Abstract
The transcription factor LexA in the cyanobacterium Synechocystis sp. strain PCC 6803 has been shown to regulate genes that are not directly involved in DNA repair but instead in several different metabolic pathways. However, the signal transduction pathways remain largely uncharacterized. The present work gives novel insights into the regulation of LexA in this unicellular cyanobacterium. A combination of Northern and Western blotting, using specific antibodies against the cyanobacterial LexA, was employed to show that this transcription regulator is under posttranscriptional control, in addition to the classical and already-described transcriptional regulation. Moreover, detailed two-dimensional (2D) electrophoresis analyses of the protein revealed that LexA undergoes posttranslational modifications. Finally, a fully segregated LexA::GFP (green fluorescent protein) fusion-modified strain was produced to image LexA's spatial distribution in live cells. The fusion protein retains DNA binding capabilities, and the GFP fluorescence indicates that LexA is localized in the innermost region of the cytoplasm, decorating the DNA in an evenly distributed pattern. The implications of these findings for the overall role of LexA in Synechocystis sp. strain PCC 6803 are further discussed.
INTRODUCTION
Besides sharing the basic cellular features of other bacteria, cyanobacteria possess unique and diagnostic characteristics. Distinctively, cyanobacteria are the only organisms ever to evolve coupled photosystems that harvest electrons from water and produce oxygen as a by-product (21). They are photosynthetic Gram-negative prokaryotes typically possessing the ability to synthesize chlorophyll a (55). Cyanobacterial ecological plasticity is remarkable, and their long evolutionary history is possibly related to some of the reasons for their success in modern habitats. Synechocystis sp. strain PCC 6803 is a unicellular cyanobacterium amenable to genetic manipulation, which makes it an attractive research model.
The protein LexA is classically associated in bacteria with the SOS response, which comprises a set of coordinated physiological responses induced by DNA damage. This response was one of the first clear networks of transcriptional regulation identified in Escherichia coli, and its mechanisms seem to be widely conserved among bacteria. However, there are several reports in the literature presenting evidence for deviations from the classical E. coli-type SOS regulation, with Synechocystis sp. strain PCC 6803 being one such case. In this cyanobacterium, LexA has been shown to directly regulate genes involved in carbon assimilation or controlled by carbon availability (8), the bidirectional hydrogenase (15), and the RNA helicase CrhR (35), but not any genes involved in DNA metabolism (8, 35). However, the signal transduction pathways directly or indirectly involved in the regulation of LexA in Synechocystis sp. strain PCC 6803 and, consequently, its downstream targets remain largely unknown. Since LexA was described in Synechocystis sp. strain PCC 6803 as being involved in regulatory networks other than the SOS response, several reports have become available describing how the lexA transcript is up- or downregulated in cells exposed to different environmental conditions (20, 37, 42, 59). However, in most of these studies, the assumption that a regulatory response on the downstream targets derives from that change in lexA transcription still prevails, without a systematic analysis of the protein levels.
In addition to the work carried out to demonstrate LexA's alternative role in Synechocystis sp. strain PCC 6803 (8, 15, 35), a careful analysis of the deduced amino acid sequence also seems to support its divergence in function. LexA in E. coli has been demonstrated to have autoproteolytic activity, which represents a crucial step in the overall SOS response (53). The autoproteolysis is dependent on two conserved protein features: a defined cleavage site and a well-characterized active site (53). However, LexA in Synechocystis sp. strain PCC 6803 does not possess the conserved cleavage site, and one of the crucial amino acids of the active site has been replaced (8, 29, 33). These changes have been suggested to exert a negative effect on the autocatalytic cleavage of this transcription factor (29). In fact, there is no indication in the literature that could suggest that LexA in Synechocystis sp. strain PCC 6803 can be autoproteolytically modified.
The proteome of Synechocystis sp. strain PCC 6803 has been extensively studied over the years, and one aspect that remains to be understood about LexA is connected to its subcellular localization. Several proteomic studies identified LexA, including two-dimensional (2D) gel analyses (11, 13, 25, 41, 44, 45, 54, 58), as well as those using more advanced techniques, such as iTRAQ (12). Despite the fact that it is a transcription factor and is predicted to be a cytoplasmic protein, LexA has also been identified in studies specifically focusing on membrane proteins, both in thylakoid (23, 45, 54) and in plasma membrane (58) fractions. Since LexA does not possess any predicted transmembrane helix, in light of these results, there is a possibility that LexA may be associated with a membrane protein and, therefore, be tethered to the membrane. However, there is still no evidence that can support this or any other hypothesis about LexA localization.
In the present work, we show for the first time evidence that LexA in Synechocystis sp. strain PCC 6803 is regulated on a posttranscriptional level, in addition to the classical transcriptional regulation. Furthermore, we present results that indicate that LexA is posttranslationally modified. Finally, we studied the subcellular localization of LexA in Synechocystis sp. strain PCC 6803 by fusing the protein with green fluorescent protein (GFP). Our results indicate that LexA has a distribution prominently restricted to the cytoplasm, specifically to the region where the DNA is located, and that it decorates the DNA in an evenly distributed pattern.
MATERIALS AND METHODS
Organisms and growth conditions.
Synechocystis sp. strain PCC 6803 wild-type cells were routinely grown in BG11 medium (46) supplemented with 10 mM HEPES, pH 7.5, bubbled with air, at 25°C and with a continuous irradiance of 40 μmol of photons m−2 s−1. The Synechocystis sp. strain PCC 6803 mutant cells were grown under the same conditions as the wild type, except that the medium was supplemented with kanamycin to a final concentration of 50 μg/ml. For total RNA and protein extractions, wild-type cells of Synechocystis sp. strain PCC 6803 were grown as described above before being transferred to dark conditions (by covering the culture with aluminum foil), supplemented with 5 mM glucose, or transferred to anaerobic (replacing the air bubbling with argon) or dark and anaerobic (bubbling with argon in addition to covering the culture with aluminum foil) conditions. E. coli strains were grown in LB liquid medium or LB agar plates supplemented with the appropriate antibiotics at 37°C.
Nucleic acid isolation and analysis.
Genomic DNA and total RNA were isolated from Synechocystis sp. strain PCC 6803 cells as described previously (3, 4, 48). Plasmid DNA was isolated from E. coli using the GenElute Plasmid Miniprep Kit (Sigma-Aldrich), and all sequencing reactions were performed at Macrogen Inc. Northern blotting was carried out as described previously (2, 43), using probes that were obtained by PCR with specific oligonucleotides (Table 1), and further labeled with [α-32P]dCTP. The even loading of the total-RNA aliquots was controlled by verification of equal abundance of the rRNA bands on the agarose gel and of the constitutive RNA component of the ribozyme RNase P (52). To identify the 3′ ends of the transcripts harboring lexA and the gene downstream (slr1735), which lies in the opposite direction to lexA, 3′ rapid amplification of cDNA ends (RACE) was carried out. Two micrograms of total RNA was reverse transcribed to cDNA using the Revert Aid H Minus M-MuLV Reverse-Transcriptase (Fermentas) with 3′ RACE adapter (Table 1) as the reaction primer. The cDNA was then subjected to nested PCR; the products obtained were analyzed by agarose gel electrophoresis before being further purified and cloned into the pCR2.1-TOPO (Invitrogen) vector. The identities of the PCR products were determined by sequencing.
Table 1.
Oligonucleotides used in this study
| Primer | Sequence (5′→3′)a | Purpose | Reference |
|---|---|---|---|
| SlexAF | GGATCCGAACCTCTCACCCGAGCCCAAAAAG | Probe for Northern blotting | 34 |
| SlexAR | AAGCTTCTAAACTCCCTGGAAATTGCGC | 34 | |
| SrnpBF | TTCTGTTCCAGGATGCGAGGCA | Probe for Northern blotting | 2 |
| SrnpBR | GAGAGTTAGGGAGGGAGTTG | 2 | |
| 3′RACE adapter | GCGAGCACAGAATTAATACGACTCACTATAGGT12CA | 3′ RACE (RT) | This work |
| 3′RACE outer | GCGAGCACAGAATTAATACGACT | 3′ RACE (1st PCR) | This work |
| 3′RACE inner | GAATTAATACGACTCACTATAGG | 3′ RACE (nested PCR) | This work |
| SlexA-3′RACE1 | CAAAGGCGGTGAACTGGTGGAA | 3′ RACE (1st PCR) | This work |
| SlexA-3′RACE2 | TGCTATCAAACGCTATTACCAAGA | 3′ RACE (nested PCR) | This work |
| slr1735-3′RACE1 | TGATAACTACCCAGACCAACT | 3′ RACE (1st PCR) | This work |
| slr1735-3′RACE2 | AGTATCCAATCGGGTCTTTTTCT | 3′ RACE (nested PCR) | This work |
| SlexAF-loc1 | GGATCCGAACCTCTCACCCGAGCCCAAAAAG | Fragment 1 (LexA::GFP fusion) | This work |
| SlexAR-loc1 | AAACTGCAGAACTCCCTGGAAATTGCGCCAA | This work | |
| SlexAF-loc2 | AAAACGCGTCCCGGCATTACCGGGAACAGCTCAAAAA | Fragment 2 (LexA::GFP fusion) | This work |
| SlexAR-loc2 | AAAAAGCTTATGACTTCCCCCACTGCTCCGTTAA | This work | |
| gfpF | AAACTGCAGTCTAGACTTGAAATGAGCAAGGGCGAGGAG | Fragment 3 (LexA::GFP fusion) | This work |
| gfpR | AAAACGCGTTTATTACTTGTACAGCTCGTCCAT | This work | |
| pUC4KF | ACGCGTTGAGGTCTGCCTCGTGAAGAA | Fragment 4 (LexA::GFP fusion) | 34 |
| pUC4KR | ACGCGTAAAGCCACGTTGTGTCTCAAA | 34 | |
| SlexAF2 | TACTCAAAGCCTCCAACAACAAA | Test chromosome segregation | This work |
| slr1735R2 | ATGCAGTTTGCCCGGGAAGTAT | This work | |
| ShoxprF | GCAATTGGGGTTGCGACTAT | DNA affinity assays | 33 |
| ShoxprR | CCTCCACAATCTTGCCCACAATAA | 33 |
The underlined nucleotides correspond to restriction sites.
Production of polyclonal antibodies against recombinant LexA. To raise antibodies against the cyanobacterial LexA, the Anabaena sp. strain PCC 7120 LexA recombinant protein, containing an amino-terminal six-histidine tag, was overexpressed in E. coli and purified as described previously (43). The fractions with the recombinant protein were further purified by excising the corresponding LexA band from SDS-polyacrylamide gels. In each lane, 160 μg His-tagged LexA was loaded, and in total, five Coomassie blue-stained bands were cut out and used for polyclonal antibody production in rabbit (Agrisera, Sweden).
Protein extraction and analysis. In the present work, proteins were extracted in different extraction buffers, depending on the downstream application. The proteins were kept under native conditions using a “native extraction buffer” (NEB) composed of 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, pH 8.0, 2 mM dithiothreitol, 0.5% Triton X-100, and 10% glycerol supplemented with a cocktail of protease inhibitors (cOmplete Mini, EDTA free, Roche). Alternatively, proteins were extracted under denaturing conditions using a “denaturing extraction buffer” (DEB) containing 7 M urea, 2 M thiourea, 4% (wt/vol) CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 40 mM dithiothreitol, and 2% IPG Buffer (GE Healthcare). In either case, the cell pellet was resuspended with an appropriate volume of extraction buffer and mixed with 0.35 g of acid-washed glass beads. The cells were then homogenized using a Precellys 24 homogenizer (Bertin Technologies). The cell debris was pelleted by centrifugation, and the proteins were stored at −20°C until further analysis.
For one-dimensional analyses, protein extracts were separated by electrophoresis on a 12% (wt/vol) SDS-polyacrylamide gel and visualized with colloidal Coomassie blue staining (Sigma) or transferred to a Hybond ECL nitrocellulose membrane (GE Healthcare). A buffer containing 192 mM glycine, 25 mM Tris, 20% (vol/vol) methanol was used as the transfer buffer in a TE 22 tank transfer unit (Amersham Biosciences). After the transfer, the membranes were blocked for 2 h with 5% (wt/vol) milk powder in TTBS (Tris-buffered saline [TBS] with 0.1% [vol/vol] Tween 20) and then incubated for 1.5 h with polyclonal rabbit anti-LexA or anti-green fluorescent protein (GFP) N-terminal antibody produced in rabbit (Sigma-Aldrich) or THE Anti-His monoclonal antibody (GenScript Corporation). After washing with TTBS, the blots were incubated for 1 h with enhanced chemiluminescence (ECL) donkey-anti-rabbit IgG (GE Healthcare) when anti-LexA or anti-GFP was used as the first antibody or goat anti-mouse IgG (GE Healthcare) when anti-His was used as the first antibody, both linked to horseradish peroxidase. The membranes were washed with TTBS before immunodetection was performed by chemiluminescence using ECL Western Blotting Analysis System detection reagents (GE Healthcare) in a ChemiDoc XRS system (Bio-Rad).
For two-dimensional analyses, 35 to 40 μg of total protein in DEB buffer was mixed with rehydration buffer before being absorbed by 7-cm-long Immobiline DryStrip gels (GE Healthcare) with a pH range of 3 to 10 (linear) or 4 to 7 (linear), according to the manufacturer's instructions. The isoelectric-focusing step was carried out on an Ettan IPGphor 3 system (GE Healthcare), after which the strips were further equilibrated with dithiothreitol and iodoacetamide. After completion of this step, the strips were applied on top of 12% (wt/vol) SDS-polyacrylamide gels, and the proteins were further separated by electrophoresis. Further analyses were performed as described above.
DNA affinity assays. DNA affinity assays were carried out as described previously (33, 34), using streptavidin-coated magnetic beads (Dynabeads M-280; Dynal Biotech, Invitrogen). A DNA fragment used in previous similar studies (33, 34), referred to as ShoxPr, was used as bait. Proteins extracted under native conditions from wild-type Synechocystis sp. strain PCC 6803, E. coli M15(pREP4) expressing a histidine-tagged version of Synechocystis sp. strain PCC 6803 LexA (33), and SFM04 cells were incubated with tagged Shoxpr. The DNA-interacting proteins were recovered from the DNA either by mixing with SDS loading buffer and further separation by ordinary SDS-polyacrylamide gel electrophoresis or by mixing with DEB, loaded on an Immobiline DryStrip gel (GE Healthcare), and separated by 2D electrophoresis as described above.
Construction of the LexA::GFP fusion strain.
Using specific oligonucleotides (Table 1), four different DNA fragments were amplified by PCR: fragment 1, a fragment of 606 bp, covering the coding sequence of sll1626 (lexA) but excluding the stop codon; fragment 2, a fragment of approximately 1 kb, covering the intergenic region downstream of lexA and the full coding sequence of slr1735 (fragments 1 and 2 were amplified using Synechocystis sp. strain PCC 6803 genomic DNA as a template); fragment 3, the gene encoding the green fluorescent protein (obtained from the vector pTHH001-GFP [T. Heidorn and P. Lindblad, unpublished data]); and fragment 4, the kanamycin resistance cassette from the plamsid pUC4K (GE Healthcare). All fragments were cloned in the vector pCR2.1-TOPO (Invitrogen), resulting in vectors pTPO1, pTPO2, pTPO3, and pTPO4, respectively. The different fragments' identities were confirmed by sequencing. Using the restriction sites introduced at both ends of the PCR fragments, fragment 3 (corresponding to the GFP gene) was cloned into pTPO2 as a PstI-MluI fragment, upstream of fragment 2, resulting in vector pTPO32. Then, the approximately 1.8-kb fragment between restriction sites PstI and HindIII (including fragments 2 and 3) was further cloned in pBluescript SK+ (Stratagene), producing the vector pSPO32. After that, fragment 1 was cut from pTPO1 using BamHI and PstI and cloned in the same restriction sites of pSPO32, producing vector pSPO132; with this step, lexA became fused to the GFP gene. The final fusion protein possesses a linker sequence composed of GVLQSRLEMS, where GV are the last 2 amino acids of LexA and MS are the first 2 residues of GFP. Finally, the kanamycin resistance cassette (fragment 4) was cloned into the MluI restriction site of pSPO132, localized between fragments 3 and 2, resulting in the vector pSPO1342.
Transformation of Synechocystis sp. strain PCC 6803 cells with the vector pSPO1342 was performed as described previously (24). Selection of mutants was carried out in plates initially supplemented with 15 μg/ml kanamycin, followed by several rounds of selection in plates with 30 μg/ml kanamycin. To analyze the extension of chromosome segregation after transforming Synechocystis sp. strain PCC 6803 with the vector pSPO1342, PCR amplifications were carried out with specific oligonucleotides (Table 1).
Sample preparation and microscopy.
Cells of wild-type Synechocystis sp. strain PCC 6803, SFM04 (harboring the LexA::GFP fusion), and Synechocystis sp. strain PCC 6803 harboring pPMQAK1-GFP (19) were grown in liquid medium in an orbital shaker under the same conditions described above but without air bubbling. In addition, SFM04 cells were also grown in liquid BG11 medium supplemented with 10 mM HEPES, pH 7.5, and kanamycin and sparged with air, either at 25 or at 30°C. To experiment with high-light conditions, SFM04 cells were grown photoautotrophically as described, with a continuous irradiance with 40 μmol of photons m−2 s−1, before being exposed to 300 μmol of photons m−2 s−1. Samples (5 μl) were loaded on a glass slide, followed by 12.5 μl of 1% low-melting-point agarose dissolved in BG11 medium, and this mixture was finally covered with a coverslip. The GFP emission (collected between 500 and 540 nm) was observed when the cells were exposed to a laser beam at 488 nm, using a Leica TCS SP5 confocal microscope. Cyanobacterial autofluorescence was collected between 640 and 720 nm after excitation at 633 nm. Alternatively, epifluorescence was analyzed using a Leica DMRXE microscope, and the filter specificities were as follows: filter cube A (for the analysis of the DNA-staining dye Hoechst 33342), excitation filter (ex.), band pass (BP) 340 to 380; dichromatic mirror (dc.), 400; emission suppression filter (em.), long pass (LP) 425; filter cube N2.1 (for cyanobacterial autofluorescence analysis), ex., BP 515 to 560; dc., 580; em. LP 590; and filter cube L5 (for GFP analysis), ex., BP 480/40; dc., 505; em., BP 527/30.
RESULTS
lexA is posttranscriptionally regulated.
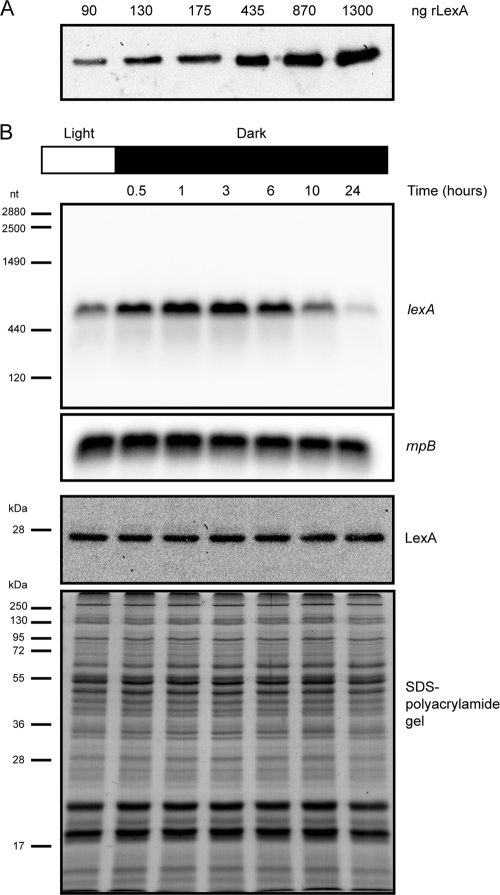
In the present work, the changes in LexA relative levels in response to up- or downregulation of lexA transcription were studied. First, cyanobacterial LexA polyclonal antibodies were raised, and their dynamic ranges were tested using recombinant histidine-tagged LexA (rLexA) (Fig. 1 A). Then, a number of environmental conditions known to induce up- or downregulation of lexA transcription were selected (darkness [35], anaerobiosis [20, 47], and presence of glucose in the medium [35]), and wild-type cells of Synechocystis sp. strain PCC 6803 were exposed to such conditions. Samples were taken at different time points, and total RNA and proteins were extracted, separated on gels, and analyzed by Northern and Western blotting, respectively. Figure 1B shows that the lexA transcript levels increase in cells exposed to 30 min of darkness after growing continuously in light. In addition, the lexA transcript levels steadily increase up to 3 h into the dark period (the time when it reaches its maximum level, as monitored in these experiments), gradually decreasing over the course of the experiment (Fig. 1B). However, the LexA levels, monitored in samples obtained at the same time points do not follow this trend; in fact, they remain unchanged throughout the experiment (Fig. 1B). Similar experiments using cells exposed to anaerobic conditions or cultures to which glucose had been added produced comparable results: despite the change in RNA levels, the protein levels remained constant (data not shown).
Fig. 1.
LexA is regulated on a posttranscriptional level. (A) The dynamic range of the anti-LexA antibodies raised in the present work was tested using purified rLexA; the amount of rLexA loaded on each well is shown. (B) Synechocystis sp. strain PCC 6803 cells were grown in BG11 under continuous light before being transferred to dark conditions. Cells were collected at different time points, and total RNA and proteins were extracted from the samples. Northern blot analyses of the relative levels of lexA and rnpB transcripts are shown (two upper blots). The numbers on the left of the upper blot indicate sizes in nucleotides as estimated from the rRNA and rnpB bands. The two lower panels depict the Western blot analysis of LexA and the Coomassie blue-stained SDS-polyacrylamide gel showing the equal loading of the samples. The molecular masses of the Fermentas protein marker are indicated on the left.
The lexA and slr1735 transcripts do not possess overlapping regions.
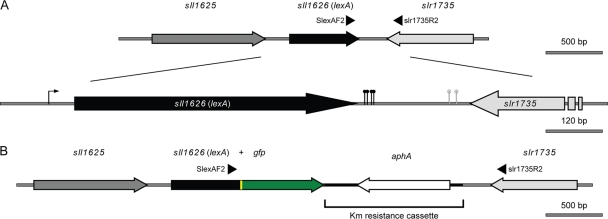
After observing the results presented in Fig. 1, it became important to further study the lexA transcript. Its start site has been identified −34 bp upstream from the ATG start codon (8), but no experiments have been done to characterize the 3′ end of the transcript. Therefore, 3′ RACE (see Fig. S1 in the supplemental material) was used to map the end of the lexA transcript, and different sites were identified in which a poly(A)-rich tail was found, as depicted in Fig. 2 A. Based on these results and on the previously identified transcription start point (8), the size of the longest lexA transcript is 676 nucleotides (nt), which matches well with the estimation of its size by Northern blotting (approximately 700 nt). Applying the same 3′ RACE technique to the gene slr1735, which lies downstream of lexA (see Fig. S1 in the supplemental material) but in the opposite direction (Fig. 2A), it was also possible to identify similar sites. From this analysis, it seems evident that the lexA and slr1735 transcripts do not possess overlapping and complementary regions at their 3′ ends, which otherwise could suggest a mechanism of antisense RNA, as previously described for the transcription regulator furA in the cyanobacterium Anabaena sp. strain PCC 7120 (17). Even though 3′ RACE does not exactly determine the point where a given cyanobacterial transcript is terminated (polyadenylation of transcripts in Synechocystis sp. strain PCC 6803 is known to occur [38], but associated with their degradation rather than with their stabilization, as in eukaryotes), it has been used successfully with cyanobacterial RNA to examine the 3′ ends of different transcripts (32, 38). Thus, even assuming that the real transcription stop points of the lexA and slr1735 transcripts are localized between 50 and 80 nucleotides downstream of each of the identified sites, these transcripts still would not overlap. Furthermore, large-scale screenings to uncover the presence of noncoding RNA and antisense RNA in the cyanobacterium Synechocystis sp. strain PCC 6803 did not identify antisense RNA in the lexA locus and its vicinity (14, 31).
Fig. 2.
Schematic representation of the lexA locus in the genome of Synechocystis sp. strain PCC 6803. (A) The lexA gene is represented in black, while the upstream and downstream genes are represented in dark and light gray, respectively. The relative positions of the oligonucleotides SlexAF2 and slr1735R2 are shown as arrowheads. The lower diagram is a magnification of the map represented above, where it is possible to recognize the transcription start point of lexA (the arrow upstream of the gene) identified by Domain et al. (8) and the transcription stop points identified in the present study by 3′ RACE (the black and gray pins are the lexA and slr1735 transcription stop points, respectively). (B) Physical map of the lexA locus in SFM04 (Synechocystis sp. strain PCC 6803 containing a LexA::GFP fusion). The gfp gene is shown in green, the linker between lexA and gfp is in yellow, and the aphA gene (aminoglycoside 3′-phosphotransferase, which confers resistance to kanamycin) is in white. The kanamycin resistance cassette taken from the vector pUC4K is highlighted.
On the other hand, transcript instability does not seem to account for the posttranscriptional control described in the present work either, since the lexA transcript is detectable as a single band of the same size irrespective of the condition tested, with no intermediate degradation products accumulating, e.g., in the form of a smear or other shorter bands (Fig. 1B).
LexA is posttranslationally modified.
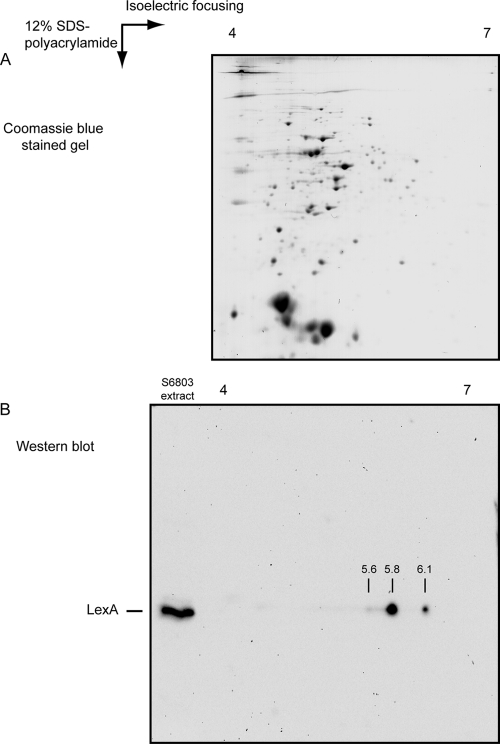
In the SOS response, the first step in the regulatory cascade consists of reducing the levels of functional LexA. Therefore, LexA undergoes autoproteolysis with the help of RecA upon induction of DNA damage, losing its affinity to DNA and consequently ceasing to work as a transcription regulator (53). Thus, whether LexA could also be subjected to posttranslational modifications in Synechocystis sp. strain PCC 6803 developed into an interesting question to examine. Based on the Western blot analyses performed, it was never possible to observe any proteolytic processing of LexA either before or after changing the growth conditions. Thus, the LexA protein was investigated, making use of 2D gels, separating the proteins according to their isoelectric points (pIs) in the first dimension and their molecular mass in the second (Fig. 3). Based on the ExPASy (Expert Protein Analysis System) calculation tool to determine the pI of a given protein (http://expasy.org/tools/pi_tool.html), LexA has an expected pI of 5.84. However, Western blots using anti-LexA antibodies revealed that LexA exists in at least 3 forms with different pIs—one coincides with the theoretical pI (∼5.8), while the other two were estimated to be approximately 5.6 and 6.1 (Fig. 3B). These spots do not seem to result from technical handling of the samples (e.g., carbamylation or deamidation), since the proteins separated by 2D gels and visible by Coomassie blue staining of the gels do not show generalized signs of such modifications (Fig. 3A). However, attempts to identify the differences between the 3 LexA forms failed to produce any results using the product of in-gel trypsin digestions of isolated LexA spots in matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) and tandem mass spectrometry (MS-MS). The reason for this may be the fact that, from these experiments, only 50% or less of the LexA sequence from the obtained peptides could be covered, which is enough for identification but leaves a significant part of the protein unexamined. The low coverage percentage may be explained by LexA's amino acid sequence, which contains 13 arginine and 16 lysine residues, together representing almost 15% of the full sequence. However, considering the minimal difference in size between the various forms of LexA, it may be hypothesized that the functional group(s) attached is rather small. Performing similar 2D runs, followed by Western blot analyses using proteins extracted from cells kept in darkness, produced results comparable to those shown in Fig. 3B (3 spots). Furthermore, no significant differences could be discerned in either the relative levels of the different forms or the estimated pIs (data not shown).
Fig. 3.
LexA in Synechocystis sp. strain PCC 6803 possesses different isoelectric points. (A) Coomassie blue-stained 2D gel with approximately 40 μg total protein from Synechocystis sp. strain PCC 6803 grown photoautotrophically in BG11 supplemented with 10 mM HEPES, pH 7.5, and bubbled with air at 25°C. As the first step, proteins were separated according to their pIs, using a strip with a pH gradient between 4 and 7, which was followed by the second separation on a 12% SDS-polyacrylamide gel. (B) Western blot analysis of the multiple LexA forms. A 2D separation of proteins was performed as shown in panel A prior to the Western blotting. On the same SDS-polyacrylamide gel, 5 μg total protein extract was loaded and separated on the left side of the gel as a control of the position of LexA. Using this approach, at least three different forms of LexA could be detected, with pIs of approximately 5.6, 5.8, and 6.1.
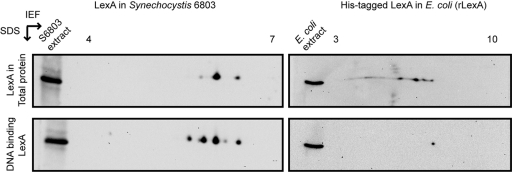
Following these results, experiments that could clarify which of these forms of LexA can interact with DNA were performed, making use of DNA affinity assays. The DNA fragment Shoxpr (covering part of the bidirectional hydrogenase promoter [33, 34]) was used to recover proteins that can interact with it, including LexA, as previously described (33). The Shoxpr-interacting proteins were further separated on 2D gels, and LexA was analyzed by Western blotting. Unexpectedly, 5 spots could now be observed (lower left panel in Fig. 4): 3 of these have apparent pIs that correspond to those of the 3 forms previously identified (Fig. 3B and upper left panel in Fig. 4), while 2 new spots could now be discerned, with apparent pIs of 5.4 and 5.9. The additional spots may be forms of LexA that are present at relatively low levels and that cannot be identified in the total extract using the method described here. However, by enriching the total amount of LexA through DNA affinity assays, they become detectable. Alternatively, these forms of LexA may also result from the handling of the samples. To rule out the latter possibility, rLexA (theoretical pI, 6.72) overexpressed in and purified from E. coli was used in similar experiments. After separation of the E. coli protein extracts expressing rLexA, Western blot analyses were carried out with THE Anti-His monoclonal antibody (GenScript Corporation), which is specific for recognizing the histidine tag. This was chosen over the anti-LexA antibody to avoid possible difficulties in the interpretation of the results, which could arise from the cross-reaction of the anti-LexA antibody with the E. coli native LexA. Four spots were identified (upper right panel in Fig. 4), while rLexA interacting with Shoxpr in DNA affinity assays appears as only one form (lower right panel in Fig. 4). These results indicate that (i) at least some modifications performed by E. coli on rLexA are different from the ones that LexA experiences in Synechocystis sp. strain PCC 6803, since not all forms of rLexA could bind to Shoxpr, and (ii) since both LexA and rLexA were extracted from Synechocystis sp. strain PCC 6803 and E. coli, respectively, with the same buffer and conditions and after being handled in similar manners throughout the various steps of the analysis, the method itself does not introduce modifications.
Fig. 4.
Isoelectric points of LexA and rLexA, both from total cell extracts and recovered from DNA affinity assays. Total protein extracts from Synechocystis sp. strain PCC 6803 and E. coli expressing rLexA were separated on 2D gels (top). Western blot analyses of LexA (top left) and rLexA (top right) were carried out using anti-LexA and anti-histidine antibodies, respectively. DNA affinity assays using the DNA fragment Shoxpr were carried out to recover LexA and rLexA with DNA binding capability and separated on 2D gels (bottom). Western blot analyses of LexA (bottom left) and rLexA (bottom right) were carried out using anti-LexA and anti-histidine antibodies, respectively. Five micrograms of total protein extract (either of Synechocystis sp. strain PCC 6803 [left] or of E. coli expressing rLexA [right]) was loaded and separated on the left side of each gel as a control of the position of LexA. IEF, isoelectric focusing.
The SFM04 mutant is fully segregated.
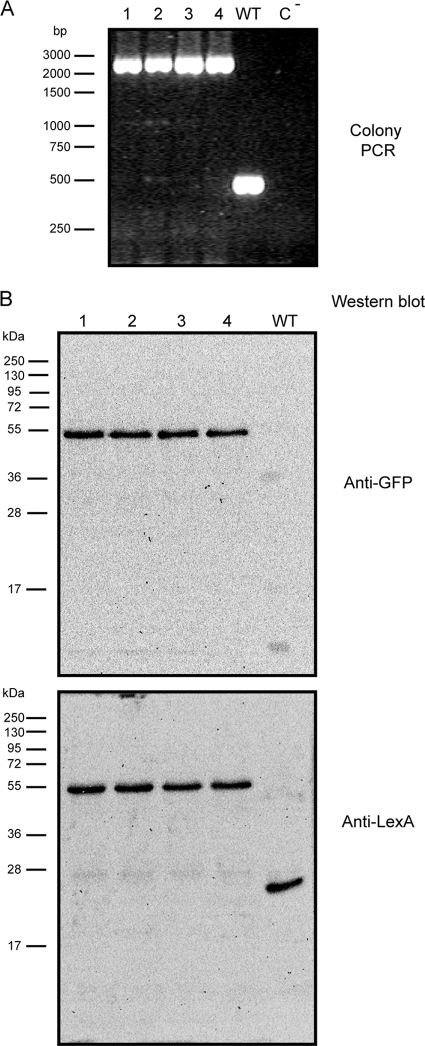
To image the spatial distribution of LexA in live cells of Synechocystis sp. strain PCC 6803, LexA was fused with GFP and its fluorescence was further monitored. This noninvasive method has been used successfully in this cyanobacterium to assess the localization of other proteins (1, 7, 22, 30, 50, 56). The LexA::GFP mutant was also created with the goal of understanding whether its localization could shed more light on the mechanisms of transcriptional regulation. The genetic modifications introduced to produce such a chimeric protein in Synechocystis sp. strain PCC 6803 were targeted to the lexA locus (Fig. 2B). The objective was to completely replace the wild-type LexA by LexA fused to GFP. Transformation of Synechocystis sp. strain PCC 6803 with the plasmid pSPO1342 (see Materials and Methods) resulted in several colonies, some of which were further spread on plates with increasing antibiotic concentrations. After a few rounds of selection, colony PCR was performed to analyze the extent of chromosome segregation. Figure 5 A depicts the analysis of 4 different colonies, showing that all of them became fully segregated. To further support these initial observations, Western blots were carried out with protein samples extracted from the above-mentioned transformants. Using two different antibodies, anti-GFP and anti-LexA, it was possible to confirm that LexA is indeed fused to GFP and that no wild-type LexA remains in the transformed cells (Fig. 5B). The transformant chosen for further experiments was named SFM04.
Fig. 5.
The mutant SFM04 is fully segregated. (A) PCR analysis of 4 different colonies (1 to 4) transformed with the vector pSPO1342 (see Materials and Methods) and the wild-type Synechocystis sp. strain PCC 6803 (WT). A control using water as a template (C−) is also shown. The oligonucleotides SlexAF2 and slr1735R2 (Fig. 2) were used to prime the reaction. Different sizes of the 1-kb marker from Fermentas are shown on the left. (B) Western blot analyses of protein samples obtained from transformants 1 to 4 and wild-type cells using an anti-GFP antibody (top) and the anti-LexA antibody (bottom) are shown. The molecular masses of the Fermentas protein marker are indicated on the left.
The fusion protein LexA::GFP retains DNA binding capability.
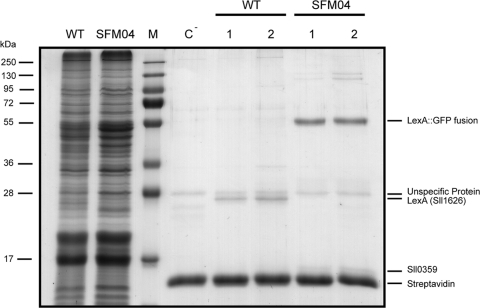
Various attempts have been made to completely knock out lexA in Synechocystis sp. strain PCC 6803; however, as described in the literature, it has never been possible to produce such a mutant, which has led to the assumption that LexA plays a vital role in the cyanobacterium (8). The mutant SFM04 is therefore the first fully segregated mutant of LexA ever to be reported, even though its design assumes that LexA should retain its activity, i.e., interacting with DNA and working as a transcription regulator. Consequently, the fact that SFM04 could be fully segregated was per se an indication that LexA::GFP was still working as the wild-type LexA. However, to further verify this possibility, DNA affinity assays were carried out: the DNA fragment Shoxpr was used to recover proteins that can interact with it. Protein samples from both wild-type Synechocystis sp. strain PCC 6803 and SFM04 were used. Figure 6 shows that, as previously described (33), LexA could be recovered when proteins extracted from wild-type cells were used; alternatively, when proteins from SFM04 were used, no band could be discerned where LexA was expected (additionally confirming the full segregation of SFM04's chromosome), but instead, a band was found with a molecular mass of approximately 53 kDa. This was further analyzed by mass spectrometry and identified as LexA and GFP. These results indicated that the fusion protein LexA::GFP possesses DNA binding capability.
Fig. 6.
The LexA::GFP fusion protein retains its DNA binding capacity. The Coomassie-stained SDS-polyacrylamide gel shows the results of DNA affinity assays performed with proteins from both the wild-type Synechocystis sp. strain PCC 6803 (WT) and the SFM04 mutant strain, using part of the hox promoter as bait. Lanes: WT, 15 μg total protein of Synechocystis sp. strain PCC 6803 wild type; SFM04, 15 μg total protein of the mutant strain SFM04; M, Fermentas protein marker (the molecular masses are indicated on the left); C−, DNA affinity assay performed with beads without DNA (negative control) and 600 μg total protein from SFM04; WT-1 and WT-2, DNA affinity assays performed with beads covered with the DNA fragment Shoxpr and 150 μg and 600 μg of total protein from wild-type Synechocystis sp. strain PCC 6803, respectively; SFM04-1 and SFM04-2, DNA affinity assays performed with beads covered with the DNA fragment Shoxpr and 150 μg and 600 μg of total protein from the mutant strain SFM04, respectively. The identities of some of the peptides are given on the right.
LexA::GFP is predominantly localized in the cytoplasm.
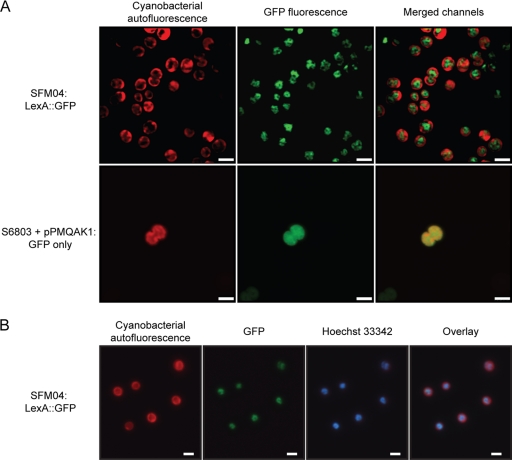
Confocal microscopy analyses of the strain SFM04 revealed that the signal from the LexA::GFP fusion protein is localized in the cytoplasm (Fig. 7), which was expected, given that LexA is predicted to be a soluble protein. However, the GFP signal could not be detected throughout the whole cytoplasm, but instead, it was confined to the most inner and central region of the cytoplasm (Fig. 7). In order to understand whether this distribution was significant and specific to this chimeric protein, a previously described strain of Synechocystis sp. strain PCC 6803 (19) expressing GFP under the control of the trc promoter (containing two operators for enhanced repression) was used as a control. In this strain, the GFP signal could be found evenly distributed in the cytoplasm, both in its central region and in the periphery, where the thylakoid membranes are located. This confirms that LexA::GFP is specifically and predominantly localized in the core region of the cytoplasm, with its diffusion toward outer regions of the cell rather restricted (Fig. 7A).
Fig. 7.
LexA::GFP in SFM04 is located in the core region of the cytoplasm in an evenly distributed pattern. (A) Confocal micrographs were obtained from cells of the strain SFM04 (possessing the LexA::GFP fusion protein) (top row) and of Synechocystis sp. strain PCC 6803 harboring the self-replicative plasmid pPMQAK1, which contains gfp expressed under the regulation of a modified trc promoter (19) (bottom row). The cyanobacterial autofluorescence is depicted in the left column, while the collected GFP signal is shown in the middle column. The result of merging the signals from both channels (cyanobacterial autofluorescence and GFP) is shown in the right column. Size bars, 2.5 μm. (B) Fluorescence micrographs were acquired from SFM04 cells, specifically analyzing the cyanobacterial autofluorescence, the LexA::GFP signal, and the fluorescence of the DNA-staining dye Hoechst 33342. An overlay of the different micrographs is shown on the right. Size bars, 2 μm.
Given the distribution of the DNA in Synechocystis sp. strain PCC 6803 analyzed by DAPI (4′,6-diamidino-2-phenylindole) staining as previously described (39) and examining the LexA::GFP signal reported in the present work, it is striking that they are similar. Therefore, it is plausible that LexA is mainly found associated with DNA. To test that possibility, the dye Hoechst 33342 was used to stain the DNA in SFM04 cells, and its distribution was compared to that of the LexA::GFP signal. The resulting micrographs show that the LexA::GFP signal does colocalize with the fluorescence signal from Hoechst 33342, confirming that this fusion protein is found associated with or in the near vicinity of the DNA (Fig. 7B). It is also possible to observe that LexA::GFP colocalizes in its full extent with the stained DNA, i.e., the localization of LexA::GFP does not seem to be preferentially in a particular region of the DNA. Furthermore, it became evident that the LexA::GFP distribution is homogeneous, i.e., no concentrated spots with stronger fluorescence could be detected. With other DNA binding proteins, e.g., RNA polymerase and the transcription elongation factors NusA, NusB, and NusG in Bacillus subtilis (26) or the response regulator OmpR in E. coli (5), it is possible to distinguish specific regions (transcription foci) where the tagged proteins preferentially concentrate. Instead, the LexA::GFP signal characterized here possesses a “hank shape” (Fig. 7; see Fig. S2 in the supplemental material), as if decorating the DNA, as opposed to the strain with GFP only, where the signal is even and smooth.
In the studies where LexA has been identified in membrane fractions (23, 45, 54, 58), when stated, Synechocystis sp. strain PCC 6803 wild-type cells were grown photoautotrophically in BG11 or AA/8 medium at 30°C, with continuous irradiance at intensities varying between 30 and 60 μmol of photons m−2 s−1. Bubbling, when performed, was carried out with air. In our initial experimental setup (Fig. 7), SFM04 was grown photoautotrophically in BG11 at 25°C, with a continuous irradiance of 40 μmol of photons m−2 s−1 without bubbling (see Materials and Methods). To address the question of whether LexA::GFP could be differently localized under other conditions, experiments were carried out with SFM04 cells, growing them at different temperature (25°C and 30°C), with air bubbling, and by exposing them to high light (300 μmol of photons m−2 s−1). Under all conditions tested, the GFP signal was monitored as described above and was consistently found in the centermost region of the cytoplasm (data not shown). In addition, when SFM04 cells were subjected to 3 h of darkness, the signal of LexA::GFP was still distributed in the same fashion as cells that had been growing in continuous light (data not shown). At least in the latter case, this suggests that no major changes occur in LexA's spatial distribution under some conditions that elicit an increase of lexA transcription.
DISCUSSION
The transcription factor LexA in the cyanobacterium Synechocystis sp. strain PCC 6803 has been suggested to regulate several genes in the most varied regulatory networks. LexA binding regions have been described in the promoters of lexA itself (36) and the hox (15, 33), the crhR (35, 36), and the sbtA (27) genes. More recently, Patterson-Fortin and Owttrim (36) presented evidence that LexA binds as a dimer to 12-bp direct repeats containing a CTA-N9-CTA sequence. Therefore, given that LexA in Synechocystis sp. strain PCC 6803 has different functions and controlling mechanisms than the LexA-mediated SOS response in E. coli, it represents an interesting model protein to understand how molecular evolution can take place in transcription factors and transcriptional networks.
Despite all the efforts to clarify the role of LexA in Synechocystis sp. strain PCC 6803, it is still not known how the overall regulation is operated, i.e., how the signal transduction pathways occur. The environmental cues that trigger LexA to up- or downregulate its regulon genes, the molecular mechanisms for perceiving the environmental change and transferring the corresponding signal and the identities of the LexA gene targets remain largely uncharacterized.
In the current work, we present novel results that show that LexA is regulated on a posttranscriptional level, in addition to the classical transcriptional regulation. In fact, the lexA transcript has been shown to respond to various environmental and growth conditions; high light (18), darkness, cold, the presence of glucose in the medium (35), and anaerobiosis (20, 47) are just a few examples. Our results suggest that simply studying the transcriptional response of lexA to different environmental cues does not necessarily provide enough information about the levels of LexA in the cell and, consequently, what sort of regulation this transcription regulator is exerting on the downstream genes. However, the mechanisms by which the cell is able to regulate the translation of the lexA transcript to protein remain unknown. Among the various possibilities that could account for the observed results are riboswitches, translation efficiency, antisense RNA, and transcript stability. This is a subject that definitely deserves further investigation, given the strong implications for an understanding of the overall LexA regulation.
In addition, here, we present evidence that the LexA protein is modified on a posttranslational level, although in a different manner than what has been described for E. coli LexA. We were not able to identify any LexA proteolytic degradation products under any of the tested conditions, in contrast to what was observed with LexA from Anabaena sp. strain PCC 7120 (P. Oliveira and P. Lindblad, unpublished results) and in agreement with what has been reported by Yano and coworkers (57). Instead, we identified different isoelectric forms of the protein. Other cyanobacterial transcription factors and signaling proteins have been shown to be modified after translation, as well, such as PII (phosphorylation) (10), the KaiC protein (phosphorylation) (49), and the AbrB-like protein in Aphanizomenon ovalisporum (N-acetylation and methylation) (40). Particularly in the last example, Shalev-Malul and coworkers identified differences between the native AbrB-like protein isolated from the cyanobacterium and the recombinant protein produced in and purified from E. coli (40). It was suggested by the authors of that work that the posttranslational modifications may affect the structural organization of the protein and thus its affinity for DNA (40). In this work, we also provide evidence of differences between the LexA protein isolated from Synechocystis sp. strain PCC 6803 and the LexA protein overexpressed and purified from E. coli (rLexA); furthermore, we show that these differences do affect the binding properties of the protein (Fig. 4). Even though we were not able to uncover the chemical nature of the posttranslational modifications that LexA is subjected to in Synechocystis sp. strain PCC 6803 in the present work, there are indications of how LexA may be modified. The soluble protein ferredoxin is the primary electron acceptor at the end of the photosynthetic electron transport chain known to directly deliver electrons to a wide range of proteins involved in various metabolic and signaling mechanisms (16). A recent screen for ferredoxin electron transfer partners in Synechocystis sp. strain PCC 6803 identified LexA as one of the interacting partners whose interaction was shown to be redox dependent (16). Hanke et al. (16) showed that LexA could interact with reduced ferredoxin, but not when it was in its oxidized form, further suggesting that the redox state of ferredoxin may play a role in regulating LexA-dependent transcriptional repression and activation (16). However, more investigations need to be carried out to support this hypothesis and to reveal the identity of LexA's modifications.
Another aspect whose study was justified was the localization of LexA in Synechocystis sp. strain PCC 6803, not only to address the question of its subcellular positioning, but also to understand whether its localization could shed light on its mechanisms of regulation. In this work, we report that a LexA::GFP protein fusion is mainly located in the inner region of the cytoplasm, as monitored by GFP fluorescence. These results suggest that the identification of LexA in different membrane fractions reported in various proteomic studies (23, 45, 54, 58) is most likely the result of DNA-contaminated membrane fractions. On the other hand, LexA was found to be evenly distributed on the DNA. A bioinformatics analysis of the genome sequence of Synechocystis sp. strain PCC 6803 revealed that the sequence CTA-N9-CTA (LexA has been reported to bind to 12-bp direct repeats containing this motif [36]) is present 1,389 times. It is highly unlikely that all of these correspond to genuine binding sites, but it points to the possibility that LexA has multiple binding sites in the genome of Synechocystis sp. strain PCC 6803. In agreement with this, multiple LexA binding sites have been bioinformatically predicted in the genome of the cyanobacterium Anabaena sp. strain PCC 7120 (43). This could mean that LexA has a widespread distribution on the DNA, and it could account for the evenly distributed GFP signal.
However, the question remains, how does LexA regulate the transcription of the genes within its regulatory network? The DNA binding functions of many transcription factors are regulated by multiple and varied mechanisms: small-molecule effectors referred to as corepressors and inducers are one of those mechanisms (6). Classical examples of these allosteric transcription factors are the lactose and purine repressor proteins (28). Binding of small molecules to the respective proteins modulates their affinities for their target sites in transcription initiation regions, as a consequence of an alteration in the orientation of the DNA binding molecules (6). In contrast to these allosteric transcription factors, the DNA binding properties of a number of transcription-regulatory proteins are modulated by small ligands via their effects on the self-assembly of the protein (6). In these systems, the small molecule functions in altering DNA binding affinity by modulating the self-assembly of the transcription regulator (6). In addition to small-ligand binding, transcription factor assembly can also be influenced by a number of other processes. First, posttranslational modification may promote either assembly or dissociation of an oligomeric transcription factor or even affect its DNA binding capacity, as mentioned above. Second, DNA binding can enhance the assembly of homo-oligomeric and hetero-oligomeric transcription-regulatory complexes. Finally, other protein-protein interactions may inhibit or enhance the assembly of transcription factors, thereby altering their functions in transcription initiation (for a review, see reference 6). In cyanobacteria, the reported examples of such transcription factor DNA binding regulation are scarce. Still, it is known that 2-oxoglutarate increases the DNA binding affinity of the global nitrogen regulator NtcA to its various target sites (51). In addition, Espinosa et al. (9) reported that PipX interacts with both PII and NtcA, providing a mechanistic link between the two factors. Moreover, the authors showed that PipX is required for NtcA-dependent transcriptional activation, implying that it may function as a prokaryotic transcriptional coactivator (9). Given all these possible ways by which the cell can control the DNA binding function of a transcription factor, it becomes quite possible that one or more of these mechanisms is crucial for LexA regulation.
To the best of our knowledge, this is the first time that a cyanobacterial transcription regulator has been reported to possess different levels of regulation (transcriptional, posttranscriptional, and posttranslational) and has been characterized in terms of its subcellular localization. Consequently, this study opens up the possibility for further investigations. How is the lexA transcript controlled to tune the LexA levels in the cell? What are the modifications LexA undergoes, and what is their physiological importance? Why is LexA so homogeneously distributed on the DNA? How can all these possible regulatory points be coordinated to produce a transcriptional response of the downstream genes? Based on these data, further investigations will definitely clarify the role of the LexA-related protein in Synechocystis sp. strain PCC 6803 and uncover the molecular mechanisms by which different environmental cues trigger a transcriptional response via LexA.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniel Camsund for the initial steps of the anti-LexA antibody production, Åsa Söderberg for the LexA localization studies in Synechocystis protein fractions, and Saw Yen Ow and Åke Engström for their technical help in trying to identify LexA's posttranslational modifications. We also thank the reviewers for improving the quality of the paper.
The work was supported by the Swedish Energy Agency; the Knut and Alice Wallenberg Foundation; the Royal Norwegian Embassy in New Delhi, India (project BioCO2); and the EU/Energy FP7 project SOLAR-H2 (contract 212508).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Aldridge C., Spence E., Kirkilionis M. A., Frigerio L., Robinson C. 2008. Tat-dependent targeting of Rieske iron-sulphur proteins to both the plasma and thylakoid membranes in the cyanobacterium Synechocystis PCC6803. Mol. Microbiol. 70:140–150 [DOI] [PubMed] [Google Scholar]
- 2. Antal T. K., Oliveira P., Lindblad P. 2006. The bidirectional hydrogenase in the cyanobacterium Synechocystis sp. strain PCC 6803. Int. J. Hydrogen Energ. 31:1439–1444 [Google Scholar]
- 3. Argueta C., Yuksek K., Summers M. 2004. Construction and use of GFP reporter vectors for analysis of cell-type-specific gene expression in Nostoc punctiforme. J. Microbiol. Methods 59:181–188 [DOI] [PubMed] [Google Scholar]
- 4. Axelsson R., Lindblad P. 2002. Transcriptional regulation of Nostoc hydrogenases: effects of oxygen, hydrogen, and nickel. Appl. Environ. Microbiol. 68:444–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Batchelor E., Goulian M. 2006. Imaging OmpR localization in Escherichia coli. Mol. Microbiol. 59:1767–1778 [DOI] [PubMed] [Google Scholar]
- 6. Beckett D. 2001. Regulated assembly of transcription factors and control of transcription initiation. J. Mol. Biol. 314:335–352 [DOI] [PubMed] [Google Scholar]
- 7. Birungi M., et al. 2010. Possibilities of subunit localization with fluorescent protein tags and electron microscopy exemplified by a cyanobacterial NDH-1 study. Biochim. Biophys. Acta 1797:1681–1686 [DOI] [PubMed] [Google Scholar]
- 8. Domain F., Houot L., Chauvat F., Cassier-Chauvat C. 2004. Function and regulation of the cyanobacterial genes lexA, recA, and ruvB: LexA is critical to the survival of cells facing inorganic carbon starvation. Mol. Microbiol. 53:65–80 [DOI] [PubMed] [Google Scholar]
- 9. Espinosa J., Forchhammer K., Burillo S., Contreras A. 2006. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 61:457–469 [DOI] [PubMed] [Google Scholar]
- 10. Forchhammer K., Tandeau de Marsac N. 1995. Functional analysis of the phosphoprotein PII (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 177:2033–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fulda S., et al. 2006. Proteome analysis of salt stress response in the cyanobacterium Synechocystis sp. strain PCC 6803. Proteomics 6:2733–2745 [DOI] [PubMed] [Google Scholar]
- 12. Gan C. S., Reardon K. F., Wright P. C. 2005. Comparison of protein and peptide prefractionation methods for the shotgun proteomic analysis of Synechocystis sp. PCC 6803. Proteomics 5:2468–2478 [DOI] [PubMed] [Google Scholar]
- 13. Gao Y., et al. 2009. Identification of the proteomic changes in Synechocystis sp. PCC 6803 following prolonged UV-B irradiation. J. Exp. Bot. 60:1141–1154 [DOI] [PubMed] [Google Scholar]
- 14. Georg J., et al. 2009. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol. Syst. Biol. 5:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gutekunst K., et al. 2005. LexA regulates the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803 as a transcription activator. Mol. Microbiol. 58:810–823 [DOI] [PubMed] [Google Scholar]
- 16. Hanke G. T., Satomi Y., Shinmura K., Takao T., Hase T. 2011. A screen for potential ferredoxin electron transfer partners uncovers new, redox dependent interactions. Biochim. Biophys. Acta 1814:366–374 [DOI] [PubMed] [Google Scholar]
- 17. Hernández J. A., et al. 2006. Identification of a furA cis antisense RNA in the cyanobacterium Anabaena sp. PCC 7120. J. Mol. Biol. 355:325–334 [DOI] [PubMed] [Google Scholar]
- 18. Hihara Y., Kamei A., Kanehisa M., Kaplan A., Ikeuchi M. 2001. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13:793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang H.-H., Camsund D., Lindblad P., Heidorn T. 2010. Design and characterization of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 38:2577–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kiss È., Kós P. B., Vass I. 2009. Transcriptional regulation of the bidirectional hydrogenase in the cyanobacterium Synechocystis 6803. J. Biotechnol. 142:31–37 [DOI] [PubMed] [Google Scholar]
- 21. Knoll A. H. 2008. Cyanobacteria and earth history, p. 1–19 In Herrero A., Flores E. (ed.), The cyanobacteria: molecular biology, genomics and evolution. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 22. Komenda J., et al. 2006. The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis sp. PCC 6803. J. Biol. Chem. 281:1145–1151 [DOI] [PubMed] [Google Scholar]
- 23. Kubota H., et al. 2010. Purification and characterization of photosystem I complex from Synechocystis sp. PCC 6803 by expressing histidine-tagged subunits. Biochim. Biophys. Acta 1797:98–105 [DOI] [PubMed] [Google Scholar]
- 24. Kufryk G. I., Sachet M., Schmetterer G., Vermaas W. F. J. 2002. Transformation of the cyanobacterium Synechocystis sp. PCC 6803 as a tool for genetic mapping: optimization of efficiency. FEMS Microbiol. Lett. 206:215–219 [DOI] [PubMed] [Google Scholar]
- 25. Kurian D., Jansèn T., Mäenpää P. 2006. Proteomic analysis of heterotrophy in Synechocystis sp. PCC 6803. Proteomics 6:1483–1494 [DOI] [PubMed] [Google Scholar]
- 26. Lewis P. J., Doherty G. P., Clarke J. 2008. Transcription factor dynamics. Microbiology 154:1837–1844 [DOI] [PubMed] [Google Scholar]
- 27. Lieman-Hurwitz J., et al. 2009. A cyanobacterial AbrB-like protein affects the apparent photosynthetic affinity for CO2 by modulating low-CO2-induced gene expression. Environ. Microbiol. 11:927–936 [DOI] [PubMed] [Google Scholar]
- 28. Matthews K. S., Falcon C. M., Swint-Kruse L. 2000. Relieving repression. Nat. Struct. Mol. Biol. 7:184–187 [DOI] [PubMed] [Google Scholar]
- 29. Mazón G., et al. 2004. LexA-binding sequences in Gram-positive and cyanobacteria are closely related. Mol. Genet. Genomics 271:40–49 [DOI] [PubMed] [Google Scholar]
- 30. Mazouni K., Domain F., Cassier-Chauvat C., Chauvat F. 2004. Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN and MinCDE. Mol. Microbiol. 52:1145–1158 [DOI] [PubMed] [Google Scholar]
- 31. Mitschke J., et al. 2011. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U. S. A. 108:2124–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oliveira P., Leitão E., Tamagnini P., Moradas-Ferreira P., Oxelfelt F. 2004. Characterization and transcriptional analysis of hupSLW in Gloeothece sp. ATCC 27152: an uptake hydrogenase from a unicellular cyanobacterium. Microbiology 150:3647–3655 [DOI] [PubMed] [Google Scholar]
- 33. Oliveira P., Lindblad P. 2005. LexA, a transcription regulator binding in the promoter region of the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 251:59–66 [DOI] [PubMed] [Google Scholar]
- 34. Oliveira P., Lindblad P. 2008. An AbrB-like protein regulates the expression of the bidirectional hydrogenase in Synechocystis sp. strain PCC 6803. J. Bacteriol. 190:1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patterson-Fortin L. M., Colvin K. R., Owttrim G. W. 2006. A LexA-related protein regulates redox-sensitive expression of the cyanobacterial RNA helicase, crhR. Nucleic Acids Res. 34:3446–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patterson-Fortin L. M., Owttrim G. W. 2008. A Synechocystis LexA-orthologue binds direct repeats in target genes. FEBS Lett. 582:2424–2430 [DOI] [PubMed] [Google Scholar]
- 37. Prakash J. S. S., et al. 2009. DNA supercoiling regulates the stress-inducible expression of genes in the cyanobacterium Synechocystis. Mol. Biosyst. 5:1904–1912 [DOI] [PubMed] [Google Scholar]
- 38. Rott R., Zipor G., Portnoy V., Liveanu V., Schuster G. 2003. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli. J. Biol. Chem. 278:15771–15777 [DOI] [PubMed] [Google Scholar]
- 39. Schneider D., Fuhrmann E., Scholz I., Hess W., Graumann P. 2007. Fluorescence staining of live cyanobacterial cells suggest non-stringent chromosome segregation and absence of a connection between cytoplasmic and thylakoid membranes. BMC Cell Biol. 8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shalev-Malul G., et al. 2008. An AbrB-like protein might be involved in the regulation of cylindrospermopsin production by Aphanizomenon ovalisporum. Environ. Microbiol. 10:988–999 [DOI] [PubMed] [Google Scholar]
- 41. Shin B.-J., et al. 2008. Cyanobacterial hybrid kinase Sll0043 regulates phototaxis by suppressing pilin and twitching motility protein. J. Microbiol. 46:300–308 [DOI] [PubMed] [Google Scholar]
- 42. Singh A. K., et al. 2008. Integration of carbon and nitrogen metabolism with energy production is crucial to light acclimation in the cyanobacterium Synechocystis. Plant Physiol. 148:467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sjöholm J., Oliveira P., Lindblad P. 2007. Transcription and regulation of the bidirectional hydrogenase in the cyanobacterium Nostoc sp. strain PCC 7120. Appl. Environ. Microbiol. 73:5435–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Slabas A. R., Suzuki I., Murata N., Simon W. J., Hall J. J. 2006. Proteomic analysis of the heat shock response in Synechocystis PCC6803 and a thermally tolerant knockout strain lacking the histidine kinase 34 gene. Proteomics 6:845–864 [DOI] [PubMed] [Google Scholar]
- 45. Srivastava R., Pisareva T., Norling B. 2005. Proteomic studies of the thylakoid membrane of Synechocystis sp. PCC 6803. Proteomics 5:4905–4916 [DOI] [PubMed] [Google Scholar]
- 46. Stanier R. Y., Kunisawa R., Mandel M., Cohen-Bazire G. 1971. Purification and properties of unicellular blue-green algae (order Chroococcales). Microbiol. Mol. Biol. Rev. 35:171–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Summerfield T. C., Nagarajan S., Sherman L. 2011. Gene expression under low oxygen conditions in the cyanobacterium Synechocystis sp. PCC 6803 demonstrates Hik31-dependent and Hik31-independent responses. Microbiology 157:301–312 [DOI] [PubMed] [Google Scholar]
- 48. Tamagnini P., Troshina O., Oxelfelt F., Salema R., Lindblad P. 1997. Hydrogenases in Nostoc sp. strain PCC 73102, a strain lacking a bidirectional enzyme. Appl. Environ. Microbiol. 63:1801–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Terauchi K., Kondo T. 2008. The cyanobacterial circadian clock and the KaiC phosphorylation cycle, p. 199–216 In Herrero A., Flores E. (ed.), The cyanobacteria: molecular biology, genomics and evolution. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 50. van Thor J. J., Gruters O. W. M., Matthijs H. C. P., Hellingwerf K. J. 1999. Localization and function of ferredoxin:NADP+ reductase bound to the phycobilisomes of Synechocystis. EMBO J. 18:4128–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vázquez-Bermúdez M. F., Herrero A., Flores E. 2002. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 512:71–74 [DOI] [PubMed] [Google Scholar]
- 52. Vioque A. 1992. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 20:6331–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wagner R. 2000. Transcription regulation in prokaryotes. Oxford University Press, New York, NY [Google Scholar]
- 54. Wang Y., Sun J., Chitnis P. R. 2000. Proteomic study of the peripheral proteins from thylakoid membranes of the cyanobacterium Synechocystis sp. PCC 6803. Electrophoresis 21:1746–1754 [DOI] [PubMed] [Google Scholar]
- 55. Whitton B. A., Potts M. 2000. Introduction to the Cyanobacteria. In Whitton B. A., Potts M. (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 56. Wilson A., et al. 2006. A soluble carotenoid protein involved in phycobilisome-related energy dissipation in cyanobacteria. Plant Cell 18:992–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yano F., et al. 2004. Analysis of LexA protein of two cyanobacteria; Anabaena and Synechocystis. Plant Cell Physiol. 45:s146–s473 [Google Scholar]
- 58. Zhang L.-F., et al. 2009. Proteomic analysis of plasma membranes of cyanobacterium Synechocystis sp. strain PCC 6803 in response to high pH stress. J. Proteome Res. 8:2892–2902 [DOI] [PubMed] [Google Scholar]
- 59. Zhang Z., Pendse N., Phillips K., Cotner J., Khodursky A. 2008. Gene expression patterns of sulfur starvation in Synechocystis sp. PCC 6803. BMC Genomics 9:344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.