Abstract
Anabaenopeptins (AP) are bioactive cyclic hexapeptides synthesized nonribosomally in cyanobacteria. APs are characterized by several conserved motifs, including the ureido bond, N-methylation in position 5, and d-Lys in position 2. All other positions of the AP molecule are variable, resulting in numerous structural variants. We have identified a nonribosomal peptide synthetase (NRPS) operon from Planktothrix agardhii strain CYA126/8 consisting of five genes (apnA to apnE) encoding six NRPS modules and have confirmed its role in AP synthesis by the generation of a mutant via insertional inactivation of apnC. In order to correlate the genetic diversity among adenylation domains (A domains) with AP structure variation, we sequenced the A domains of all six NRPS modules from seven Planktothrix strains differing in the production of AP congeners. It is remarkable that single strains coproduce APs bearing either of the chemically divergent amino acids Arg and Tyr in exocyclic position 1. Since the A domain of the initiation module (the ApnA A1 domain) has been proposed to activate the amino acid incorporated into exocyclic position 1, we decided to analyze this domain both biochemically and phylogenetically. Only ApnA A1 enzymes from strains producing AP molecules containing Arg or Tyr in position 1 were found to activate these two chemically divergent amino acids in vitro. Phylogenetic analysis of apn A domain sequences revealed that strains with a promiscuous ApnA A1 domain are derived from an ancestor that activates only Arg. Surprisingly, positive selection appears to affect only three codons within the apnA A1 gene, suggesting that this remarkable promiscuity has evolved from point mutations only.
INTRODUCTION
Cyclic peptides of both ribosomal and nonribosomal biosynthetic origin from bacteria and fungi possess a wide range of biological activities. Among bacteria, the cyanobacteria are prolific producers of biologically active compounds that in most cases arise via the nonribosomal peptide synthesis (NRPS) pathway, sometimes in combination with the polyketide synthesis (PKS) pathway via the thio-template mechanism (48). NRPS enzymes have a modular structure, and except for the initiation module, each module contains specific functional domains for activation (aminoacyl adenylation domains [A domains]), thioesterification (thiolation domains [T domains]) of the activated monomer, and elongation (condensation domains [C domains]) of the growing natural product. In addition, a number of tailoring enzymes, such as methyltransferases, epimerases, and thioesterases (TEs), lead to the modification of the synthesized product (11). It is a characteristic feature of cyanobacterial NRPS pathways that they often produce entire families of structurally related compounds co-occurring in one specific isolate, e.g., the cryptophycins (15), laxaphycins (2, 13), and nostopeptolides (16). In such NRPS peptide families, structurally related amino acids, such as Val, Ile, and Leu, are found in equivalent positions of the peptides, suggesting that the A domains of these NRPS enzymes may possess a relaxed substrate specificity (19). Since in principle each A domain involved in biosynthesis could have such a relaxed specificity, the producer organism may be able to generate a true natural combinatorial library with a diversity limited by the number of A domains that display relaxed substrate specificity for structurally related amino acids, e.g., as observed for the insulapeptolides (29).
In contrast to these semiconservative substitutions of related amino acids, in some other metabolite classes, e.g., the anabaenopeptins (APs) and the microcystins (MCs), chemically distinct amino acids occupy equivalent positions in congeners retrieved from one strain. For example, we have recently described the structures of APs 908 and 915 (Fig. 1) from Planktothrix agardhii strain CYA126/8, which differ in the exocyclic ureido-bound amino acids (Arg or Tyr) that are attached to a common cyclopentapeptide core (34). An analogous difference between Leu and Arg is found in position 2 within MCs produced by the same isolate, i.e., MC-[Asp3]-RR and MC-[Asp3]-LR (4). Therefore, one can postulate that the first A domain of McyB (the McyB A1 domain), which is responsible for the activation of amino acids in position 2 of the MC molecule, is able to activate Arg and Leu (4). The polymorphism within the mcyB A1 domain, as observed for the genera Microcystis, Planktothrix, and Anabaena, has been investigated in detail (10, 26, 30, 45). Within each genus, the genetic variation within the mcyB A1 domain could be correlated with a specific structural variation in position 2 of the MC molecules found in the genus. For example, among Planktothrix strains, mcyB A1 genotypes that produce MC variants bearing either Arg or Leu, or either homotyrosine (Hty) or Leu, in position 2 were identified (26). Unfortunately, efforts to characterize the specific substrate activation profiles of McyB A1 domains biochemically have not been successful to date.
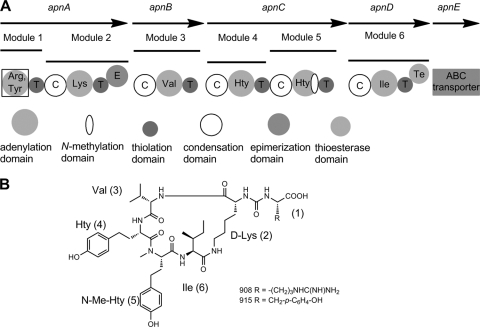
Fig. 1.
(A) Scheme of the structural organization of the anabaenopeptin (apn) biosynthetic gene cluster from Planktothrix agardhii strain CYA126/8. The ApnA A1 domain (boxed) was probed using the ATP-pyrophosphate exchange assay. (B) Chemical structures of anabaenopeptin variants (AP 908 and AP 915) produced by the same strain (34).
Recently, the AP gene cluster of Anabaena strain 90, comprising seven genes (aptA to aptF), was characterized, and the biosynthetic pathway for the three AP structural variants AP A (m/z 844), AP B (m/z 837), and AP C (m/z 809), containing either Tyr, Arg, or Lys, respectively, in the exocyclic position, was proposed (39). Interestingly, this AP gene cluster includes two aptA genes encoding two alternative NRPS starter bimodular proteins that putatively arose from duplication and subsequent intragenomic recombination. While the A domain of the initiation module (the AptA1 A1 domain) was not characterized biochemically, it was postulated that this module is responsible for the activation of Arg or Lys. In contrast, the A domain of the alternative starter module (the AptA2 A1 domain) was expressed and displayed high substrate selectivity for l-Tyr. The authors concluded that the presence of two aptA genes enables Anabaena strain 90 to incorporate chemically distinct amino acids into the equivalent position during the biosynthesis of the three different AP variants. The same authors described the apt gene clusters of Nostoc PCC73102 and Nodularia spumigena, neither of which shows evidence of duplication of the starter module.
In the present study, we identified an NRPS gene cluster putatively responsible for AP synthesis in the genus Planktothrix and confirmed its role by insertional inactivation of the apnC gene. In order to elucidate the influence of the genetic diversity among A domains on AP structure variation, we compared the A domains of the apn gene cluster among seven Planktothrix sp. strains that differ in AP production in that they contain AP metabolites with Arg only or with either Arg or Tyr in exocylic position 1 (22). In contrast to the findings for the apt gene cluster from Anabaena strain 90 (39), we found no evidence for duplication of a bimodular protein carrying two alternative initiation modules that could explain the co-occurrence of AP metabolites bearing Arg or Tyr in exocylic position 1. Therefore, we expressed representative A domains of the apt gene cluster initiation module from seven Planktothrix strains as His6-tagged fusion proteins and conducted assays for adenylation activity in vitro using the standard ATP-pyrophosphate exchange assay (27). The results show that only strains producing AP molecules bearing Arg or Tyr in position 1 possess ApnA A1 domains that activate both amino acids. In contrast, ApnA A1 domains from strains producing AP molecules bearing solely Arg in position 1 are specific for Arg only.
MATERIALS AND METHODS
Bacterial strains and AP composition.
Planktothrix sp. strains were grown in BG11 medium under sterile conditions at 20°C and 40 to 60 μmol m−2 s−1 as described previously (22). The strains did not show genetic variation within the 16S rRNA gene region (301 bp). However, they were polymorphic within other housekeeping gene regions, i.e., in spacer regions between the 16S rRNA and 23S rRNA, cpcB and cpcA, and psaA and psaB genes (5). Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) was used to determine AP structural composition either directly from harvested biomass (23) or from aqueous-methanol extracts fractionated by high-performance liquid chromatography with diode array detection (HPLC-DAD) (22, 47). This analysis revealed the occurrence of two groups of AP structural variants among the strains investigated: (i) AP 908 (m/z 909) and AP 915 (m/z 916), produced by strain CYA126/8 (Fig. 1), and (ii) AP B (m/z 837), AP F (m/z 851), AP A (m/z 844), and oscillamide Y (m/z 858), produced by the other strains (Table 1).
Table 1.
Proportions of AP variants in seven Planktothrix sp. strains used in this study to analyze A domain variability within the AP gene cluster
| AP varianta | m/z | Amino acid at position: |
% of AP variant (avg ± SE)b in: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | CYA126/8 (Langsjön, Finland, 1984)c | No. 3 (Mondsee, Austria, 2001) | CCAP1459/36 (Gjersjön, Norway, 1968) | PCC7821 (Gjersjön, Norway, 1971) | No. 39 (Wannsee, Germany, 2001) | No. 66 (Jägerteich, Austria, 2001) | No. 80 (Schwarzensee, Austria, 2001) | ||
| AP 908 | 909 | Arg | Lys | Val | Hty | MHty | Ile | 38 ± 2 | ||||||
| AP 915 | 916 | Tyr | Lys | Val | Hty | MHty | Ile | 62 ± 2 | ||||||
| AP B | 837 | Arg | Lys | Val | Hty | MAla | Phe | 38 ± 2 | 22 ± 2 | 6 ± 0 | 29 ± 1 | 64 ± 6 | 100 ± 0 | |
| AP F | 851 | Arg | Lys | Ile | Hty | MAla | Phe | 22.2 ± 1 | 33 ± 1 | 12 ± 1 | 71 ± 1 | 36 ± 6 | ||
| AP A | 844 | Tyr | Lys | Val | Hty | MAla | Phe | 20 ± 1 | 17 ± 1 | 21 ± 2 | ||||
| Osc Y | 858 | Tyr | Lys | Ile | Hty | MAla | Phe | 21 ± 1 | 29 ± 2 | 61 ± 2 | ||||
Results from six replicates are shown. The average concentrations of APs (given in equivalents of AP B [22]) ± standard errors in strains CYA126/8, no. 3, CCAP1459/36, PCC7821, no. 39, no. 66, and no. 80 are 2 ± 0, 6 ± 1, 2 ± 0, 3 ± 1, 5 ± 0, 7 ± 1, and 6 ± 0 μg mg−1 (dry weight), respectively.
The origin and year of isolation of each strain are given in parentheses.
apnC insertional inactivation.
DNA was extracted by the procedure of Franche and Damerval (12). Since all APs contain a strictly conserved N-methylated amino acid in position 2, we performed PCR from DNA of Planktothrix strain CYA126/8 using forward primer MTF2 (33), binding in the conserved A2 motif of A domains, and a degenerated reverse primer designed for binding to the conserved region encoding the S-adenosylmethionine (SAM)-binding motif of the N-methyltransferase (NMT) in A domains (see Table S1 in the supplemental material). The resulting amplicon of approximately 1.5 kbp was cloned, and 10 colonies were isolated and analyzed by restriction fragment length polymorphism (RFLP) after transformation into Escherichia coli. Restriction analysis with MboI revealed only two different restriction types, and one representative of each group was sequenced. One clone represented the mcyA A1 domain (4), while the closest homolog for the second RFLP type was an uncharacterized A domain from Nostoc punctiforme PCC73102. This fragment, named A2-Nmt2, was the starting point for plasmid apnCKO, subsequently used in an insertional gene inactivation experiment according to our established technique (4). The A2-Nmt2 nucleotide sequence was then used to search a draft genome of P. agardhii strain CYA126/8 (R. Kurmayer and G. Christiansen, unpublished results), and an NRPS gene cluster putatively coding for AP synthesis was identified (EMBL/GenBank/DDBJ accession no. EF672686).
Analysis of APs.
Cells of wild-type (WT) strain CYA126/8 and the mutant culture were harvested on preweighed glass fiber filters (BMC; Ederol, Vienna, Austria). The cell mass was freeze-dried, and the filters were reweighed for the determination of the dry weight (6 mg each for wild type and the mutant). Filters were extracted in 50% (vol/vol) aqueous methanol (MeOH) and were analyzed by HPLC-DAD at 210 nm using a linear gradient of aqueous acetonitrile (0.05% [vol/vol] trifluoroacetic acid) starting with 20% acetonitrile (vol/vol) and increasing to 50% acetonitrile within 45 min with a flow rate of 1 ml min−1 using a LiChrospher 100 octyldecyl silane (ODS) (5-μm particle size) LiChroCART 250-4 cartridge system (Merck, Darmstadt, Germany), (20). Under the conditions described here, the detection limit was 10 ng for the AP B standard (22).
Amplification and cloning of A domains for DNA sequencing.
Oligonucleotide primers (see Table S1 in the supplemental material) were derived from the apn gene operon of strain CYA126/8 (EMBL/GenBank/DDBJ accession no. EF672686). The different loci were amplified by PCR under the following conditions: 50 μl (final volume) containing 50 ng of the chromosomal DNA template, 1× PCR buffer, 200 μM each deoxynucleotide, 0.25 μM each primer, and 2.5 U of QiaTaq DNA polymerase (Qiagen, Hilden, Germany). The following conditions were used: 95°C (3 min); 35 cycles of 94°C (30 s), 60°C (30 s), and 72°C (1 min per kb of the expected amplicon); and 95°C (3 min). The products of successful PCRs (45 μl) were purified and ligated into the pDRIVE vector (Qiagen). For each A domain, PCR amplicons were cloned and sequenced in triplicate.
Oligonucleotide primers ApnCA2+/−(see Table S1 in the supplemental material) failed to amplify a DNA fragment with DNA isolated from all Planktothrix strains except that of CYA126/8. Therefore, primers (ApnCA1+Aus60.9 LS NonCYA and ApnDA1-Inn60.2 LS NonCYA), binding further upstream at the 5′- and downstream at the 3′-end-flanking regions, were used. The resulting amplicon of 6,449 bp was cloned and sequenced.
Phylogenetic analysis.
The A domain sequences (1,441 bp) from all six NRPS modules were aligned (ClustalW2). Neighbor-joining (NJ) analysis, maximum parsimony (MP), and maximum likelihood (ML) were used to construct a phylogenetic tree (PHYLIP, version 3.6 alpha) (9). In general, sites were not weighted, and gaps in the alignment were not removed. For NJ, the DNA distance matrix was calculated using the F84 model (DNADIST), and a tree was constructed by successive clustering of lineages (NEIGHBOR). For MP, the method of Fitch as employed in the DNAPARS package was used to count the number of nucleotide changes needed on a phylogenetic tree. For ML, the default parameters of the DNAML package, such as a constant rate variation among sites, as well as a fixed transition/transversion ratio, were used. The statistical significance of the branches was estimated by bootstrap analysis generating 1,000 replicates of the original data set. Finally, consensus trees were computed by following the 50% majority rule. For all of the A domain genes, phylogenetic trees were congruent, and the NJ tree and the significant bootstrap values for all three methods are presented. The runs test implemented in the GENECONV program (version 1.81) (42) was used to investigate whether substitutions were significantly clustered and whether gene conversion (recombination) events occurred within the six apn A domains. The settings used were the default (/g0; mismatches within fragments were not allowed).
For the A domain sequences (excluding the NMT of the apnC A2 sequence) of the six NRPS modules, the ratio of nonsynonymous (dN) to synonymous (dS) substitutions per codon site was determined using maximum-likelihood estimates as implemented in the CODEML program of the PAML package (version 4.2b) (52). The “one-ratio” model, estimating one dN/dS ratio for all the A domain sequences, and a “six-ratio” model assuming that the A domain dN/dS ratios differ between the six NRPS modules, were statistically compared by constructing a likelihood ratio test (LRT) (51). The difference between the two models was found to be significant (twice the log likelihood difference [2Δl] = 29.7; df = 5; P < 0.001), implying that the overall A domain dN/dS ratio (0.19) differs significantly from the A domain dN/dS ratios determined for each NRPS module separately (see Results).
To test for positive selection within the A domain sequences of an NRPS module, an LRT (df = 2) was again constructed to compare the likelihoods of the phylogenetic trees calculated with two different types of models: (i) the null models M1 (nearly neutral), M7 (beta), and M8a (beta & dN/dS = 1), which do not allow for positively selected sites (dN/dS ≤ 1), and (ii) the alternative models M2 (positive selection) and M8 (beta & dN/dS > 1), with an additional site class that accounts for positive selection (dN/dS > 1). If the LRT was found significant, the Bayes empirical Bayes (BEB) approach was used to calculate the posterior probabilities that a site comes from the site class with dN/dS > 1 (53).
Heterologous overexpression and protein purification.
PCR amplicons obtained with the primer pair ApnAA1fwd(54/67)-ApnAA1rev(55/67), which were used for protein overexpression, were digested with NdeI and XhoI (MBI Fermentas, St. Leon-Rot, Germany). The digested and purified amplicons were ligated into the identically digested vector pET28b (Novagen, Merck, Darmstadt, Germany). Plasmids pET28bApnA-A1CYA126/8, -No3, -CCAP1459/36, -PCC7821, -No39, -No66, and -No80 were transformed into E. coli BL21(DE3) (Invitrogen, Lofer, Austria) cells, which were plated on LB agar with kanamycin. A single colony was inoculated into 25 ml LB medium with kanamycin and was grown overnight to saturation. LB medium with kanamycin (1 liter) was inoculated with 10 ml of the overnight culture, which was grown at 30°C for 3.5 h (optical density at 600 nm [OD600], ∼0.6 to 0.8), after which the cultures were cooled to 22°C (30 min), and isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM for an additional 3.5 h at 22°C. Cells were collected by centrifugation (4,000 × g, 10 min, 4°C), and the resulting pellet was stored at −20°C until use.
The collected pellet was thawed on ice and was resuspended in 8 ml of lysis buffer (50 mM Tris, 300 mM NaCl, 5 mM imidazole [pH 8.0]) with Roche Complete EDTA-free protease inhibitor cocktail added (Roche, Vienna, Austria). The cells were lysed by sonication on ice 10 times at 30-s intervals. The cellular debris was removed by centrifugation (10,000 × g, 30 min, 4°C), followed by transfer of the supernatant to Eppendorf tubes and further centrifugation (13,000 × g, 15 min, 4°C) to ensure that no insoluble debris remained. The cleared supernatant was incubated with 0.125 ml Ni-nitrilotriacetic acid (NTA) agarose (Qiagen). The agarose was then transferred to a Poly-Prep column (Bio-Rad, Munich, Germany) and drained. The resin was then washed with 1.25 ml lysis buffer with 5 mM β-mercaptoethanol (BME) added, followed first by 0.5 ml and then by 0.25 ml of wash buffer (50 mM Tris, 300 mM NaCl, 25 mM imidazole, 5 mM BME [pH 8.0]), and the protein was eluted with 0.4 ml elution buffer (50 mM Tris, 300 mM NaCl, 250 mM imidazole, 5 mM BME [pH 8.0]). The fractions were checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and fractions containing the desired protein were desalted using an Econo-Pac 10DG desalting column (Bio-Rad) equilibrated with storage buffer 2 (50 mM Tris, 50 mM NaCl, 5 mM BME, 10% [wt/vol] glycerol [pH 8.0]). The protein was flash frozen in liquid nitrogen and was stored at −80°C until use. Protein yields were typically between 0.75 and 1 mg liter−1.
ATP-pyrophosphate exchange assay.
In a 96-well plate, 75 mM Tris (pH 8.0), 10 mM MgCl2, 10 mM ATP, 2 mM NaPPi, 1.5 mM amino acid, 0.5 μCi [32P]NaPPi (Perkin-Elmer, Waltham, MA), and 10 μg of protein were added to a final volume of 100 μl (19). The reaction was initiated by the addition of protein and was allowed to proceed at room temperature for 45 min. The reaction was terminated by the addition of 30 μl stop buffer (2.5% [wt/vol] activated charcoal, 10% [wt/vol] trichloroacetic acid [TCA], and 20 mM NaPPi). The charcoal was then applied to a filter plate using a FilterMate harvester (Perkin-Elmer), and the bound charcoal was washed 10 times with 200 μl distilled water (dH2O), followed by 200 μl ethanol (EtOH) (95%, vol/vol), and was then allowed to air dry for 5 to 10 min. To each well, 60 μl Micro-Scint-20 cocktail (Perkin-Elmer) was added, and then the radiation from beta-emitting nuclides was measured using a TopCount NXT counter (Perkin-Elmer).
Nucleotide sequence accession numbers.
The sequence data obtained in this study for the apn gene cluster and the A domains of all the strains have been submitted to the DDBJ/EMBL/GenBank databases under accession no. EF67686 and HM773399 to HM773422.
RESULTS
Identification of the AP biosynthetic gene cluster.
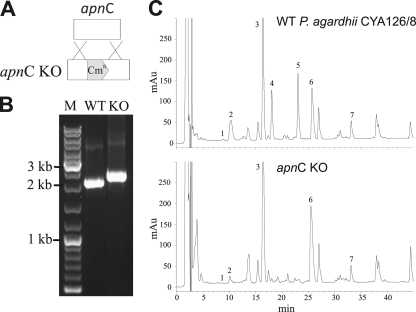
Using degenerate primers targeting the conserved A2 motif of A domains and the SAM-binding motif of NMT domains (28), we were able to amplify a DNA fragment of an uncharacterized A domain putatively involved in the activation of the strictly conserved N-methylated amino acid in position 5 (see Materials and Methods for a full description). To confirm the involvement of this DNA fragment (A2-Nmt2) in AP biosynthesis, we used this fragment for insertional inactivation by homologous recombination. PCR amplification from the apnC genes of the chloramphenicol-resistant cells showed stable integration of the construct at the expected position due to a double homologous-crossover recombination event (Fig. 2A and B): While the wild type gave the calculated PCR product of 2,000 bp, the apnC mutant showed an amplicon of 2.8 kbp (2 kbp plus the 0.8-kbp Cmr fragment). HPLC analysis of mutant clones showed that the mutant lacked the peaks corresponding to AP 908 and AP 915 (34), while production of the aeruginosides (20), microviridin K (36), and MC-LR and MC-RR (4) was unaffected (Fig. 2C). This observation demonstrated unequivocally that the cloned fragment belonged to the AP (apn) gene cluster.
Fig. 2.
(A) Inactivation of the apnC gene by insertional inactivation via homologous recombination. The transformation construct contains a selection marker (Cmr; 800 bp) (shaded) that is flanked by homologous sequences on both the 5′ and 3′ ends. KO, knockout. (B) Testing of the full segregation of the apnC mutation by PCR using primers specific to the flanking region of the construct inserted into apnC (sequence position 15074 in EMBL/GenBank/DDBJ accession no. EF672686). (C) HPLC analysis of the aqueous-methanol extracts of WT apnC (top) and KO apnC, deficient in AP synthesis (bottom). Peaks 1 and 2 represent aeruginosides 126A (m/z 691; 9.07 min) and 126B (m/z 715; 10.56 min); peak 3, microviridin K (m/z 1,771; 16.09 min); peaks 4 and 5, AP 908 (m/z 909; 18.29 min) and AP 915 (m/z 916; 22.67 min); and peaks 6 and 7, MC-RR (m/z 1,024; 26.61 min) and MC-LR (m/z 981; 33.83 min), respectively.
Characterization of the AP operon from Planktothrix agardhii.
The complete AP (apn) operon of P. agardhii CYA126/8 was identified from a draft genome (Kurmayer and Christiansen, unpublished) and spans 23,949 bp containing five open reading frames (ORFs) (apnA to apnE), which encode five distinct proteins, including four NRPS proteins (ApnA to -D) and an ATP binding cassette (ABC) transporter (ApnE) (Fig. 1). ApnA is a bimodular NRPS enzyme comprising two A domains, two thiolation domains (T domains), one C domain, and an epimerase domain (E domain). ApnB contains one NRPS elongation module. ApnC contains two NRPS elongation modules, with the second A domain bearing an NMT domain. The termination module ApnD contains one NRPS module including a thioesterase (TE) domain. Overall, the organization of the AP gene cluster is highly similar to those of the AP synthesis operons described for Anabaena, Nostoc, and Nodularia (39). In accordance with the colinearity rule (7), it is deduced that the modules act in the sequence in which they are arranged. This hypothesis is supported by bioinformatic predictions of A domain selectivity using the specificity-conferring codes defined by Stachelhaus et al (44) and the residues within 8 Å around the substrate as defined by Rausch et al (37). These signature sequences suggest that the second A domain of ApnA activates Lys in position 2, a prediction supported by the presence of an epimerization domain, while the A domains of ApnB and ApnD are predicted to activate Val and Ile for incorporation into positions 3 and 6, respectively (Table 2). In contrast, the specificity-conferring codes of the A domains in the initiation (the ApnA A1 domain) and elongation (the ApnC A1 and A2 domains) modules do not match any entries in the database (37). The two A domains of ApnC showed identical specificity-conferring codes, suggesting that they activated the same amino acid, which is in accordance with the structures of AP 908 and AP 915, both of which contain Hty in positions 4 and 5.
Table 2.
Specificity-conferring codes of A domains of the AP biosynthetic gene clustersa
| Parameter | ApnA A1 | ApnA A2 | ApnB A | ApnC A1 | ApnC A2 | ApnD A |
|---|---|---|---|---|---|---|
| Length (bp) | 1,641 | 1,608 | 1,376 | 1,387 | 2,631 | 1,532 |
| % variability | ||||||
| Nucleotide | 6.2 | 9.7 | 8.3 | 7.1 | 24.1 | 29.2 |
| aa | 6.8 | 8.4 | 9.2 | 7.9 | 30 | 34.8 |
| Code (substrate) inb: | ||||||
| CYA126/8 | DVESIGAIAK (—) | DAEDIGSVVK (Lys) | DALWIGGVFK (Val) | DLGFTGVCTK (—) | DLGFTGCVTK (—) | DAFFLGVTFK (Ile) |
| No. 3 | DVESIGAIAK (—) | DAEDIGSVVK (Lys) | DALWIGGVFK (Val) | DLGFTGVCTK (—) | DLFNNALTYK (Ala) | DAWTIAGVCK (Phe) |
| CCAP1459/36 | DVESIGVIAK (—) | DAEDIGSVVK (Lys) | DALWIGGVFK (Val) | DLGFTGVCTK (—) | DLFNNALTYK (Ala) | DAWTIAGVCK (Phe) |
| PCC7821 | DVESIGVIAK (—) | DAEDIGSVVK (Lys) | DALWIGGVFK (Val) | DLGFTGVCTK (—) | DLFNNALTYK (Ala) | DAWTIAGVCK (Phe) |
| No. 39 | DVEDIGAVEK (Arg) | DAEDIGSVVK (Lys) | DALWIGGVFK (Val) | DLGFTGVCTK (—) | DLFNNALTYK (Ala) | DAWTIAGVCK (Phe) |
| No. 66 | DVEDIGAIEK (Arg) | DAEDIGSVVK (Lys) | DALWIGGVFK (Val) | DLGFAGVCTK (—) | DLFNNALTYK (Ala) | DAWTIAGVCK (Phe) |
| No. 80 | DVEDIGAVEK (Arg) | DAEDIGSVVK (Lys) | DALWIGGVFK (Val) | DLGFTGVCTK (—) | DLFNNALTYK (Ala) | DAWTIAGVCK (Phe) |
| Position and aa in AP molecule | 1; Arg/Tyr | 2; Lys | 3; Val/Ile | 4; Hty | 5; Hty, Ala | 6; Ile, Phe |
According to the work of Stachelhaus et al. (44). Amino acid (aa) residue numbers are according to GrsA (Swissprot: P0C061; 235, 236, 239, 278, 299, 301, 322, 330, 331, 517).
Amino acid residues that differ between Planktothrix strains are boldface and underlined. The predicted substrate is in parentheses (NRPS predictor [37]). —, no precedent in the database.
Sequence variability of A domains and structural variation of APs.
Comparison of the ApnA A1 domains of the Planktothrix strains showed 6.2% variability on the nucleotide level and 6.8% variability in the deduced amino acid sequence. These sequence changes resulted in four different signature sequences (Table 2). The S278D, A322V, I330V, and A331E substitutions were observed (amino acid residues are numbered according to the GrsA sequence [SwissProt accession no. P0C061] [44]). One group (strains no. 39, no. 66, and no. 80) contained 278D and 331E, resulting in a more acidic binding pocket, predicted to activate Arg. This prediction correlates with the observation that those strains contained only AP molecules bearing Arg in the exocyclic position (Table 1). In contrast, the second group (CYA126/8, no. 3, CCAP1459/36, and PCC7821) formed a less acidic pocket containing 278S and 331A, which lack precedents in the database.
Although similar degrees of sequence variability on both the nucleotide (9.7%) and the amino acid (8.4%) level were observed for the ApnA A2 domains in the seven strains, no variation was observed within the signature sequence that predicted the activation of Lys. This fits the observation that the d-Lys residue in position 2 of the AP molecule is strictly conserved (48). A similar extent of variation on the nucleotide level was found for the apnB A domains of the seven strains (8.3%). Again, no differences were found in the specificity-conferring code (predicting the activation of Val) among the strains. Within the apnC A1 domain (7.1% nucleotide variability), only strain no. 66 showed one substitution affecting its signature sequence. In contrast, the sequence variability within the apnC A2 domain was much higher, i.e., all strains except for CYA126/8 were identical, but they differed by 24.1% (2,631 bp) from CYA126/8. Analysis of the substrate specificity codes of the ApnC A2 domain revealed that all strains except CYA126/8 were predicted to activate Ala, which is confirmed by the observation that all these isolates produce APs bearing an N-methyl Ala residue in position 5. For the apnD A domain, highly dissimilar sequences (29.2% nucleotide variability) were observed between strain CYA126/8, on the one hand, and the other six isolates, which displayed only 2.4% variability. This, again, resulted in two divergent binding pockets predicted to activate Phe instead of Ile in all the strains other than CYA126/8. Accordingly, all isolates containing this signature sequence produce AP metabolites bearing Phe at the lysine ε amino group (position 6), whereas the APs of CYA126/8 contain Ile in that position.
Evolution of A domain substrate specificity.
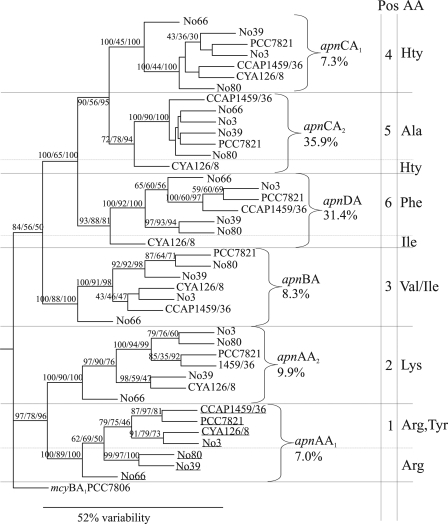
In general, phylogenetic analysis of all apn A domain sequences (1,441 bp) revealed six branches corresponding to the six different NRPS modules of the apn gene cluster. Two major branches were found: while the apnA A1 and A2 sequences were found in one distinct clade, the apnB A, apnC A1 and A2, and apnD A sequences formed a second clade. Within the latter branch, the apnC A1 and A2 domains share a recent ancestor (Fig. 3). A domain sequences also clustered according to the predicted A domain specificity in accordance with the AP metabolite profile. The large sequence variabilities observed within the apnC A2 and apnD A domains between CYA126/8 and all the other strains clearly correlate with the differences between Hty and Ala at position 5 and between Ile and Phe at position 6 of the AP molecule, respectively. A 3,235-bp fragment, including both the condensation domain (apnC C2) and the adjacent A domain (apnC A2), was exchanged through past gene conversion in all strains except CYA126/8 (P, <0.05 by GENECONV). A second potential locus of recombination, constituting only part of the A domain (795 bp), was identified in the apnD A domain. Remarkably, the promiscuous apnA A1 domain sequences found in strains incorporating Arg or Tyr evolved from an apnA A1 domain ancestor sequence found in strains incorporating exclusively Arg in position 1.
Fig. 3.
Phylogenetic tree of the six A domains (core motifs A1 to A10; 1,441 bp) of the apn gene cluster sequenced from the seven Planktothrix strains investigated. The significant bootstrap percentages were obtained from 1,000 pseudoreplicates using neighbor joining/parsimony/maximum likelihood. For each clade, the genetic dissimilarity was calculated. In addition, the position and identity of each amino acid (AA) incorporated into one of the six positions of the AP molecule are given. The NMT region (1,175 bp) of the apnC A2 domain was not included in the phylogenetic analysis. The mcyB A1 sequence of Microcystis strain PCC7806 served as an outgroup. Underlining indicates A domain genetic variants that were probed using the ATP-pyrophosphate exchange assay.
ATP-PPi exchange assay of ApnA A1 domains from different genotypes.
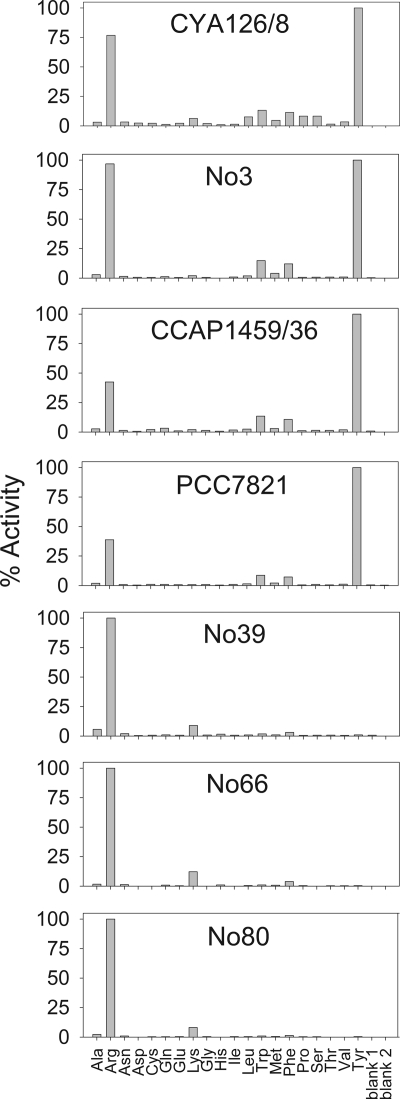
In order to analyze the functional consequences of ApnA A1 domain variability, we heterologously overexpressed the ApnA A1 domains of the strains investigated and characterized them via the ATP-PPi exchange assay. Arg, Lys, Trp, Phe, and Tyr were the only amino acids that showed significant activation rates (>10% activation) (Fig. 4). Therefore, subsequent assays focused on these amino acids and included the higher homologs of Arg (homoarginine [Har]) and Tyr (Hty). The most striking result was that the A domains of the three strains containing the more acidic binding pocket (no. 39, no. 66, and no. 80) showed activation only of the basic amino acids Arg, Har, and Lys (see Fig. S1 in the supplemental material). In contrast, the A domains of strains CYA126/8, no. 3, CCAP1459/36, and PCC7821 activated both basic and aromatic amino acids (Arg, Har, Tyr, Hty, Trp, and Phe). Within the latter group of A domains, the ratio of Arg activation to Tyr activation differed: the A domains of strains CCAP1459/36 and PCC7821, containing V322A, showed lower rates of Arg activation (39 and 42% relative to Tyr activation) than the A domain from strain no. 3 (96% relative to Tyr activation).
Fig. 4.
Comparison of amino acid activation between ApnA A1 domains expressed from seven different Planktothrix strains by using the ATP-pyrophosphate exchange assay. Blank 1 contains protein; blank 2 does not.
Ratio of nonsynonymous to synonymous substitutions.
In order to test for the presence of positive selection and to identify those selected sites, nonsynonymous (dN) and synonymous (dS) substitutions were compared using maximum likelihood (52). In general, a low dN/dS ratio (<1) is indicative of purifying selection, while a dN/dS ratio of ∼1 indicates a relaxation of selective constraints, and dN/dS ratios of >1 imply positive selection (53). For all apn A domains (1,441 bp), the dN/dS ratio was 0.19, suggesting that overall, the A domains were under purifying selection. The lowest dN/dS ratio (0.12) was observed for the apnA A2 gene, which encodes a domain activating the strictly conserved Lys in position 2 of the AP molecule. For the other A domains, the apnA A1, apnB A, apnC A1 and A2, and apnD A domains, the dN/dS ratios were more variable, i.e., 0.19, 0.18, 0.16, 0.47, and 0.17, respectively. The dN/dS ratios remained low when the 330-bp regions encoding the putative binding pocket from core motif A4 to A5 were compared: 0.17 for the apnA A1 domain, 0.08 for the apnA A2 domain, 0.17 for the apnB A domain, 0.13 for the apnC A1 domain, 0.28 for the apnC A2 domain, and 0.23 for the apnD A domain.
To increase the sensitivity of detection of positive selection, “site models” were applied (52). Only the apnA A1 gene was found to be under positive selection; none of the other A domain sequences showed statistical evidence of positive selection (Table 3). Surprisingly, within the ApnA A1 domain, positive selection affects a relatively small number of codons. Interestingly, one positively selected codon is part of the specificity-conferring code (D278S; posterior probability [P], 0.75) as defined by Stachelhaus et al (44) and the residues within 8 Å around the substrate as defined by Rausch et al. (37) (see Fig. S2 in the supplemental material). Three other positively selected sites include several amino acid substitutions located outside the putative binding pocket as defined by Stachelhaus et al (44) (Table 3). This rather small number of positively selected sites suggests that the remarkable substrate promiscuity of the ApnA A1 domains of strains CYA126/8, no, 3, CCAP1459/36, and PCC7821 evolved from point mutations only.
Table 3.
Likelihood ratio tests of positive selection in AP synthetase A domains
| apn A domain | Site modela | lnLb | LRTc | dN/dSd | Positively selected sitese |
|---|---|---|---|---|---|
| apnA A1 | M1 | −2,960.79 | 8.34** | 0.22 | M2 vs M1: 52, 117, 124, 172***, 225, 278, 366, 403, 405, 458, 466, 481, 494 |
| M2 | −2,956.62 | 0.28 | |||
| M7 | −2,960.84 | 8.56** | 0.21 | M8 vs M7: 52, 108*, 117**, 124, 167, 172***, 218, 222, | |
| M8 | −2,956.56 | 0.27 | 225, 226, 278, 366, 380, 403, 405, 458, 466, 481, 485, | ||
| M8a | −2,960.79 | 8.46*** | 0.22 | 494, 521, 525, 542 | |
| apnA A2 | M1 | −2,643.08 | 2.02 | 0.15 | |
| M2 | −2,642.07 | 0.17 | |||
| M7 | −2,643.51 | 2.86 | 0.15 | ||
| M8 | −2,642.09 | 0.17 | |||
| M8a | −2,643.09 | 2.02 | 0.15 | ||
| apnB A1 | M1 | −2,311.49 | 1.1 | 0.19 | |
| M2 | −2,310.94 | 0.25 | |||
| M7 | −2,311.54 | 1.38 | 0.19 | ||
| M8 | −2,310.85 | 0.25 | |||
| M8a | −2,311.49 | 1.28 | 0.19 | ||
| apnC A1 | M1 | −2,225.77 | 1.72 | 0.20 | |
| M2 | −2,224.91 | 0.24 | |||
| M7 | −2,225.77 | 1.72 | 0.20 | ||
| M8 | −2,224.91 | 0.24 | |||
| M8a | −2,225.77 | 1.72 | 0.20 | ||
| apnC A2 | M1 | −3,511.91 | Not applicablef | ||
| M2 | −3,515.25 | ||||
| M7 | −3,513.69 | ||||
| M8 | −3,509.95 | ||||
| M8a | −3,511.91 | ||||
| apnD A | M1 | −3,608.7 | Not applicablef | ||
| M2 | −3,608.7 | ||||
| M7 | −3,601.1 | ||||
| M8 | −3,600.54 | ||||
| M8a | −3,601.03 |
M1, nearly neutral; M2, positive selection; M7, beta; M8, beta & dN/dS > 1; M8a, beta & dN/dS = 1.
Log likelihood.
Likelihood ratio test. Asterisks indicate significance levels as follows: *, 90%; **, 95%; ***, 99%.
Ratio of nonsynonymous to synonymous substitutions per codon site.
Amino acid residues numbered according to the GrsA (SwissProt accession no. P0C061) sequence. Identification of positive selection was based on the Bayes empirical Bayes approach.
Due to recombination of the entire A domain (apnC A2) or the partial A domain (apnD A).
DISCUSSION
Substrate specificity of ApnA A1 domains and AP structural variation.
In this study an effort was made to elucidate the genetic basis responsible for the structural variation of APs among isolates of the genus Planktothrix. While this approach has been applied previously (10, 24, 25, 30, 45), until now none of the reported genetic variants has been probed biochemically. Therefore, the biochemical consequences of genetic variation within A domains in toxin-producing cyanobacteria have remained elusive. In the present study, we observed a significant correlation between AP structure variation and apn A domain genetic variation among the producing strains. The biochemical analysis of the catalytic activities of ApnA A1 domains matches the observation of the AP structural variation in position 1: only strains bearing AP structural variants with Arg or Tyr contained ApnA A1 domains specifically activating Arg and Tyr. The Arg-specific A domains of strains no. 39, no. 66, and no. 80 were found to activate the higher homolog (Har) as well. APs bearing Har in exocyclic position 1 have not been found in Planktothrix, although they are known from Microcystis (1). Reasoning that it is likely that Har is not available in the cells, we fed synthetic Har to cultures of strain no. 80. We observed that Har was indeed incorporated into exocyclic position 1 of the AP molecule, resulting in the detection of AP B (m/z 837) and a new AP molecule bearing Har (m/z 851) (G. Christiansen, T. Hemscheidt, and R. Kurmayer, unpublished data). During the biochemical characterization of the ApnA A1 domain, small amounts of Trp (9 to 15%) and Phe (7 to 12%) activation were observed beside the major activation of Arg and Tyr (Fig. 4), suggesting that APs bearing either Trp or Phe in exocyclic position 1 might occur as well. Indeed, AP structural variants with Trp in position 1 have been described as ferintoic acids A (m/z 867) and B (m/z 881) from Microcystis sp. (49), and several AP variants bearing Phe in position 1 have been described for Anabaena (17, 39). To date, such AP structural variants have not been found in CYA126/8 (34). Nevertheless, the possibility cannot be excluded that AP variants bearing either Trp or Phe in position 1 are produced by Planktothrix strains no. 3, CCAP1459/36, and PCC7821 in amounts below the limit of detection, which is estimated to be about 3% of the major AP metabolite in the isolates.
Structural variation and microevolution of the substrate specificity of ApnA A1 domains.
A plethora of AP-type metabolites have been isolated from several cyanobacterial genera and other organisms (46, 48). All APs are characterized by a cyclic pentapeptide backbone containing a strictly conserved d-Lys residue in position 2 and N-methylation on the amino acid located in the i+1 site from the ε amino group of Lys. Like MCs, AP B and oscillamides B and C have been described as inhibitors of protein phosphatase 1 (PP1) and PP2A (14, 41). In contrast, oscillamide Y and schizopeptin have been described as effective inhibitors of serine proteases such as chymotrypsin and trypsin (38, 40). Numerous variants of AP (APs G, H, T, I, and J) and brunsvicamide A have been described as potent inhibitors of carboxypeptidase A (CPD A) (21, 32, 46). Most recently, AP B, AP F, and the brunsvicamides were reported to inhibit porcine pancreatic elastase (PPE) and human leukocyte elastase (HLE) in the micromolar range (3, 46). The range of enzyme inhibition activities of APs is impressive, and studies of structure-activity relationships have confirmed the essential role of the strictly conserved d-Lys residue in the inhibition of CPD A (46). Furthermore, an alanine scan of brunsvicamide A changing all six positions except the exocyclic amino acid (Ile) revealed that substitutions affecting the amino acids in the cyclic backbone have no effect on bioactivity. In contrast, the exchange in exocyclic position 1 reduced the bioactivity of brunsvicamide A significantly.
In our study, we have identified three codons in the apnA A1 domain that are under positive selection. They are not located only within the putative binding pocket but are distributed over the entire A domain. In fact, the sequence variability within the binding pocket of the apnA A1 domain was indistinguishable from that affecting the entire A domain. Correspondingly, Tooming-Klunderud et al. (45) identified only a small number of positively selected codons that were also distributed over the entire A domains of the mcyB and mcyC genes in Anabaena, Planktothrix, and Microcystis. Recently, only two substitutions located within the mcyC A domain (1,344 bp) were observed to correlate with the replacement of Arg by Hty in position 4 of the MC molecule in Planktothrix (6). We conclude that under the maximum-parsimony criterion, a rather small number of substitutions are related to the change in the substrate specificity of the ApnA A1 domain as observed in the in vitro experiments. For terpene cyclases in higher plants (Nicotiana, Hyoscyamus), detailed mutational analysis has been performed in order to characterize the catalytic landscape underlying the evolution of sesquiterpene chemical diversity (35). The authors concluded that novel catalytic specificities require as little as a single nucleotide polymorphism, not necessarily located on the active-site surface of the protein. In that study, a large fraction of mutants was found to be able to produce several classes of sesquiterpenes. The authors postulated that from those promiscuous enzymes, new specific enzyme activities arise by selective pressure on the producers of these compounds. Therefore, we suggest that the ApnA A1 domains showing Arg/Tyr substrate promiscuity actually represent a transitional state in the evolutionary process of reshaping APs as enzyme inhibitors, which may result in a Tyr-specific ApnA A1 domain. If an aromatic amino acid residue in exocyclic position 1 has a selective advantage, anabaenopeptins with solely Phe, Tyr, or Trp in position 1 will become more abundant in the future. At present, only two strains (Anabaena NZ-3-1 and Nostoc PCC73102) that bear exclusively Phe in position 1 have been isolated (17, 39). It is notable that the Arg/Tyr dichotomy demonstrated here is also found at certain positions in other cyanobacterial metabolite families (in MCs at position 2 and in cyanopeptolins at position 2 adjacent to the 3-amino-6-hydroxy-2-piperidone [Ahp] moiety) and could have functional implications. For example, Yamaki et al. (50) have shown that among cyanopeptolins, the dichotomy of basic versus aromatic amino acids is decisive for either trypsin or chymotrypsin inhibition.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for the comments of three anonymous referees on an earlier version of this article.
This work was supported by the Austrian Science Fund (FWF-P20231) and in part by funds from the National Science Foundation (OCE04-32479) and the NIEHS (P50ES012740).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 27 May 2011.
REFERENCES
- 1. Beresovsky D., et al. 2006. Toxins and biologically active secondary metabolites of Microcystis sp. isolated from Lake Kinneret. Isr. J. Chem. 46:79–87 [Google Scholar]
- 2. Bonnard I., et al. 2007. Total structure and inhibition of tumor cell proliferation of laxaphycins. J. Med. Chem. 50:1266–1279 [DOI] [PubMed] [Google Scholar]
- 3. Bubik A., Sedmak B., Novinec M., Lenarcic B., Lah T. T. 2008. Cytotoxic and peptidase inhibitory activities of selected non-hepatotoxic cyclic peptides from cyanobacteria. Biol. Chem. 389:1339–1346 [DOI] [PubMed] [Google Scholar]
- 4. Christiansen G., Fastner J., Erhard M., Börner T., Dittmann E. 2003. Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation. J. Bacteriol. 185:564–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Christiansen G., Molitor C., Philmus B., Kurmayer R. 2008. Nontoxic strains of cyanobacteria are the result of major gene deletion events induced by a transposable element. Mol. Biol. Evol. 25:1695–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christiansen G., et al. 2008. Isolation and structure determination of two microcystins and sequence comparisons of McyABC adenylation domains in Planktothrix species. J. Nat. Prod. 71:1881–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Conti E., Stachelhaus T., Marahiel M. A., Brick P. 1997. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 16:4174–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Erhard M., von Döhren H., Jungblut P. 1999. Rapid identification of the new anabaenopeptin G from Planktothrix agardhii HUB 011 using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 13:337–343 [DOI] [PubMed] [Google Scholar]
- 9. Felsenstein J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 10. Fewer D. P., et al. 2007. Recurrent adenylation domain replacement in the microcystin synthetase gene cluster. BMC Evol. Biol. 7:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fischbach M. A., Walsh C. T. 2006. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106:3468–3496 [DOI] [PubMed] [Google Scholar]
- 12. Franche C., Damerval T. 1988. Test on nif probes and DNA hybridizations. Methods Enzymol. 167:803–808 [Google Scholar]
- 13. Frankmölle W. P., Knübel G., Moore R. E., Patterson G. M. L. 1992. Antifungal cyclic peptides from the terrestrial blue-green alga Anabaena laxa. II. Structures of laxaphycins A, B, D and E. J. Antibiot. (Tokyo) 45:1458–1466 [DOI] [PubMed] [Google Scholar]
- 14. Gkelis S., Lanares T., Sivonen K. 2006. The presence of microcystins and other cyanobacterial bioactive peptides in aquatic fauna collected from Greek freshwaters. Aquat. Toxicol. 78:32–41 [DOI] [PubMed] [Google Scholar]
- 15. Golakoti T., et al. 1995. Structure determination, conformational analysis, chemical stability studies, and antitumor evaluation of the cryptophycins. Isolation of 18 new analogs from Nostoc sp. strain GSV 224. J. Am. Chem. Soc. 117:12030–12049 [Google Scholar]
- 16. Golakoti T., Yoshida W. Y., Chaganty S., Moore R. E. 2000. Isolation and structures of nostopeptolides A1, A2 and A3 from the cyanobacterium Nostoc sp. GSV224. Tetrahedron 56:9093–9102 [Google Scholar]
- 17. Grach-Pogrebinsky O., Carmeli S. 2008. Three novel anabaenopeptins from the cyanobacterium Anabaena sp. Tetrahedron 64:10233–10238 [Google Scholar]
- 18. Harada K., Fujii K., Shimada T., Suzuki M. 1995. Two cyclic peptides, anabaenopeptins, a third group of bioactive compounds from the cyanobacterium Anabaena flos-aquae NRC 525-17. Tetrahedron Lett. 36:1511–1514 [Google Scholar]
- 19. Hoffmann D., Hevel J. M., Moore R. E., Moore B. S. 2003. Sequence analysis and biochemical characterization of the nostopeptolide A biosynthetic gene cluster from Nostoc sp. GSV224. Gene 311:171–180 [DOI] [PubMed] [Google Scholar]
- 20. Ishida K., et al. 2007. Biosynthetic pathway and structure analysis of aeruginoside 126A and B, cyanobacterial peptide glycosides bearing an unusual 2-carboxy-6-hydroxyoctahydroindole moiety. Chem. Biol. 14:565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Itou Y., Suzuki S., Ishida K., Murakami M. 1999. Anabaenopeptins G and H, potent carboxypeptidase A inhibitors from the cyanobacterium Oscillatoria agardhii (NIES-595). Bioorg. Med. Chem. Lett. 9:1243–1246 [DOI] [PubMed] [Google Scholar]
- 22. Kosol S., Schmidt J., Kurmayer R. 2009. Variation in peptide net production and growth among strains of the toxic cyanobacterium Planktothrix spp. Eur. J. Phycol. 44:49–62 [Google Scholar]
- 23. Kurmayer R., Christiansen G., Fastner J., Börner T. 2004. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ. Microbiol. 6:831–841 [DOI] [PubMed] [Google Scholar]
- 24. Kurmayer R., Christiansen G., Gumpenberger M., Fastner J. 2005. Genetic identification of microcystin ecotypes in toxic cyanobacteria of the genus Planktothrix. Microbiology 151:1525–1533 [DOI] [PubMed] [Google Scholar]
- 25. Kurmayer R., Dittmann E., Fastner J., Chorus I. 2002. Diversity of microcystin genes within a population of the toxic cyanobacterium Microcystis spp. in Lake Wannsee (Berlin, Germany). Microb. Ecol. 43:107–118 [DOI] [PubMed] [Google Scholar]
- 26. Kurmayer R., Gumpenberger M. 2006. Diversity of microcystin genotypes among populations of the filamentous cyanobacteria Planktothrix rubescens and Planktothrix agardhii. Mol. Ecol. 15:3849–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lipmann F. 1971. Attempts to map a process evolution of peptide biosynthesis. Science 173:875–884 [DOI] [PubMed] [Google Scholar]
- 28. Marahiel M. A., Stachelhaus T., Mootz H. D. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651–2674 [DOI] [PubMed] [Google Scholar]
- 29. Mehner C., et al. 2008. New peptolides from the cyanobacterium Nostoc insulare as selective and potent inhibitors of human leukocyte elastase. Chembiochem 9:2692–2703 [DOI] [PubMed] [Google Scholar]
- 30. Mikalsen B., et al. 2003. Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J. Bacteriol. 185:2774–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murakami M., Shin H. J., Matsuda H., Ishida K., Yamaguchi K. 1997. A cyclic peptide, anabaenopeptin B, from the cyanobacterium Oscillatoria agardhii. Phytochemistry 44:449–452 [Google Scholar]
- 32. Murakami M., Suzuki S., Itou Y., Kodani S., Ishida K. 2000. New anabaenopeptins, potent carboxypeptidase-A inhibitors from the cyanobacterium Aphanizomenon flos-aquae. J. Nat. Prod. 63:1280–1282 [DOI] [PubMed] [Google Scholar]
- 33. Neilan B. A., et al. 1999. Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J. Bacteriol. 181:4089–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Okumura H., Philmus B., Portmann C., Hemscheidt T. 2009. Homotyrosine-containing cyanopeptolins 880 and 960 and anabaenopeptins 908 and 915 from Planktothrix agardhii CYA 126/8. J. Nat. Prod. 72:172–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O'Maille P. E., et al. 2008. Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat. Chem. Biol. 4:617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Philmus B., Christiansen G., Yoshida W., Hemscheidt T. 2008. Posttranslational modification in microviridin biosynthesis. Chembiochem 9:3066–3073 [DOI] [PubMed] [Google Scholar]
- 37. Rausch C., Weber T., Kohlbacher O., Wohlleben W., Huson D. 2005. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 33:5799–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reshef V., Carmeli S. 2002. Schizopeptin 791, a new anabaenopeptin-like cyclic peptide from the cyanobacterium Schizothrix sp. J. Nat. Prod. 65:1187–1189 [DOI] [PubMed] [Google Scholar]
- 39. Rouhiainen L., Jokela J., Fewer D. P., Urmann M., Sivonen K. 2010. Two alternative starter modules for the non-ribosomal biosynthesis of specific anabaenopeptin variants in Anabaena (Cyanobacteria). Chem. Biol. 17:265–273 [DOI] [PubMed] [Google Scholar]
- 40. Sano T., Kaya K. 1995. Oscillamide Y, a chymotrypsin inhibitor from toxic Oscillatoria agardhii. Tetrahedron Lett. 36:5933–5936 [DOI] [PubMed] [Google Scholar]
- 41. Sano T., Usui T., Ueda K., Osada H., Kaya K. 2001. Isolation of new protein phosphatase inhibitors from two cyanobacteria species, Planktothrix spp. J. Nat. Prod. 64:1052–1055 [DOI] [PubMed] [Google Scholar]
- 42. Sawyer S. 1999. GENECONV: a computer package for the statistical detection of gene conversion. Distributed by the author Department of Mathematics, Washington University, St. Louis, MO: http://www.math.wustl.edu/∼sawyer/geneconv/index.html [Google Scholar]
- 43. Shin H. J., Matsuda H., Murakami M., Yamaguchi K. 1997. Anabaenopeptins E and F, two new cyclic peptides from the cyanobacterium Oscillatoria agardhii (NIES 204). J. Nat. Prod. 60:139–141 [Google Scholar]
- 44. Stachelhaus T., Mootz H. D., Marahiel M. A. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493–505 [DOI] [PubMed] [Google Scholar]
- 45. Tooming-Klunderud A., et al. 2008. Evidence for positive selection acting on microcystin synthetase adenylation domains in three cyanobacterial genera. BMC Evol. Biol. 8:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walther T., Renner S., Waldmann H., Arndt H. D. 2009. Synthesis and structure-activity correlation of a brunsvicamide-inspired cyclopeptide collection. Chembiochem 10:1153–1162 [DOI] [PubMed] [Google Scholar]
- 47. Welker M., Erhard M. 2007. Consistency between chemotyping of single filaments of Planktothrix rubescens (Cyanobacteria) by MALDI-TOF and the peptide patterns of strains determined by HPLC-MS. J. Mass Spectrom. 42:1062–1068 [DOI] [PubMed] [Google Scholar]
- 48. Welker M., von Döhren H. 2006. Cyanobacterial peptides—nature′s own combinatorial biosynthesis. FEMS Microbiol. Rev. 30:530–563 [DOI] [PubMed] [Google Scholar]
- 49. Williams D. E., Craig M., Holmes C. F. B., Andersen R. J. 1996. Ferintoic acids A and B, new cyclic hexapeptides from the freshwater cyanobacterium Microcystis aeruginosa. J. Nat. Prod. 59:570–575 [Google Scholar]
- 50. Yamaki H., Sitachitta N., Sano T., Kaya K. 2005. Two new chymotrypsin inhibitors isolated from the cyanobacterium Microcystis aeruginosa NIES-88. J. Nat. Prod. 68:14–18 [DOI] [PubMed] [Google Scholar]
- 51. Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555–556 [DOI] [PubMed] [Google Scholar]
- 52. Yang Z., Nielsen R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19:908–917 [DOI] [PubMed] [Google Scholar]
- 53. Yang Z. H., Wong W. S. W., Nielsen R. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22:1107–1118 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.