reduction at 30°C. The reaction dependence on cofactor concentration was assessed using purified BceC incubated with increasing concentrations of NAD+ in the presence of saturating UDP-Glc substrate. Similarly, dependence of reaction kinetics on substrate concentration was obtained using increasing UDP-Glc concentrations in the presence of saturating NAD+ cofactor. Experimental data were plotted using GraphPad Prism, version 5, software, and Km and Vmax were calculated by fitting data to a modified form of the Michaelis-Menten equation: V = Vmax[S]n/([S]n + Kmn), where n is the Hill coefficient and S is the substrate.
Reaction transition state enthalpy and entropy differences and their estimated uncertainties were obtained using the Eyring-Polanyi equation (18, 20) by linear regression of ln(k/T) versus 1/T, where k is the reaction rate constant and T is absolute temperature, using the program LINEST in Excel from Microsoft Office.
Crystallization, diffraction data collection, and processing.
Native BceC crystallization was described elsewhere (47). In spite of extensive crystallization screens, no crystals of the Y10F mutant were ever obtained, but suitable crystals of the Y10S and Y10K mutants were readily produced by optimization of the native BceC sitting-drop vapor diffusion crystallization conditions (47). The best Y10S and Y10K crystals were obtained by equilibration of 1 μl of mutant protein solution and 1 μl of a precipitant solution of 100 mM sodium acetate buffer, pH 4.5, 200 mM ammonium sulfate, and 11 to 14% (wt/vol) polyethylene glycol (PEG) 4000. Octahedral crystals appeared and grew up to dimensions of 0.06 to 0.1 mm within 24 h.
Native crystals were cryo-protected by direct transfer to a mother-liquor drop including 25% (vol/vol) glycerol. Y10S and Y10K mutant crystal cryo-protection required consecutive crystal transfers between drops of mother liquor including glycerol at increasing concentrations up to 25% (vol/vol). Crystals were stored upon a swift plunge in liquid nitrogen. Diffraction data were measured at beam line ID14-EH1 of the European Synchrotron Radiation Facility (ESRF), Grenoble, France. Diffraction images were processed with MOSFLM (30), intensities without a sigma cutoff were merged and scaled with SCALA (19), and the corresponding structure factors were calculated using TRUNCATE (22) from the CCP4 suite (8) (crystal and diffraction statistics are given in Table 1).
Table 1.
Crystallographic data and refinement statistics of native BceC and Y10 mutants
| Parameter | Value for the proteina |
||
|---|---|---|---|
| BceC | Y10S | Y10K | |
| Wavelength (Å) | 0.9334 | 0.9334 | 0.9334 |
| Resolution range (Å) | 47.56–1.75 | 47.40–1.70 | 57.83–2.80 |
| (1.84–1.75) | (1.79–1.70) | (2.95–2.80) | |
| Space group | P212121 | P212121 | P212121 |
| a (Å) | 97.63 | 97.36 | 101.62 |
| b (Å) | 108.93 | 108.62 | 109.25 |
| c (Å) | 187.71 | 187.46 | 183.44 |
| No. of observations | 750,258 (109,058) | 803,863 (115,360) | 206,820 (30,278) |
| No. of unique observations | 197,721 (29,063) | 217,359 (31,490) | 50,832 (7,331) |
| Redundancy | 3.8 (3.8) | 3.7 (3.7) | 4.1 (4.1) |
| Completeness (%) | 98.4 (99.7) | 99.8 (100.0) | 99.8 (100.0) |
| I/σ(I) | 6.7 (2.0) | 7.6 (2.0) | 2.8 (1.7) |
| Rpimb | 0.038 (0.188) | 0.044 (0.283) | 0.119 (0.241) |
| Rrimc | 0.076 (0.369) | 0.087 (0.554) | 0.249 (0.501) |
| Rsymd | 0.066 (0.315) | 0.074 (0.474) | 0.217 (0.436) |
| Mosaicity (°) | 0.26 | 0.25 | 0.8 |
| Solvent content (%) | 49.5 | 47.5 | 48.9 |
| No. molecules in AU | 4 | 4 | 4 |
| Wilson B (Å2) | 18.3 | 17.7 | 42.2 |
| Rwork (%)f | 16.3 | 16.3 | 22.8 |
| Rfree (%)f | 19.7 | 19.7 | 26.3 |
| R (%)e | 16.5 | 16.5 | 22.2 |
| RMSD bonds (Å) | 0.013 | 0.010 | 0.010 |
| RMSD angles (°) | 1.4 | 1.3 | 1.2 |
| Chain <ADP> (Å2)g | 21, 22, 29, 21 | 21, 22, 29, 20 | 28, 29, 28, 30 |
| Main chain <ADP> RMSD (Å2) | 1.5, 1.6, 1.6, 1.5 | 1.5, 1.5, 1.7, 1.5 | 4.2, 4.1, 4.2, 4.3 |
| Side chain <ADP> RMSD (Å2) | 2.8, 2.8, 2.9, 2.7 | 2.8, 2.8, 2.7, 2.6 | 6.3, 6.3, 6.6, 6.3 |
| Glucuronic acid <ADP> (Å2) | 12, 11, 12, 11 | 12, 10, 14, 11 | 28, 27, 21, 25 |
| No. of residues per chain (total) | 461, 451, 455, 454 (1,821) | 461, 450, 458, 454 (1,823) | 455, 449, 451, 453 (1,808) |
| No. of solvent waters | 1,374 | 1,304 | |
| No. of solvent sulfates | 14 | 14 | 9 |
| No. of solvent acetates | 4 | 5 | |
| No. of solvent glycerols | 6 | 6 | |
| No. of solvent Tris molecules | 1 | ||
| No. of atoms modeled in multiple conformations | 331 | 320 | |
| Residue distribution in preferred, allowed, and outlier regions of Ramachandran diagram (%) | 96.9, 3.0, 0.1 | 97.5, 2.4, 0.1 | 95.5, 3.8, 0.7 |
| PDB code | 2Y0E | 2Y0C | 2Y0D |
Values within parentheses correspond to highest-resolution shells (except when specified otherwise); four values in a series separated by commas correspond to each of the four crystallographically independent molecules.
Rpim = Σh [1/(N − 1)]1/2 Σi |Ii(h) − <I(h)>|/Σh Σi Ii (h), where N is the data redundancy, I is the observed intensity, and <I> is the average intensity of multiple observations from symmetry-related reflections. It is an indicator of the precision of the final merged and averaged data set (55).
Rrim = Rmeas = Σh [N/(N − 1)]1/2 Σi |Ii(h) − <I(h)>|/Σh Σi Ii(h), where N is the data redundancy, I is the observed intensity, and <I> is the average intensity of multiple observations of symmetry-related reflections. It is an indicator of the average spread of the individual measurements (14, 55).
Rsym = Σh Σi |Ii(h) − <I(h)>|/Σh Σi Ii (h), where I is the observed intensity and <I> is the average intensity of multiple observations from symmetry-related reflections.
R = Σ |Fobs − Fcalc|/Σ Fobs, where Fobs and Fcalc are the amplitudes of the observed and the model calculated structure factors, respectively. It is a measure of the agreement between the experimental X-ray diffraction data and the crystallographic model.
Rwork refers to the actual working data set used in refinement, while Rfree refers to a cross validation set that is not directly used in refinement and is therefore free from refinement bias.
<ADP>, average for atomic displacement parameters (also called B factors).
Structural determination and refinement.
Molecular replacement trials with native BceC as a target and the Streptococcus pyogenes UGD (Protein Data Bank [PDB] code 1dli) (6), which was available at the time, as a search structure were unsuccessful, possibly due to their low homology, with sequence identities of ca. 23%. However, once a preliminary three-dimensional (3D) model of UGD from Sphingomonas elodea ATCC 31461 was obtained in our laboratory (46), with a sequence identity of 42% against BceC, the molecular replacement solution was readily obtained (47). Using the initial diffraction data set to 2.09-Å resolution, the phases of the native BceC molecular replacement solution were improved (47) with the program RESOLVE using its automatically implemented 2-fold noncrystallographic symmetry (NCS) averaging and “prime and switch” procedures (54). Interpretable electron density maps were produced with a figure of merit of 0.48 and examined using COOT (17). Maps showed well-defined densities for a UDP-GlcA molecule nestled in each BceC monomer, in a model with 1,358 out of 1,912 possible residues in the asymmetric unit (AU). Nevertheless, at that stage only 349 residues were docked in sequence. Using the obtained 1.75-Å resolution diffraction data set of native BceC and the program ARP/wARP (42), the four-chain model was extended to 1,604 residues, of which 1,530 were docked in sequence. The protein and solvent structures were further examined with COOT against their σA maps (43), and the model was improved accordingly and refined with PHENIX (1). Upon determination of the polypeptide model translation, libration, and screw (TLS) motions using TLSMD (http://skuld.bmsc.washington.edu/∼tlsmd) (41), four homologous sequential domains were assigned to each chain and used both for TLS refinement and to define NCS restraints among chains in atomic positional and isotropic atomic displacement parameters. Steps of protein and solvent refinement and modeling were iteratively repeated until convergence of Rfree, where 5% of the reflections were consistently set aside for cross-validation.
Phases for the mutants were obtained from the BceC native structure upon truncation of Tyr10 into Ala. As native BceC and its Y10S mutant produced isomorphous crystals, cell dimensions differed by no more than 0.3%, and a rigid-body refinement of the four chains produced suitable phases. The Y10S model was refined using TLS and NCS domains similar to those of the native BceC. For the nonisomorphous Y10K mutant crystal, cell lengths differ up to 4% in the a direction, and phases were obtained by molecular replacement using PHASER (35). Refinement was performed with specific TLS and NCS domain definitions. For both mutants, the iterative map inspection with protein and solvent modeling and refinement were performed as described for the native case, with 5% of the reflections used for Rfree monitoring purposes.
Structural analysis.
Molecular stereochemistry was checked within COOT (17), intermolecular contacts and crystallographic packing was checked with PISA (28), and structural comparisons were performed with SSM (29). Colored figures were produced with PYMOL (13).
Protein structure accession numbers.
The atomic coordinates and structure factors of native BceC and its Y10S and Y10K single mutants have been deposited in the PDB, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://rcsb-deposit.rutgers.edu/), under accession codes 2Y0E, 2Y0C, and 2Y0D, respectively.
RESULTS AND DISCUSSION
Biochemical characterization of BceC.
Under denaturing conditions, the purified fusion protein migrated as a single polypeptide with a molecular mass of approximately 52.6 kDa (data not shown), corresponding to 51.3 kDa from the native plus 1.3 kDa from the His6 tag. In order to show that BceC is a UGD, the bceC gene from plasmid pBceC was overexpressed by IPTG induction, and the UGD activity was assayed in cell crude extracts harboring either pBceC or the cloning vector pWH844 (control). No UGD activity was detected in the extract prepared from the control cells, while the extract from cells overexpressing the bceC gene exhibited strong UGD activity (data not shown). Vmax values for the most active batch of purified BceC relative to UDP-Glc or NAD+ were virtually identical, with 8.0 μmol·min−1·mg−1 or 7.8 μmol·min−1·mg−1, respectively. The estimated apparent Km values for UDP-Glc and NAD+ were 0.23 and 0.53 mM, respectively. The values of the Hill coefficient for UDP-glucose and NAD+ were 0.89 and 1.5, respectively. BceC had a Kcat of 413 and 401 per min and a catalytic efficiency of 1,796 min−1·mM−1 and 757 min−1·mM−1 for UDP-glucose and NAD+, respectively.
The Km values for UDP-glucose and NAD+ are higher than those indicated for two characterized B. cenocepacia K56-2 UGD proteins (0.02 and 0.17 mM for BCAL2946 and 0.02 and 0.21 mM for BCAM0855, respectively) (32). BceC shares 76% and 97% amino acid identity with BCAL2946 and BCAM0855, respectively. Thus, we could expect that orthologous enzymes have identical kinetic parameters. Nevertheless, since each enzyme assay conditions differed for the tested UGDs, conclusions should be made carefully. BceC characteristics were also compared with other published UGDs, revealing that Kms for UDP-Glc and NAD+ are within the range reported for the UGD of P. aeruginosa PAO1 (PA2022) (27) or UgdG of S. elodea ATCC31461 (24).
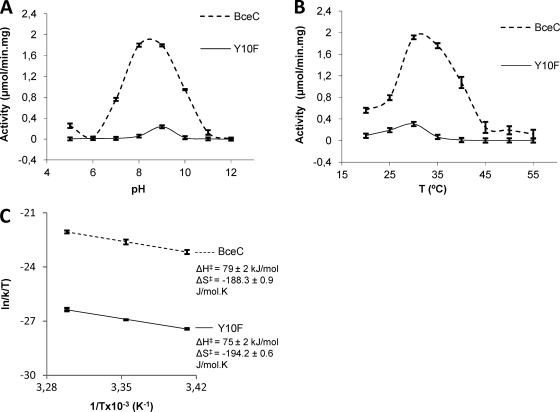
The optimal pH at 30°C for the UGD activity of purified BceC was 8.7 (Fig. 1 A). At pH 7.0, BceC exhibits only 37% of its maximal activity. The optimal temperature for the activity of BceC was 30°C. Between 30°C and 40°C, more than 70% of the activity was maintained, but the activity decreased significantly when the temperature was below 25°C (Fig. 1B). Further enzymatic assays of BceC and its mutants were performed at 30°C.
Fig. 1.
Dehydrogenase activities of BceC and its Y10F mutant. (A) Activity relative to the pH of the medium at 30°C. (B) Activity relative to the temperature of the medium at pH 8.7. (C) Plot of the first three values of ln(k/T) versus 1/T (the ignored remaining values correspond to degraded conformations of the enzyme at higher temperatures) allows an estimation of ΔH‡ (enthalpy of activation) and ΔS‡ (entropy of activation) according to the Eyring-Polanyi equation: ln(k/T) = −ΔH‡/R × 1/T + ln(kB/h) + ΔS‡/R, where kB is the Boltzmann constant, h is Planck's constant, and R is the gas constant (18, 20). Bars represent maximal intervals of (two or three) the measurements replicates.
Crystal data and the reliability of BceC and its mutants models.
Upon successive crystal optimizations and data collection sessions at ESRF, data sets of native BceC and its Y10S and Y10K mutants reached final resolutions of 1.75, 1.70, and 2.80 Å, respectively. All crystals belong to space group P212121, showing similar cell dimensions, and contained four independent molecules in the asymmetric unit (AU), corresponding to calculated solvent contents in the range of 47.5 to 49.5%.
Structure refinement was performed using the application phenix.refine in PHENIX (1), whereby the program was set automatically to optimize the X-ray weights that minimized Rfree; except in the final refinement step, all data were used, and the X-ray weight was manually adjusted so that both geometrical and atomic displacement parameter (ADP) root mean square deviations (RMSDs) replicated those previously obtained in the presence of cross-validation Rfree sets. Native BceC was refined to Rwork/Rfree of 16.3/19.7%. Based on all data, the final R factor was 16.5%. The four independent molecules of the AU contained 96.9, 3.0, and 0.1% of their residues within the preferred, allowed, or outlier regions of the Ramachandran diagram, respectively. Y10S and Y10K models were refined to Rwork/Rfree values of 16.3/19.7% and 22.8/26.3% and to final R values of 16.5 and 22.2%, respectively. The final models contained 97.5, 2.4, and 0.1% of Y10S residues and 95.5, 3.8, and 0.7% of Y10K residues within the preferred, allowed, and outlier regions of the Ramachandran diagram, respectively. Y10F could not be crystallized; Y10K crystals were produced but, compared with those of Y10S or BceC, they showed over 1-Å lower resolution limit and weaker diffraction power though the structure could be refined to acceptable parameters. No obvious differences were detected between BceC and its mutants, except for the mutated resi due 10.
Crystal contents of native BceC and its 10Y mutants.
While the two residues of the linker between the His tag (47) and the first methionine of BceC could be modeled as Gly2 and Ser1 (except for the Y10K mutant, where only the serine was visible), the six histidines of the tag and the last 12 residues (up to 17 residues in the case of the Y10K mutant) of the C termini were not visible in 1 σ electron density maps and were therefore omitted from the models. While chain A in the native form and Y10S mutant could be uninterruptedly modeled, two solvent-exposed loops (by residues 88 to 94 or 142 to 145) in other chains were eventually invisible in the maps and were omitted from the models. Similarly, some residues were modeled in multiple conformations. Globally, 1,821, 1,823, and 1,808 out of 1,912 possible residues (four chains with His6-Gly-Ser plus 470 residues) were modeled in native BceC, Y10S, and Y10K mutant structures, respectively. Additionally, a UDP-GlcA molecule was found docked to each protein chain, and whenever characteristic positive (mFo − DFc) density (43) was found in a chemically suitable environment, sulfate, acetate, glycerol, or Tris solvent molecules were modeled in addition to solvent waters, in particular for the higher-resolution crystals of the native form and Y10S mutant. Globally, a total of 1,374 or 1,304 solvent molecules were modeled in the native or Y10S structure, respectively (find further details in Table 1).
BceC structural features.
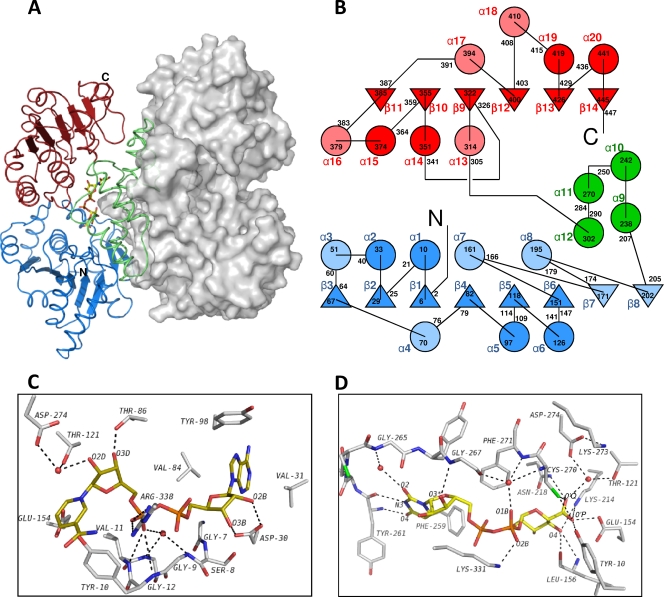
BceC is consisted of Rossmann-like fold domains at the N-terminal (residues 1 to 205) and C-terminal regions (residues 305 to 459), linked together by an intermediate which belongs entirely to the α subdomain (residues 206 to 304). This subdomain, similar to other dehydrogenases, harbors the catalytic center and is the main contributor to the enzyme's homodimerization. A fit of the four crystallographically independent BceC chains against the S. pyogenes UGD of 402 residues with 23% sequence identity showed that 332 to 337 residues get superposed within a 1.75-Å radius cutoff and within 1.3- to 1.5-Å Cα RMSDs. This was relatively high in comparison with the Cα RMSD values between the BceC chains, which were within 0.4 to 0.7 Å. The low sequence identity between the two proteins is reflected in their 3D dissimilarities, which hindered PHASER in solving the phase problem by molecular replacement.
In solution, BceC forms particles of ca. 100 kDa, according to dynamic light scattering (DLS) or gel filtration chromatography assays (data not shown), similar to E. coli (49) or P. aeruginosa (27) UGDs, indicating that homodimers (two chains of 470 residues) are the BceC functional units. A crystal packing analysis also confirms such dimeric arrangement, with four molecules in the asymmetric unit associated in two dimers. Their formation implies a solvent occlusion of about 2.6 × 103 Å2, which corresponds to 13% solvent-accessible surface per monomer. Each of the remaining crystal packing-interacting regions does not exceed 3.5% solvent-accessible area per monomer. Dimerization involves interactions mainly between the intermediating helical subdomains (Fig. 2A and B), with particular relevance to an antiparallel coupling of the two dimer partners long central helices α9. The dimerization interface involves 65 residues (of the 99 that compose each helical subdomain) and ca. 20 hydrogen bond interactions.
Fig. 2.
BceC 3D structure. (A) View of the BceC homodimer, with the left monomer as a cartoon and the right one represented by its solvent-accessible surface. BceC domains are highlighted in blue and red for the N- and C-terminal dinucleotide binding Rossmann-like folds, respectively, and in green for the intermediate α-helical subdomain. (B) BceC topology with β-strands as triangles and α-helices as circles. Dark blue and dark red items represent topological features common to both Rossmann-like folds, in contrast to light blue and light red that represent unique secondary structure elements. The intermediate α-helical subdomain is shown in green. Secondary structure elements are labeled alphanumerically, with delimiting residue numbers in black. (C) BceC cofactor binding pocket showing peptide bonds as sticks, with atoms shown in gray for carbon, in blue for nitrogen, in red for oxygen, in orange for phosphorus, and in green for sulfur and including an in silico docked NAD(H) model with carbon shown yellow. (D) Representation similar to that in panel C of the BceC substrate/product binding pocket. While the glucosidic moiety interacts mainly with conserved BceC residue side chains, including the catalytic amino acids, pyrophosphate and uridine groups interact essentially with the peptide main chain, often via bridging water molecules. A lower sequence conservation is found for these loci than with, e.g., the cofactor NAD+ binding pocket (see Table S1 in the supplemental material for further details).
Cofactor binding pocket.
Crystals of BceC were obtained only if both the cofactor, NAD+, and the reaction product, UDP-GlcA, were present in the initial protein solution (47). However, no cofactor was bound in the crystals that were obtained, as indicated by the electron density map. The absence (or degradation) of the cofactor in UGD crystal structures seems to correlate with low crystallization pHs (3, 34, 37). Examples include the UGD structures from Porphyromonas gingivalis (pH 5.4; PDB code 3gg2), Caenorhabditis elegans (pH 5.5; PDB code 2o3j), Homo sapiens (pH 6.0; PDB code 3itk), and S. elodea (pH 6.0; showing NAD degradation) (J. Rocha, unpublished data). In contrast, structures of UGD where the NAD cofactor was observed at higher pHs include structures from H. sapiens (pH 6.5; PDB code 2qg4) and S. pyogenes (pH 7.8) (6).
In an attempt to compare the BceC cofactor binding pocket with those of the human and S. pyogenes UGDs, whose structures had been experimentally obtained with docked NAD, a superposition of their 3D models was performed using SSM (29) within COOT (17). The observed high local homology in the pocket allowed a manual fit of NAD also in BceC, as shown in Fig. 2C. The UGD cofactor lies at an accessible cleft near the carboxy ends of strands β1, β4, and β5 in an extended conformation similar to several NAD+-enzyme complexes (3), leaving most of its atoms exposed to the solvent. This is in agreement with a “Bi-Uni-Uni-Bi ping-pong” mechanism, whereby the alcohol UDP-Glc binds first before the acid UDP-GlcA is released (7, 23, 40). The cofactor molecules then readily access the enzyme, being reduced and released into the medium without requirement of major conformational changes of the enzyme molecule. Several conserved residues in the UDP-glucose/GDP-mannose 6-dehydrogenase family (UGD/GMD) are involved in cofactor-protein interactions. At the beginning of the first Rossmann fold, a stretch of 30 to 35 amino acid residues shows a high degree of similarity, reminiscent of different types of dehydrogenases such as alcohol or aldehyde dehydrogenases, and is named the fingerprint region (3). The side chain of Asp30, a conserved residue in this region, establishes hydrogen bonds with the adenine ribose hydroxyls O2B and O3B (Fig. 2C). Also located in the fingerprint region is the glycine-rich consensus sequence, GXGYXG (conserved Tyr10 residue in italics), also known as a phosphate-binding loop, where main-chain amides donate hydrogens, either directly or via bridging water molecules, for H bonding with the pyrophosphate moiety. It is noteworthy that conservation in the UGD/GMD family of proteins seems to go beyond the three glycine residues, as the fourth position of this motif, Tyr10 in BceC, is also strictly conserved in the family. Surprisingly, the nicotinamide carboxamide group of the in silico docked NAD was found overlapping the side chains of Tyr10 and Glu154 (Fig. 2C; see also discussion in the next section).
Substrate binding pocket.
In contrast to the cofactor cleft, the substrate-binding pocket, where the reaction product was found (Fig. 2D), is deeply buried, requiring protein structural changes for the access and release of reactant and product (6). Almost all N or O atoms of the substrate are engaged in H-bonding interactions with the enzyme. Main-chain interactions are prevalent with uracil or ribose moieties, while the interactions with glucose, usually in multiple hydrogen bonds, occur mainly with side chains of the residues. Also noteworthy is the higher residue conservation in the vicinity of the active site than in the remaining substrate binding pocket. Six residues are within van der Waals distances to UDP-GlcA, which establishes hydrogen bonds with another 11 residues and 11 solvent water molecules. A detailed list of BceC hydrogen bond interactions in the substrate binding pocket is presented in Table S1 in the supplemental material, together with those found in the UGD structures of H. sapiens (PDB code 2q3e) and S. pyogenes structures (6), evidencing a highly conserved hydrogen-bonding pattern within the substrate binding pocket in the three structures. The UDP moiety is flanked by a coiled stretch comprising residues 259 to 267, with the uracil and Phe259 rings showing a π-edge stacking similar to that observed in homologous Phe265 and Tyr249 from H. sapiens and S. pyogenes UGDs, respectively. As mentioned earlier, the enzyme functions as a homodimer. The Arg254 from one monomer extends its side chain into its partner monomer's active site; the guanido group forms hydrogen bonds with the O2′ and O3′ hydroxyls of the sugar (data not shown). Additionally, the Lys214 ε-amino group forms a hydrogen bond with O4′ and, together with the Asn218 side chain amide, also hydrogen bonds to O′P of the UDP-GlcA carboxylic group. Its O′Q is, in turn, involved in hydrogen bonds with the sulfhydryl of catalytic Cys270, with the hydroxyl of Tyr10, and with the carboxylic of Glu154. These two last interactions are absent in both S. pyogenes and in human UGDs because their structures were obtained from ternary complexes including both reactants while in the present study no NAD+ is present. The superposition of the three structures showed that Tyr10 and Glu154 side chains have space to adopt the conformations of their homologues in S. pyogenes or in human UGDs, but as BceC contains no NAD, they access instead the cofactor binding pocket. The O1B atom of the pyrophosphate is hydrogen bonded to a solvent water molecule that, in turn, forms several hydrogen bonds in the immediate vicinity of the catalytic site, namely, with Gly267 carbonyl and main-chain amides from Cys270 and Phe271, and thus helps to set the catalytic positioning of substrate and Cys270.
Catalytic mechanism.
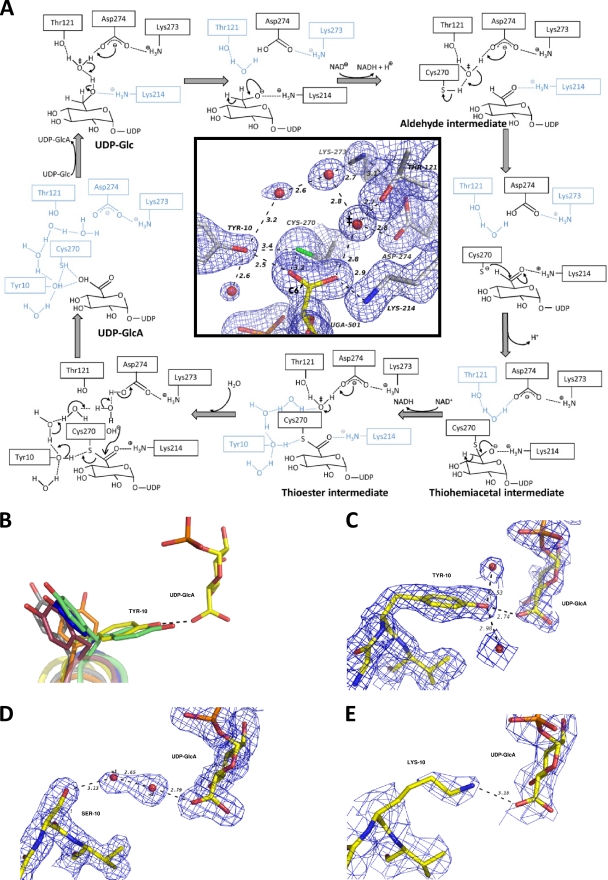
The UGD/GMD conversion of an alcohol into a carboxylic acid requires a four-electron oxidation of the pyranose C-6′ center during its transformation from a methylene into an acyl group. The reaction engages two successive hydride transfers to NAD+ and a final irreversible hydrolysis of a thioester intermediate at the covalent bonded sulfur and C-6′ atoms. The scheme of the reaction mechanism was initially determined based on bovine liver UGD studies (40) but has been successively fine-tuned using kinetic, point mutational, and crystallographic studies also with the S. pyogenes and human enzymes. The reaction (Fig. 3A) is initiated with deprotonation of a water molecule by the carboxylate group of Asp274 (15, 23), in which the base capacity and catalytic positioning are influenced by hydrogen bonds with side chains of Asn218 and Lys273 (52). As this carboxylate group is solvent accessible (when no NAD+/NADH is docked), it works as a direct exit route for the two protons produced during the overall reaction. The hydroxide anion formed is kept in place by a hydrogen bond with the side chain of Thr121, whose positioning is in turn ensured by hydrogen bonding with Ser120 and Pro123 residues (23). Upon deprotonation of the UDP-Glc O6 hydroxyl by the hydroxide, the formed oxyanion is stabilized by electrostatic interactions with the nearby ε-amino group of Lys214, at hydrogen-bonding distance (6), until the first C-6′ pro-R hydride of UDP-Glc is transferred onto NAD+ (6, 45). The resulting aldehyde intermediate (7) remains sequestered by the enzyme (23, 39). The increase in the oxidation state with the electron withdrawal effect of the double-bonded oxygen in C-6′ makes it susceptible to a nucleophilic attack. The catalytic water/hydroxide site also neighbors the sulfhydryl group of Cys270 and may extract its proton. Thus, the thiolate formed proceeds with a nucleophilic attack to the previously formed acyl center at C-6′, leading to the formation of a covalently bonded thiohemiacetal intermediate (PDB 3KHU) (23, 45, 52). The resulting oxyanion is again stabilized by electrostatic interactions with the nearby ε-amino group of Lys214 (6) until transfer of the second C-6′ pro-S hydride (6, 45) into a freshly docked NAD+, and the global oxidation of C-6′ is accomplished with the resulting thioester. The final catalytic step, the irreversible hydrolysis of the thioester (40, 45), involves a second nucleophilic attack to C-6′, most possibly again by a water/hydroxyl activated by Asp274, which will lead to the disruption of the thioester sulfur-carbon bond and will produce the final carboxylic group at C-6′. This is the unique irreversible and rate-limiting step of the overall reaction (40) and requires the stabilization of the departing thiolate. However, as the ε-amino group of Lys214 is at the opposite side relative to C-6′ (Fig. 3A, inset), it is too far away to be invoked as a key residue in this step, and the relevant catalytic agent of the hydrolysis step thus still remains elusive (15, 23).
Fig. 3.
BceC Tyr10 is a key residue in the UGD/GMD family mechanism. (A) Enzymatic mechanism of BceC in black (catalytic resting components in cyan), by homology with the UGD mechanism reported in the literature (see text). The insert shows a view of the BceC active site, with the reaction center (C-6′), the catalytic water/hydroxide ion (‡), the Cys270 side chain, and the Tyr10 side chain, among other active site components. The Tyr10 hydroxyl group is located at the opposite side of Lys214 relative to the acyl group and may work as a proton conveyer from the aqueous medium to a forming thiolate, making it thus capable of catalyzing the hydrolysis of the intermediate thioester. Bonds are shown as sticks (color codes are as described in the legend of Fig. 2); blue mesh represents 1.0 σ Fourier synthesis map (43), and black dashes represent hydrogen bonds. (B) The side chain of the conserved Tyr within the UGD glycine-rich motif was found adopting various Chi1 values, with a gauche(+) G+ conformation only for BceC and for one of the alternating side chains in P. gingivalis, when no cofactor occupied its binding pocket. Bonds are shown as sticks but with carbon shown in yellow for BceC, green for P. gingivalis (PDB code 3gg2), pink for H. sapiens (PDB code 2q3e), blue for S. pyogenes (PDB code 1dli), gray for C. elegans (PDB code 2o3j), and orange for S. elodea (J. Rocha et al., unpublished data). (C) In the native BceC structure the Tyr10 hydroxyl group forms hydrogen bonds with UDP-GlcA carboxylate and two solvent water molecules (the interaction with thiol of Cys270 is not shown here for clarity). (D) In the Y10S mutant two solvent water molecules are involved in a hydrogen-bonding network between Ser10 Oγ and UDP-GlcA carboxylate. (E) In the Y10K mutant a salt bridge is formed between the Lys10 ε-amino group and UDP-GlcA.
UGD/GMD final hydrolysis step is catalyzed by the conserved Y of the GXGYXG consensus motif.
In BceC, and similar to P. gingivalis UGD structure (PDB code 3gg2), the hydroxyl group of the conserved Tyr10 establishes hydrogen bonds with the thiol group of Cys270, the carboxylic group of UDP-GlcA, and to solvent water molecules (Fig. 3A, inset, and B). We performed mutagenic studies on BceC, whereby we mutated Tyr10 into phenylalanine, serine, and lysine. These mutants showed severely impaired activities, with decreases down to 9%, 3%, and 2% of BceC activity for Y10F, Y10S, and Y10K, respectively. We followed the activity of BceC and Y10F (the other mutants display activity that is too low) under saturating concentrations of substrate and cofactor at pH 8.7 (data not shown), and we compared their activity profiles against temperature (Fig. 1B). It is noteworthy that latent activity was detected, similar to that reported for the Y10F mutation in B. subtilis UGD, where the critical importance of the Tyr in catalysis was anticipated but not further discussed (36). The structures and kinetic data that we present in this study imply that Tyr10 plays a catalytic role in the final hydrolysis step. Upon release of NADH after the second oxidation step, Tyr10 may work as a proton conveyer from the aqueous hydrogen-bonding proton “wire” system (10, 25) to the hydrolytic site. In fact, the hydroxyl of Tyr10 is not only correctly positioned to promote stabilization of the forming thiolate upon direct proton transfer but also has its two electron doublets establishing hydrogen bonds with the surrounding water-solvent molecules (Fig. 3A, inset, and C). The three inactive mutants lack this capacity at residue 10: Y10F has no proton donor group; Y10S has an OG hydroxyl, but the side chain is too short for its direct assistance to the proton transfer (Fig. 3D); and Y10K, at biological pH, has the ε-amino group in the protonated ammonium form, and thus there is no electron doublet free to convey protons from the aqueous medium (Fig. 3E). Interestingly, although Tyr and Lys side chains show similarly high pKa values—above 10 (9)—they display contrasting catalytic capacities because the transfer of aqueous protons to the forming thiolate via the tyrosine hydroxyl group does not result from its general acid-base properties. This is due to the two hydrogen-bonded water molecules at Tyr10 (Fig. 3A, inset, and C): a proton always remains attached to the OH of the tyrosine throughout the catalytic process. In the absence of Tyr, hydrolysis proceeds only when a solvent water molecule happens to be in place for donation of the stabilizing proton to the forming thiolate. Such a difference in the mechanisms of native versus Y10 mutated BceC implies a similar catalytic activation state but two different ground states, particularly in the readiness to provide a correctly positioned proton to stabilize the thiolate. In fact, the dependence of their activities on temperature (Fig. 1B) confirms that the decreased activity of the Y10F mutant relative to BceC should be ascribed mainly to a higher activation entropy difference, ΔS‡ (more negative), of Y10F versus that of BceC, as shown by their ln (k/T) versus 1/T plots, with similar slopes but different origins (Fig. 1C).
In contrast with earlier structures, where the presence of NAD excluded the Tyr10 side chain from the active site, the BceC structure allowed the visualization of Tyr10 side chain interactions with its catalytic partners and thus prompted the final elucidation of the irreversible and rate-determining UGD catalytic step.
Because of its crucial role on cepacian synthesis, it has been suggested that BceC could be an attractive target for drug development against Burkholderia strains in cystic fibrosis patients. However, the fact that Burkholderia species have multiple copies of this gene makes this approach less attractive. On the other hand, the available structural data indicate that the residues involved in the catalytic mechanisms are conserved among all prokaryotic and human UGDs (16). Taken together, these data could indicate that UGD might not be an appropriate target for the development of anti-Burkholderia drugs. Nevertheless, Egger and coworkers (16) have recently performed a sequence-based analysis and a structural classification of prokaryotic and eukaryotic UGDs available in the databases. Interestingly, this study reveals that UGD enzymes can be categorized in two groups (UGD-I and UGD-II) according to their origins, molecular sizes, and quaternary structures. UGD-I includes prokaryotic enzymes, whereas UGD-II represents the members from eukaryotic organisms. These findings may open new possibilities for further investigations of the use of UGD as a new therapeutic target for drug development against Burkholderia strains in cystic fibrosis patients.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff at ESRF in Grenoble, France, for the use of synchrotron radiation.
Fundação para a Ciência e Tecnologia (FCT) granted Ph.D. fellowships to J.R. (SFRH BD/24216/2005), to A.O.P. (SFRH/BD/30430/2006), and to D.M.-H. (SFRH/BD/41760/2007). This work was funded by FCT grants POCTI/BME/38859/2001, POCI/BIO/58401/2004, PTDC/QI/67925/2006, and PTDC/SAU-MII/69591/2006.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Adams P. D., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agodi A., et al. 2001. Burkholderia cepacia complex infection in Italian patients with cystic fibrosis: prevalence, epidemiology, and genomovar status. J. Clin. Microbiol. 39:2891–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bellamacina C. R. 1996. The nicotinamide dinucleotide binding motif: a comparison of nucleotide binding proteins. FASEB J. 10:1257–1269 [DOI] [PubMed] [Google Scholar]
- 4. Brisse S., et al. 2004. Species distribution and ribotype diversity of Burkholderia cepacia complex isolates from French patients with cystic fibrosis. J. Clin. Microbiol. 42:4824–4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Campana S., et al. 2005. Transmission of Burkholderia cepacia complex: evidence for new epidemic clones infecting cystic fibrosis patients in Italy. J. Clin. Microbiol. 43:5136–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Campbell R. E., Mosimann S. C., van De Rijn I., Tanner M. E., Strynadka N. C. 2000. The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation. Biochemistry 39:7012–7023 [PubMed] [Google Scholar]
- 7. Campbell R. E., Sala R. F., van de Rijn I., Tanner M. E. 1997. Properties and kinetic analysis of UDP-glucose dehydrogenase from group A streptococci. Irreversible inhibition by UDP-chloroacetol. J. Biol. Chem. 272:3416–3422 [DOI] [PubMed] [Google Scholar]
- 8. Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 9. Creighton T. E. 1993. Proteins, 2nd ed. W. E. Freeman and Company, New York, NY [Google Scholar]
- 10. Cukierman S. 2006. Et tu, Grotthuss! and other unfinished stories. Biochim. Biophys. Acta 1757:876–885 [DOI] [PubMed] [Google Scholar]
- 11. Cunha M. V., et al. 2003. Molecular analysis of Burkholderia cepacia complex isolates from a Portuguese cystic fibrosis center: a 7-year study. J. Clin. Microbiol. 41:4113–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cunha M. V., et al. 2007. Exceptionally high representation of Burkholderia cepacia among B. cepacia complex isolates recovered from the major Portuguese cystic fibrosis center. J. Clin. Microbiol. 45:1628–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DeLano W. L. 2008. The Pymol molecular graphics system. DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 14. Diederichs K., Karplus P. A. 1997. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 4:269–275 [DOI] [PubMed] [Google Scholar]
- 15. Easley K. E., Sommer B. J., Boanca G., Barycki J. J., Simpson M. A. 2007. Characterization of human UDP-glucose dehydrogenase reveals critical catalytic roles for lysine 220 and aspartate 280. Biochemistry 46:369–378 [DOI] [PubMed] [Google Scholar]
- 16. Egger S., Chaikuad A., Kavanagh K. L., Oppermann U., Nidetzky B. 2010. UDP-glucose dehydrogenase: structure and function of a potential drug target. Biochem. Soc. Trans. 38:1378–1385 [DOI] [PubMed] [Google Scholar]
- 17. Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 18. Evans M. G., Polanyi M. 1935. Some applications of the transition state method to the calculation of reaction velocities, especially in solution. Trans. Faraday Soc. 31:875–894 [Google Scholar]
- 19. Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62:72–82 [DOI] [PubMed] [Google Scholar]
- 20. Eyring H. 1935. The activated complex in chemical reactions. J. Chem. Phys. 3:107–115 [Google Scholar]
- 21. Ferreira A. S., et al. 2010. Distribution of cepacian biosynthesis genes among environmental and clinical Burkholderia strains and role of cepacian exopolysaccharide in resistance to stress conditions. Appl. Environ. Microbiol. 76:441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. French G. S., Wilson K. S. 1978. On the treatment of negative intensity observations. Acta Crystallogr. A 34:517–525 [Google Scholar]
- 23. Ge X., Penney L. C., van de Rijn I., Tanner M. E. 2004. Active site residues and mechanism of UDP-glucose dehydrogenase. Eur. J. Biochem. 271:14–22 [DOI] [PubMed] [Google Scholar]
- 24. Granja A. T., Popescu A., Marques A. R., Sa-Correia I., Fialho A. M. 2007. Biochemical characterization and phylogenetic analysis of UDP-glucose dehydrogenase from the gellan gum producer Sphingomonas elodea ATCC 31461. Appl. Microbiol. Biotechnol. 76:1319–1327 [DOI] [PubMed] [Google Scholar]
- 25. Grotthuss C. J. T. 1806. Sur la décomposition de l'eau et des corps q'uelle tient en dissolution à l'aide de l'électricité galavnique. Ann. Chim. LVIII:54–74 [Google Scholar]
- 26. Hempel J., et al. 1994. UDP-glucose dehydrogenase from bovine liver: primary structure and relationship to other dehydrogenases. Protein Sci. 3:1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hung R. J., et al. 2007. Comparative analysis of two UDP-glucose dehydrogenases in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 282:17738–17748 [DOI] [PubMed] [Google Scholar]
- 28. Krissinel E., Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 [DOI] [PubMed] [Google Scholar]
- 29. Krissinel E., Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60:2256–2268 [DOI] [PubMed] [Google Scholar]
- 30. Leslie A. G. W. 1992. Recent changes to the MOSFILM package for processing film and image plate data. Joint CCP4 ESF-EACBM Newsl. Protein Crystallogr. 26:27–33 [Google Scholar]
- 31. LiPuma J. J., et al. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 164:92–96 [DOI] [PubMed] [Google Scholar]
- 32. Loutet S. A., Bartholdson S. J., Govan J. R., Campopiano D. J., Valvano M. A. 2009. Contributions of two UDP-glucose dehydrogenases to viability and polymyxin B resistance of Burkholderia cenocepacia. Microbiology 155:2029–2039 [DOI] [PubMed] [Google Scholar]
- 33. Mahenthiralingam E., et al. 2001. Infection with Burkholderia cepacia complex genomovars in patients with cystic fibrosis: virulent transmissible strains of genomovar III can replace Burkholderia multivorans. Clin. Infect. Dis. 33:1469–1475 [DOI] [PubMed] [Google Scholar]
- 34. Margolis S. A., Howell B. F., Schaffer R. 1976. Purification and analysis of the purity of NADH. Clin. Chem. 22:1322–1329 [PubMed] [Google Scholar]
- 35. McCoy A. J., et al. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mijakovic I., Petranovic D., Deutscher J. 2004. How tyrosine phosphorylation affects the UDP-glucose dehydrogenase activity of Bacillus subtilis YwqF. J. Mol. Microbiol. Biotechnol. 8:19–25 [DOI] [PubMed] [Google Scholar]
- 37. Miksic J. R., Brown P. R. 1978. Reactions of reduced nicotinamide adenine dinucleotide in acid: studies by reversed-phase high-pressure liquid chromatography. Biochemistry 17:2234–2238 [DOI] [PubMed] [Google Scholar]
- 38. Moreira L. M., et al. 2003. Identification and physical organization of the gene cluster involved in the biosynthesis of Burkholderia cepacia complex exopolysaccharide. Biochem. Biophys. Res. Commun. 312:323–333 [DOI] [PubMed] [Google Scholar]
- 39. Nelsestuen G. L., Kirkwood S. 1971. The mechanism of action of uridine diphosphoglucose dehydrogenase. Uridine diphosphohexodialdoses as intermediates. J. Biol. Chem. 246:3828–3834 [PubMed] [Google Scholar]
- 40. Ordman A. B., Kirkwood S. 1977. UDPglucose dehydrogenase. Kinetics and their mechanistic implications. Biochim. Biophys. Acta 481:25–32 [DOI] [PubMed] [Google Scholar]
- 41. Painter J., Merritt E. A. 2006. TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39:109–111 [Google Scholar]
- 42. Perrakis A., Harkiolaki M., Wilson K. S., Lamzin V. S. 2001. ARP/wARP and molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 57:1445–1450 [DOI] [PubMed] [Google Scholar]
- 43. Read R. J. 1986. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A 42:140–149 [Google Scholar]
- 44. Reik R., Spilker T., Lipuma J. J. 2005. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. J. Clin. Microbiol. 43:2926–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ridley W. P., Houchins J. P., Kirkwood S. 1975. Mechanism of action of uridine diphosphoglucose dehydrogenase. Evidence for a second reversible dehydrogenation step involving an essential thiol group. J. Biol. Chem. 250:8761–8767 [PubMed] [Google Scholar]
- 46. Rocha J., Granja A. T., Sa-Correia I., Fialho A., Frazao C. 2010. Cloning, expression, purification, crystallization and preliminary crystallographic studies of UgdG, an UDP-glucose dehydrogenase from Sphingomonas elodea ATCC 31461. Acta Crystallogr Sect. F Struct. Biol. Cryst. Commun. 66:69–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rocha J., Popescu A. O., Sa-Correia I., Fialho A. M., Frazao C. 2010. Cloning, expression, purification, crystallization and preliminary crystallographic studies of BceC, a UDP-glucose dehydrogenase from Burkholderia cepacia IST 408. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66:269–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roman E., Roberts I., Lidholt K., Kusche-Gullberg M. 2003. Overexpression of UDP-glucose dehydrogenase in Escherichia coli results in decreased biosynthesis of K5 polysaccharide. Biochem. J. 374:767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sieberth V., Rigg G. P., Roberts I. S., Jann K. 1995. Expression and characterization of UDPGlc dehydrogenase (KfiD), which is encoded in the type-specific region 2 of the Escherichia coli K5 capsule genes. J. Bacteriol. 177:4562–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sist P., et al. 2003. Macromolecular and solution properties of Cepacian: the exopolysaccharide produced by a strain of Burkholderia cepacia isolated from a cystic fibrosis patient. Carbohydr. Res. 338:1861–1867 [DOI] [PubMed] [Google Scholar]
- 51. Snook C. F., Tipton P. A., Beamer L. J. 2003. Crystal structure of GDP-mannose dehydrogenase: a key enzyme of alginate biosynthesis in P. aeruginosa. Biochemistry 42:4658–4668 [DOI] [PubMed] [Google Scholar]
- 52. Sommer B. J., Barycki J. J., Simpson M. A. 2004. Characterization of human UDP-glucose dehydrogenase. CYS-276 is required for the second of two successive oxidations. J. Biol. Chem. 279:23590–23596 [DOI] [PubMed] [Google Scholar]
- 53. Sousa S. A., et al. 2007. Virulence of Burkholderia cepacia complex strains in gp91phox−/− mice. Cell Microbiol. 9:2817–2825 [DOI] [PubMed] [Google Scholar]
- 54. Terwilliger T. C. 2003. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 59:38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weiss M. S. 2001. Global indicators of X-ray data quality. J. Appl. Crystallogr. 34:130–135 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.