Abstract
Cell-free extracts prepared from Kingella kingae colony biofilms were found to inhibit biofilm formation by Aggregatibacter actinomycetemcomitans, Klebsiella pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Candida albicans, and K. kingae. The extracts evidently inhibited biofilm formation by modifying the physicochemical properties of the cell surface, the biofilm matrix, and the substrate. Chemical and biochemical analyses indicated that the biofilm inhibition activity in the K. kingae extract was due to polysaccharide. Structural analyses showed that the extract contained two major polysaccharides. One was a linear polysaccharide with the structure →6)-α-d-GlcNAcp-(1→5)-β-d-OclAp-(2→, which was identical to a capsular polysaccharide produced by Actinobacillus pleuropneumoniae serotype 5. The second was a novel linear polysaccharide, designated PAM galactan, with the structure →3)-β-d-Galf-(1→6)-β-d-Galf-(1→. Purified PAM galactan exhibited broad-spectrum biofilm inhibition activity. A cluster of three K. kingae genes encoding UDP-galactopyranose mutase (ugm) and two putative galactofuranosyl transferases was sufficient for the synthesis of PAM galactan in Escherichia coli. PAM galactan is one of a growing number of bacterial polysaccharides that exhibit antibiofilm activity. The biological roles and potential technological applications of these molecules remain unknown.
INTRODUCTION
Biofilm is the predominant mode of growth for bacteria in most environments. Biofilms typically contain millions of tightly packed bacterial cells encased in a polymeric matrix and attached to a tissue or surface. The biofilm mode of growth protects the bacterial cells from cell stressors such as desiccation, predators, and antibiotics. Biofilms cause corrosion and biofouling of industrial equipment and chronic infections in clinical settings (24).
Many bacterial biofilms secrete molecules such as quorum-sensing signals (28, 29), surfactants (7), enzymes (17, 21), and polysaccharides (20, 26, 30) that function to regulate biofilm architecture or mediate the release of cells from biofilms during the dispersal stage of the biofilm life cycle (15). These compounds often exhibit broad-spectrum biofilm-inhibiting or biofilm-detaching activity when tested against biofilms cultured in vitro. Such compounds may represent a novel source of antibiofilm compounds for technological development.
In a screen for novel antibiofilm compounds, we observed potent biofilm inhibition activity in cell-free extracts prepared from colony biofilms produced by the Gram-negative oral bacterium Kingella kingae. Preliminary characterization of this extract indicated that it contained abundant DNA, and it was therefore named poly-DNA-containing anti-adhesive material extract (referred to here as PAM extract). In this report we show that PAM extract exhibits surfactant-like properties, inhibits biofilm formation by phylogenetically diverse bacteria and fungi, and contains a novel galactan (referred to here as PAM galactan) as one of its active components.
MATERIALS AND METHODS
Strains and culture conditions.
The strains used in this study were K. kingae PYKK081, PYKK181, PYP8, and PYKK129 (19), Staphylococcus aureus SH1000 (11), S. aureus Xen29 (33), Staphylococcus epidermidis NJ9709 (18), Aggregatibacter actinomycetemcomitans CU1000 (9), Klebsiella pneumoniae 1840 (14), Escherichia coli JM109 (35), and Candida albicans CAF2-1 (10). K. pneumoniae and E. coli were cultured in LB, C. albicans was cultured in tryptic soy broth (TSB) supplemented with 10% fetal bovine serum, and all other bacteria were cultured in TSB supplemented with 6 g/liter yeast extract and 8 g/liter glucose. All cultures were incubated at 37°C. K. kingae and A. actinomycetemcomitans cultures were incubated in 10% CO2.
Preparation of K. kingae colony biofilm extracts.
Extracts were prepared from bacterial lawns (colony biofilms) cultured on tryptic soy agar plates. Lawns were prepared by spreading 109 to 1010 CFU directly onto the surface of the plate and incubating the plate for 2 days. The cell paste was scraped from the surface of the plate with a cell scraper, transferred to a polycarbonate thick-walled centrifuge tube (Sorvall no. 03237), and subjected to ultracentrifugation at 115,000 × g for 15 min. The supernatant was transferred to a new tube and sterilized by passage through a 0.22-μm cellulose acetate centrifuge tube filter (Costar no. 8160). The resulting extract, termed PAM extract, was stored at 4°C.
Biofilm inhibition assay.
Biofilms were cultured in 96-well polystyrene microtiter plates as previously described (13). Briefly, cells were diluted to 104 to 105 CFU/ml in TSB supplemented with 0.1 volume of PAM extract or 0.1 volume of water as a control. Aliquots of cells (200 μl each) were transferred to the wells of a microtiter plate, and the plate was incubated at 37°C for 18 h. Biofilms were rinsed with water and stained for 1 min with 200 μl of Gram's crystal violet. Stained biofilms were rinsed with water and dried. The amount of crystal violet binding was quantitated by destaining the biofilms for 10 min with 200 μl of 33% acetic acid and then measuring the absorbance of the crystal violet solution at 595 nm. All assays were performed 3 to 5 times with similarly significant decreases in absorbance values.
Surface attachment assays.
Single-cell suspensions of S. aureus strain SH1000 were prepared using a filtration protocol as previously described (16). Aliquots of cells (0.5 ml each, ca. 107 to 108 CFU/ml) were transferred to 1.5-ml polypropylene microcentrifuge tubes. Cell suspensions were supplemented with 0.03 volume of PAM extract or with 0.03 volume of water as a control. Stainless steel rods (0.6-mm diameter by 13-mm length) or polystyrene rods (1.4-mm diameter by 15-mm length) were then placed in the tubes, and the tubes were incubated at 37°C. After 30 or 60 min, the rods were removed from the tubes, rinsed three times with phosphate-buffered saline (PBS), and transferred to a 15-ml conical centrifuge tube containing 1 ml of PBS. The rods were sonicated on ice (twice for 30 s each) using an IKA Labortechnik sonicator set to 50% power and 50% duty cycle. CFU in the sonicate were quantitated by dilution plating. In another experiment, S. aureus single-cell suspensions were supplemented with 0.01 or 0.1 volume of PAM extract, and then 1-ml aliquots of cells were transferred to 15-ml polycarbonate tubes. After 15 min, the cells were removed from the tubes, and the tubes were rinsed three times with PBS. One milliliter of PBS was then added to each tube, the tubes were sonicated as described above, and CFU in the sonicate were quantitated by dilution plating. All assays were performed 2 or 3 times with similarly significant reductions in CFU values.
Surface coating assay.
A volume of 10 μl of PAM extract (or 10 μl of water as a control) was transferred to the center of a well of a 24-well tissue culture-treated polystyrene microtiter plate (Falcon no. 353047). The plate was incubated at 37°C for 30 min to allow complete evaporation of the liquid. The wells were then filled with 1 ml of TSB containing 104 to 105 CFU/ml of S. epidermidis or A. actinomycetemcomitans. After 18 h, biofilms were rinsed with water and stained with 1 ml of Gram's crystal violet. Stained biofilms were rinsed with water and dried, and the wells were photographed.
Biochemical and chemical analyses of PAM extract.
For enzymatic treatments, PAM extract was incubated for 1 h at 37°C with 100 μg/ml DNase I, RNase A, or proteinase K (Sigma-Aldrich). For ether extraction, an equal volume of ether was added to the PAM extract, and the tube was mixed by vortex agitation for 15 s. The ether layer was removed with a pipette and discarded, and the residual ether was allowed to evaporate for 15 min at room temperature. For sodium metaperiodate treatment, 0.1 volume of 100 mM sodium metaperiodate was added to the PAM extract, and the extract was incubated at 37°C for 1 h. Molecular masses were determined using Microcon centrifugal concentrators (Millipore) with 10-kDa, 30-kDa, and 100-kDa-molecular-mass-cutoff filters.
Analytical techniques.
PAM extract was treated with DNase I and RNase A (100 μg/ml each; 1 h, 37°C) and passed through a 100-kDa-molecular-mass-cutoff cellulose centrifugal filter (Amicon no. UFC81008). The retentate was lyophilized, and the residue was dissolved in D2O. The precipitate was removed by centrifugation, and the resulting supernatant was analyzed by nuclear magnetic resonance (NMR) spectroscopy as previously described (31). Galactofuranose residues were identified on the basis of 13C NMR chemical shifts as described by Bock and Pedersen (3). Structures were confirmed by spectrum simulation using the Bacterial Carbohydrate Structure database v.3 (www.glyco.ac.ru/bcsdb3/). Linkage positions were identified on the basis of nuclear Overhauser effect (NOE) correlation and heteronuclear multiple-bond correlation (HMBC) and by methylation analysis. The identity of galactose was confirmed by monosaccharide analysis.
Purification of PAM galactan.
PAM extract was treated with DNase and RNase, filtered, and lyophilized as described above. The residue was resuspended in 2% acetic acid and heated at 100°C for 3 h. The precipitate was removed by ultracentrifugation at 100,000 × g for 3 h. The supernatant was fractionated by Sephadex G-50 column chromatography in pyridine-acetic acid buffer (4 ml/liter of pyridine and 1% acetic acid). The eluate was monitored by refractive index, and collected fractions (10 ml) were analyzed by thin-layer chromatography on SiO2 plates. Spots were visualized with 5% H2SO4 in ethyl alcohol.
Cloning and sequencing of the K. kingae pam genes.
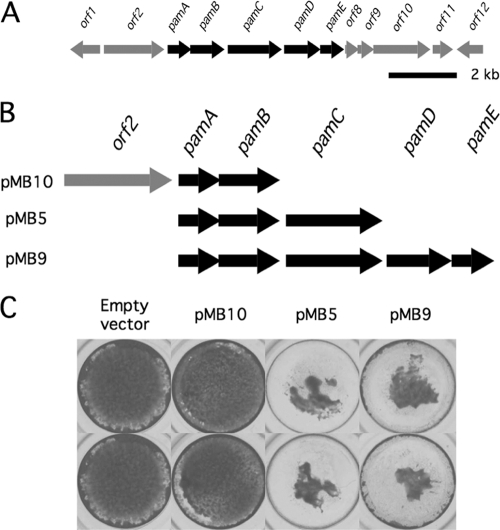
Genomic DNA was isolated from K. kingae strain PYKK181 using a DNeasy blood and tissue kit (Qiagen). The DNA was amplified by PCR using primers that amplify orf2-pamAB, pamABC, and pamABCDE (see Fig. 7B). The primer pairs used were F1 (5′-TGGTAGGCAAAGTTTGCG-3′) and R1 (5′-CGTACCACCCCAATAACG-3′), F2 (5′-TTGATGAATGCAGTGGCG-3′) and R2 (5′-ATGCCGCCTGTTATTAGCC-3′), and F2 and R3 (5′-TGCCTTCGGTGTGAAAGG-3′). The PCR products were cloned into plasmid vector pGEM-T (Promega), resulting in plasmids pMB10, pMB5, and pMB9, respectively. The nucleotide sequences of both strands of the plasmid inserts were determined using an ABI 3130xl Genetic Analyzer.
Fig. 7.
Expression of the K. kingae pam genes in E. coli. (A) Genetic map of the K. kingae pam genes and surrounding region. Arrows indicate open reading frames and direction of transcription. Scale bar, 2 kb. (B) Genetic maps of expression plasmid inserts. (C) Inhibition of S. aureus biofilm formation by colony biofilm extracts isolated from plasmid-harboring E. coli strains. Biofilms were stained with crystal violet and photographed. Duplicate wells are shown.
Expression of the K. kingae pam genes in E. coli.
Plasmids pGEM-T, pMB10, pMB5, and pMB9 were transformed into E. coli strain JM109. Transformants were selected on agar supplemented with 50 μg/ml ampicillin. Lawns of transformants were grown on agar plates supplemented with 50 μg/ml ampicillin and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Lawns were harvested, and cell-free extracts were prepared by ultracentrifugation as described above. Extracts were sterilized by passage through a 0.22-μm filter, incubated at 100°C for 15 min, and then tested for activity in the biofilm inhibition assay described above.
Nucleotide sequence accession numbers.
The nucleotide sequences of the pamABC gene clusters from K. kingae strains PYKK081 and PYKK181 were deposited into GenBank under accession numbers JF441233 and JF441234, respectively.
RESULTS
K. kingae PAM extract inhibits S. aureus biofilm formation.
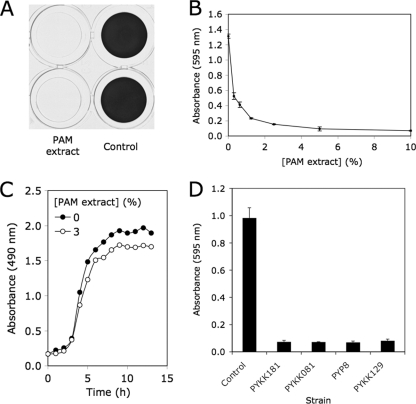
A cell-free extract prepared from colony biofilms of K. kingae clinical strain PYKK181 (referred to here as K. kingae PAM extract) was tested for its ability to inhibit biofilm formation by S. aureus strain SH1000 using a 96-well microtiter plate crystal violet binding assay. When S. aureus cultures were supplemented with PAM extract at a concentration of 10% (by volume), nearly complete inhibition of biofilm formation was observed (Fig. 1A). PAM extract inhibited S. aureus biofilm formation in a dose-dependent manner (Fig. 1B). Greater than 50% reduction in crystal violet binding was observed at concentrations of as low as 0.03%. PAM extract did not inhibit the growth of S. aureus strain SH1000 (Fig. 1C) or of S. aureus Xen29, E. coli JM109, or K. pneumoniae 1840 (data not shown). Colony biofilm extracts prepared from three other K. kingae clinical strains (PYKK081, PYKK129, and PYP8) also exhibited biofilm inhibition activity against S. aureus (Fig. 1D). Biofilm inhibition activity could also be detected in the supernatant of K. kingae PYKK181 broth cultures, but the activity was much weaker than the activity in colony biofilm extracts (data not shown).
Fig. 1.
Inhibition of S. aureus biofilm formation by K. kingae PAM extract. (A) Biofilm formation by S. aureus SH1000 in 96-well microtiter plate wells in the presence or absence of 10% PAM extract. Biofilms were stained with crystal violet. Duplicate wells are shown. (B) Biofilm formation by S. aureus in the presence of increasing concentrations of PAM extract. Biofilm biomass was quantitated by destaining the biofilm and measuring the absorbance of the crystal violet solution. Values show mean absorbance and range for duplicate wells. (C) Growth of S. aureus strain SH1000 in the presence or absence of 3% PAM extract. Growth was monitored by measuring the absorbance of the culture at 490 nm. Values show mean absorbance for duplicate tubes. Error bars were omitted for clarity. (D) Inhibition of S. aureus SH1000 biofilm formation by colony biofilm extracts prepared from four K. kingae clinical strains. Extracts were tested at a concentration of 10% by volume. Values show mean absorbance and range for duplicate wells.
K. kingae PAM extract exhibits surfactant-like activity.
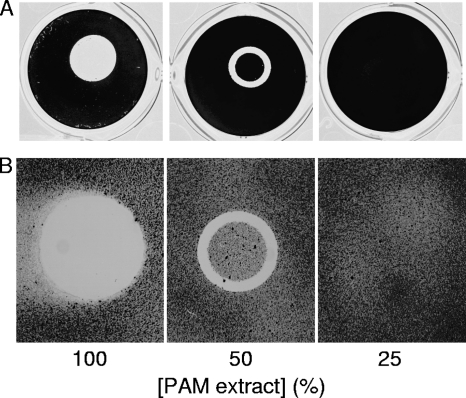
To determine whether PAM extract can modify the surface properties of an abiotic substrate, we used evaporation coating to deposit PAM extract onto the surface of polystyrene wells and then tested the ability of the coated surfaces to resist biofilm formation by S. epidermidis and A. actinomycetemcomitans (Fig. 2). When PAM extract was applied to polystyrene surfaces, the coated surfaces efficiently repelled biofilm formation by both species of bacteria (Fig. 2, left panels). When PAM extract was diluted with an equal volume of water, the evaporative coating produced a ring of biofilm inhibition (Fig. 2, middle panels). This ring probably develops as a consequence of surface tension-driven flow resulting from evaporation of liquid at the edge of the drop (8). The second contact line that develops toward the center of the drop may result from suppression of Marangoni flow due to the presence of organic compounds in the extract (12) or from capillary forces (32). When PAM extract was diluted with 3 volumes of water, the evaporative coating did not efficiently inhibit biofilm formation (Fig. 2, right panels). These findings demonstrate that PAM extract can alter the physiochemical properties of a surface and suggest that a minimum thickness of coating is necessary for efficient surface modification and biofilm inhibition.
Fig. 2.
Polystyrene surfaces coated with PAM extract inhibit biofilm formation. Drops of PAM extract were pipetted onto the surface of a 24-well microtiter plate and allowed to evaporate. In some wells the PAM extract was diluted with 1 or 3 volumes of water. Wells were then inoculated with S. epidermidis (A) or A. actinomycetemcomitans (B) and incubated at 37°C. After 18 h, wells were rinsed with water and stained with crystal violet.
K. kingae PAM extract interferes with cell-substrate interactions and disrupts preformed biofilms.
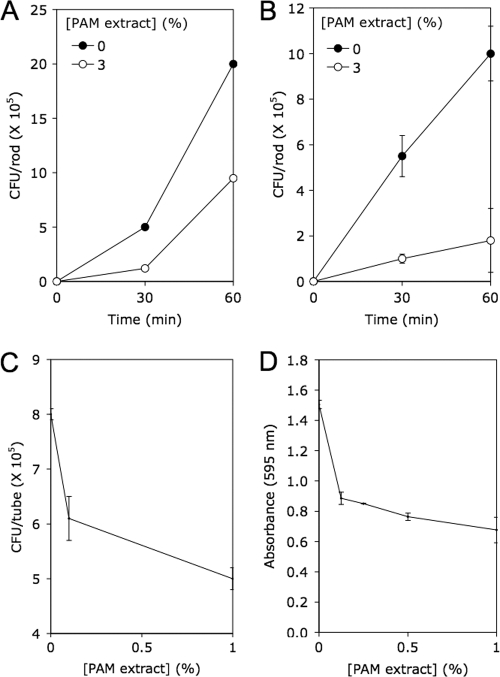
To determine whether PAM extract inhibits binding of S. aureus cells to abiotic surfaces, we placed stainless steel or polystyrene rods in tubes containing S. aureus single-cell suspensions supplemented with 3% PAM extract and counted the numbers of cells attached to the rods after 30 or 60 min (Fig. 3A and B). For both materials and both time points, the numbers of cells bound to the rods were significantly less in tubes containing PAM extract than in control tubes (P < 0.05 by Student's t test). PAM extract also significantly inhibited binding of S. aureus cells to polycarbonate tubes at concentrations of 0.1 and 1% (P < 0.05) (Fig. 3C) and significantly detached preformed S. aureus biofilms at concentrations of 0.1, 0.5, and 1% (P < 0.05) (Fig. 3D). These results suggest that PAM extract may modify the physicochemical properties of the cell surface or substrate or decrease the cohesiveness of the biofilm matrix. Alternately, PAM extract may act as a signal that downregulates production of the adhesive components of the cell surface or biofilm matrix.
Fig. 3.
PAM extract inhibits S. aureus surface attachment and disperses preformed S. aureus biofilms. (A and B) Binding of S. aureus cells to stainless steel (A) and polystyrene (B) rods in the presence or absence of 3% PAM extract. Values show mean CFU per rod and range for duplicate rods. (C) Binding of S. aureus cells to polycarbonate tubes in 15 min in the presence of 0, 0.1, or 1% PAM extract. Values show mean CFU per tube and range for duplicate tubes. (D) Detachment of 18-h-old S. aureus biofilms by PAM extract. Biofilms were rinsed with water and treated with the indicated concentration of PAM extract for 1 h. Biofilms were then rinsed with water and quantitated by crystal violet staining. Values show mean absorbance and range for duplicate wells.
K. kingae PAM extract exhibits broad-spectrum biofilm inhibition activity.
We measured the ability of PAM extract to inhibit biofilm formation by K. kingae, K. pneumoniae, S. epidermidis, and C. albicans in a 96-well microtiter plate assay (Fig. 4). PAM extract inhibited biofilm formation by all four species in a dose-dependent manner.
Fig. 4.
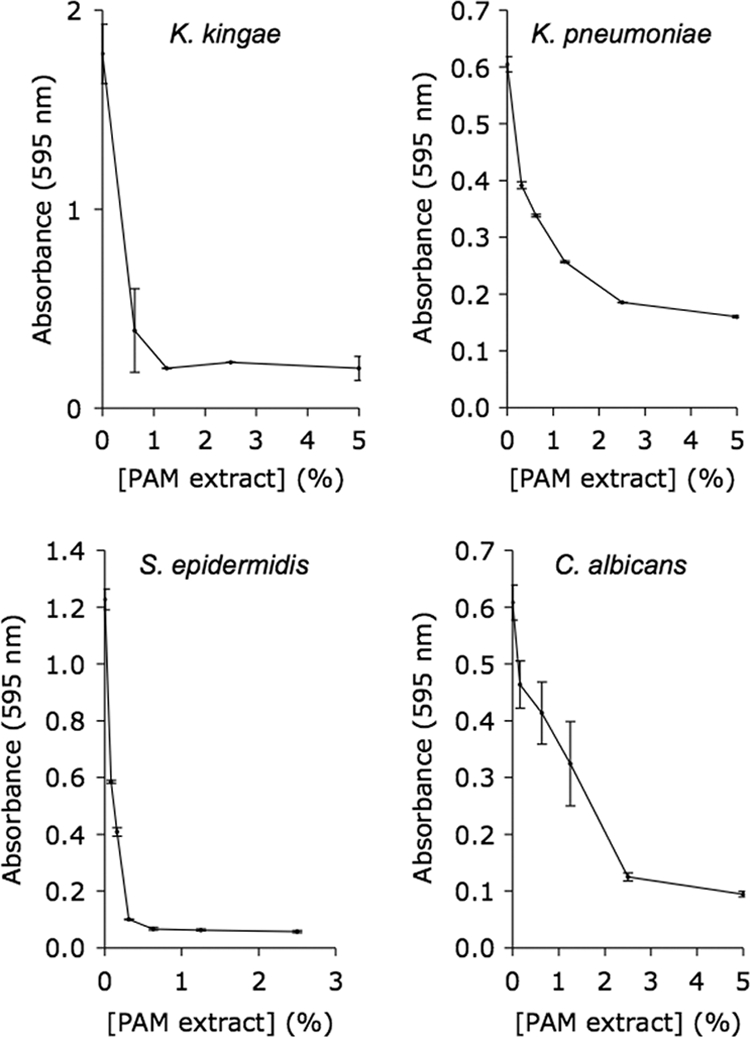
PAM extract inhibits biofilm formation by K. kingae, K. pneumoniae, S. epidermidis, and C. albicans. Biofilms were cultured in 96-well microtiter plates and stained with crystal violet. Values shown mean absorbance and range for duplicate wells.
The biofilm-inhibiting activity of K. kingae PAM extract is due to polysaccharide.
To determine the chemical nature of the biofilm-inhibiting compound in PAM extract, we treated the extract with DNase I, RNase A, and proteinase K. None of these enzymes affected the biofilm-inhibiting activity of the extract (data not shown). Similarly, extraction of PAM extract with ether, which removes lipids and fatty acids, had no effect on its antibiofilm activity (data not shown). In contrast, treatment of PAM extract with the carbohydrate active compound sodium metaperiodate or treatment of the extract with 2 M HCl at 100°C for 2 h significantly reduced its biofilm-inhibiting activity (data not shown). In addition, the biofilm-inhibiting material was heat stable (100°C, 15 min) and >100 kDa in size as determined by size exclusion filtration (data not shown). These findings suggest that the biofilm-inhibiting activity of the PAM extract is due to a high-molecular-mass polysaccharide.
Structural analysis of K. kingae exopolysaccharides.
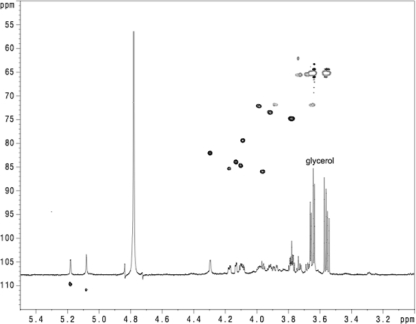
NMR analysis of K. kingae PAM extract treated with DNase and RNase revealed the presence of galactan (Fig. 5 and Table 1). 13C NMR chemical shift patterns and spectrum simulations were consistent with the structure →3)-β-d-Galf-(1→6)-β-d-Galf-(1→. This polysaccharide is referred to here as PAM galactan.
Fig. 5.
1H-13C heteronuclear multiple-quantum coherence (HMQC) spectrum of PAM extract from K. kingae strain PYKK181.
Table 1.
NMR data for K. kingae PAM galactan
| Glycose residue | Chemical shift (ppm) |
|||||
|---|---|---|---|---|---|---|
| H/C-1 | H/C-2 | H/C-3 | H/C-4 | H/C-5 | H/C-6 | |
| →6)-Galf-(1→ | ||||||
| H | 5.18 | 4.13 | 4.09 | 3.96 | 3.98 | 3.65, 3.88 |
| C | 108.10 | 82.40 | 77.80 | 84.40 | 70.70 | 70.30 |
| →3)-Galf-(1→ | ||||||
| H | 5.08 | 4.30 | 4.17 | 4.10 | 3.92 | 3.67, 3.73 |
| C | 109.40 | 80.40 | 83.80 | 83.20 | 71.90 | 63.90 |
Weaker and broader signals of another polysaccharide (PS2) were also identified (Fig. 5 and data not shown). NOE correlation between H-1 of GlcNAc and H-5 of 3-deoxy-d-manno-octulosonic acid (Kdo), as well as low-field shift of the C-5 signal of Kdo, indicated that GlcNAc was connected to O-5 of Kdo in PS2. Low-field shift of GlcNAc C-6 (65.5 ppm) was consistent with Kdo linked to this position. Alkaline deacylation of PAM extract with subsequent separation on a Sephadex G-50 column gave a product with decreased molecular mass (PS3). Deamination of this polysaccharide produced OS1. NMR data for OS1 (data not shown) allowed us to reliably identify the anomeric configuration of Kdo as β by comparing the positions of all 1H and 13C NMR signals to published data. These NMR data are consistent with the structure →5)-β-Kdo-(2→6)-α-GlcNAc-(1→ for PS2.
Hydrolysis of PAM extract with acetic acid resulted in a precipitate and a soluble fraction, which contained both PAM galactan and PS2. Additional PS2 could be recovered from the precipitate by 5% NH4OH extraction. Monosaccharide analysis of PAM extract showed the presence of Glc (3% of sample mass), Gal (15%), and GlcN (10%). Kdo was identified after methanolysis and acetylation.
Bradford protein assay using bovine serum albumin (BSA) as a standard showed that PAM extract contained approximately 12% protein. A large quantity of nonaromatic peptides was obtained after strong alkaline deacylation of PAM extract. Fatty acid analysis of PAM extract showed C14 and C16 acids, as well as unsaturated C16 and C18. The C14 acids were the main component. A small amount of lipid was also present. A disaccharide (OS2) was obtained after deacylation of the precipitate, indicating the presence of lipopolysaccharide (LPS). However, no characteristic 3-OH fatty acids for LPS were detected by gas chromatography-mass spectrometry (GC-MS) analysis.
Together, PAM galactan and PS2 constituted 30 to 50% of the PAM extract sample. PAM galactan is a novel polysaccharide. PS2 is identical to a capsular polysaccharide produced by Actinobacillus pleuropneumoniae serotype 5 (1).
Purified K. kingae PAM galactan exhibits biofilm inhibition activity.
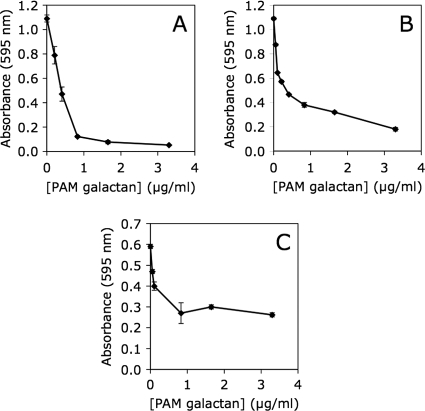
PAM extract was fractionated on a Sephadex G-50 column (data not shown). PAM galactan eluted after the void volume but spread significantly. Pooled PAM fractions yielded a homogeneous high-molecular-mass polysaccharide that was free of PS2 as determined by NMR spectroscopy (data not shown). Purified PAM galactan exhibited biofilm-inhibiting activity against S. aureus, K. kingae, and K. pneumoniae (Fig. 6), demonstrating that PAM galactan is an active biofilm-inhibiting component of PAM extract.
Fig. 6.
Inhibition of S. aureus SH1000 (A), K. kingae PYKK181 (B), and K. pneumoniae 1840 (C) biofilm formation by purified K. kingae PAM galactan. Biofilm biomass was quantitated by crystal violet staining. Graphs show mean crystal violet absorbance values and range for duplicate wells.
Identification of a putative K. kingae pam gene cluster.
In bacteria, the biosynthesis of galactofuranose is catalyzed by the enzyme UDP-galactopyranose mutase, which converts UDP-galactopyranose to UDP-galactofuranose (25). We searched a whole-genome shotgun sequence assembly of K. kingae strain PYKK081 for homologues of E. coli ugm, which encodes UDP-galactopyranose mutase (22). We identified an open reading frame that encodes a protein exhibiting 64% amino acid identity to E. coli UDP-galactopyranose mutase. The K. kingae ugm homologue was located in a cluster of five genes designated pamABCDE, where pamB encodes the UDP-galactopyranose mutase and pamA, pamC, pamD, and pamE encode putative glycosyltransferases. The G+C content of pamABCDE is less than that of the flanking regions, suggesting that these genes may have been acquired by horizontal transfer (data not shown). Among the flanking genes, orf2 encodes the β subunit of glycyl-tRNA synthetase, and orf8 encodes a conserved hypothetical protein that is present in the genomes of many members of the family Neisseriaceae. We hypothesized that the pamABCDE gene cluster is involved in the production of PAM galactan. A genetic map of the K. kingae pamABCDE gene cluster and flanking regions is shown in Fig. 7A.
Expression of the K. kingae pam genes in E. coli.
To confirm that the K. kingae pamABCDE genes are involved in the production of PAM galactan, we amplified by PCR orf2-pamAB, pamABC, and pamABCDE from K. kingae strain PYKK181, cloned the PCR products into a high-copy-number plasmid downstream from an IPTG-inducible tac promoter, and transformed the plasmids into E. coli. The nucleotide sequence of pamABCDE from K. kingae strain PYKK181 was 97% identical to the orthologous sequence from K. kingae strain PYKK081. Schematic diagrams of the genes carried on the expression plasmids are shown in Fig. 7B.
Extracts were prepared from colony biofilms of plasmid-harboring E. coli transformants and tested for their ability to inhibit biofilm formation by S. aureus in the microtiter plate assay (Fig. 7C). The biofilm inhibition activity of extracts prepared from the pMB10-harboring transformant (containing orf2-pamAB) was not significantly different from that of extracts prepared from the plasmid vector control. In contrast, extracts prepared from pMB5- and pMB9-harboring strains (containing pamABC and pamABCDE, respectively) inhibited S. aureus biofilm formation as evidenced by a reduction in the amount of crystal violet binding (Fig. 7C). In addition, NMR spectra revealed anomeric signals corresponding to PAM galactan in colony biofilm extracts prepared from pMB5-haboring E. coli but not in those from the plasmid vector control (data not shown). These results demonstrate that the K. kingae pamABC genes are sufficient for the production of bioactive PAM galactan in E. coli.
DISCUSSION
Kingella kingae is a Gram-negative bacterium that is a member of the family Neisseriaceae and is closely related to Neisseria meningitidis. K. kingae colonizes the posterior pharynges of young children (12 to 24 months) and is a common etiology of pediatric infections, including septic arthritis, osteomyelitis, and endocarditis (34). K. kingae is also a member of the clinically important HACEK group of bacteria (Haemophilus parainfluenzae, Aggregatibacter spp., Cardiobacterium hominus, Eikenella corrodens, and Kingella kingae) (2). These fastidious, Gram-negative organisms are considered normal flora of the human oral cavity but can infrequently enter the submucosa and cause extraoral infections, including endocarditis, bacteremias, and abscesses (6).
In this study we found that K. kingae forms biofilms in an in vitro microtiter plate assay (Fig. 4 and 6B). This is the first report of biofilm formation in this bacterium. Biofilm formation is likely to play an important role in the ability of K. kingae to colonize the pharynx and oral cavity (23) and in the pathogenesis of septic arthritis, osteomyelitis, endocarditis, and other localized infections caused by K. kingae (24).
In this study we also found that K. kingae produces an extracellular galactan (termed PAM galactan) that inhibits its own biofilm formation, as well as biofilm formation by phylogenetically diverse bacteria and C. albicans. Previous studies showed that other bacterial exopolysaccharides also exhibit biofilm inhibition activity. E. coli strains that produce group II capsular polysaccharide, for example, release a soluble polysaccharide of unknown structure that prevents biofilm formation by a wide range of Gram-positive and Gram-negative bacteria (30). Like PAM galactan, the E. coli polysaccharide induces physicochemical surface alterations and inhibits surface colonization when applied to abiotic surfaces. Similarly, Pseudomonas aeruginosa Pel and Psl exopolysaccharides have been shown to inhibit S. epidermidis biofilm formation and disrupt preformed S. epidermidis biofilms (26). Psl consists of a repeating pentasaccharide containing d-mannose, d-glucose, and l-rhamnose (4), whereas Pel is a glucose-rich polysaccharide whose exact structure remains unknown (5). In addition, exopolysaccharides of unknown structure produced by Lactobacillus acidophilus A4 were shown to inhibit initial attachment and autoaggregation of E. coli O157:H7 during biofilm development and to inhibit biofilm formation by phylogenetically diverse Gram-negative and Gram-positive bacteria (20). At least part of the antibiofilm activity of the L. acidophilus exopolysaccharides was due to their surface-modifying properties. These findings demonstrate that diverse bacterial exopolysaccharides can exhibit surfactant-like activity. Previous studies showed that other bacterial polysaccharides also exhibit surfactant-like properties. For example, Proteus mirabilis capsular polysaccharide has been shown to influence cell-cell interactions during population migration, increase surface fluidity, and create a fluid environment necessary for swarming motility (27).
We found that K. kingae PAM galactan consisted of a linear polymer of galactofuranose residues in alternating β(1→3)-β(1→6) linkages. Galactofuranose is a component of several bacterial glycoconjugates, including O antigens and mycobacterial cell wall polysaccharides, and its biosynthesis may be a potential target for antimicrobial chemotherapy (25). The biosynthesis of galactofuranose is catalyzed by the enzyme UDP-galactopyranose mutase (25). In the present study we identified a homologue of ugm, which encodes UDP-galactopyranose mutase, in a whole-genome shotgun sequence assembly of K. kingae strain PYKK081. We further showed that K. kingae ugm is the second gene in a cluster of five genes (pamABCDE), the other four of which encode putative glycosyl transferases. We further showed that three of these genes (pamABC) were sufficient for the biosynthesis of K. kingae PAM galactan in E. coli (Fig. 7 and data not shown). Homologues of pamABC are present in the genome of the oral bacterium Simonsiella mulleri, and a modified pamABC gene cluster containing one additional gene located between pamA and pamB is present in the genome of the swine respiratory pathogen Actinobacillus pleuropneumoniae. Preliminary results indicate that extracts prepared from S. mulleri and A. pleuropneumoniae colony biofilms also exhibit broad-spectrum antibiofilm activity (unpublished data), although it is not known whether these bacteria also produce PAM galactan.
In conclusion, we isolated a surfactant-like galactan from K. kingae colony biofilms that exhibits broad-spectrum antibiofilm activity. The biological role of this galactan and other biofilm-inhibiting bacterial polysaccharides is unknown. These molecules may function to regulate biofilm architecture or mediate the release of cells from biofilms during the dispersal stage of the biofilm life cycle (15). It is also possible that these polysaccharides function as a defense mechanism to prevent surface attachment and biofilm formation by competing bacteria. Antibiofilm bacterial polysaccharides may represent a novel strategy for mitigating biofilms in clinical and industrial settings.
ACKNOWLEDGMENTS
We thank Pablo Yagupsky (Soroka University Medical Center, Beer-Sheva, Israel) for providing K. kingae strains and Neeraj Chauhan (Public Health Research Institute, Newark, NJ) for providing C. albicans CAF2-1. We also thank Jia Rao, Yongyi Mei, Layla Farid, Babar Khan, and William F. Scott IV for technical assistance.
This work was supported in part by grants from the National Institute of Allergy and Infectious Diseases to J.B.K. (AI82392), S.C.K. (AI80606), and N.V.B. (AI080844), by a grant from the American Heart Association to N.V.B. (9SDG2310194), and by a UMDNJ Foundation faculty research grant to D.E.K. and J.B.K.
Footnotes
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Altman E., Brisson J.-R., Perry M. B. 1987. Structure of the capsular polysaccharide of Haemophilus pleuropneumoniae serotype 5. Eur. J. Biochem. 170:185–192 [DOI] [PubMed] [Google Scholar]
- 2. Berbari E. F., Cockerill F. R., Stickelberg J. M. 1997. Infective endocarditis due to unusual or fastidious microorganisms. Mayo Clin. Proc. 72:532–542 [DOI] [PubMed] [Google Scholar]
- 3. Bock K., Pedersen C. 1983. Carbon-13 nuclear magnetic resonance spectroscopy of monosaccharides. Adv. Carbohydr. Chem. Biochem. 41:27–66 [Google Scholar]
- 4. Byrd M. S., et al. 2009. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73:622–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Colvin K. M., et al. 2011. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7:e1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Das M., Badley A. D., Cockerill F. R., Steckelberg J. M., Wilson W. R. 1997. Infective endocarditis caused by HACEK microorganisms. Annu. Rev. Med. 48:25–33 [DOI] [PubMed] [Google Scholar]
- 7. Davey M. E., Caiazza N. C., O'Toole G. A. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deegan R. D., et al. 1997. Capillary flow as the cause of ring stains from dried liquid drops. Nature 389:827–829 [Google Scholar]
- 9. Fine D. H., Furgang D., Kaplan J., Charlesworth J., Figurski D. H. 1999. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch. Oral Biol. 44:1063–1076 [DOI] [PubMed] [Google Scholar]
- 10. Fonzi W. A., Irwin M. Y. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horsburgh M., et al. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu H., Larson R. G. 2006. Marangoni effect reverses coffee-ring depositions. J. Phys. Chem. B 110:7090–7094 [DOI] [PubMed] [Google Scholar]
- 13. Izano E. A., Amarante M. A., Kher W. B., Kaplan J. B. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kadouri D., Venzon N. C., O'Toole G. A. 2007. Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Appl. Environ. Microbiol. 73:605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaplan J. B. 2010. Biofilm dispersal: mechanisms, clinical implications and potential therapeutic uses. J. Dent. Res. 89:205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaplan J. B., Fine D. H. 2002. Biofilm dispersal of Neisseria subflava and other phylogenetically diverse oral bacteria. Appl. Environ. Microbiol. 68:4943–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaplan J. B., Ragunath C., Ramasubbu N., Fine D. H. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J. Bacteriol. 185:4692–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaplan J. B., Ragunath C., Velliyagounder K., Fine D. H., Ramasubbu N. 2004. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 48:2633–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kehl-Fie T. E., et al. 2010. Examination of type IV pilus expression and pilus-associated phenotypes in Kingella kingae clinical isolates. Infect. Immun. 78:1692–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim Y., Oh S., Kim S. H. 2009. Released exopolysaccharide (r-EPS) produced from probiotic bacteria reduce biofilm formation of enterohemorrhagic Escherichia coli O157:H7. Biochem. Biophys. Res. Commun. 379:324–329 [DOI] [PubMed] [Google Scholar]
- 21. Mann E. E., et al. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4:e5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nassau P. M., et al. 1996. Galactofuranose biosynthesis in Escherichia coli K-12: identification and cloning of UDP-galactopyranose mutase. J. Bacteriol. 178:1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neil R. B., Shao J. Q., Apicella M. A. 2009. Biofilm formation on human airway epithelia by encapsulated Neisseria meningitidis serogroup B. Microbes Infect. 11:281–287 [DOI] [PubMed] [Google Scholar]
- 24. Parsek M. P., Singh P. K. 2003. Bacterial biofilms: and emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677–701 [DOI] [PubMed] [Google Scholar]
- 25. Pedersen L. L., Turco S. J. 2003. Galactofuranose metabolism: a potential target for antimicrobial chemotherapy. Cell. Mol. Life Sci. 60:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qin Z., Yang L., Qu D., Molin S., Tolker-Nielsen T. 2009. Pseudomonas aeruginosa extracellular products inhibit staphylococcal growth, and disrupt established biofilms produced by Staphylococcus epidermidis. Microbiology 155:2148–2156 [DOI] [PubMed] [Google Scholar]
- 27. Rahman M. M., Guard-Petter J., Asokan K., Hughes C., Carlson R. W. 1999. The structure of the colony migration factor from pathogenic Proteus mirabilis: a capsular polysaccharide that facilitates swarming. J. Biol. Chem. 274:22993–22998 [DOI] [PubMed] [Google Scholar]
- 28. Rice S. A., et al. 2005. Biofilm formation and sloughing in Serratia marcescens are controlled by quorum sensing and nutrient cues. J. Bacteriol. 187:3477–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schooling S. R., Charaf U. K., Allison D. G., Gilbert P. 2004. A role for rhamnolipid in biofilm dispersion. Biofilms 1:91–99 [Google Scholar]
- 30. Valle J., et al. 2006. Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc. Natl. Acad. Sci. U. S. A. 103:12558–12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vinogradov E., Conlan W. J., Gunn J. S., Perry M. B. 2004. Characterization of the lipopolysaccharide O-antigen of Francisella novicida. Carbohydr. Res. 339:649–654 [DOI] [PubMed] [Google Scholar]
- 32. Weon B. M., Je J. H. 2010. Capillary force repels coffee-ring effect. Phys. Rev. E 82:015305. [DOI] [PubMed] [Google Scholar]
- 33. Xiong Y. Q., et al. 2005. Real-time in vivo bioluminescent imaging for evaluating the efficacy of antibiotics in a rat Staphylococcus aureus endocarditis model. Antimicrob. Agents Chemother. 49:380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yagupsky P., Porsch E., St. Geme J. W., III 2011. Kingella kingae: an emerging pathogen in youg children. Pediatrics 127:557–565 [DOI] [PubMed] [Google Scholar]
- 35. Yanisch-Perron C., Vieira J., Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]