Abstract
The bacterial pathogen Chromobacterium violaceum uses a LuxIR-type quorum-sensing system to detect and respond to changes in cell population density. CviI synthesizes the autoinducer C10-homoserine lactone (C10-HSL), and CviR is a cytoplasmic DNA binding transcription factor that activates gene expression following binding to C10-HSL. A number of behaviors are controlled by quorum sensing in C. violaceum. However, few genes have been shown to be directly controlled by CviR, in part because the DNA motif bound by CviR is not well characterized. Here, we define the DNA sequence required for promoter recognition by CviR. Using in vivo data generated from a library of point mutations in a CviR-regulated promoter, we find that CviR binds to a palindrome with the ideal sequence CTGNCCNNNNGGNCAG. We constructed a position weight matrix using these in vivo data and scanned the C. violaceum genome to predict CviR binding sites. We measured direct activation of the identified promoters by CviR and found that CviR controls the expression of the promoter for a chitinase, a type VI secretion-related gene, a transcriptional regulator gene, a guanine deaminase gene, and cviI. Indeed, regulation of cviI expression by CviR generates a canonical quorum-sensing positive-feedback loop.
INTRODUCTION
Quorum sensing is a process of bacterial cell-cell communication in which cells produce, detect, and respond to extracellular signal molecules called autoinducers. Using quorum sensing, bacteria change their gene expression patterns and, in turn, their behavior in response to changes in cell density. Canonical Gram-negative quorum-sensing systems consist of LuxI-type autoinducer synthases that produce specific acylated homoserine lactone (AHL) autoinducers and cognate LuxR-type receptors (5). At low cell density, the AHL signal concentration is low and unliganded LuxR receptors are intrinsically unstable and rapidly degraded (28). As cell density increases, the AHL concentration likewise increases. Accumulated AHL binds the LuxR-type receptor, leading to stabilization of the protein-ligand complex (16, 17, 28). The LuxR:AHL complex subsequently binds DNA at promoters driving genes regulated by quorum sensing (26, 27).
Quorum sensing controls collective behaviors, including bioluminescence, biofilm formation, and DNA exchange (6, 8, 9, 21, 24). Bacterial pathogens rely heavily on quorum-sensing systems to control the expression of genes required for virulence (13, 15). One such pathogen, and the focus of the present work, is Chromobacterium violaceum, an aquatic bacterium that can infect humans and cause abscesses and bacteremia (20). The C. violaceum quorum-sensing system consists of the LuxI/LuxR homologues CviI/CviR. The CviI/CviR circuit controls virulence, as evidenced by the fact that antagonist molecules that bind in place of the natural AHL ligand and induce a CviR conformation that prevents DNA binding protect the nematode Caenorhabditis elegans from C. violaceum-mediated killing (2, 19). These findings demonstrate the importance of quorum sensing in C. violaceum pathogenesis and suggest that quorum-sensing inhibitors could be valuable in battling virulent bacteria.
The only well-studied trait controlled by quorum sensing in C. violaceum is production of the hallmark purple pigment violacein (12). Violacein is synthesized from tryptophan by the products of the vioABCD operon (1). The vioA promoter is controlled by CviR both in C. violaceum and in recombinant Escherichia coli, demonstrating that regulation is direct (19). Other C. violaceum phenotypes that are known to depend on AHL include biofilm formation and chitinase production (3). However, whether this is through direct or indirect regulation is not known, in part because the CviR operator DNA binding site has not been well defined.
Here, we have engineered and screened a comprehensive library of vioA promoter mutations, allowing us to define an ideal CviR binding site (CTGNCCNNNNGGNCAG). This analysis coupled with genome scanning enabled prediction of CviR-regulated genes in C. violaceum. Our findings reveal a number of promoters containing predicted CviR binding sites. We show that these genes are directly regulated by CviR. Furthermore cviI, the gene encoding the C10-HSL autoinducer synthase, is, not surprisingly, controlled by CviIR and thus regulated by positive feedback.
MATERIALS AND METHODS
Bacterial strains and protein purification.
The strains used in this study are listed in Table 1. Wild-type Chromobacterium violaceum strain 12472 has been described previously (4). Escherichia coli strain Top10 (Invitrogen) was used for plasmid constructions. E. coli BL21(DE3) (Novagen) was used for recombinant protein production. Expression and purification of the CviR:C10-HSL complex were carried out as described previously (2).
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Reference or source | Comment |
|---|---|---|
| C. violaceum strain 12472 (wild type) | 4 | C10-homoserine lactone-producing strain |
| E. coli strains | ||
| Top10 | Invitrogen | Strain used for all cloning and reporter assays |
| BL21(DE3) | Novagen | Strain used for all protein expression |
| E. coli plasmids | ||
| pET23b | Novagen | E. coli overexpression plasmid |
| pET23cviR | This study | cviR from C. violaceum strain 12472 inserted into pET23b |
| pvioA-gfp | 19 | |
| pvioA-gfpΔ25-35 | This study | pvioA-gfp with −35 site deleted |
| pvioA-gfpΔ37-46 | This study | pvioA-gfp with first half of IR deleted |
| pvioA-gfpΔ47-56 | This study | pvioA-gfp with second half of IR deleted |
| pvioA-gfpΔ57-66 | This study | pvioA-gfp with 10-bp deletion upstream of IR |
| pvioA-gfpΔ67-76 | This study | pvioA-gfp with 10-bp deletion upstream of IR |
| pvioA-gfpΔ77-86 | This study | pvioA-gfp with 10-bp deletion upstream of IR |
| pvioA-gfpscram | This study | pvioA-gfp with scrambled IR |
| pvioA-gfpΔ49-58 | This study | pvioA-gfp with 10-bp deletion upstream of IR |
| pvioA-gfpΔ51-60 | This study | pvioA-gfp with 10-bp deletion upstream of IR |
| pvioA-gfpΔ53-62 | This study | pvioA-gfp with 10-bp deletion upstream of IR |
| pvioA-gfpΔ55-64 | This study | pvioA-gfp with 10-bp deletion upstream of IR |
| pvioA-gfpΔ79-88 | This study | pvioA-gfp with 10-bp deletion upstream of IR |
| pvioA-gfpΔ81-90 | This study | pvioA-gfp with 10-bp deletion upstream of IR |
| pvioA-gfpΔ83-92 | This study | pvioA-gfp with 10-bp deletion upstream of IR |
| pvioA-gfpΔ85-94 | This study | pvioA-gfp with 10-bp deletion upstream of IR |
| pEVSlux | This study | luxCDABE from pBBRlux cloned into plasmid pEVS141 |
| pcviI-gfp | This study | C. violaceum strain 12472 cviI promoter fused to gfp |
| pcviI*-gfp | This study | pcviI-gfp containing mutant CviR binding site |
| pcviIR | This study | pBBR322 carrying cviIR |
| pcviI*R | This study | pBBR322 carrying cviIR with mutant CviR binding site |
| pccviIRstop | This study | pBBR322 carrying cviIR with stop codon insertion in cviR |
| p4091-lux | This study | CV_4091 promoter fused to luxCDABE |
| p4240-lux | This study | CV_4240 promoter fused to luxCDABE |
| p1432-lux | This study | CV_1432 promoter fused to luxCDABE |
| p0577-lux | This study | CV_0577 promoter fused to luxCDABE |
| p0578-lux | This study | CV_0578 promoter fused to luxCDABE |
| pcviI(31532)-lux | This study | C. violaceum 31532 cviI promoter fused to luxCDABE |
Plasmid construction.
The plasmids used in this study are listed in Table 1. A plasmid expressing C. violaceum 12472 CviR in E. coli was constructed by inserting the cviR gene between the NdeI and XhoI sites of plasmid pET23b (Novagen). This plasmid was designated pET23cviR. A reporter plasmid harboring pvioA-gfp (19) was used as the template for Pfu mutagenesis (23) of the vioA promoter. Each base between positions −62 and −79 of the vioA promoter was subjected to site-directed mutagenesis to engineer a defined library of potential CviR DNA binding site mutants. Synthetic, complementary 31-nucleotide oligonucleotides consisting of a 1:1:1 mixture of every non-wild-type base at a particular position in this region were used to generate mutant pools. The primers contained 15 nucleotides flanking each side of the lesion for annealing purposes. Mutant vioA-gfp fusions were sequenced (GeneWiz, Inc.) to identify one mutant containing each non-wild-type base at each position.
The cviR and cviI genes are located adjacent to one another on the chromosome and are transcribed toward one another. Plasmid pcviIR was constructed by cloning the entire cviIR coding sequence as well as flanking DNA (starting 142 base pairs upstream of the cviR start codon, through cviR and cviI, and ending 117 base pairs upstream of the cviI start codon) into pBBR322 using NheI and HindIII. Mutations in pcviIR were incorporated by Pfu mutagenesis (23) as follows. For generation of plasmid pcviIR containing a nonsense mutation in cviR (designated pcviIRstop), a stop codon was introduced at cviR residue Q21. For generation of pcviIR lacking a CviR binding site in the cviI promoter (designated pcviI*R), residues CTG of the CviR binding palindrome were mutated to GAC. Promoter-luciferase fusions were engineered by cloning the luxCDABE genes from plasmid pBBRlux into plasmid pEVS141 between the NheI and BamHI sites to make plasmid pEVSlux. Next, candidate C. violaceum 12472 promoters containing putative CviR binding sites were amplified and cloned into pEVSlux using EcoRI and NheI. All plasmids were confirmed by DNA sequencing (GeneWiz, Inc.).
Position weight matrix construction.
Each plasmid from the library of CviR binding site mutants in pvioA-gfp was transformed into E. coli Top10(pET23cviR), and the fold increase in green fluorescent protein (GFP) in response to 1 μM C10-HSL was measured. The C10-HSL-induced GFP production was compared to wild-type production and designated Fbn, where n is the position within the sequence and b is the base (normal activation equals 1, and no activation equals zero). At each position, the sum of the four fractions corresponding to each possible base was calculated and designated FNn. For example, at position 2, where C is the base in the wild type, FA2 = 0.14, FT2 = 0.29, FG2 = 0.07, and FC2 = 1.0; in this case, FN2 = 1.5. The relative importance of each residue expressed as a normalized fraction was calculated by dividing Fbn by FNn (Fbn/FNn = F′bn). F′ factors were rounded, and 10 arbitrary sequences containing residue b at position n with a frequency corresponding to F′bn were constructed. For example, at position 2, F′A2 = 0.10, FT2 = 0.19, FG2 = 0.04, and FC2 = 0.67; one of the 10 sequences would have an A at position two, two would have a T, none would have a G, and seven would have a C. These sequences were entered into a position weight matrix calculator (25) that was subsequently used to scan the C. violaceum 12472 genome for potential CviR binding sites.
GFP and bioluminescence analyses.
Cultures for GFP measurements were grown in the presence of 1 μM C10-HSL unless otherwise noted. GFP production was measured as described previously (19). Cultures for bioluminescence measurements were grown overnight in LB broth at 37°C with shaking. Cells were subcultured at 1:100 in triplicate into black-walled 96-well plates containing fresh medium or medium plus 1 μM C10-HSL. Plates were incubated at 37°C with shaking for 4 h. Bioluminescence was measured using a Perkin-Elmer 2103 plate reader. To calculate the fold increase in light emission in response to C10-HSL, bioluminescence units from wells of cells grown in the presence of C10-HSL were divided by bioluminescence units from wells of cells grown in the absence of C10-HSL.
Autoinducer production assays.
E. coli Top10 cultures harboring pcviIR or pcviIR mutants were grown overnight in triplicate, pelleted by centrifugation at 10,000 × g for 1 min, washed twice in LB broth, and subcultured 1:2,000 in 25 ml fresh LB broth. Samples of 500 μl were removed and pelleted for 1 min at 16,000 × g, and culture fluids were collected and frozen at −80°C. Autoinducer levels were subsequently determined using an autoinducer bioassay as follows. Ninety-six-well autoinducer bioassay plates were loaded with 120 μl LB broth plus a 1:50 subculture of E. coli(pvioA-gfp, pET23cviR). Cell-free culture fluids (60 μl) from autoinducer-producing strains or 60 μl of LB broth plus 6 μM C10-HSL was added, the contents of the wells were mixed, and 1:3 serial dilutions were performed. Plates were incubated at 37°C with shaking for 7 h, and GFP levels were measured on a Perkin-Elmer 2103 plate reader. GFP production in response to autoinducer produced by E. coli(pcviIR) was compared to the C10-HSL standard curve to yield culture fluid autoinducer concentrations.
Gel mobility shift assays.
For competition gel shifts, 100-base-pair probes containing candidate promoter DNA were amplified by PCR from C. violaceum genomic DNA using primers containing NotI sites. DNA was digested with NotI and gel purified, and Klenow radiolabeling reaction mixtures were assembled (20-μl reaction mixtures containing 100 ng/μl NotI-digested DNA, 10 mM Tris-HCl [pH 7.9], 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol [DTT], 4 μl of 6,000-Ci/mmol [α-32P]dGTP [Perkin-Elmer], and 2.5 U Klenow [exo−, New England BioLabs]). Klenow radiolabeling reaction mixtures were incubated at 30°C for 30 min followed by 75°C for 15 min. DNA was purified using a Zymoclean gel DNA recovery kit (Zymo Research). Competitor DNA (100 base pairs) was PCR amplified using pvioA-gfp mutant templates and gel purified as described above. Double-stranded oligonucleotide probes were synthesized as follows. Complementary pairs of 26-mer oligonucleotides were synthesized. T4 polynucleotide kinase (PNK) labeling reaction mixtures were assembled (50-μl reaction mixtures containing 1 μM oligonucleotide, 70 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 5 mM DTT, 2 μl of 3,000-Ci/mmol [γ-32P]ATP [Perkin-Elmer], and 20 U T4 PNK [New England BioLabs]). T4 PNK labeling reaction mixtures were incubated at 37°C for 30 min followed by 80°C for 20 min. DNA was purified using ProbeQuant G-50 columns (GE Healthcare). For all gel shift assays, 10-μl binding reaction mixtures were assembled [20 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 10 mM CaCl2, 1 mM DTT, 100 μg/ml bovine serum albumin (BSA), 10% glycerol, 170 mM KCl, 50 μg/ml poly(dI-dC), 4,000 cpm radiolabeled DNA, 1 μl CviR:C10-HSL complex in 10 mM imidazole (pH 8.0), 300 mM NaCl, 1 mM EDTA, and 1 mM DTT]. CviR:C10-HSL was used at concentrations of below 10 μM due to protein insolubility at higher concentrations. Binding reaction mixtures were incubated at room temperature for 30 min and resolved on Tris-borate-EDTA (TBE)-polyacrylamide gels (6% for 100-bp probes and 10% for double-stranded oligonucleotides). Gels were dried and analyzed using a phosphorimager.
RESULTS
CviR binds to and regulates the vioA promoter.
To investigate quorum sensing in C. violaceum, we have previously employed genetic, biochemical, and structural analyses to examine autoinducer and antagonist binding to the receptor CviR (2, 19). These studies exploited reporters using the vioA promoter to drive expression of GFP. It is well known that CviR activates vioA expression both in vivo and in recombinant E. coli (12, 19); however, the native CviR binding site has not been well characterized. To address this deficit in our understanding, we analyzed the vioA promoter for putative CviR binding sites. Initially, we focused on an inverted repeat (IR) upstream of the vioA transcription start site (Fig. 1 A, positions −56 to −36 with respect to the transcription start site; see Fig. S1 in the supplemental material for sequence information) because LuxR-type receptors homologous to CviR are reported to bind to similar, so-called “lux boxes” containing palindromic sequences (7).
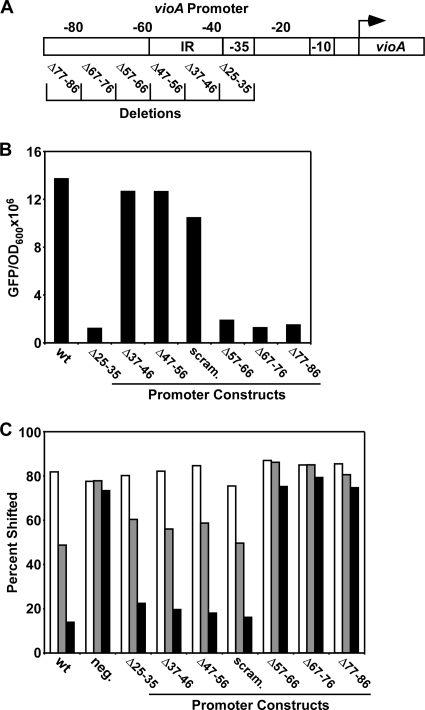
Fig. 1.
An inverted repeat in the vioA promoter is dispensable for binding and activation by CviR. (A) Schematic of the CviR-regulated vioA promoter (IR, inverted repeat; −35 and −10, RNA polymerase binding sites; arrow, transcription start site). Numbers above the boxes indicate positions with respect to the transcription start site. Deletions analyzed in panel B are shown at the bottom. (B) Effect of mutations on vioA promoter activity. Production of GFP in response to 1 μM C10-HSL was measured in E. coli expressing CviR and containing the vioA-gfp fusions shown in panel A. wt, vioA promoter from position −108 to +192; scram., vioA promoter from position −108 to +192 with the IR randomized. The numbers below the other bars indicate the deleted regions. (C) CviR competition gel shift assay. Electrophoretic mobility shift assays were performed on a radiolabeled 100-base-pair vioA promoter probe (spanning bases −108 to −8) incubated with 1 μM CviR:C10-HSL and 10 nM (white bars), 40 nM (gray bars), or 150 nM (black bars) unlabeled DNA. neg., intragenic DNA used as a control; scram., 100-base-pair vioA promoter probe (spanning bases −108 to −8) with the IR randomized. Complexes were resolved by polyacrylamide gel electrophoresis, and the percentage of shifted radiolabeled probe was calculated using a phosphorimager. All results are representative of three independent experiments.
The wild-type level of GFP production by vioA-gfp in response to C10-HSL is shown in Fig. 1B. We found that deletion of the region from position −25 to −35 of the vioA promoter, containing the RNA polymerase (RNAP) binding site, completely eliminated GFP production. To our surprise, we found that the IR sequence is not required for CviR activation of the vioA promoter. Indeed, deletion of either IR half-site (Δ37-46 and Δ47-56 in Fig. 1B) or randomization of the entire IR (scrambled IR [scram. in Fig. 1B]) did not eliminate GFP production in response to C10-HSL.
To support these in vivo data, we tested whether the IR is required for binding of the purified CviR:C10-HSL complex to vioA promoter DNA. Using competition-based gel shift assays, we found that the unlabeled vioA promoter (bases −108 to −8 with respect to the transcription start site) competes with labeled vioA promoter DNA of the same sequence for binding to CviR:C10-HSL (wild type [wt in Fig. 1C]). Intragenic vioA DNA used as the negative control (neg. in Fig. 1C), however, is not capable of competing for CviR binding, presumably because there is no CviR binding site. In addition, deletion of the −35 site (Δ25-35) does not influence competition because RNA polymerase, not CviR, binds at this site. Consistent with our in vivo data, competitor DNAs containing deletions in either IR half-site (Δ37-46 and Δ47-56) or randomization of the entire IR (scram.) fully compete for CviR binding to the vioA promoter (Fig. 1C). Thus, the IR is not bound by CviR or required for control of the vioA promoter by CviR, and we suggest that it could instead encode a binding site for some unknown regulator (e.g., a repressor or another activator) of violacein synthesis that exists in C. violaceum.
In light of the above results, we engineered a series of additional vioA promoter deletions upstream and downstream of the IR sequence to locate the CviR binding site. This analysis revealed a 30-base-pair region upstream of the IR that is required for CviR-directed activation of vioA (Δ57-66, Δ67-76, and Δ77-86 in Fig. 1B). Deletion of regions further upstream of this 30-base-pair sequence had no effect on vioA activation in response to CviR (see “Delineation of the CviR binding site” below). Consistent with this, in competition gel shift assays, unlabeled vioA promoter DNA harboring deletions that render the vioA promoter incapable of responding to CviR in vivo are also unable to compete for the CviR-vioA promoter interaction in vitro (Δ57-66, Δ67-76, and Δ77-86 in Fig. 1C). Together, these results show that the region from position −57 to −86 must contain a CviR recognition motif.
Delineation of the CviR binding site.
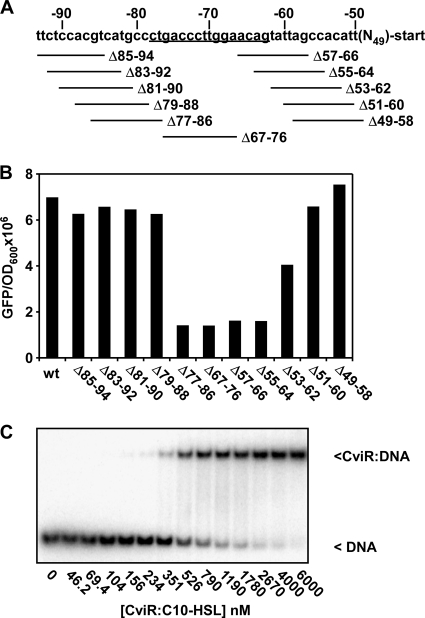
To pinpoint the CviR binding site within the candidate 30-base-pair region defined in Fig. 1, a series of 10-base-pair deletions that span this region starting at every second base were constructed (Fig. 2A). This analysis identified a 16-base-pair region (−63 to −78) required for vioA promoter activation (Fig. 2B). Specifically, deletion of base pairs 77 to 86 or base pairs 55 to 64 eliminated vioA promoter activation, while adjacent deletions did not. This result suggests that CviR binds to the vioA promoter at a sequence within the 16-base-pair region between base pairs −63 and −78. CviR:C10-HSL binds to gel shift probes containing this 16-base-pair region with a binding affinity (Kd) of 652 ± 116 nM (Fig. 2C). Importantly, this 16-base-pair region (CTGACCCTTGGAACAG) is similar (43.8% identity) to the binding site for the LuxR homologue LasR (CTATCTCATTTGCTAG; underlined residues are critical for LasR recognition) (7). These results indicate that the conserved mechanism for DNA recognition known for LuxR homologues is used by CviR. This 16-base-pair region can form a palindrome with five complementary base pairs across the symmetry axis (Fig. 3A).
Fig. 2.
Identification of the site required for CviR binding to the vioA promoter. (A) Schematic of the vioA promoter. Numbers indicate the base position with respect to the transcription start site; deletions are shown below the sequence. The region required for activation by CviR is underlined. (B) Production of GFP in response to 1 μM C10-HSL for the constructs shown in panel A was measured in E. coli expressing CviR and harboring the vioA-gfp plasmid. (C) CviR:C10-HSL electrophoretic mobility shift assay on a double-stranded oligonucleotide (CCGCCCTGACCCTTGGAACAGTATCC) containing the sequence shown to be required for activation by CviR in panel B. Numbers below the gel indicate the concentration of CviR:C10-HSL in each binding reaction mixture.
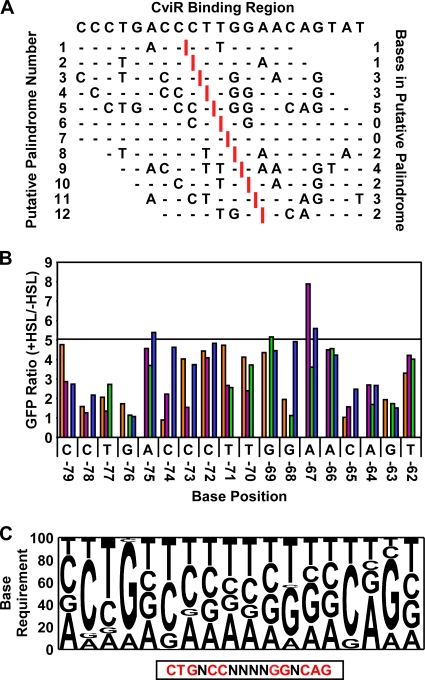
Fig. 3.
A mutagenesis-based position weight matrix for predicting CviR-regulated genes. (A) Prediction of palindromes in the CviR binding region of the vioA promoter (base pairs −80 to −59 with respect to the transcription start site). Putative axes of symmetry (indicated by vertical red bars) were sequentially moved through the CviR binding region in the vioA promoter to generate putative palindromes 1 to 12 (left column). Sites that contain palindromic residues surrounding the putative symmetry axes are shown with base designations. Sites with residues that do not make a palindrome half-site are indicated by horizontal dashes. The number of complementary residues per putative palindrome is indicated in the right column. (B) Fold GFP production from vioA-gfp point mutants. Eighteen bases in the CviR binding site in the vioA promoter (labeled with base designation and position with respect to the transcription start site) were mutated to each non-wild-type residue (orange, A; purple, G; green, C; blue, T). The fold change in production of GFP in response to 1 μM C10-HSL was measured in E. coli expressing CviR. The horizontal line indicates GFP production when the wild-type site is present (5-fold). (C) Position weight matrix constructed from the in vivo data shown in panel B. The relative effect of mutating each base in the CviR binding site to a non-wild-type residue was calculated. The palindromic sequence corresponding to the ideal CviR site is shown below the box.
Construction of a defined CviR binding site library.
To determine which bases in the palindrome are critical for CviR binding, we constructed a defined pool of mutant vioA-gfp promoters in which each base in the putative 16-base-pair CviR binding region as well as one base on either side of the site was mutated to every other non-wild-type residue. We next assayed CviR-dependent GFP production from this library of vioA-gfp point mutants (Fig. 3B). We used the in vivo data in Fig. 3B to construct a position weight matrix defining the nucleotides in the CviR binding site that are most important for CviR activation of vioA (Fig. 3C). A clear palindrome emerged from these studies (CTGNCCNNNNGGNCAG). Importantly, this palindrome has the same axis of symmetry as the palindrome that we have shown in Fig. 3A to contain the greatest number of complementary bases across the symmetry axis (palindrome number 5).
To validate that the position weight matrix produced the correct CviR binding site, we analyzed CviR:C10-HSL binding to altered CviR binding sites in vitro. To do this, we synthesized double-stranded oligonucleotides containing the wild-type CviR binding site or CviR binding sites harboring single-base-pair alterations at each position in the CviR binding site. We performed quantitative gel shift assays on these probes. Consistent with the in vivo results, mutations in the palindrome that impair CviR activation of vioA also show reduced binding to CviR:C10-HSL (Table 2; compare to Kd of 652 ± 116 nM for the wild-type CviR binding site [Fig. 2C]). Quite to the contrary, mutation of the base at position 4 or 12 of the CviR site (positions −75 and −67 with respect to the transcription start site) results in higher-than-average binding affinity and increased expression of vioA-gfp (Table 2 and Fig. 3B). These results indicate that the vioA promoter has not evolved to promote maximal activation in response to CviR.
Table 2.
CviR DNA binding affinities
| Oligonucleotide sequencea |
Kd (nM) |
|
|---|---|---|
| Mean | SD | |
| CCGCCGTGACCCTTGGAACAGTATCC | >10,000 | |
| CCGCCCAGACCCTTGGAACAGTATCC | >10,000 | |
| CCGCCCTCACCCTTGGAACAGTATCC | >10,000 | |
| CCGCCCTGTCCCTTGGAACAGTATCC | 547 | 110 |
| CCGCCCTGAGCCTTGGAACAGTATCC | 5,836 | 2,805 |
| CCGCCCTGACGCTTGGAACAGTATCC | 7,214 | 2,576 |
| CCGCCCTGACCGTTGGAACAGTATCC | 939 | 219 |
| CCGCCCTGACCCGTGGAACAGTATCC | 1,034 | 303 |
| CCGCCCTGACCCTGGGAACAGTATCC | 1,369 | 224 |
| CCGCCCTGACCCTTGGAACAGTATCC | 975 | 228 |
| CCGCCCTGACCCTTGCAACAGTATCC | >10,000 | |
| CCGCCCTGACCCTTGGTACAGTATCC | 538 | 30 |
| CCGCCCTGACCCTTGGATCAGTATCC | 1,505 | 281 |
| CCGCCCTGACCCTTGGAAGAGTATCC | >10,000 | |
| CCGCCCTGACCCTTGGAACTGTATCC | >10,000 | |
| CCGCCCTGACCCTTGGAACACTATCC | 7,451 | 2,976 |
Underlining indicates a mutated nucleotide.
Identification of putative CviR-regulated genes using position weight matrices constructed from in vivo data.
In most bacteria that engage in cell-cell communication, quorum sensing regulates an array of genes that are critical for group behaviors (13, 22). We reasoned that CviR also regulates multiple genes in C. violaceum. Accordingly, we scanned the C. violaceum genome for potential CviR binding sites using the position weight matrix generated as described above (Fig. 3C). Using a stringent cutoff score (7.9 out of 8.4, with an average score across the genome of 5.0), we identified 53 potential CviR binding sites (see Table S1 in the supplemental material). Twenty-two of these potential CviR sites reside in intergenic or promoter regions.
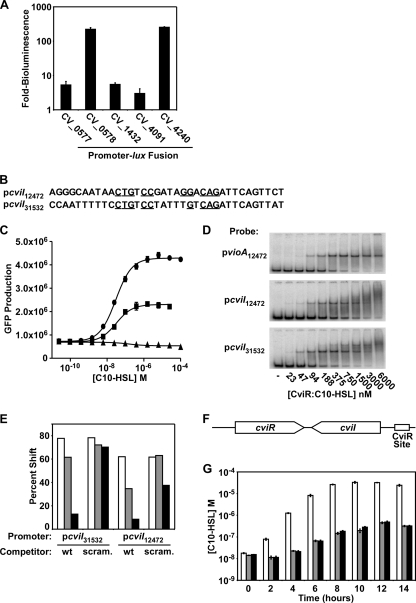
To test whether this method accurately predicts CviR-regulated genes, we fused a subset of the promoters that we identified to the lux operon, introduced these fusions into E. coli expressing CviR, and analyzed bioluminescence in response to C10-HSL. For the initial experiment, we selected promoters upstream of genes with predicted functions or those that drive known quorum-sensing genes (CV_0577, encoding a transcriptional regulator; CV_0578, encoding a guanine deaminase; CV_1432, a gene with a putative role in type VI secretion; CV_4091, the autoinducer synthase gene cviI; and CV_4240, encoding a chitinase). All of these promoters are activated by CviR (Fig. 4 A), suggesting that that we can indeed predict CviR binding sites and, in turn, CviR-regulated genes.
Fig. 4.
Identification of targets of CviR regulation. (A) CviR activation of candidate promoters. Fusions between the indicated C. violaceum promoters and luxCDABE were tested for autoinducer-dependent activation of bioluminescence in E. coli expressing CviR. Shown are averages for four replicates; error bars correspond to one standard deviation from the mean. (B) Predicted CviR-binding region in the cviI promoter from C. violaceum strain 31532 (pcviI31532) and C. violaceum strain 12472 (pcviI12472). Residues identical to those in the ideal CviR binding site are indicated with underlines. (C) Dependence of cviI activation on the predicted CviR binding site. The C. violaceum strain 12472 vioA promoter (circles), the cviI promoter (squares), and the cviI promoter containing a scrambled CviR binding site (triangles) were fused to the GFP gene. GFP production in response to various concentrations of C10-HSL was measured. (D) CviR binding to the cviI promoter. PCR-amplified vioA or cviI promoter DNA from C. violaceum strain 12472 or 31532 was radiolabeled and added to the indicated concentration of purified CviR:C10-HSL. Complexes were resolved by PAGE and analyzed using a phosphorimager. (E) CviR competitive gel shift. CviR:C10-HSL (1 μM) was added to radiolabeled cviI promoter DNA from either C. violaceum strain 12472 or 31532. Competitor wild-type (wt) cviI promoter DNA or cviI promoter DNA containing a randomized CviR binding site (scram.) was added to binding reaction mixtures to a final concentration of 5 nM (white bars), 30 nM (gray bars), or 150 nM (black bars) and analyzed as for panel D. All results are typical of at least three independent experiments. (F) Schematic of the cviI-cviR locus, encoding the autoinducer synthase (cviI), the autoinducer receptor (cviR), and the CviR binding site in the cviI promoter. (G) Role of positive feedback in autoinducer synthesis. Plasmid pcviIR (white bars), pcviIRstop (pcviIR containing a stop codon in cviR) (gray bars), or cviI*R (pcviIR containing a mutant CviR binding site in the cviI promoter) (black bars) was introduced into E. coli. The concentration of C10-HSL in cell-free culture fluids was determined. Shown are averages for three replicates; error bars correspond to one standard deviation from the mean.
CviR binds to and regulates the C. violaceum cviI promoter.
One of the promoters we identified as activated by CviR is upstream of cviI, encoding the C. violaceum autoinducer synthase. Two different strains of C. violaceum have available cviIR sequence information: strain 12472 (used here) and strain 31532. We identified a CviR binding site in the cviI promoters of both strains, suggesting that CviR regulation of cviI expression is a conserved feature among the Chromobacteria (Fig. 4B).
To examine quorum-sensing regulation of cviI, we constructed a C. violaceum 12472 cviI-gfp reporter plasmid and randomized the putative CviR binding site. In E. coli, CviR activated cviI expression, and activity was eliminated when the CviR binding site was randomized (Fig. 4C). Consistent with these results, CviR:C10-HSL bound the cviI promoters from both C. violaceum strains 12472 and 31532 in gel shift assays (Fig. 4D), and in both cases the CviR-cviI promoter interaction was abolished by addition of excess unlabeled wild-type cviI promoter DNA (Fig. 4E). Excess unlabeled cviI promoter DNA containing a randomized CviR binding site (scrambled CviR site [scram.]) was impaired in its ability to compete (Fig. 4E).
A positive-feedback loop controls C. violaceum quorum sensing.
In many bacteria, quorum sensing is controlled by positive feedback in which signaling in response to autoinducer upregulates expression of the autoinducer receptor, the autoinducer synthase, or both (6, 14, 18). To test whether quorum sensing is regulated by positive feedback in C. violaceum, we constructed an E. coli strain carrying the native cviI-cviR locus (Fig. 4F). If positive feedback operates, we predict that in this E. coli strain, CviI will produce autoinducer, which will accumulate at high cell density, leading to the activation of CviR DNA binding activity. In turn, the CviR:C10-HSL complex will activate cviI expression.
To test this, we used an autoinducer bioassay to measure autoinducer production by the recombinant E. coli strain harboring cviI-cviR. We found that in E. coli carrying wild-type cviI-cviR, autoinducer synthesis per cell increases dramatically once the extracellular concentration of C10-HSL reaches 100 nM (Fig. 4G, white bars), consistent with the concentration of C10-HSL required to activate expression of the cviI promoter (Fig. 4C). Increased autoinducer synthesis required CviR, as E. coli carrying cviIR with a null mutation in cviR produced less C10-HSL than the wild type (Fig. 4G, gray bars). A similar decrease occurred when CviR was wild type but the CviR binding site in the cviI promoter was mutated (Fig. 4G, black bars). These data demonstrate that CviR binding to the cviI promoter is required for high-level production of autoinducer at high cell density, suggesting that, as in other canonical LuxIR-type quorum-sensing systems, a positive-feedback loop operates.
DISCUSSION
We have characterized the interaction between the C. violaceum quorum-sensing receptor CviR and the DNA element required for promoter recognition. Using in vivo data generated from a comprehensive library of binding site mutations, we defined the CviR binding site as CTGNCCNNNNGGNCAG and deciphered the relative importance of each base. This approach has allowed us to predict CviR sites in the C. violaceum genome and thereby identify genes that are members of the quorum-sensing regulon.
The quorum-sensing networks of many bacteria include positive feedback loops in which activation of the autoinducer receptor by ligand binding leads to induction of the gene encoding the autoinducer receptor, synthase, or both (6, 14, 18). Furthermore, positive-feedback loops are found in analogous but unrelated quorum-sensing systems that operate by other mechanisms (13), suggesting that this strategy for increasing production of quorum-sensing components in response to autoinducers has emerged independently as an important feature of quorum-sensing circuits. In principle, positive feedback could be used to impose homogeneity in the quorum-sensing response over the population, which could maximize the effects of group behavior. We have previously shown that quorum-sensing genes are required for killing of C. elegans by C. violaceum and that quorum-sensing inhibitors protect nematodes (19). We speculate that positive feedback may be involved in the interaction between C. violaceum and its host, where bacteria need to collectively regulate virulence in order to overcome host defenses.
Interrogation of the C. violaceum genome for putative CviR binding sites revealed over 20 promoters potentially regulated by CviR. These promoters are predicted to drive genes with a variety of functions, including gene regulation, motility, coenzyme synthesis, nutrient utilization, and virulence. We measured positive, direct regulation by CviR for a subset of these promoters, including CV_0577, encoding a transcriptional regulator; CV_0578, encoding a guanine deaminase; CV_1432, a gene with a putative role in type VI secretion; and CV_4091, the autoinducer synthase gene cviI. Of particular interest is that positive regulation also occurred for CV_4240, which is predicted to encode an extracellular chitinase. Chitin is the major structural component of arthropod and fungal cell walls, and it provides an abundant carbon source in the environment (10, 11). Our observation of direct regulation of chitinase production by CviR supports previous studies showing that chitinase activity is regulated by quorum sensing in a related strain of C. violaceum (3). Chernin et al. hypothesized that quorum-sensing control of chitinolytic enzymes may be important for blocking the growth of fungi in soil or water or during colonization of plants, giving C. violaceum a competitive advantage (3). Interestingly, quorum sensing in C. violaceum strain 31532, which recognizes C6-HSL, is antagonized by long-chain AHLs produced by other bacterial species, including C10-HSL produced by C. violaceum strain 12472 (2, 12, 19). It is possible that quorum sensing controls competition in the environment between C. violaceum and fungi (through chitinase secretion). It is also possible that quorum sensing is itself controlled by competition between different strains of C. violaceum or other bacteria that produce autoinducers that act as quorum-sensing antagonists. Antagonism may give particular strains of C. violaceum competitive advantages in specific niches.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Howard Hughes Medical Institute, National Institutes of Health (NIH) grant 5R01GM065859, NIH grant 5R01AI054442, National Science Foundation grant MCB-0343821 to B.L.B., and Ruth L. Kirschstein NIH/NRSA fellowship F32 GM096537-1 to D.L.S.
We thank Lark Perez for synthesis of acylated homoserine lactone derivatives, Guozhou Chen for protein purification, and Suhyun Kim for assistance with plasmid construction.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 27 May 2011.
REFERENCES
- 1. August P. R., et al. 2000. Sequence analysis and functional characterization of the violacein biosynthetic pathway from Chromobacterium violaceum. J. Mol. Microbiol. Biotechnol. 2:513–519 [PubMed] [Google Scholar]
- 2. Chen G., et al. A strategy for antagonizing quorum sensing. Mol. Cell 42:199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chernin L. S., et al. 1998. Chitinolytic activity in Chromobacterium violaceum: substrate analysis and regulation by quorum sensing. J. Bacteriol. 180:4435–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davis P. J., Gustafson M., Rosazza J. P. 1975. Metabolism of N-carbobenzoxyl-l-tryptophan by Chromobacterium violaceum. Biochim. Biophys. Acta 385:133–144 [DOI] [PubMed] [Google Scholar]
- 5. Fuqua C., Parsek M. R., Greenberg E. P. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439–468 [DOI] [PubMed] [Google Scholar]
- 6. Fuqua W. C., Winans S. C. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gilbert K. B., Kim T. H., Gupta R., Greenberg E. P., Schuster M. 2009. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol. Microbiol. 73:1072–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Havarstein L. S., Coomaraswamy G., Morrison D. A. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 92:11140–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Henke J. M., Bassler B. L. 2004. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 186:6902–6914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kurita K. 2006. Chitin and chitosan: functional biopolymers from marine crustaceans. Mar. Biotechnol. (New York, N.Y.) 8:203–226 [DOI] [PubMed] [Google Scholar]
- 11. Lenardon M. D., Munro C. A., Gow N. A. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 13:416–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McClean K. H., et al. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703–3711 [DOI] [PubMed] [Google Scholar]
- 13. Ng W. L., Bassler B. L. 2009. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43:197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Novick R. P., et al. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446–458 [DOI] [PubMed] [Google Scholar]
- 15. Parsek M. R., Greenberg E. P. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. U. S. A. 97:8789–8793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pinto U. M., Winans S. C. 2009. Dimerization of the quorum-sensing transcription factor TraR enhances resistance to cytoplasmic proteolysis. Mol. Microbiol. 73:32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sappington K. J., Dandekar A. A., Oinuma K., Greenberg E. P. 2011. Reversible signal binding by the Pseudomonas aeruginosa quorum-sensing signal receptor LasR. MBio 2:1 e00011-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seed P. C., Passador L., Iglewski B. H. 1995. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J. Bacteriol. 177:654–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Swem L. R., et al. 2009. A quorum-sensing antagonist targets both membrane-bound and cytoplasmic receptors and controls bacterial pathogenicity. Mol. Cell 35:143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Teoh A. Y., et al. 2006. Fatal septicaemia from Chromobacterium violaceum: case reports and review of the literature. Hong Kong Med. J. 12:228–231 [PubMed] [Google Scholar]
- 21. Ulitzur S., Hastings J. W. 1978. Growth, luminescence, respiration, and the ATP pool during autoinduction in Beneckea harveyi. J. Bacteriol. 133:1307–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Waters C. M., Bassler B. L. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319–346 [DOI] [PubMed] [Google Scholar]
- 23. Weiner M. P., et al. 1994. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 151:119–123 [DOI] [PubMed] [Google Scholar]
- 24. Yarwood J. M., Bartels D. J., Volper E. M., Greenberg E. P. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186:1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu J. 2003. A teaching tool for position-specific scoring matrices. http://coding.plantpath.ksu.edu/profile/
- 26. Zhang R. G., et al. 2002. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417:971–974 [DOI] [PubMed] [Google Scholar]
- 27. Zhu J., Winans S. C. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. U. S. A. 96:4832–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu J., Winans S. C. 2001. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. U. S. A. 98:1507–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.