Abstract
The hexuronate metabolism in Escherichia coli is regulated by two related transcription factors from the FadR subfamily of the GntR family, UxuR and ExuR. UxuR controls the d-glucuronate metabolism, while ExuR represses genes involved in the metabolism of all hexuronates. We use a comparative genomics approach to reconstruct the hexuronate metabolic pathways and transcriptional regulons in gammaproteobacteria. We demonstrate differences in the binding motifs of UxuR and ExuR, identify new candidate members of the UxuR/ExuR regulons, and describe the links between the UxuR/ExuR regulons and the adjacent regulons UidR, KdgR, and YjjM. We provide experimental evidence that two predicted members of the UxuR regulon, yjjM and yjjN, are the subject of complex regulation by this transcription factor in E. coli.
INTRODUCTION
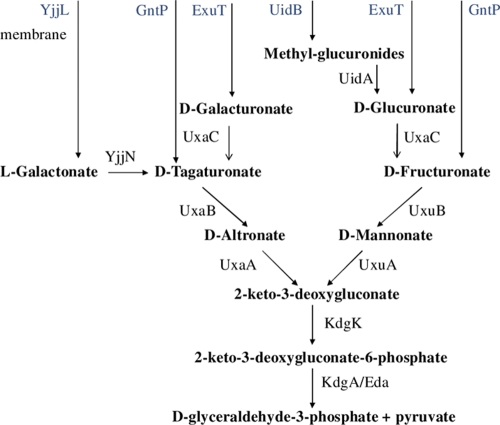
d-Glucuronate and d-galacturonate can each serve as a sole carbon source for the growth of Escherichia coli. They are utilized via the Ashwell catabolic pathway (27). Hexuronates enter the cell using two different transport systems, GntP and ExuT (from the GntP and MFS families of transporters, respectively), which have dual specificity, importing d-tagaturonate/d-fructuronate and d-galacturonate/d-glucuronate, respectively. In addition, β-d-glucuronides can be utilized to form d-glucuronate using the UidB transporter and the UidA glucuronidase (Fig. 1).
Fig. 1.
Hexuronate metabolism in E. coli. Transport proteins are indicated in blue, and enzymes are indicated in black.
d-Galacturonate and d-glucuronate are converted to the same metabolite, 2-keto-3-deoxy-d-gluconate, via parallel pathways, which involve one enzyme common for both pathways, d-glucuronate/d-galacturonate isomerase (UxaC), and two pairs of enzymes, d-mannonate and d-altronate hydrolases (UxuA and UxaA, respectively) and oxidoreductases (UxuB and UxaB, respectively), in each pathway (Fig. 1). The UxuB and UxaB proteins of E. coli are homologous (identity, 26%; positives, 42%; coverage, 95%) but may be confidently distinguished by the genome context analysis. The UxuA and UxaA demonstrate no discernible homology.
The hexuronate metabolism in E. coli is regulated by two related transcription factors (TFs) from the FadR subfamily of the GntR family, UxuR and ExuR, whose amino acid sequences are 46% identical. UxuR controls the d-glucuronate metabolism by repressing uxuAB, uidABC, gntP, and its own gene uxuR (3, 7, 17, 24–27). ExuR negatively controls most of the genes involved in the metabolism of both d-galacturonate and d-glucuronate, including exuT, uxaCA, uxaB, uxuAB, and exuR (6, 22, 24–27).
Understanding the regulation of the uidABC genes may have practical applications. Indeed, inhibiting β-glucuronidase produced by the gut flora can prevent the intestinal metabolism of the anticancer drug irinotecan, thereby diminishing life-threatening toxicity and conceivably allowing dose escalation that will enhance the drug's efficacy (33). It is also known that bacterial β-glucuronidase inhibitors can prevent a side effect of the common colon cancer chemotherapeutic CPT-11, severe diarrhea caused by β-glucuronidases from symbiotic bacteria that reactivate the drug in the gut. These inhibitors were highly effective against the enzyme target in aerobic and anaerobic bacteria but did not kill the bacteria and had no effect on the mammalian enzyme, thus causing no harm to mammalian cells (21).
Comparative genomics is a powerful approach for the prediction of gene regulation and the annotation of the bacterial genome (28). Previous in silico analysis of the UxuR/ExuR regulon revealed a common DNA-binding motif with consensus AAATTGGTATACCAATTT in upstream regions of many known UxuR/ExuR-regulated genes from E. coli and their orthologs in three other gammaproteobacteria (30). Here, we report a detailed analysis of the d-glucuronate and d-galacturonate pathways and their regulation by the UxuR and ExuR TFs in a large number of available genomes of gammaproteobacteria. We characterize evolutionary restructuring of these metabolic pathways, genomic loci, and regulatory interactions and identify new candidate members of these regulons. The detailed comparative analysis of candidate DNA-binding sites allowed us to build two distinct recognition rules for UxuR- and ExuR-binding sites. Using overexpression of UxuR and correlation analysis, we validated the regulation of two novel regulon members, yjjM and yjjN, by UxuR in E. coli.
MATERIALS AND METHODS
Computational analysis of regulons.
The genomic sequences of analyzed gammaproteobacteria that possess either uxuR or exuR genes have been obtained from GenBank (4) and are listed in Table S1 in the supplemental material (redundant species were excluded). Orthologs of UxuR and ExuR were identified by PSI-BLAST (1) searches (E-value cutoff, e−20) and confirmed by construction of the phylogenetic tree for identified homologs and by analysis of the respective gene neighborhoods on the chromosome (colocalization with the hexuronate metabolism genes). Amino acid sequence alignment was performed using the MUSCLE package (default parameters) (9). Phylogenetic trees were constructed with the PHYLIP package, using the protdist program for the computation of distance and the maximum-likelihood method for the tree construction (default parameters) (11).
For all bacterial species that had either uxuR and/or exuR, a comparative genomics-based reconstruction of the UxuR and ExuR regulons was performed. Nucleotide position weight matrices (profiles) for the UxuR- and ExuR-binding sites (see Table S2 in the supplemental material) were constructed by the SignalX program as previously described (13), using two training sets of upstream regions of genes from the studied gammaproteobacteria. The UxuR training set included known UxuR-regulated genes from E. coli (uxuR, uxuAB, gntP, and uidABC) and their orthologs from uxuR-containing genomes (PLU, STM, VPA, VVU, PPR, HSM, HIT, MSU, and ASU). The ExuR training set included known ExuR-regulated genes from E. coli (exuR, exuT, uxaCA, and uxaB) and their orthologs from exuR-containing genomes (CKO, ENT, ECA, KPN, SPE, and YPK). Computational search for candidate regulator-binding sites in upstream gene regions (for all genes in genomes, −400 nucleotides (nt) upstream and +50 nt downstream relative the gene start) was performed using the GenomeExplorer program package (15) and the RegPredict web server (19). The threshold scores for putative UxuR- and ExuR-binding sites were set to 4.0 and 5.0, respectively. Weaker sites with scores in the interval 3.7 to 4.0 and 4.5 to 5.0, respectively, were also taken into account, if their position was similar to positions of stronger sites upstream of orthologous genes and there were no stronger competing sites in the same intergenic region.
Candidate binding sites were confirmed by phylogenetic footprinting (28). In summary, we manually analyzed alignments of upstream regions of orthologous genes and identified groups of consecutive conserved positions, relying on the assumption that binding sites are more conserved than adjacent intergenic regions. New candidate members of the UxuR and ExuR regulons were attributed to the regulon if they were preceded by candidate regulator binding sites in at least three genomes. The reconstructed regulons were extended to include all genes in putative operons, the latter defined as strings of genes transcribed in the same direction with intergenic distances not exceeding 200 nt and also when such organization persisted in several (at least three) genomes. Motif logos were constructed using WebLogo (8).
The inferred regulons have been deposited to the RegPrecise database for computationally reconstructed microbial regulons (18) and are available at http://regprecise.lbl.gov/RegPrecise/collection_regspec.jsp?collection_id=104.
Experimental validation of novel UxuR regulon members.
Quantitative PCR (qPCR) assays were used to study expression pattern for genes from the UxuR regulon. At the first step, predicted dependence on UxuR was verified in E. coli K-12 MG1655 grown in mineral medium M9 supplied with either 0.2% d-glucose or d-glucuronate as a carbon source. Alternatively, the gene uxuR was cloned into the plasmid pGEMA (20) instead of rpoA under the control of the T7 promoter using the XbaI site. The resultant plasmid pGEMU was used to transform E. coli cells BL21(DE3) (experimental culture). Cells transformed by the plasmid pGEM with a deleted rpoA gene were used as a control sample. Both cultures were grown under constant shaking at 37°C in the standard Luria-Bertani (LB) medium supplied with 100 μg of ampicillin/ml. These conditions provide a moderate level of transcription for the recombinant uxuR gene, thus permitting the expression analysis for putative genes of the UxuR-regulated genes both with or without specific induction by IPTG (isopropyl-β-d-thiogalactopyranoside; 0.1 mM).
For qPCR, total cellular RNA was extracted as described in reference 10 and treated with DNase I (New England Biolabs). Primers for uxuR, uxuB, yjjM, yjjN, and hns genes used for reverse transcription (RT) and subsequent cDNA amplification are listed in Table S3 in the supplemental material.
For RT, the mixture (15 μl) containing 5 μg of total RNA and 4 pmol of RT primer was denatured by heating at 70°C for 10 min, and then deoxynucleoside triphosphates (dNTPs; 1.7 mM each) and reaction buffer (Fermentas) were added (total 8 μl). Probes were chilled on ice for 2 min, briefly spun down (1 to 2 s) and, after the addition of M-MuLV revertase (20 U; Fermentas, Lithuania), placed on 42°C for 40 min. Revertase was inactivated by 5 min of heating at 85°C.
qPCR was run on DT-322 thermal cycler (DNA Technology, Russia). The reaction mixture (20 μl) contained 2 to 4 μl of cDNA, 4 pmol of each primer, 0.1 mM concentrations of each dNTP, standard buffer (Evrogen, Russia), 1 μl of SYBR green I (diluted 1:3,000; Invitrogen), and 1 U of Taq DNA polymerase (Evrogen). The amplification program was the same for all target genes and included initial denaturation at 95°C for 2 min, followed by 30 cycles of 95°C for 20 s, annealing at 56°C for 20 s, and synthesis at 72°C for 25 s. The fluorescence of intercalating into double-stranded DNA fluorescent dye SYBR green I was measured at the end of each cycle during the last 15 s. Samples containing genomic DNA of E. coli instead of cDNA were used as positive controls, while probes with RNA passed through the reaction of primer extension without revertase were used as negative ones. mRNA of hns, encoding the histone-like nucleoid protein of E. coli, was used as a reference product. Quantitative data were computed using qPCR software (DNA Technology). The quality of PCR products was tested by 5% polyacrylamide gel electrophoresis (210 V, 100 mA). Only products of the expected size were registered in all cases.
Correlation in the gene expression for the UxuR regulon.
Fifteen sets of the microarray data deposited in NCBI GEO database (reference numbers GSE1121, GSE1154, GSE1981, GSE2037, GSE2928, GSE3591, GSE4344, GSE4706, GSE6425, GSE7885, GSE9814, GSE10345, GSE12797, GSE15059, and GSE18935) (2) were used to estimate correlation in the expression pattern for members of UxuR regulon (a total of 80 experiments). The analyzed data set of expression profiles includes experiments for the wild-type E. coli K-12 (mostly MG1655 strain) cells, grown either in LB or minimal medium (M9 or morpholinepropanesulfonic acid) in aerobic or anaerobic conditions in the presence of different carbon supplies (glucose, glycerol, succinate, acetate, l-alanine, and l-proline) and harvested at logarithmic phase. The only data sets selected were those for which information on the significant fluorescence signals was provided by the authors or could be obtained using raw data across the whole chip. In the latter case, background correction was achieved using the fluorescence intensity in the chip regions surrounding the spotted areas. Spots containing unrelated or randomized DNA were used to estimate average efficiency of nonspecific hybridization. Only signals whose fluorescence intensity exceeded the average efficiency of nonspecific hybridization were used for further analysis. To compare expression profiles obtained by different research groups on different microarray platforms, the abundance of each mRNA was estimated as a percentage of all significant mRNA products. Pearson correlation coefficients in the expression pattern of target genes were quantified by using Excel.
RESULTS AND DISCUSSION
Identification of transcription factors and binding motifs.
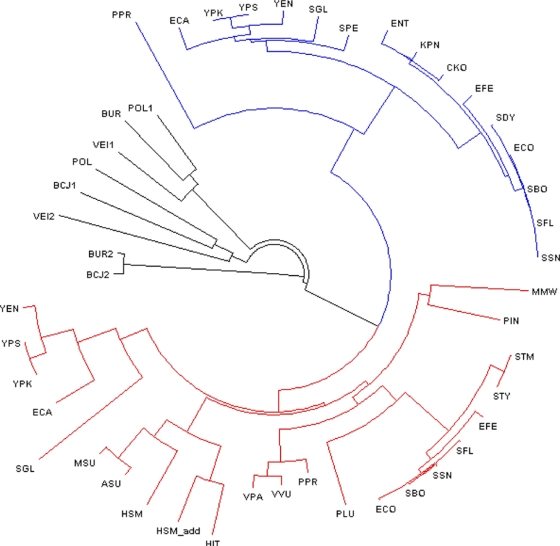
Orthologs of ExuR and UxuR were found only in the gammaproteobacteria. The phylogenetic tree of these TFs is shown in Fig. 2. The phylogenetic analysis shows that ExuR is present only in the Enterobacteriales and in one Vibrionales genome, Photobacterium profundum, whereas UxuR is present in several orders of the gammaproteobacteria (Enterobacteriales, Pasteurellales, Vibrionales, Alteromonadales, and Oceanospirillales). Separation of closely related transcription factors into the ExuR and UxuR groups was confirmed by their chromosomal colocalization with specific genes of the respective regulons.
Fig. 2.
Phylogenetic tree of the UxuR and ExuR transcription factors from the gammaproteobacteria. Colors: red, UxuR; blue, ExuR; outgroups, black. The “add” denotes a second, additional copy of a TF in the genome. The TF labels correspond to the abbreviations given in Table S1 in the supplemental material.
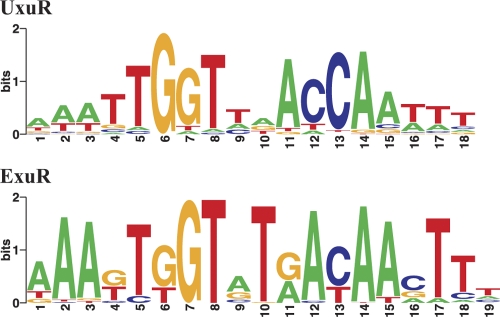
UxuR- and ExuR-specific profiles were made using binding sites upstream of known regulon members in E. coli and orthologous genes in related gammaproteobacteria (for more details, see Materials and Methods). Although previous studies assumed that candidate UxuR- and ExuR-binding sites both match the consensus sequence AAATTGGTATACCAATTT and are too similar to be distinguished by sequence analysis (30), we observed that if the larger set of sites from different related genomes is used and sites are attributed to the TFs based on the function of the regulated genes, it is possible to separate UxuR and ExuR binding motifs and detect subtle differences between the regulators. We found two distinct motifs (Fig. 3), differing in the 3′-box, ACCA for UxuR and AYAA for ExuR, and in the length of the spacer between the boxes (2 and 3 bp, respectively). The ExuR motif is not strictly palindromic in positions 6/14, 7/13, and 9/11 (Fig. 3). This creates a possibility of cross-recognition of the same area by both ExuR and UxuR: the AACAA sequence can play the role of ACCA with one substituted C (the first or the second one, respectively), complementary to TGGT in both 18-bp UxuR and 19-bp ExuR binding motifs. Hence, we propose that after duplication ExuR developed a new DNA-binding specificity.
Fig. 3.
Sequence logo for the UxuR and ExuR binding motifs. Horizontal axis, position in the binding site; vertical axis, information content in bits. The height of each column is proportional to the positional information content in the given position; the height of each individual symbol reflects its prevalence at the given position.
Regulon composition.
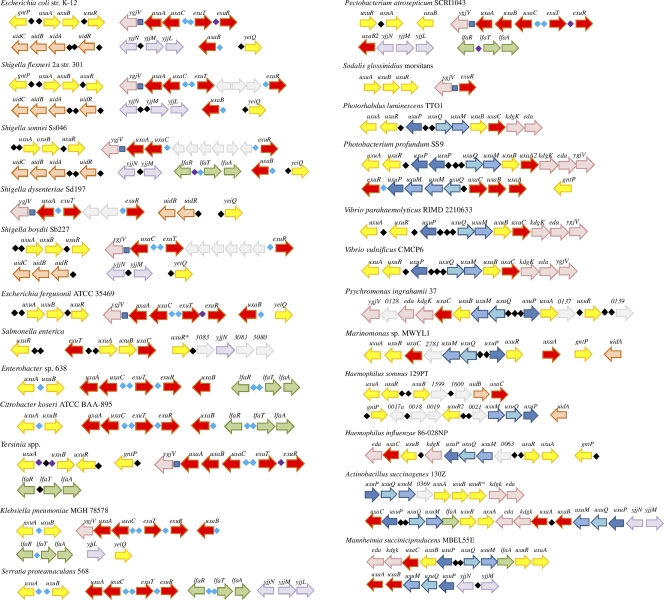
The reconstructed UxuR and ExuR regulons in all studied genomes are shown in Fig. 4. The majority of the Enterobacteriales have both uxu and uxa genes and thus are able to utilize both d-glucuronate and d-galacturonate. In Escherichia, Shigella (except S. dysenteriae that lacks UxuR), Yersinia, and Pectobacterium spp., both the UxuR and the ExuR regulators are present and control the d-glucuronate and d-galacturonate catabolic pathways, respectively. Citrobacter koseri, Enterobacter sp. strain 638, Klebsiella pneumoniae, and Serratia proteamaculans have only ExuR TFs that regulate genes of both catabolic pathways. Two bacterial genera in this group (Photorhabdus luminescens and Salmonella spp.) have only the d-glucuronate utilization pathways controlled by UxuR and lack the d-galacturonate pathway. Interestingly, the composition of the UxuR regulons in the latter two genera is significantly different: the uxuAB-uxaC, uxuPQM, kdgK, and eda genes in P. luminescens (similar to UxuR regulons in the Vibrionales, see below) and uxuABC-exuT in Salmonella spp.
Fig. 4.
UxuR/ExuR regulon organization in various gammaproteobacteria. UxuR binding sites are indicated as black diamonds, ExuR binding sites are indicated as light blue diamonds, sites predicted to be recognized by both UxuR and ExuR are indicated as violet diamonds, and KdgR binding sites (upstream of ygjV) are indicated as dark blue squares. Numbers denote the locus_tags of unknown genes. Genes are colored according to the locus organization in E. coli.
Most Enterobacteriaceae have multiple paralogous genes encoding d-mannonate oxidoreductase, for example, uxuB, yeiQ, and ydfI in E. coli. There are putative UxuR binding sites upstream of yeiQ in E. coli and the Shigella spp. At that, S. dysenteriae that lacks UxuR nevertheless has strong UxuR binding sites upstream of the uidR and yeiQ genes, indicating that the loss of the TF gene has been very recent.
Unlike the Enterobacteriales, other gammaproteobacteria from the Pasteurellales (four species), Vibrionales (three species), Alteromonadales (Psychromonas spp.), and Oceanospirillales (Marinomonas spp.) orders have only UxuR TFs that typically control the glucuronate utilization pathway, but not the galacturonate one (Fig. 1). As an exception, P. profundum has both the UxuR and the ExuR regulators controlling the d-glucuronate and d-galacturonate catabolic gene loci, probably as a result of horizontal gene transfer. A typical feature of the d-glucuronate/d-galacturonate utilization gene clusters in bacteria from these taxa (and P. luminescens from the Enterobacteriales) is the replacement of exuT by a novel hypothetical transport system from the tripartite ATP-independent periplasmic (TRAP) transporter family (12, 14, 31). Since this UxuR-regulated TRAP system is similar to the C4-dicarboxylate transport system DctPQM, we name it UxuPQM and propose its involvement in transport of hexuronates (see the next section).
A. succinogenes has an additional d-glucuronate utilization locus containing paralogs of the uxuAB, kdgK, eda, and uxuPQM genes and a gene encoding a different GntR family TF that is related but not orthologous to UxuR (Asuc_0372, named uxuR* in Fig. 4). Haemophilus somnus has two uxuR paralogs, HSM_0414 and HSM_1913, that are clustered with the uxuAB-uxaC and uxuPQM genes, respectively. There is also an additional GntR family TF in Salmonella enterica serovar Typhimurium strain LT2, related to UxuR and ExuR, but not close enough to be considered an orthologous repressor (STM3084.S, named uxuR* in Fig. 4). It is transcribed divergently to an operon that consists of d-mannonate oxidoreductase, yjjN, dehydrogenase with unknown specificity, and putative mannitol dehydrogenase.
A common product of the d-glucuronate and d-galacturonate utilization pathways, 2-keto-3-deoxy-d-gluconate (KDG), is further utilized by a consequent action of the KdgK kinase and the Eda/KdgA aldolase. In E. coli, the downstream KDG metabolism is controlled by the KdgR repressor from the IclR family, which is also regulating the pectin utilization in plant-pathogenic species of the Enterobacteriales (16). Here we found that the UxuR regulons in the Vibrionales, Pasteurellales, P. luminescens, and P. ingrahamii include the kdgK and eda genes. All of these species except Vibrio spp. lack KdgR TFs, thus their UxuR regulons are expanded to include genes from the downstream pathway. In contrast, Vibrio spp. have two copies of both kdgK and eda genes, one of which is regulated by UxuR, whereas the other one is under the KdgR control (29).
The ygjV gene encoding a hypothetical transporter was found immediately downstream of the galacturonate catabolic cluster uxaCBA/uxaCA in most enterobacteria (P. atrosepticum, Escherichia spp., K. pneumoniae, Shigella spp., and Yersinia spp.) and downstream uxaC-kdgK-eda in Vibrio spp., Photobacterium profundum SS9, and Psychromonas ingrahamii 37. It was considered to be a part of the KdgR regulon and has strong upstream candidate KdgR-binding sites in all enterobacteria, except K. pneumoniae (29). A candidate KdgR-binding site is located between uxaA and ygjV, immediately after the predicted rho-independent transcriptional terminator of the uxa operon (see Fig. 4) (29). There are several paralogs of ygjV in the Vibrio spp. possessing the KdgR TF: one copy is located in the uxaC-kdgK-eda-ygjV cluster (not regulated by KdgR), another one belongs to the kduD-ygjV-kdgF-spiX cluster preceded by two candidate KdgR sites. The Vibrio spp. also have additional copies of the kdgK and eda genes that are members of the KdgR regulon (29). This duplication may be a result of a recent specialization of the paralogs toward catabolism of either pectin (regulated by KdgR) or hexuronate, and its localization suggests that yjgV could be a part of the UxuR regulon, involved in the transport of some intermediates of the hexuronate catabolic pathway.
New members of the regulons.
We have identified several new candidate members of the UxuR/ExuR regulons, including the hypothetical carbohydrate catabolic genes lfaR-lfaTA and yjjN-yjjML in the Enterobacteriales and the transporters uxuPMQ in other studied gammaproteobacteria.
The divergently transcribed lfaR-lfaTA genes encode, respectively, a LacI family repressor, an MFS-type transporter (homologous to ExuT), and a family 31 glycosylhydrolase similar to α-glucosidase. This catabolic gene cluster is involved in the phytopathogenicity of Erwinia chrysanthemi 3937, being required for utilization of an as-yet-unknown carbohydrate of plant origin (32). Accordingly, it was detected in other pathogenic enterobacteria studied here, such as the Citrobacter, Yersinia, Enterobacter, Serratia, Shigella, and Pectobacterium spp., where it was always found under the control of either UxuR or ExuR.
The novel UxuR-regulated transport system named UxuPQM belongs to the TRAP family of ATP-independent transporters that are present in many prokaryotes. TRAP systems are known to transport various carbohydrates such as l-xylulose, gluconate, mannonate, succinate, etc. (12, 14, 31). Since the occurrence pattern of the uxuPQM genes in gammaproteobacteria correlates with the absence of the known hexuronate transport gene exuT, we propose that the former plays a role in the hexuronate transport. Most bacteria (Haemophilus spp., Photorhabdus luminescens, Psychromonas ingrahamii, and Vibrio spp.) have only one set of the uxuPQM genes in the hexuronate regulons. In these cases, the uxuPQM genes are colocalized with the genes of the glucuronate utilization pathway and thus are likely involved in d-glucuronate/d-fructuronate uptake rather than the uptake of d-galacturonate/d-tagaturonate. Several bacteria (Mannheimia succiniciproducens, A. succinogenes, and P. profundum) have two or more sets of uxuPQM genes in the UxuR/ExuR regulons, suggesting that some of these paralogs are involved in the d-galacturonate uptake, in accordance with their colocalization with the d-galacturonate catabolic genes.
The yjjN/yjjML gene cluster encodes l-galactonate oxidoreductase (YjjN converts its substrate to d-tagaturonate) (23), a GntR family TF (YjjM), and a putative l-galactonate transporter (YjjL). Previously, it was shown that in E. coli YjjM is involved in the l-galactonate utilization, since the yjjM mutant strain failed to grow on l-galactonate (23). Orthologs of yjjM are present in several other enterobacteria (Shigella, Serratia, and Pectobacterium spp.) and in two Pasteurellales (M. succiniciproducens and A. succinogenes). In the studied genomes, the yjjM gene forms a divergon with yjjN and is often colocalized with yjjL, presumably forming an yjjML operon. We propose that YjjM can regulate the transcription of the yjjM/yjjN genes; however, the comparative analysis of their intergenic region did not reveal any conserved motif. Among enterobacteria, candidate UxuR-binding sites with low similarity to the consensus UxuR motif were detected between the yjjN and yjjM genes in E. coli, Shigella, and Serratia but not in Pectobacterium.
Experimental validation of the yjjM and yjjN modulation by UxuR.
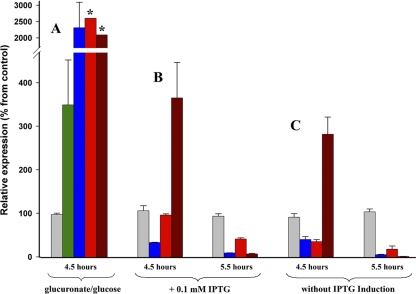
In order to validate the predicted UxuR regulation for the yjjM and yjjN genes in E. coli, the content of corresponding mRNAs was first estimated in bacteria grown in conditions requiring expression of UxuR-dependent genes. If d-glucose is used as a sole carbon source, expression of the uxa/uxu operons is repressed by UxuR. The presence of d-fructuronate (the first intermediate in glucuronate catabolism) relieves this repression (3). Thus, we grew E. coli K-12 cells in a mineral medium supplemented with either d-glucuronate (experimental culture) or d-glucose (control sample) and harvested them after 4.5 h of cultivation (Fig. 5 A). The abundance of the hns mRNA, which was used as a control RNA product in qPCR experiments, showed no dependence on carbohydrate source. An expected increase in the expression pattern of uxuB (positive control), was very high, which agrees with previously published data (3). The responses of yjjM and yjjN appeared to be very similar (Fig. 5A). These data imply the involvement of corresponding proteins in the metabolic switch between the d-glucose and d-glucuronate pathways and led to further experiments aimed at determining whether this participation is mediated by UxuR.
Fig. 5.
Effect of UxuR on the expression of target genes as measured by qPCR. Bars represent the average abundance of the hns (gray), uxuB (blue), yjjM (bright red), yjjN (dark red), and uxuR (green) mRNAs in experimental bacterial cultures compared to the control samples. (A) Relative expression of genes within cells grown in minimal M9 broth, supplemented with 0.2% d-glucose (control sample) or d-glucuronate (experimental culture). Assays indicated by asterisks in four biological replications (8 to 10 technical repeats) vary in the ranges of 250 to 6,900% (yjjM) and 150 to 4,200%. (B and C) Cells transformed by either pGEMU (experimental culture) or pGEM (control sample) were grown in the presence (B) or absence (C) of IPTG and harvested after 4.5 or 5.5 h of cultivation. (B) IPTG (0.1 mM) was added to bacterial cultures after 3.5 h (optical density at 600 nm = 0.4). Induction was allowed for 1 or 2 h, and the cells were harvested as described in panel C. Bars represent average values of changes registered in three independent experiments, with two concentration variables in each experiment. High UxuR-specific bars (see the text) are eliminated from the panels B and C so as to increase the resolution of the plot.
UxuR blocks the transcription of the target genes, including its own gene, by specific interaction with corresponding operator sites and can do so without any cofactor (3). Although d-fructuronate is required to relieve this repression (3), the expression of UxuR regulon should be dependent on intracellular concentration of UxuR. Thus, alteration in the cellular content of yjjM and yjjN mRNAs was tested in the condition of UxuR overproduction (Fig. 5B). BL21(DE3) cells were transformed by either pGEMU (uxuR-containing) or pGEM (control) plasmid. In pGEMU, uxuR was cloned under the control of the T7 promoter, avoiding a possibility of negative feedback since it lacks an UxuR-binding site. IPTG induction yielded a 98- to 120-fold difference between experimental and control samples. The abundance of the hns mRNA within the pGEMU-transformants appeared to be the same as in the cells transformed by pGEM, while the amount of the uxuB mRNA significantly decreased, confirming its negative regulation by UxuR (Fig. 5B). A weaker response was registered for the yjjM mRNA, but the yjjN gene displayed a different behavior: the initial 3.7-fold increase was followed by a dramatic decrease in the amount of its RNA product (Fig. 5A).
Bacteria grown in the absence of IPTG demonstrated practically the same time dependence in the expression pattern of analyzed genes (Fig. 5C). In this case, moderate transcription of the recombinant gene was achieved by the presence of lactose among the nutrients of LB medium. Lactose, just like its synthetic analog IPTG, abolishes LacI repression of the lacUV5 promoter, which controls production of T7 RNA polymerase in BL21(DE3) cells. A low concentration of this sugar provides an opportunity to observe cellular response under more physiological conditions. The abundance of the uxuR mRNA within the pGEMU transformants after 5.5 h of cultivation appeared to be only (6.75 ± 2.4)-fold higher than in the control cells. As in the previous case, this was accompanied by alterations in the amount of intracellular yjjM and yjjN mRNAs, thus supporting predictions made on the basis of comparative genomics.
A peculiar increase in the abundance of yjjN has been observed at the same time point (Fig. 5C), excluding simple dosage effects and assuming certain links with a particular growth phase. Its amplitude appeared to be smaller (2.8-fold) than in the previous set of experiments (compare Fig. 5B and C). The same tendency for a decrease has been observed for other examined mRNAs (except uxuB at the 4.5-h time point). That assumes positive correlation in the amount of UxuR and RNA products of genes negatively regulated by this transcription factor. Semidirectional changes in the abundance of uxuR, uxuB, yjjM, and yjjN were also registered upon metabolic switch (Fig. 5A). Although increased transcription of uxuR in this case is explained by the relieved autorepression (due to the presence of fructuronate) and by the functionality of cyclic AMP-CRP (due to the absence of glucose), this assumes “unreasonable” interference with the required induction of UxuR regulon. Since all analyzed mRNAs have comparable half-lives (4.2 to 7.9 min) (5), correlation analysis was undertaken to determine whether a larger set of experimental data also supports this revealed tendency.
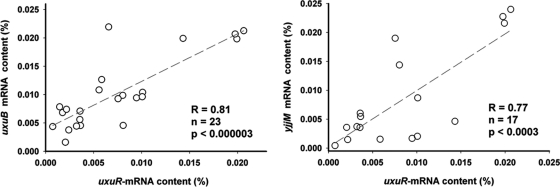
A total of 80 microarray experiments deposited in the NCBI GEO database were used for comparative analysis. These data reflect gene expression profiles in the wild-type E. coli K-12 cells grown in various media and harvested at the logarithmic stage. Even though the cellular abundance of the uxuR mRNA was measured only in 24 experiments, the available data indeed demonstrate positive correlations in the expression level of uxuR and almost all members of the UxuR regulon, including yjjM and yjjN (examples are given in Fig. 6). The most significant correlations were observed for uxuAB (Fig. 6), uxaC (R = 0.81, P < 0.000005), and uxaB (R = 0.77, P < 0.00007). Only the expression of uidB demonstrated some negative correlation with that of uxuR; however, the value of R = −0.51 obtained on a small number of available points (n = 13) is statistically not significant (P < 0.07). The observed correlation does not seem to be an artifact caused by a limited number of successful experiments estimating the abundance of the uxuR mRNA, since the expression of exuR, measured in 73 assays, demonstrates the same concordance. The most significant correlations were observed with the uxuAB operon (for uxuA, R = 0.63, P < 0.000001), uxaC (R = 0.54, P < 0.000002), and uxuR (R = 0.629, P < 0.0013). Thus, we conclude that the physiological conditions yielding the hexuronate metabolism also increase the expression of regulators UxuR and ExuR. Even though a detailed molecular mechanisms underlying UxuR/ExuR regulatory action remains to be elucidated, it seems likely that yjjM and yjjN are regulated by at least UxuR.
Fig. 6.
Correlation in the cellular abundance of the uxuR mRNA with that of the uxuB (left) and yjjM (right) mRNAs. The first-order regression lines are added to trace the points. The Pearson correlation coefficients (R), numbers of available points (n), and corresponding P values are indicated.
Conclusions.
Orthologs of ExuR and UxuR were found only among the gammaproteobacteria. ExuR is present in the Enterobacteriales and P. profundum, while UxuR is present in several orders of the gammaproteobacteria. The diversity of the UxuR regulon organization in different taxonomic groups leads to the question about the evolution of the hexuronate metabolism. The phylogenetic tree of the transcription factors (Fig. 2) demonstrates that the duplication into the UxuR and ExuR branches occurred before diversification of the UxuR factors into taxon-specific branches. Hence, a plausible scenario seems to be that the ancestral form of the regulatory system included both transcription factors with subsequent loss of ExuR in all groups but the Enterobacteriales and P. profundum. An alternative scenario is the duplication in the common ancestor of the Enterobacteriales, with a subsequent relatively rapid sequence change in ExuR and horizontal transfer to P. profundum. Both scenarios have a number of problems. The former one presumes numerous, independent losses of the exuR gene, whereas the latter contradicts the position of the root in the phylogenetic tree and requires a highly nonuniform evolution rate.
We have distinguished UxuR and ExuR candidate binding sites by positional and sequence analysis. The UxuR binding motif is a 18-bp palindromic sequence; the 19-bp palindromic ExuR binding motif is rather similar. The ExuR binding motif seems to have evolved from the UxuR one, and now we observe early stages of the TFs' diversification. Overall, the reconstructed UxuR and ExuR regulons are very flexible in the studied gammaproteobacteria. Among the Enterobacteriales, UxuR mostly represses the uxuABR and gntP genes, whereas ExuR mostly represses the exuR, exuT, and uxaABC genes, as well as the uxuAB genes in some genomes that lack UxuR. We have also identified new candidate members of the UxuR/ExuR regulons, a novel predicted TRAP transporter for hexuronates (uxuPQM), hypothetical carbohydrate catabolic genes (lfaR/lfaTA), and the l-galactonate utilization gene cluster (yjjN/yjjML). In several genomes, the UxuR regulon is expanded to include the genes of the downstream KDG catabolic pathway, kdgK and eda.
We experimentally validated the UxuR-dependent control of the yjjM and yjjN transcription in E. coli. The obtained data revealed a complex response of yjjN on the induced production of UxuR, as well as unexpected, positive correlations in the expression pattern of genes encoding repressors UxuR/ExuR and members of the corresponding regulons. It may be speculated that, along with the simple repression of genes encoding temporarily unneeded enzymes, these TFs perform another important biological function, that of keeping the expression of target genes on a controllable level when their transcription is strongly induced by the global catabolic activator protein CRP.
Supplementary Material
ACKNOWLEDGMENTS
This study was partially supported by the Russian Foundation of Basic Research under grants 09-04-92745, 10-04-00431, 10-04-01768, and 10-04-01218; State Contract 2.740.11.0101; and the Russian Academy of Sciences via the program Molecular and Cellular Biology. D.A.R. was also supported by the Department of Energy under award DE-Se 0004999.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 27 May 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barrett T., et al. 2011. NCBI GEO: archive for functional genomics data sets −10 years on. Nucleic Acids Res. 39:D1005–D1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bates-Utz C., Nguyen A. B., Smalley D. J., Anderson A. B., Conway T. 2004. GntP is the Escherichia coli fructuronic acid transporter and belongs to the UxuR regulon. J. Bacteriol. 186:7690–7696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benson D. A., et al. 1999. GenBank. Nucleic Acids Res. 27:12–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bernstein J. A., Khodursky A. B., Lin P. H., Lin-Chao S., Cohen S. N. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. U. S. A. 99:9697–9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blanco C., Mata-Gilsinger M., Ritzenthaler P. 1983. Construction of hybrid plasmids containing the Escherichia coli uxaB gene: analysis of its regulation and direction of transcription. J. Bacteriol. 153:747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blanco C., Ritzenthaler P., Mata-Gilsinger M. 1986. Negative dominant mutations of the uidR gene in Escherichia coli: genetic proof for a cooperative regulation of uidA expression. Genetics 112:173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edgar R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Favre-Bonte S., Joly B., Forestier C. 1999. Consequences of reduction of Klebsiella pneumoniae capsule expression on interactions of this bacterium with epithelial cells. Infect. Immun. 67:554–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Felsenstein J. 1996. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 266:418–427 [DOI] [PubMed] [Google Scholar]
- 12. Forward J. A., Behrendt M. C., Wyborn N. R., Cross R., Kelly D. J. 1997. TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria. J. Bacteriol. 179:5482–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gelfand M. S., Koonin E. V., Mironov A. A. 2000. Prediction of transcription regulatory sites in Archaea by a comparative genomic approach. Nucleic Acids Res. 28:695–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kelly D. J., Thomas G. H. 2001. The tripartite ATP-independent periplasmic (TRAP) transporters of bacteria and archaea. FEMS Microbiol. Rev. 25:405–424 [DOI] [PubMed] [Google Scholar]
- 15. Mironov A. A., Vinokurova N. P., Gel'fand M. S. 2000. Software for analyzing bacterial genomes. Mol. Biol. (Mosk.) 34:253–262 [PubMed] [Google Scholar]
- 16. Murray E. L., Conway T. 2005. Multiple regulators control expression of the Entner-Doudoroff aldolase (Eda) of Escherichia coli. J. Bacteriol. 187:991–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Novel M., Novel G. 1976. Regulation of beta-glucuronidase synthesis in Escherichia coli K-12: pleiotropic constitutive mutations affecting uxu and uidA expression. J. Bacteriol. 127:418–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Novichkov P. S., et al. 2010. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38:D111–D118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Novichkov P. S., et al. 2010. RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res. 38:W299–W307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ozoline O. N., Fujita N., Ishihama A. 2001. Mode of DNA-protein interaction between the C-terminal domain of Escherichia coli RNA polymerase alpha subunit and T7D promoter UP element. Nucleic Acids Res. 29:4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patel A. G., Kaufmann S. H. 2010. Cancer Targeting bacteria to improve cancer therapy. Science 330:766–767 [DOI] [PubMed] [Google Scholar]
- 22. Portalier R., Robert-Baudouy J., Stoeber F. 1980. Regulation of Escherichia coli K-12 hexuronate system genes: exu regulon. J. Bacteriol. 143:1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reed J. L., et al. 2006. Systems approach to refining genome annotation. Proc. Natl. Acad. Sci. U. S. A. 103:17480–17484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ritzenthaler P., Blanco C., Mata-Gilsinger M. 1985. Genetic analysis of uxuR and exuR genes: evidence for ExuR and UxuR monomer repressors interactions. Mol. Gen. Genet. 199:507–511 [DOI] [PubMed] [Google Scholar]
- 25. Ritzenthaler P., Mata-Gilsinger M. 1982. Use of in vitro gene fusions to study the uxuR regulatory gene in Escherichia coli K-12: direction of transcription and regulation of its expression. J. Bacteriol. 150:1040–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robert-Baudouy J., Portalier R., Stoeber F. 1981. Regulation of hexuronate system genes in Escherichia coli K-12: multiple regulation of the uxu operon by exuR and uxuR gene products. J. Bacteriol. 145:211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robert-Baudouy J. M., Portalier R. C., Stoeber F. R. 1974. Regulation of hexuronate metabolism in Escherichia coli K-12. Kinetics of enzyme induction of the hexuronate system. Eur. J. Biochem. 43:1–15 [DOI] [PubMed] [Google Scholar]
- 28. Rodionov D. A. 2007. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem. Rev. 107:3467–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rodionov D. A., Gelfand M. S., Hugouvieux-Cotte-Pattat N. 2004. Comparative genomics of the KdgR regulon in Erwinia chrysanthemi 3937 and other gammaproteobacteria. Microbiology 150:3571–3590 [DOI] [PubMed] [Google Scholar]
- 30. Rodionov D. A., Mironov A. A., Rakhmaninova A. B., Gelfand M. S. 2000. Transcriptional regulation of transport and utilization systems for hexuronides, hexuronates, and hexonates in gamma purple bacteria. Mol. Microbiol. 38:673–683 [DOI] [PubMed] [Google Scholar]
- 31. Thomas G. H., Southworth T., Leon-Kempis M. R., Leech A., Kelly D. J. 2006. Novel ligands for the extracellular solute receptors of two bacterial TRAP transporters. Microbiology 152:187–198 [DOI] [PubMed] [Google Scholar]
- 32. Van Gijsegem F., Wlodarczyk A., Cornu A., Reverchon S., Hugouvieux-Cotte-Pattat N. 2008. Analysis of the LacI family regulators of Erwinia chrysanthemi 3937, involvement in the bacterial phytopathogenicity. Mol. Plant-Microbe Interact. 21:1471–1481 [DOI] [PubMed] [Google Scholar]
- 33. Wallace B. D., et al. 2010. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330:831–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.