Abstract
Nickel and cobalt are both essential trace elements that are toxic when present in excess. The main resistance mechanism that bacteria use to overcome this toxicity is the efflux of these cations out of the cytoplasm. RND (resistance-nodulation-cell division)- and MFS (major facilitator superfamily)-type efflux systems are known to export either nickel or cobalt. The RcnA efflux pump, which belongs to a unique family, is responsible for the detoxification of Ni and Co in Escherichia coli. In this work, the role of the gene yohN, which is located downstream of rcnA, is investigated. yohN is cotranscribed with rcnA, and its expression is induced by Ni and Co. Surprisingly, in contrast to the effect of deleting rcnA, deletion of yohN conferred enhanced resistance to Ni and Co in E. coli, accompanied by decreased metal accumulation. We show that YohN is localized to the periplasm and does not bind Ni or Co ions directly. Physiological and genetic experiments demonstrate that YohN is not involved in Ni import. YohN is conserved among proteobacteria and belongs to a new family of proteins; consequently, yohN has been renamed rcnB. We show that the enhanced resistance of rcnB mutants to Ni and Co and their decreased Ni and Co intracellular accumulation are linked to the greater efflux of these ions in the absence of rcnB. Taken together, these results suggest that RcnB is required to maintain metal ion homeostasis, in conjunction with the efflux pump RcnA, presumably by modulating RcnA-mediated export of Ni and Co to avoid excess efflux of Ni and Co ions via an unknown novel mechanism.
INTRODUCTION
Nickel and cobalt are both essential trace elements. Co is incorporated into a subset of proteins; in Escherichia coli, it is present in vitamin B12, which is involved in many crucial biological functions (28). Ni is a catalytic cofactor of eight prokaryotic enzymes involved in both redox and nonredox reactions, the best-characterized enzymes being hydrogenase and urease (27). However, using improved predictive and analytical methods, the assignment or reassignment of the metal content in proteins is still in progress. For instance, two new Ni-containing enzymes have been identified recently: a cupin/putative sugar-binding protein and an alanyl-tRNA editing hydrolase (8).
In eubacteria, Co import occurs via NiCoT secondary transporters (16) or via recently characterized ECF-type ATP binding cassette (ABC) importers, such as the cobalt uptake system CbiMNQO (41). E. coli lacks a specific Co importer but possesses a vitamin B12 (or cobalamin) uptake system. BtuB is the outer membrane protein that, by a TonB-dependent mechanism, imports cobalamin into the periplasm (5). The transmembrane component of the BtuCDF ABC importer, BtuC, and the nucleotide binding protein, BtuD (30), function together with the periplasmic solute-binding protein BtuF (4) to import cobalamin into the cytoplasm.
Ni import is mediated either by ABC transporters or by NiCoT permease (15). In E. coli, specific Ni uptake occurs via the NikABCDE system of the ABC family. NikBC are channel-forming transmembrane proteins, NikDE are nucleotide-binding proteins (34), and NikA is a periplasmic solute binding protein (13). In some organisms, the same importer accomplishes the uptake of both Ni and Co. A member of the NiCoT family, NhlF from Rhodococcus rhodochrous J1, has been shown to import both metals, although with different affinities, with Co being a better substrate than Ni (11). Mutagenesis studies have shown that the transport mechanism is conserved among NiCoT members and that the specificity relies on a few residues, some being shared for Ni and Co coordination (10). Similarly, the FecDE and CeuE ABC transporter has been shown recently to import both Ni and Co in Helicobacter mustelae (49), in parallel with the NiCoT Ni importer NixA. Apart from these dedicated specific uptake systems, under conditions where the concentration of metals is high, nonspecific import is an important source of metal entry in bacteria. In E. coli, both Ni and Co are transported through the Mg(II) transporter CorA (35). For Ni, mutation of the nik genes results in a complete loss of hydrogenase activity (54). In this case, the enzyme is synthesized normally, but it lacks the Ni required for maturation (42) and eventual translocation to the periplasm (43). Addition of excess Ni to the growth medium restores hydrogenase activity (53), owing to nonspecific Ni uptake by CorA. However, when the nik and corA genes are mutated, nickel sensitivity is not fully abolished, suggesting the existence of an as-yet-unidentified supplementary transporter (34). Similarly, Co uptake can occur via an alternative transporter, ZupT, which is primarily involved in Zn uptake (20), and active Co uptake can be detected in the absence of CorA and ZupT (29).
Like many other transition metals, Ni and Co are toxic in excess, causing growth arrest and cell death (19, 55). The molecular mechanisms of nickel toxicity need to be reevaluated, but Ni mainly causes oxidative stress (52). A recent transcriptomic study in E. coli revealed that the toxicity of Co occurs primarily by impairing the biogenesis of Fe/S clusters and secondarily by decreasing iron availability (17). The most widespread mechanism that bacteria use to resist metal toxicity is the efflux of the ions out of the cell (36), although alternative mechanisms, such as biomineralization (22) or biofilm formation (38), have also been described. Ni and Co are often expelled by the same efflux pumps. In E. coli, the only identified mechanism of resistance to Ni or Co is executed by the RcnA efflux pump. RcnA (formerly YohM) is a six-transmembrane-domain protein of a novel family, and it is thought to function as a secondary transporter (44). The expression of rcnA is induced by Ni and Co and not by other divalent cations (44). Other members of the RcnA family have been identified in Pseudomonas syringae, RcnAPsy, and in Pseudomonas putida, MdrH (23). RcnAPsy confers resistance to Ni and Co, whereas MdrH confers resistance to Ni, Zn, and Cd, but not to Co, and its expression is induced by Ni, Zn, Cd, and Co (23). Interestingly, MdrH exhibits a chimeric structure encompassing a domain similar to that of CzcB of the resistance-nodulation-cell division (RND) efflux family.
In order to better understand the efflux mechanism of the RcnA family, in the present work, we have investigated the role of the yohN gene in E. coli. yohN lies directly downstream of rcnA, and we show here that it is coexpressed with rcnA and that its expression is induced by Ni and Co via RcnR. Surprisingly, in contrast to the effect of deleting rcnA, deletion of the yohN gene confers increased resistance to Ni and Co, which is accompanied by reduced intracellular Ni and Co pools. We show that yohN is not involved in Ni uptake and that it encodes a periplasmic protein. Finally, we find that in the yohN mutant, the efflux of Ni is considerably increased. These results indicate that YohN is essential for maintaining a proper Ni and Co homeostasis in E. coli, most likely in connection with the efflux pump RcnA. Based on these results, yohN has been renamed rcnB.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains used in this work are summarized in Table S1A in the supplemental material and are all derivatives of Escherichia coli K-12. Bacterial cells were grown in LB medium or M63 minimal medium supplemented with 0.4% glucose. Anaerobic conditions were obtained with liquid cultures by filling bottles to the top or by placing petri dishes in a GasPak jar, according to the manufacturer's instructions (BD). Cultures were incubated at 37°C unless otherwise stated. Where required, antibiotics purchased from Sigma were used at the following concentrations: chloramphenicol at 20 μg/ml, kanamycin at 50 μg/ml, ampicillin at 100 μg/ml, and spectinomycin at 100 μg/ml.
Construction of mutant and gene reporter strains.
Standard genetic methods were used (46). Insertional inactivation of yohN was accomplished using the one-step mutagenesis method in strain BW25113(pKD46) (9), using the primers yoNFRTup and yoNFRTdwn2 (see Table S1B in the supplemental material) to amplify the cat cassette from plasmid pKD3. The cat gene was inserted antisense to yohN. The yohN-cat fusion was transferred from strain BW25113 into other strains via generalized phage transduction using P1 vir, as described by Miller (32). The same procedure was used to construct a ΔrcnA-cat strain using primers rcnAFRTup1 and rcnAFRTdwn2. The transcriptional rcnA-uidA fusion, carried by strain ARY023 (44), was moved into W3110 or NM522 strains using P1 phage transduction.
RT-PCR assays.
Total RNA was isolated from cultured cells with the RNeasy kit (Qiagen) and treated twice with DNase (Ambion) for 30 min at 37°C. The absence of DNA contamination was verified by direct PCR. RNA was quantified by measuring the optical density at 260 nm (OD260), and its integrity was confirmed by agarose gel electrophoresis. RNA was reverse transcribed using the Access reverse transcriptase PCR (RT-PCR) kit (Promega) using the primers RTMup and RTMdwn, YoN2up and Yodwn2, and Yodwn2 and RTMup for amplification of the yohM, yohN, and yohMN transcripts, respectively. Controls were treated as samples, but reverse transcriptase (RT) was omitted. PCR and RT-PCR products were visualized on a 2% agarose gel.
cDNA synthesis and quantitative real-time PCR (qRT-PCR).
Total RNA was extracted, as described above, from 10 ml of log-phase cultures. cDNA synthesis was performed using 2 μg of RNA with Superscript II RT (Invitrogen), according to the manufacturer's instructions. Real-time PCR was performed using Master Plus SYBR green I (Roche Applied Science) to quantify the expression level of the rcnA and rcnB genes. The rpoB and rpoA genes were chosen as reference genes for data normalization. Primers were designed with the Primer 3 software. Amplification and detection of the specific products were conducted with the LightCycler system, and data analysis was performed with the LightCycler relative quantification software (Roche Applied Science). Relative expression was calculated as the ratio of the normalized value of each sample to that of the corresponding untreated control, with a correction for individual PCR efficiency according to the work of Pfaffl (39). All real-time PCRs were performed on cDNA obtained from at least three independent cultures.
Nickel and cobalt susceptibility testing.
Metal sensitivity assays were conducted as follows. First, bacteria were grown to mid-log phase in LB medium. A 10-fold serial dilution of the cultures in M63 medium was performed, and 5 μl was spotted onto 0.4% glucose-M63 minimal medium plates containing increasing concentrations of each metal. Plates were incubated at 37°C for 24 to 48 h under aerobic conditions or for 48 to 72 h under anaerobic conditions.
Enzymatic assays.
β-Glucuronidase and β-galactosidase activities were measured on toluenized extracts by monitoring the degradation of synthetic substrates (Sigma): orthonitrophenyl-β-d-galactoside (ONPG) for β-galactosidase assays or paranitrophenyl-β-d-glucuronide (PNPU) for β-glucuronidase assays.
For fluorescence quantification, cells were grown to stationary phase, and fluorescence was measured with an SFM 23/B spectrofluorometer (Kontron Instruments).
For hydrogenase activity assays, 25 ml of anaerobic cultures (OD600 = 0.5) in LBSM medium (LB supplemented with 2 μM sodium selenite and 2 μM sodium molybdate) at 37°C was harvested by centrifugation and washed twice with a wash buffer consisting of 50 mM sodium phosphate buffer, pH 6.8 (NaH2PO4/Na2HPO4). Bacteria were resuspended in 1/10 volume of the wash buffer. Toluenized extracts were assayed for the oxidation of benzyl viologen as previously described (42).
Overexpression and purification of the RcnB protein.
Strain BL21, harboring the pETRCB plasmid, was grown to mid-log phase in LB medium. Next, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to induce the expression of RcnB. After 4 h at 30°C, cells were harvested by centrifugation and either frozen at −80°C for later use or diluted in 50 mM Tris-HCl, pH 7, supplemented with 1.5 M ammonium sulfate, and lysed by 3 passages through a French press in the presence of Halt protease inhibitor (Thermo). The supernatant was filtered through a 0.45-μm filter and loaded onto a phenyl HP column (HiTrap; GE Healthcare). Elution followed a linear gradient, from 1.5 M to 0 M ammonium sulfate in 50 mM Tris-HCl, pH 7, and fractions containing the RcnB protein (0.7 M ammonium sulfate) were pooled and dialyzed against 50 mM Tris-HCl, pH 7, to remove the ammonium sulfate. The fraction containing RcnB was loaded onto an anion-exchange UnoQ1 column (Bio-Rad). Under these conditions, RcnB was not retained on the column, in contrast to the contaminating proteins present in the loaded fraction. The purity of RcnB was verified on a 15% Tris-Tricine polyacrylamide gel. When necessary, purified RcnB was concentrated using 6-kDa-cutoff Vivaspin 6 concentrators (Sartorius). RcnB was quantified either by measuring OD278 using an ε of 21,430 M−1 cm−1 or by using the Bradford protein assay kit (Bio-Rad).
Immunodetection of proteins.
Polyclonal antibodies against RcnB were obtained from Covalab (Villeurbanne, France) by immunization of a rabbit with the purified protein. For Western blot analysis, periplasmic extracts of cells grown in the presence of 0.1% lactose were fractionated from whole cells using the chloroform extraction procedure (13). To control for contamination of the periplasmic fraction with cytoplasmic proteins, β-galactosidase activity was measured on crude and periplasmic extracts. After migration on a 15% Tris-Tricine PAGE gel, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane. Anti-RcnB antibodies were used at a dilution of 1/3,000, and anti-BlaM antibodies were used at a 1/5,000 dilution. Detection was performed using horseradish peroxidase (HRP)-conjugated secondary antibodies (Sigma) and Immobilon Western chemiluminescent substrate (Millipore).
Metal-protein interactions.
Filter binding assays were conducted as described in reference 14. Purified RcnB, in 50 mM Tris-HCl, pH 7, was incubated overnight with increasing quantities of radiolabeled 63NiCl2 and was subsequently immobilized on a buffer-equilibrated 0.2-μm Westran PVDF membrane (Millipore). The membrane was washed thoroughly with buffer, and radioactivity was quantified by immersing dried filters in 5 ml of OCS scintillation liquid (Amersham Biosciences). Background radioactivity (in the absence of protein) was subtracted.
ICP-OES assays.
Strains were grown to mid-log phase in 250 ml LB medium supplemented with 500 μM NiSO4 or 50 μM CoCl2 or in minimal medium containing 50 μM NiSO4 or 15 μM CoCl2; cells were subsequently harvested by centrifugation, washed once with phosphate buffer plus 1 mM EDTA, and then washed twice with phosphate buffer alone. The bacterial pellet was vacuum dried and weighed (typically circa 0.100 g was obtained) before being dissolved in 2 ml of 10 M nitric acid (Traceselect; Fluka). Digestion was conducted in a Mars XPress microwave device (CEM Corporation) by applying a linear temperature gradient from room temperature to 180°C over 15 min, followed by 30 min at 180°C. After cooling, mineralized samples were diluted to 8 ml with ultrapure water and analyzed by inductively coupled plasma optical emission spectrometry (ICP-OES) using an axial JY 138 Ultrace prototype spectrometer (Horiba, Longjumeau, France) operating at 1,075 W and equipped with a Meinhard-type nebulizer and a Scott spray chamber. Compressed air was used as shear gas. Flow rates were as follows: plasma gas, 12 liters min−1; nebulizer gas, 0.7 liter min−1; sheath gas, 0.1 liter min−1. Integration time was set at 2 s, and the wavelength was set to 221.65 nm for Ni and 228.62 nm for Co.
63Ni uptake assay.
Cells were grown in LB medium in the presence of 500 μM NiSO4 at 37°C under aerobic conditions to mid-log phase (OD600 = 0.6); cells were subsequently harvested and washed once with a buffer consisting of 66 mM KH2PO4-K2HPO4 (pH 6.8) and 0.4% glucose. Bacteria were resuspended in 1/5 volume of the wash buffer. Transport assays were conducted by adding 63Ni2+ to a final concentration of 5 μM, with filtration performed at the indicated times. Nitrocellulose filters (0.45-μm pore size; Millipore) were washed twice with the same buffer containing 10 mM EDTA. Filters were dried and immersed in 5 ml scintillation fluid (OCS; Amersham Biosciences). Radioactivity was counted with a Tri-Carb Packard Bell scintillation counter. 63NiCl2 (396 Ci/mol) was acquired from Amersham Biosciences.
63Ni efflux assay.
The 63Ni efflux assay procedure was adapted from the work of Pimentel et al (40). Cells were grown in M63 medium containing 0.4% glucose (medium A) in the presence of 5 μM NiSO4 at 37°C under aerobic conditions until early log phase (OD600 = 0.4); cells were subsequently harvested and washed twice with prewarmed medium A and resuspended in 1/10 volume of the same medium. 63NiCl2 (5 μM) was added, and the cells were incubated for 15 min at 37°C. Aliquots of 200 μl were diluted in 20 ml 50 mM Na2HPO4-NaH2PO4, pH 7.2, and incubated at 37°C. At various time points, 1-ml aliquots were filtered, and the filters were washed and radioactivity was counted as described above.
RESULTS
rcnB is part of the rcn operon.
We have previously reported on the role of RcnA as an efflux pump involved in Co and Ni detoxification (44). In E. coli, rcnA is followed by the yohN gene, which is transcribed in the same direction. In order to assess the possible cotranscription of rcnA and yohN, mRNA was isolated from strain NM522 after growth in LB medium supplemented with 0.25 mM NiSO4, conditions that are known to induce the expression of rcnA (44). After a reverse transcription step, amplification of cDNA using rcnA- and yohN-specific primers (see Table S1B in the supplemental material) yielded 720-bp and 300-bp products, respectively, indicating that yohN is transcribed under the growth conditions used (see Fig. S1A in the supplemental material). Using primers complementary to the 5′ end of rcnA and the 3′ end of yohN, a 1,300-bp PCR product was detected, indicating that rcnA and yohN are cotranscribed. It has been previously shown that the transcription of rcnA is induced by Ni or Co (44) via the metalloregulator RcnR (3, 26). Quantitative real-time PCR assays were performed to monitor the expression of the yohN mRNA in NM522 cells grown in LB medium alone or medium supplemented with 1 mM NiSO4 or 0.1 mM CoCl2. These metal ion concentrations induced the highest expression of the rcnA::uidA gene reporter fusion (44). Under these conditions, the expression of yohN was similarly induced by Ni or Co, leading to a 10-fold increase in expression, compared to cells grown in LB medium alone (see Fig. S1B). When assayed in the rcnR mutant strain, DB150, the level of expression of yohN was 8-fold higher than that of the wild-type (WT) strain, and the addition of Ni or Co to the growth medium had no significant effect (P > 0.5), indicating that yohN is negatively regulated by rcnR. The same pattern of mRNA expression has been previously obtained for rcnA (3), reaffirming the fact that rcnA and yohN are coexpressed and coregulated. For these reasons, yohN has been renamed rcnB.
rcnB is involved in Ni and Co resistance.
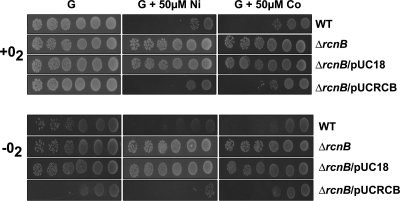
Because rcnB forms an operon with rcnA, we naturally investigated its function in Ni and Co metabolism. For that purpose, an rcnB deletion mutant was constructed from the WT strain, W3110, giving rise to strain WRCB1. Serial dilutions of fresh cultures grown in LB medium were spotted on M63 minimal medium plates supplemented with 0.4% glucose and, as required, with 50 μM NiSO4 or CoCl2, followed by incubation at 37°C under aerobic or anaerobic growth conditions. After aerobic culture, growth of the WT strain was observed only for the two most concentrated bacterial dilutions when either Ni or Co was present in the medium (Fig. 1, upper panel). In contrast, the growth of the rcnB mutant was not influenced by Ni or Co because growth could be observed throughout the dilution range, similar to the growth observed in the absence of metals. This metal resistance phenotype is fully linked to the absence of rcnB because complementation of the rcnB mutant by a plasmid-borne copy of the gene restored the wild-type phenotype. Under anaerobic growth conditions, similar results were obtained, with the exception that growth of the complemented mutant on glucose-supplemented medium was impaired, which will be discussed below. Therefore, rcnB plays a role in Ni and Co metabolism. Surprisingly, this phenotype is the opposite of what has been observed for the rcnA mutant cells, which are sensitive to Ni and Co (44).
Fig. 1.
rcnB is involved in Ni and Co resistance. The wild-type strain (W3110), the ΔrcnB isogenic mutant (WRCB1), WRCB1 × pUC18, and WRCB1 × pUCRCB (rcnB in pUC18) were grown in LB medium to mid-log phase. A 10-fold serial dilution of the cultures was performed, and 5 μl was spotted on M63 minimal medium supplemented with 0.4% glucose (G), 0.4% glucose plus 50 μM NiSO4, or 0.4% glucose plus 50 μM CoCl2 and grown aerobically (top) or anaerobically (bottom) at 37°C in petri dishes. Spots corresponding to 107 (rightmost column) to 102 (leftmost column) bacteria are shown.
rcnB mutants show reduced intracellular Ni or Co levels.
As rcnB mutants are more resistant to Ni and Co, one possible explanation is the reduced accumulation of these metals in the bacteria. Metal quantification by ICP-OES was first performed on whole, dried bacteria after growth in LB medium supplemented with subinhibitory concentrations of Ni or Co (0.5 mM Ni or 0.05 mM Co). The bacteria were harvested and washed with EDTA-supplemented buffer prior to analysis in order to minimize nonspecific interactions between metals and the bacterial cell envelope. Ni or Co was not detectable after growth in LB medium alone. Compared to the WT strain, the Ni or Co content in the rcnB mutant was reduced by 20% and 40%, respectively (Table 1), under the different conditions tested. Small peptides present in LB medium are known to chelate metals and to lower free metal concentrations. The same assays were thus repeated in minimal medium in the presence of 0.05 mM NiSO4 or 0.015 mM CoCl2; these conditions inhibited growth of the WT cells by approximately 10-fold (final OD), whereas no change was observed for the rcnB mutant. The metal content in the rcnB mutant was reduced by 68% for Co and 84% for Ni, compared to WT cells (Table 1). These results are consistent with those of the metal susceptibility test, and they support the proposed role of rcnB in Ni and Co metabolism. Complementation of the rcnB mutant by rcnB hosted on the pUC18 plasmid restored the cellular Ni and Co content to a level higher than what was measured for WT cells, confirming the function of rcnB in modulating Ni and Co accumulation. The increased quantity of the two metals present in this case is probably due to the high copy number of the pUC18 plasmid.
Table 1.
Ni and Co contents of a ΔrcnB mutanta
| Growth medium | Metal assayed | Mean (SD) for strain: |
||
|---|---|---|---|---|
| WT | ΔrcnB mutant | ΔrcnB/rcnB mutant | ||
| LB + Ni | Ni | 14.3 (0.2) | 11.8 (0.2) | 18.6 (0.1) |
| LB + Co | Co | 11.9 (0.5) | 6.9 (0.1) | 16.2 (0.1) |
| M63 + Ni | Ni | 88 (13) | 14 (0.5) | ND |
| M63 + Co | Co | 114 (25) | 37 (0.4) | ND |
The wild-type strain (W3110), the ΔrcnB isogenic mutant (WRCB1), and the ΔrcnB mutant complemented by rcnB (WRCB1/pUCRCB) were grown in LB medium supplemented with 500 μM NiSO4 or 50 μM CoCl2 or in 0.4 % glucose-M63 minimal medium supplemented with 50 μM Ni or 15 μM Co. Values (μg/g dry weight) are the means of at least three independent ICP-OES measurements performed on independent cultures. ND, not determined.
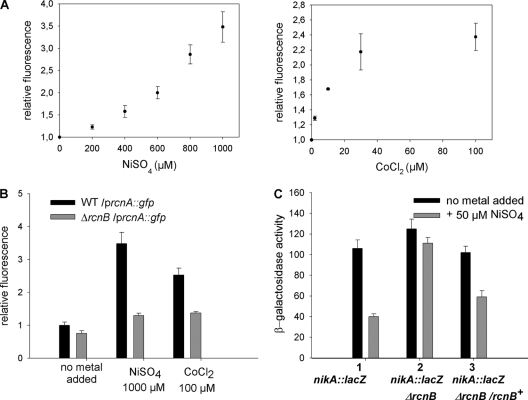
Because rcnB affects the profile of resistance to Ni and Co, several hypotheses regarding metal sequestration or compartmentalization in the bacterial cell may explain this phenomenon. To gain insights into the mode of action of rcnB, we investigated the consequences of changes in intracellular Ni and Co levels in E. coli. To achieve this, we took advantage of the genetic systems utilized in our laboratory, specifically the rcn and nik systems. rcnA expression is induced by Ni and Co (3, 44) via the control of the metalloregulator RcnR (26, 29). A plasmid-borne gene fusion of rcnA and green fluorescent protein (rcnA-gfp; p157) (38) was used to monitor the level of expression of rcnA in LB medium supplemented with increasing concentrations of Ni or Co (Fig. 2A). Within the range of 0 to 1,000 μM NiSO4 and 0 to 30 μM CoCl2, green fluorescent protein (GFP) levels increased as a function of the concentration of added metal, and thus, this fusion can be used as an in vivo sensor of intracellular Ni or Co concentrations. The Ni and Co concentrations chosen thereafter were those providing the maximum response from the reporter gene. When introduced into the ΔrcnB strain, WRCB1, the level of fluorescence was sharply reduced in the presence of 1,000 μM NiSO4 or 100 μM CoCl2; the specific activity was 50% of that of the WT strain and was not significantly different from the background level (Fig. 2B). The same types of assays were conducted using a nikA::lacZ gene fusion. The nikABCDE operon involved in nickel uptake is expressed under anaerobic growth conditions (53), and its expression is specifically repressed by nickel via the metalloregulator NikR (12). As previously shown (54), the expression of the nikA::lacZ gene reporter fusion is inversely proportional to the concentration of Ni present. When assayed in the nikA::lacZ mutant, the β-galactosidase specific activity was reduced by more than half when 0.05 mM NiSO4 was added to the growth medium (Fig. 2C, plot 1), and consequently, this concentration was used for further assays. Interestingly, in the ΔrcnB derivative, the expression of the fusion gene was the same in the absence and in the presence of Ni. Finally, when rcnB was expressed in trans, the expression profile was restored to the pattern exhibited by the nikA mutant. Taken together, these results demonstrate that when rcnB is impaired, the total cellular levels of Ni or Co are decreased and, more precisely, that it is the intracellular Ni and Co pools that are reduced.
Fig. 2.
Ni and Co intracellular concentrations are modified by rcnB. (A) Expression of the rcnA::gfp fusion in the presence of increasing concentrations of Ni (left) or Co (right). W3110 harboring plasmid p157 (rcnA::gfp) was grown aerobically in LB supplemented with the indicated metal concentrations, and the relative fluorescence of the rcnA-gfp gene fusion was calculated as follows: the specific fluorescence (F/OD600), recorded in the presence of metals, was divided by the specific fluorescence assayed in the absence of metals. Results are the means of at least three independent experiments. The increases in fluorescence as a function of Ni or Co concentration can be represented by a hyperbolic function (48) with the following parameters: fluorescence of Co [0 to 30 μM] = −0.0014 [Co]2 + 0.0777 [Co] + 1.0587, R2 = 0.9935; fluorescence of Ni [0 to 1,000 μM] = 1.781 × 10−6 [Ni]2 + 0.0007 [Ni] + 0.9975, R2 = 0.9946. (B) The wild-type strain (W3110) or the ΔrcnB isogenic mutant (WRCB1)-carrying plasmid p157 (WT/prcnA::gfp [black bars] or ΔrcnB/prcnA::gfp [gray bars]) was grown in LB, LB plus 1,000 μM NiSO4, or LB plus 100 μM CoCl2, and fluorescence was assayed as described for panel A. (C) Expression of the nikA::lacZ fusion. Cells were grown anaerobically in LB (black bars) or in LB plus 50 μM NiSO4 (gray bars). β-Galactosidase activity is in nmol ONP min−1 mg bacterial dry weight−1. Strains: 1, nikA::lacZ mutant (HYD72K1); 2, nikA::lacZ ΔrcnB mutant (HYRB1); 3, nikA::lacZ ΔrcnB/rcnB mutant (HYRB1 × pUCRCB). Data are the means of at least three independent experiments. Standard deviations are presented in the figure.
RcnB is a soluble periplasmic protein.
rcnB encodes a 112-amino-acid (aa) protein of 12,466 Da. A Sec-type signal peptide is predicted at the N terminus, with a cleavage site located after residue 21, leading to a mature polypeptide of 10,238 Da that presumably localizes to the periplasm. RcnB was overexpressed in E. coli and purified. Polyclonal antibodies raised against the purified protein were used for detection by immunoblotting. Periplasmic extracts were prepared and assayed for contamination by cytoplasmic proteins by measuring β-galactosidase activity before analysis by Western blotting. β-Lactamase, BlaM, encoded by vector pUC18, was used as a control for the periplasmic fractionation. RcnB could be detected in the periplasmic fraction of wild-type cells, demonstrating its periplasmic localization (see Fig. S2A, lane 1, in the supplemental material) and was clearly absent in the rcnB mutant (Fig. S2A, lane 3). Periplasmic localization was confirmed by N-terminal sequencing. RcnB was purified from the periplasmic fraction, and its N terminus was determined by Edman sequencing. The sequence of the first six amino acids was AGSTNT, corresponding to residues 26 to 31 of the predicted amino acid sequence. Thus, RcnB possesses a cleavable N-terminal signal sequence of 25 aa, leading to the 9,898-Da mature form of RcnB. The experimental cleavage site is 4 aa downstream of the predicted one.
Sequence comparison retrieved more than 150 homologs of RcnB among alpha-, beta-, and gammaproteobacteria, but none are previously described proteins (see Fig. S2B in the supplemental material). Similarly, no motifs could be detected in the sequence of RcnB using Interproscan (25). Alignment of the RcnB sequence demonstrated that the second half of the sequence is quite well conserved; however, there was no obvious conservation of residues potentially involved in metal liganding, except for H82 and D53 and, to a lesser extent, D95 and H111. Notably, RcnB lacks Cys residues. This is the case for all of the sequences considered, except for Q88FZ8 (P. putida) and Q882W6 (P. syringae), which do bear Cys within their predicted signal sequences and, consequently, are predicted to be lipoproteins. However, the latter prediction is tentative because the canonical sequence usually shared by lipoproteins is absent. The first half of the alignment exhibits no sequence conservation and a great variability in sequence length, which is due, in part, to the presence of a signal sequence. Concerning synteny, it is very diverse; the rcnB gene may lie close to rcnA, close to hypothetical efflux pumps, or close to uncharacterized hypothetical open reading frames (ORFs) or as an orphan gene. Some organisms possess several copies of rcnB, for example, entries Q6FEG8 and B5XP92 from Acinetobacter sp., D0FUH8 and D0FV83 from Erwinia pyrifoliae, B5Y075 and B5XP92 from Klebsiella pneumoniae, and Q88FZ8 and Q88RS9 from P. putida.
RcnB does not bind Ni or Co directly.
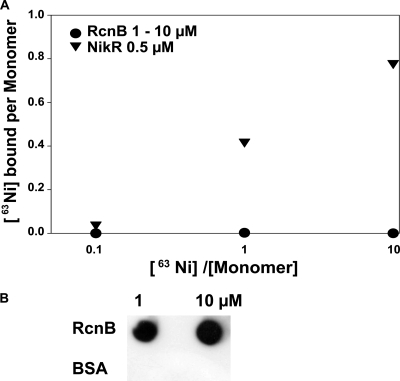
In view of its role and its cellular localization, the binding of Ni or Co ions by RcnB needed to be tested, and several different techniques were employed. The metalation of purified RcnB was first monitored by tryptophan fluorescence assays, as RcnB possesses two tryptophan residues that are well conserved in the sequence alignment. No changes in the spectra were observed following the addition of Ni or Co (data not shown). More direct methods were then applied using a 63Ni radioactive isotope. Equilibrium dialysis and flow dialysis demonstrated no nickel binding by RcnB (data not shown). Filter binding assays, which had previously been used successfully to determine the affinity constant of NikR for Ni, were performed (14). When assayed in parallel, purified NikR bound 63Ni, as observed in the previous study, whereas purified RcnB exhibited no binding (Fig. 3A). Dot blot and immunodetection showed that RcnB is retained well on the PVDF membrane after filtration (Fig. 3B). The protein and metal concentrations were varied, which indicates that the Kd (dissociation constant) of RcnB for Ni is much greater than 100 μM. Thus, we conclude that RcnB does not bind Ni or Co ions directly under the tested conditions.
Fig. 3.
Interaction of RcnB with Ni. (A) Filter binding assays: 1 μM or 10 μM RcnB (black circles) or 0.5 μM NikR (black triangles) was incubated with the indicated 63NiCl2 concentrations and filtered onto PVDF membranes. 63Ni bound to the membrane is plotted against total Ni concentration. (B) Dot blot assays: 10 μl of 1 or 10 μM RcnB or bovine serum albumin was filtered onto PVDF membranes and detected using anti-RcnB antibodies.
RcnB operates independently of Ni entry.
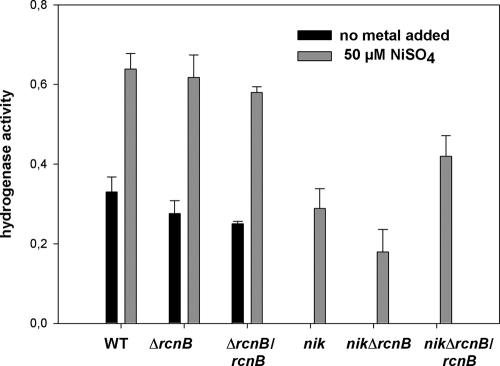
When RcnB is absent, E. coli strains are more resistant to Ni and Co. This correlates with a decrease in intracellular metal concentration (Fig. 2 and Table 1). As RcnB cannot bind Ni or Co directly, a role for RcnB in the storage of Ni and Co is unlikely. Therefore, we examined the possibility that RcnB acts on Ni and Co entry into the cytoplasm. In E. coli, specific Ni uptake occurs via the NikABCDE transporter, which is expressed under anaerobic growth conditions (34). nik mutants are defective in Ni-dependent hydrogenase activity, which can be restored when excess Ni is added to the growth medium (42, 54). Apart from the high-capacity magnesium transport system CorA, it has been suggested that another nickel import pathway operates in E. coli (34, 45). Therefore, we measured the hydrogenase activity of WT and mutant bacteria that were grown anaerobically in LB medium, either without added Ni or with 50 μM NiSO4. The restoration of hydrogenase activity in the nik mutants by the addition of nickel has been shown to reach roughly 50% of the level of the WT strain under these conditions (54). As expected, a similar pattern of hydrogenase recovery could be seen here (Fig. 4). In contrast, both the ΔrcnB mutant and the ΔrcnB mutant complemented by the plasmid-borne rcnB allele exhibited levels of hydrogenase activity similar to that of the WT strain, both in the absence and in the presence of nickel. The nik ΔrcnB double mutant was constructed by transferring the ΔrcnB deletion into the nik mutant. Interestingly, the nik ΔrcnB mutant exhibited half the level of hydrogenase activity of the nik mutant, and complementation by the rcnB allele produced a strong enhancement of hydrogenase activity over that of the nik and nik ΔrcnB mutants. This result was consistent with the nickel content analysis (Table 1), confirming that RcnB modulates Ni homeostasis and demonstrating that it functions independently of the NikABCDE uptake system. This result also suggests that RcnB interacts poorly with auxiliary Ni uptake systems because no Ni would enter the cell in a nik ΔrcnB mutant. As a consequence, the reduction in intracellular Ni concentration associated with RcnB is unlikely to be due to altered Ni acquisition by the cell.
Fig. 4.
Effect of rcnB on Ni acquisition. Total hydrogenase activity was measured after anaerobic growth in LBSM (black bars) or in LBSM supplemented with 50 μM NiSO4 (gray bars). Specific hydrogenase activity is in μmol benzyl viologen reduced min−1 mg bacterial dry weight−1. Strains: wild type (MC4100), ΔrcnB isogenic mutant (MRCB1), ΔrcnB/rcnB mutant (MRCB1/pUCRCB), nikA::lacZ isogenic mutant (HYD72K1), nikA::lacZ ΔrcnB isogenic mutant (HYRB1), and nikA::lacZ ΔrcnB/rcnB mutant (HYRB1 × pUCRCB). Data are the means of at least three independent experiments. Standard deviations are presented in the figure.
Absence of rcnB results in enhanced efflux of Ni.
In order to clarify the function of rcnB in the dynamics of Ni and Co accumulation by the cell, the accumulation of 63Ni was monitored in a time course assay. Unloaded cells were incubated with 63Ni, and the kinetics of Ni uptake confirmed that a lower level of Ni was accumulated in the rcnB mutant than in the WT cells (Fig. 5A). We also analyzed the ΔrcnA mutant. In this mutant, RcnB is synthesized, as confirmed by Western blot experiments (see Fig. S2A in the supplemental material). In this case, 63Ni accumulation was 2-fold higher than that in the parental strain (Fig. 5A), which is in agreement with previously published results (44). We previously used an rcnA::uidA insertion mutant, which was revealed by qRT-PCR to be an rcnAB double mutant because the uidA-Kanr cassette has a polar effect on the downstream gene rcnB (data not shown). When tested, the ΔrcnA and rcnA::uidA mutants showed similar behaviors, with an increased sensitivity to Ni and Co and similar Ni uptake properties (data not shown). This concurs with RcnA being a Ni and Co efflux pump, as previously established (44), and suggests that rcnA is epistatic to rcnB, as there appears to be no difference between the rcnA and rcnAB mutants. These data imply that RcnB does not facilitate Ni uptake but, rather, has a possible role in the efflux of Ni.
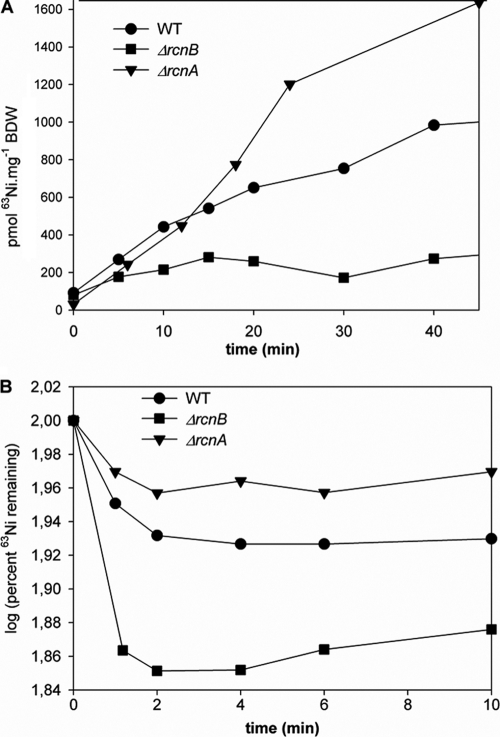
Fig. 5.
rcnB acts on Ni efflux. (A) 63Ni accumulation kinetics in the wild-type strain and its rcn derivatives. Strains: wild type (NM522, circles), ΔrcnB mutant (NRCB1, squares), and ΔrcnA mutant (NRCA1, triangles). (B) Effect of rcnB mutation on Ni efflux. Efflux was measured at 37°C in E. coli strains: wild type (W3110, circles), ΔrcnB mutant (WRCB1, squares), and ΔrcnA isogenic mutant (WRCA1, triangles). The experiments were repeated at least three times. The data shown are from a single, representative experiment.
The role of RcnB was further investigated with regard to Ni efflux. 63Ni-loaded cells were incubated in unlabeled, fresh medium, and the cellular 63Ni content was assayed at various time points. In this context, the rate of Ni efflux in the rcnB mutant was considerably higher than that of the WT strain (Fig. 5B). This result clearly demonstrates that RcnB acts on Ni efflux. The ΔrcnA mutant exhibited an efflux rate much lower than that of the WT strain (Fig. 5B), which is in agreement with RcnA being an efflux pump. Furthermore, this confirms the phenotypic dominance of RcnA over RcnB in the efflux of Ni. As the absence of rcnB facilitates the efflux of Ni, the role of RcnB is most likely to regulate or balance the efflux of Ni mediated by RcnA.
DISCUSSION
The present work identifies a protein of a new type that influences Ni and Co homeostasis in E. coli. The absence of RcnB clearly affected the physiology of the cells by dramatically increasing the resistance of E. coli to Ni and Co. We showed that this phenotype is linked to decreased intracellular concentrations of Ni or Co. As previously mentioned, metal trafficking is quite complex, and this may explain the observed differences in the resistance phenotypes between the aerobic and anaerobic growth conditions. The expression in trans of rcnB rendered the cells more sensitive to the depletion of oxygen (Fig. 1), likely due to increased metal accumulation. This phenomenon is not linked to RcnA, as we have previously shown that rcnA is expressed under both conditions (44). However, the existence of multiple import and export systems makes it difficult to demonstrate the role of an isolated protein. For this reason, we used several E. coli K-12 strains in this work, and we obtained similar results for all strains. The experimental conditions (medium compositions and metal concentrations) are also important and may explain the discrepancy observed here and in another study, where no phenotype was associated with rcnB (29). We also verified our data by performing the experiments on strain JW5346 (BW25113 ΔrcnB Cmr), obtained from the Keio collection (1), which fully confirmed our conclusions (data not shown).
To date, the only known periplasmic Ni-binding protein in E. coli is NikA, whereas none are known to bind Co. RcnB is localized to the periplasm, and there is no evidence that RcnB binds Ni or Co ions directly in the present study. Interestingly, the resolution of several NikA structures has provided breakthroughs in the understanding of metal liganding. An initial report presented the NikA structure as a typical periplasmic binding protein of the ABC family, though the position of Ni in this structure did not take into account its environment (24). A second study resolved the structure of NikA in complex with Fe(III)-EDTA (H2O) bound at the substrate site, which suggested that NikA binds Ni in complex with a metallophore (7). Finally, the resolution of NikA crystallized after a periplasmic extraction in the absence of EDTA showed that the nickelophore was best modeled as butane-1,2,4-tricarboxylate (6), although its exact nature remains to be established. RcnB could interact with metals in a similar way, by liganding Ni or Co complexed to a prosthetic group.
RcnB could be involved in Ni and Co homeostasis, either by increasing metal import or by decreasing export. We have shown, by monitoring hydrogenase activity (Fig. 4), that RcnB is certainly not involved in importing Ni. This conclusion is further supported by the fact that the expression of rcnB is induced by Ni or Co (see Fig. S1C in the supplemental material), which would conflict with a role in importing these toxic compounds.
Homeostasis is primarily the result of a balance between the entry and exit of metals. Hence, the regulation of transporters is under the control of metallosensory regulatory proteins (31). To avoid excess removal of Ni or Co from the cytoplasm, the transcription of the efflux pump RcnA is under the control of the metallorepressor RcnR (26). The transcription of the NikABCDE importer is repressed by the NikR metalloregulator (12). Induction of rcnA and repression of the nik operon by Ni have been shown to be sequential, i.e., PnikA is repressed by Ni concentrations that are lower than those needed for the activation of PrcnA (26). In this study, we demonstrate that RcnB is a soluble periplasmic protein that influences Ni and Co homeostasis. RcnB could be a transmitter in a signal transduction pathway across the cytoplasmic membrane, as is observed for other systems (51). This signal transduction cascade would lead to modifications in the transcription of Ni or Co efflux systems. More precisely, it could be supposed that RcnA is the target because we have shown that the action of RcnB is dependent on the presence of rcnA (Fig. 5); in view of the phenotype associated with rcnB, when rcnB is impaired, rcnA would be upregulated. Nevertheless, when assayed in the ΔrcnB mutant, the rcnA-gfp gene fusion exhibited induction levels that were lower than those in the WT strain in the presence of Ni or Co (Fig. 2B), which contradicts the above hypothesis.
Finally, we clearly show that RcnB is involved in the efflux of Ni. The fact that RcnB does not bind Ni or Co ions and does not influence the transcription of rcnA argues in favor of RcnB exerting its function via protein-protein interactions, especially with RcnA. This is further indicated by the fact that, in the absence of rcnA and the presence of RcnB, more Ni is accumulated by the cells (Fig. 5). Moreover, rcnA and rcnB are cotranscribed (see Fig. S1 in the supplemental material). Efflux in bacteria can occur via a wide variety of structurally diverse systems. Members of the CDF, MFS, P-type ATPase, and RND families (for a review, see reference 36) have been shown to export metals with narrow or broad substrate specificities. However, none of the metal efflux systems and, even more generally, none of the efflux systems seem to function with a soluble periplasmic protein, with the possible exception of the RND CusCFBA system of E. coli. RND protein complexes transport their substrates from the periplasm to the extracellular space. The cusCFBA operon in E. coli encodes an RND copper efflux system made up of CusC (OMF), CusB (MFP), and CusA (RND) (37). This RND system comprises an additional small periplasmic protein, CusF (21). Mutation of any of the cusCFBA genes leads to enhanced copper sensitivity, although to a lesser extent for cusF (18). CusF and CusB interact in a metal-dependent way, and Cu(I) is transferred between CusF and CusB (2). The precise mechanism of efflux is not understood, but this example emphasizes that export is also mediated by periplasmic proteins. A striking difference with the RcnAB efflux system is that mutations in cusF or cusB diminish Cu export, whereas mutations in rcnB enhance Ni and Co efflux. Moreover, no RND system involved in Ni or Co efflux has been identified in E. coli. Another interesting feature of RND systems is that they mediate efflux predominantly from the periplasmic space, outside the cell, as emerging evidence suggests that the detoxification of toxic compounds occurs in two steps. The compounds are first transported from the cytoplasm to the periplasm by an inner-membrane efflux system, and then, an RND-type exporter, with overlapping specificity, expels the drugs (50) or the metals (47) from the periplasm to the extracellular milieu. These examples highlight the importance of the step conducted in the periplasmic space in efflux-mediated resistance mechanisms, and further studies are necessary to establish a role for RcnB as a possible intermediate in these networks.
Another interesting example regarding the pivotal role of a periplasmic transport protein has been recently reported for the sialic acid-specific uptake system in Haemophilus influenzae. SiaP is the periplasmic substrate binding protein of the TRAP (tripartite ATP-independent periplasmic transporter) importer SiaPQM. In vitro experiments confirmed that SiaP is an essential component of the import system; intriguingly, under some experimental conditions, SiaP was able to trigger the reverse event, i.e., the export of sialic acid by the inner-membrane complex SiaQM (33).
The present work provides compelling evidence of a new biological function and demonstrates the existence of alternative efflux mechanisms. Indeed, from the data collected here, RcnB can be proposed to function as a valve that regulates Ni and Co efflux by RcnA. Further work is required to investigate the interactions between RcnB and other proteins and to understand its control over metal movements.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by CNRS, IFCPAR grant no. 3709, INSA Lyon BQR. C.B. was supported by a grant from the French Ministry of Research.
We thank G. Condemine for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Baba T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bagai I., Rensing C., Blackburn N. J., McEvoy M. M. 2008. Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry 47:11408–11414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blaha D., et al. 2011. The Escherichia coli metallo-regulator RcnR represses rcnA and rcnR transcription through binding on a shared operator site: insights into regulatory specificity towards nickel and cobalt. Biochimie 93:434–439 [DOI] [PubMed] [Google Scholar]
- 4. Cadieux N., et al. 2002. Identification of the periplasmic cobalamin-binding protein BtuF of Escherichia coli. J. Bacteriol. 184:706–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cadieux N., Kadner R. J. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. U. S. A. 96:10673–10678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cherrier M. V., Cavazza C., Bochot C., Lemaire D., Fontecilla-Camps J. C. 2008. Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA. Biochemistry 47:9937–9943 [DOI] [PubMed] [Google Scholar]
- 7. Cherrier M. V., et al. 2005. Crystallographic and spectroscopic evidence for high affinity binding of FeEDTA(H2O)- to the periplasmic nickel transporter NikA. J. Am. Chem. Soc. 127:10075–10082 [DOI] [PubMed] [Google Scholar]
- 8. Cvetkovic A., et al. 2010. Microbial metalloproteomes are largely uncharacterized. Nature 466:779–782 [DOI] [PubMed] [Google Scholar]
- 9. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Degen O., Eitinger T. 2002. Substrate specificity of nickel/cobalt permeases: insights from mutants altered in transmembrane domains I and II. J. Bacteriol. 184:3569–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Degen O., Kobayashi M., Shimizu S., Eitinger T. 1999. Selective transport of divalent cations by transition metal permeases: the Alcaligenes eutrophus HoxN and the Rhodococcus rhodochrous NhlF. Arch. Microbiol. 171:139–145 [DOI] [PubMed] [Google Scholar]
- 12. De Pina K., Desjardin V., Mandrand-Berthelot M. A., Giordano G., Wu L. F. 1999. Isolation and characterization of the nikR gene encoding a nickel-responsive regulator in Escherichia coli. J. Bacteriol. 181:670–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Pina K., et al. 1995. Purification and characterization of the periplasmic nickel-binding protein NikA of Escherichia coli K-12. Eur. J. Biochem. 227:857–865 [DOI] [PubMed] [Google Scholar]
- 14. Diederix R. E., Fauquant C., Rodrigue A., Mandrand-Berthelot M. A., Michaud-Soret I. 2008. Sub-micromolar affinity of Escherichia coli NikR for Ni(II). Chem. Commun. (Camb.) 15:1813–1815 [DOI] [PubMed] [Google Scholar]
- 15. Eitinger T., Rodionov D. A., Grote M., Schneider E. 2011. Canonical and ECF-type ATP-binding cassette importers in prokaryotes: diversity in modular organization and cellular functions. FEMS Microbiol. Rev. 35:3–67 [DOI] [PubMed] [Google Scholar]
- 16. Eitinger T., Suhr J., Moore L., Smith J. A. 2005. Secondary transporters for nickel and cobalt ions: theme and variations. Biometals 18:399–405 [DOI] [PubMed] [Google Scholar]
- 17. Fantino J. R., Py B., Fontecave M., Barras F. 2010. A genetic analysis of the response of Escherichia coli to cobalt stress. Environ. Microbiol. 12:2846–2857 [DOI] [PubMed] [Google Scholar]
- 18. Franke S., Grass G., Rensing C., Nies D. H. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185:3804–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gikas P. 2008. Single and combined effects of nickel (Ni(II)) and cobalt (Co(II)) ions on activated sludge and on other aerobic microorganisms: a review. J. Hazard. Mater. 159:187–203 [DOI] [PubMed] [Google Scholar]
- 20. Grass G., et al. 2005. The metal permease ZupT from Escherichia coli is a transporter with a broad substrate spectrum. J. Bacteriol. 187:1604–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grass G., Rensing C. 2001. Genes involved in copper homeostasis in Escherichia coli. J. Bacteriol. 183:2145–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haferburg G., Kloess G., Schmitz W., Kothe E. 2008. “Ni-struvite”—a new biomineral formed by a nickel resistant Streptomyces acidiscabies. Chemosphere 72:517–523 [DOI] [PubMed] [Google Scholar]
- 23. Haritha A., et al. 2009. MrdH, a novel metal resistance determinant of Pseudomonas putida KT2440, is flanked by metal-inducible mobile genetic elements. J. Bacteriol. 191:5976–5987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heddle J., Scott D. J., Unzai S., Park S. Y., Tame J. R. 2003. Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli. J. Biol. Chem. 278:50322–50329 [DOI] [PubMed] [Google Scholar]
- 25. Hunter S., et al. 2009. InterPro: the integrative protein signature database. Nucleic Acids Res. 37:D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iwig J. S., Rowe J. L., Chivers P. T. 2006. Nickel homeostasis in Escherichia coli—the rcnR-rcnA efflux pathway and its linkage to NikR function. Mol. Microbiol. 62:252–262 [DOI] [PubMed] [Google Scholar]
- 27. Kaluarachchi H., Chan Chung K. C., Zamble D. B. 2010. Microbial nickel proteins. Nat. Prod. Rep. 27:681–694 [DOI] [PubMed] [Google Scholar]
- 28. Kobayashi M., Shimizu S. 1999. Cobalt proteins. Eur. J. Biochem. 261:1–9 [DOI] [PubMed] [Google Scholar]
- 29. Koch D., Nies D. H., Grass G. 2007. The RcnRA (YohLM) system of Escherichia coli: a connection between nickel, cobalt and iron homeostasis. Biometals 20:759–771 [DOI] [PubMed] [Google Scholar]
- 30. Locher K. P., Lee A. T., Rees D. C. 2002. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296:1091–1098 [DOI] [PubMed] [Google Scholar]
- 31. Ma Z., Jacobsen F. E., Giedroc D. P. 2009. Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 109:4644–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller J. H. 1992. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 33. Mulligan C., et al. 2009. The substrate-binding protein imposes directionality on an electrochemical sodium gradient-driven TRAP transporter. Proc. Natl. Acad. Sci. U. S. A. 106:1778–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Navarro C., Wu L. F., Mandrand-Berthelot M. A. 1993. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol. Microbiol. 9:1181–1191 [DOI] [PubMed] [Google Scholar]
- 35. Niegowski D., Eshaghi S. 2007. The CorA family: structure and function revisited. Cell. Mol. Life Sci. 64:2564–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nies D. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313–339 [DOI] [PubMed] [Google Scholar]
- 37. Outten F. W., Huffman D. L., Hale J. A., O'Halloran T. V. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276:30670–30677 [DOI] [PubMed] [Google Scholar]
- 38. Perrin C., et al. 2009. Nickel promotes biofilm formation by Escherichia coli K-12 strains that produce curli. Appl. Environ. Microbiol. 75:1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pimentel B. E., Moreno-Sánchez R., Cervantes C. 2002. Efflux of chromate by Pseudomonas aeruginosa cells expressing the ChrA protein. FEMS Microbiol. Lett. 212:249–254 [DOI] [PubMed] [Google Scholar]
- 41. Rodionov D. A., Hebbeln P., Gelfand M. S., Eitinger T. 2006. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 188:317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rodrigue A., et al. 1996. Involvement of the GroE chaperonins in the nickel-dependent anaerobic biosynthesis of NiFe-hydrogenases of Escherichia coli. J. Bacteriol. 178:4453–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rodrigue A., Chanal A., Beck K., Muller M., Wu L. F. 1999. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial tat pathway. J. Biol. Chem. 274:13223–13228 [DOI] [PubMed] [Google Scholar]
- 44. Rodrigue A., Effantin G., Mandrand-Berthelot M. A. 2005. Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. J. Bacteriol. 187:2912–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rowe J. L., Starnes G. L., Chivers P. T. 2005. Complex transcriptional control links NikABCDE-dependent nickel transport with hydrogenase expression in Escherichia coli. J. Bacteriol. 187:6317–6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd edition Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 47. Scherer J., Nies D. H. 2009. CzcP is a novel efflux system contributing to transition metal resistance in Cupriavidus metallidurans CH34. Mol. Microbiol. 73:601–621 [DOI] [PubMed] [Google Scholar]
- 48. Stocker J., et al. 2003. Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environ. Sci. Technol. 37:4743–4750 [DOI] [PubMed] [Google Scholar]
- 49. Stoof J., Kuipers E. J., Klaver G., van Vliet A. H. M. 2010. An ABC transporter and a TonB ortholog contribute to Helicobacter mustelae nickel and cobalt acquisition. Infect. Immun. 78:4261–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tal N., Schuldiner S. 2009. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc. Natl. Acad. Sci. U. S. A. 106:9051–9056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tetsch L., Jung K. 2009. How are signals transduced across the cytoplasmic membrane? Transport proteins as transmitter of information. Amino Acids 37:467–477 [DOI] [PubMed] [Google Scholar]
- 52. Valko M., Morris H., Cronin M. T. 2005. Metals, toxicity and oxidative stress. Curr. Med. Chem. 12:1161–1208 [DOI] [PubMed] [Google Scholar]
- 53. Wu L. F., Mandrand-Berthelot M. A. 1986. Genetic and physiological characterization of new Escherichia coli mutants impaired in hydrogenase activity. Biochimie 68:167–179 [DOI] [PubMed] [Google Scholar]
- 54. Wu L. F., et al. 1989. Nickel deficiency gives rise to the defective hydrogenase phenotype of hydC and fnr mutants in Escherichia coli. Mol. Microbiol. 3:1709–1718 [DOI] [PubMed] [Google Scholar]
- 55. Wu L. F., Navarro C., de Pina K., Quenard M., Mandrand M. A. 1994. Antagonistic effect of nickel on the fermentative growth of Escherichia coli K-12 and comparison of nickel and cobalt toxicity on the aerobic and anaerobic growth. Environ. Health Perspect. 102(Suppl. 3):297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.