Abstract
The RecU protein from Mycoplasma genitalium, RecUMge, is a 19.4-kDa Holliday junction (HJ) resolvase that binds in a nonspecific fashion to HJ substrates and, in the presence of Mn2+, cleaves these substrates at a specific sequence (5′-G/TC↓C/TTA/GG-3′). To identify amino acid residues that are crucial for HJ binding and/or cleavage, we generated a series of 16 deletion mutants (9 N- and 7 C-terminal deletion mutants) and 31 point mutants of RecUMge. The point mutations were introduced at amino acid positions that are highly conserved among bacterial RecU-like sequences. All mutants were purified and tested for the ability to bind to, and cleave, HJ substrates. We found the five N-terminal and three C-terminal amino acid residues of RecUMge to be dispensable for its catalytic activities. Among the 31 point mutants, 7 mutants were found to be inactive in both HJ binding and cleavage. Interestingly, in 12 other mutants, these two activities were uncoupled; while these proteins displayed HJ-binding characteristics similar to those of wild-type RecUMge, they were unable to cleave HJ substrates. Thus, 12 amino acid residues were identified (E11, K31, D57, Y58, Y66, D68, E70, K72, T74, K76, Q88, and L92) that may play either a direct or indirect role in the catalysis of HJ resolution.
INTRODUCTION
Mycoplasma genitalium is a human pathogen that causes a range of urogenital tract infections in both men and women. Like all other members of the bacterial class Mollicutes, M. genitalium has a small genome (580 kb) that contains a limited number of genes (10). In spite of its restricted size, a considerable part of the M. genitalium genome (∼4%) consists of repeated DNA elements (20). These elements, which were termed MgPa repeats or MgPar sequences (19), were also found to be homologous to parts of two adjacent open reading frames within the MgPa operon (mgpB and mgpC, respectively) that encode surface-exposed antigenic proteins (MgPa and P110, respectively) (6, 28). Both MgPa and P110 are associated with the terminal tip structure of M. genitalium, which is responsible for attachment of the bacteria to human cells (2, 8–9, 12). Because the different MgPar sequences are similar but not identical in sequence, it has been hypothesized that these sequences could serve as a source of variation of the mgpB and mgpC genes by means of homologous DNA recombination between the MgPar portions within the genes and MgPar sequences at distal sites in the genome. In turn, the sequence variation within mgpB and mgpC could lead to antigenic variation of the MgPa and P110 proteins at the bacterial surface (7, 10, 14, 19). Indeed, Iverson-Cabral and coworkers reported that the sequence variation that is observed within the mgpB and mgpC genes of an M. genitalium strain can be explained by the occurrence of recombination events among MgPar sequences (14, 15).
Despite the important role that homologous DNA recombination may play in antigenic variation of M. genitalium and thereby in the immune-evasive strategies of the bacterium, little is known about its mechanism and dynamics. In order to try to delineate this mechanism, we initiated studies to identify the bacterial enzymes putatively involved in homologous DNA recombination in M. genitalium, as well as in its closest known relative, i.e., Mycoplasma pneumoniae. Several open reading frames (ORFs) have been identified in both M. genitalium and M. pneumoniae that putatively encode enzymes involved in DNA recombination processes. Among these are the ORFs encoding RuvA and single-stranded DNA-binding protein (SSB) from M. pneumoniae (13, 25) and the RecA and RecU proteins from both M. genitalium and M. pneumoniae (26, 27). While the RecA proteins from these species were found to have similar in vitro activities (27), their RecU proteins (designated RecUMge and RecUMpn, respectively) displayed dramatic differences (26). RecUMge was found to be a potent Holliday junction (HJ)-resolving enzyme (resolvase) that specifically cleaves HJ substrates at the sequence 5′-G/TC↓C/TTA/GG-3′ in the presence of Mn2+ (26). In contrast, RecUMpn, which shares significant sequence similarity with RecUMge (67% identity), did not possess obvious DNA-binding or cleavage activities. This apparent inactivity of RecUMpn appeared to be caused primarily by the presence of a glutamic acid residue at position 67 of the protein, a residue that is not conserved in RecUMge (26). Based on the important role that HJ resolvases generally play in homologous DNA recombination, it was speculated that the relatively low level of homologous DNA recombination that is observed in M. pneumoniae could be due to the inactivity of RecUMpn (26).
To further define the activities of RecUMge and identify crucial amino acids, as well as regions of the protein, here we describe the generation of a series of 16 deletion mutants (including 9 N- and 7 C-terminal deletion mutants) and 31 RecUMge point mutants. The point mutations were introduced at amino acid positions that are highly conserved among bacterial RecU-like sequences. We describe the identification of two groups of debilitating mutations in RecUMge: (i) mutations of amino acid residues that are essential for sequence-specific HJ resolution and (ii) mutations in residues that play a role in both HJ binding and resolution. The data demonstrate that the HJ-binding activity of RecUMge can be separated biochemically from its HJ cleavage activity.
MATERIALS AND METHODS
Generation of plasmid constructs expressing wild-type (wt) and mutant RecUMge.
The cloning of the M. genitalium G37 (ATCC 33530) MG352 gene, which encodes RecUMge, has been described previously (26). Plasmid pMALc-RecUMge was used for the expression of maltose-binding protein (MBP)-fused RecUMge protein. This fusion protein has activities that are indistinguishable from that of the nonfused, native RecUMge (26). The RecUMge mutants that we generated were also expressed as fusions to MBP. The plasmids used for the production of the point mutants, as well as the N- and C-terminal deletion mutants, were derived from plasmid pMALc-RecUMge. The appropriate mutations were introduced into this plasmid using a PCR-based mutagenesis protocol, similar to that used for the replacement of TGA codons (which code for tryptophan in Mycoplasma species) by TGG codons (which code for tryptophan in Escherichia coli) (26, 27). Thus, a set of plasmid constructs was generated expressing 9 N-terminal deletion mutants, 7 C-terminal deletion mutants, and 31 point mutants of RecUMge. The specific mutations that were made are listed in Table 1. The sequences of the mutagenesis primers used for the generation of the expression constructs are available upon request. The integrity of all DNA constructs used in this study was verified by dideoxy sequencing, following standard procedures (27).
Table 1.
RecUMge mutants and their activities
| Mutant | HJ-binding activityb | HJ cleavage activityc | Mutant groupd |
|---|---|---|---|
| Point mutantsa | |||
| N5A | +++ | +++ | A |
| G7A | +++ | + | B |
| M8A | +++ | +++ | A |
| L10A | + | + | B |
| E11A | +++ | − | C |
| N15A | ++ | ++ | A |
| K31A | ++ | − | C |
| V46A | +++ | + | B |
| S54A | ++ | ++ | A |
| D57A | +++ | − | C |
| Y58A | + | − | C |
| G60A | − | − | D |
| Y62A | ++ | +++ | A |
| G64A | ++ | +++ | A |
| Y66A | + | − | C |
| D68A | +++ | − | C |
| F69A | − | − | D |
| E70A | +++ | − | C |
| K72A | + | − | C |
| T74A | + | − | C |
| K76A | + | − | C |
| F79A | − | − | D |
| H87A | +++ | + | B |
| Q88A | +++ | − | C |
| H91A | − | − | D |
| L92A | ++ | − | C |
| G100A | − | − | D |
| F103A | − | − | D |
| F108A | − | − | D |
| D112A | +++ | +++ | A |
| L122A | +++ | ++ | A |
| Deletion mutantse | |||
| N terminal | |||
| NΔ2 | +++ | +++ | |
| NΔ3 | +++ | +++ | |
| NΔ5 | ++ | + | |
| NΔ8 | ++ | − | |
| NΔ11 | − | − | |
| NΔ18 | − | − | |
| NΔ31 | − | − | |
| NΔ54 | − | − | |
| NΔ79 | − | − | |
| C terminal | |||
| CΔ3 | ++ | + | |
| CΔ9 | − | − | |
| CΔ13 | − | − | |
| CΔ19 | − | ||
| CΔ23 | − | − | |
| CΔ35 | − | − | |
| CΔ112 | − | − |
The names of the point mutants indicate the amino acid that was changed, using the one-letter amino acid code, followed by its position within the RecUMge protein and an A, which indicates the alanine into which the original amino acid was changed.
HJ-binding reactions were performed as described in Materials and Methods, using 400 nM protein. The listed activities represent the percentage of activity as opposed to that of wild-type (wt) RecUMge. These activities are defined as follows: −, <5%; +, 5% to 20%; ++, 20% to 70%; +++, >70%.
HJ cleavage reactions were performed as described in Materials and Methods, using 400 nM protein. The percent cleavage (as opposed to wt protein) refers to cleavage at the specific site within substrate HJ 1.1, resulting in a 23-nucleotide product (26). The activities are defined as follows: −, <5%; +, 5% to 20%; ++, 20% to 70%; +++, >70%.
The classification of mutant proteins in groups A to D is explained in the text.
The names of the deletion mutants contain either N, for N terminus, or C, for C terminus, followed by Δ and the number of amino acid residues that were deleted from either of the two RecUMge termini.
Expression and purification of RecUMge-MBP fusion proteins.
The expression of MBP fusion proteins in E. coli BL21(DE3) was performed as follows. Each plasmid-bearing strain was grown overnight at 37°C in LB medium containing 100 μg/ml ampicillin. The cultures were diluted 1:100 in 50 ml LB medium with ampicillin and grown at 37°C to an optical density at 600 nm of 0.5. Protein expression was then induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.3 mM. After 1 h of incubation at 37°C, the bacteria were harvested by centrifugation and stored at −20°C. The frozen bacterial pellets were resuspended in 5 ml of buffer A (20 mM Tris [pH 7.5], 1 mM dithiothreitol [DTT], and 0.1 mM EDTA) containing 1 M NaCl and 0.5 mg/ml of lysozyme. The suspension was sonicated on ice and clarified by centrifugation for 20 min at 12,000 × g (4°C). All subsequent purification steps were carried out either on ice or at 4°C. To the supernatant, which contained the majority of the expressed MBP-fused proteins, 5 ml of buffer A was added. This solution was then mixed with 200 μl amylose resin (New England BioLabs), which was previously equilibrated in buffer B (buffer A plus 0.5 M NaCl). After binding of the MBP-fused proteins to the resin for 1 h (by incubation on a rotary shaker), the resin was washed three times with 1 ml of buffer B and once with 1 ml of buffer C (40 mM Tris [pH 7.5], 2 mM DTT, 0.2 mM EDTA, 0.4 M NaCl). The fusion proteins were then eluted from the amylose resin using 200 μl of buffer C containing 10 mM maltose. The purified protein was mixed with an equal volume of 100% glycerol and stored at −20°C.
DNA-binding assays.
The binding of the wild-type and mutant RecUMge proteins to HJ substrates was performed in 10-μl volumes and included 20 mM Tris-HCl, pH 7.5, 1 mM DTT, 50 ng/μl bovine serum albumin (BSA), 12.3 nM synthetic HJ substrate (HJ 1.1), and various concentrations of protein. Substrate HJ 1.1 (26) is composed of the following four annealed DNA strands: 5′-GCGACGTGATCACCAGATGATTGCTAGGCATGCTTTCCGCAAGAGAAGC-3′ (HJ11), 5′-GGCTTCTCTTGCGGAAAGCATGC↓CTAGCAATCCTGTCAGCTGCATGGAAC-3′ (5′ 6-carboxyfluorescein [FAM] labeled; HJ12), 5′-GGTTCCATGCAGCTGACAGGATTGCTAGGCTCAAGGCGAACTGCTAACGG-3′ (HJ13), and 5′-ACCGTTAGCAGTTCGCCTTGAGC↓CTAGCAATCATCTGGTGATCACGTCGC-3′ (HJ14). In these oligonucleotides, the mobile core is indicated in boldface. The RecUMge cleavage site (in strands HJ12 and HJ14) is indicated with a downward-pointing arrow. After incubation for 20 min at room temperature, 1 μl of loading dye (40% glycerol, 0.25% bromophenol blue) was added, followed by electrophoresis of the reaction mixtures through 6% polyacrylamide gels in 1× TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA). The FAM-labeled DNA was then visualized by fluorometry using a Typhoon Trio 9200 Variable Mode Imager (GE Healthcare). Digital images were recorded using Typhoon Scanner Control v4.0 software (Amersham Bioscience) and processed (visualized and quantified) using Quantity One 1-D Analysis Software (Bio-Rad). The binding activity of the wt and mutant proteins was measured as a percentage of bound DNA. The relative HJ-binding activity of each protein was expressed as a percentage of the binding activity of wild-type RecUMge.
HJ resolution assays.
Standard HJ resolution assays (10 μl) were performed with 12.3 nM HJ 1.1 (FAM labeled; see above) and (mutants of) RecUMge (at various concentrations) in the presence of 20 mM Tris-HCl, pH 7.5, 1 mM DTT, 50 ng/μl BSA, and 5 mM MnCl2. After incubation for 15 min at 37°C, the reactions were terminated by the addition of 10 μl formamide loading dye (95% formamide, 0.05% bromophenol blue). Then, the samples were incubated for 3 min at 80°C, and the samples were loaded on 15% polyacrylamide gels containing 8 M urea and 1× TBE. Following electrophoresis, the gels were analyzed by fluorometry using the Typhoon Trio imager as described above for the DNA-binding assays. The cleavage (or resolution) activities of the wt and mutant proteins, i.e., the generation of a specific 23-nucleotide resolution product (26), was measured as the percentage of cleaved product as opposed to the total HJ substrate in the reaction. The relative HJ cleavage activity of each protein was expressed as a percentage of the activity of wild-type RecUMge.
Protein structure prediction.
Three-dimensional (3D) models of RecUMge were constructed using the protein homology/analogy recognition engine Phyre (16; http://www.sbg.bio.ic.ac.uk/∼phyre/). Using this engine, the lowest E values (3.4e−22 and 3e−21), with 100% estimated precision, were generated with the structures of the RecU proteins of Bacillus stearothermophilus and Bacillus subtilis, respectively. The generated Protein Data Bank (PDB) files were analyzed using DeepView/Swiss-PdbViewer v4.0.1 (11; http://www.expasy.org/spdbv/).
RESULTS
Generation and purification of point mutants of RecUMge.
A series of 31 point mutants of the M. genitalium RecUMge protein were expressed in and purified from E. coli. The amino acids that were targeted for mutagenesis included 18 amino acid residues that are invariable among RecU-like sequences from members of the bacterial class Mollicutes, as well as from B. subtilis; these residues are indicated by black dots below the multiple RecU sequence alignment in Fig. 1A. The other 13 residues that were targeted are also highly conserved among this group of sequences, albeit not completely.
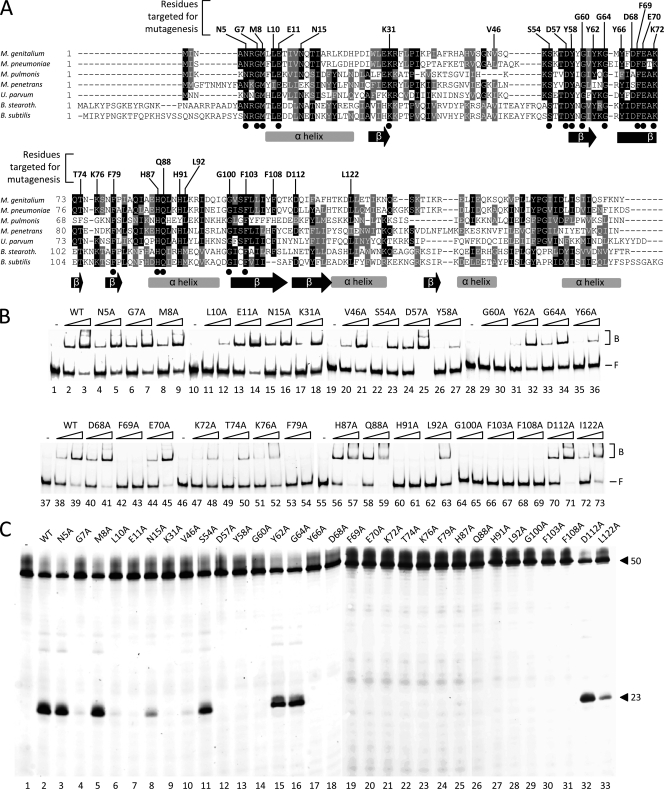
Fig. 1.
Multiple alignment of RecU sequences and activities of single point mutants of RecUMge. (A) A multiple alignment was generated with amino acid sequences predicted to be encoded by the following ORFs (with GenBank accession numbers in parentheses), M. genitalium G37 MG352 (Q49422-1), M. pneumoniae MAC MPN528a, Mycoplasma pulmonis recU (Q98RC3), Mycoplasma penetrans recU (Q8EWG0), Ureaplasma parvum recU (Q9PQJ4), B. stearothermophilus recU (Q5KXY4), and B. subtilis recU (P39792-1). The amino acid residues indicated above the sequences represent the RecUMge residues that were targeted by site-directed mutagenesis. The dots below the sequences indicate residues that are completely conserved among the RecU sequences. The gray bars and black arrows below the sequences represent α-helices and β-sheets, respectively, as determined previously for the RecUBsu protein (18). The multiple alignment was performed using Clustal W (http://www.ebi.ac.uk/Tools/clustalw/index.html). The program BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html) was used to generate white letters on black boxes (for residues that are identical in at least four out of seven sequences) and white letters on gray boxes (for similar residues). (B) HJ DNA-binding activities of point mutants of RecUMge. The binding of the proteins to the FAM-labeled HJ substrate HJ 1.1 was performed as indicated in Materials and Methods. In short, reactions were performed in volumes of 10 μl and contained 12.3 nM DNA substrate and either 0 nM protein (the lanes marked −) or two different concentrations of protein (100 nM or 400 nM), as indicated above the lanes. The protein-DNA mixtures were electrophoresed through native 6% polyacrylamide gels and visualized by fluorometry. The positions of the free, unbound DNA substrates in the gels are indicated by F on the right; the protein-bound substrates are indicated by B. (C) Holliday junction resolution by point mutants of RecUMge. The resolution reactions were performed as described in Materials and Methods, using the FAM-labeled HJ substrate HJ 1.1 and 400 nM protein. After the reaction (15 min at 37°C), the samples were analyzed by denaturing 15% polyacrylamide gel electrophoresis, followed by fluorometry. The lengths of the FAM-labeled strands from the DNA substrate (HJ12; 50 nucleotides) and the specific, major product from cleavage of the substrate by the mutants of RecUMge (23 nucleotides [26]) are indicated on the right. The names of the mutant proteins are shown above the lanes.
In each mutant protein, the targeted amino acid residue was changed into a (nonpolar) alanine residue. This alteration was incorporated in the nomenclature of the mutants (Table 1), in which the original amino acid is indicated by the one-letter amino acid code, followed by its position within the RecUMge amino acid sequence and an A, which indicates the alanine into which the original residue was changed. All mutants were produced as fusions to MBP, which allowed the application of a universal small-scale purification protocol for each mutant. This approach was possible because we previously found the activities of MBP-fused RecUMge to be indistinguishable from those of native RecUMge (26).
RecUMge point mutants can be separated into four functional groups.
The RecUMge point mutants were tested by means of electrophoretic mobility shift assays (Fig. 1B) and HJ resolution assays, respectively, for the capacity to bind to, as well as cleave, HJ substrates (Fig. 1C). When the HJ-binding and cleavage activities of the point mutants were combined, four groups of proteins could be functionally distinguished (group A to D) (Table 1).
The mutants from the first group (group A) were able to bind and cleave HJs with an efficiency similar to that of wt RecUMge. This group consists of eight mutants, i.e., N5A, M8A, N15A, S54A, Y62A, G64A, D112A, and L122A (Fig. 1 and Table 1). Obviously, the amino acids that were targeted in these mutants do not exert a crucial function in HJ binding or the catalysis of HJ resolution by RecUMge.
Group B mutants display reduced resolution activities.
The proteins from the second group of mutants (group B, consisting of four mutants) displayed significantly reduced HJ cleavage activities (5 to 20% of the wt level) and, in most cases, reduced binding activities (Fig. 1 and Table 1). Only two of the mutants from this group, G7A and H87A, were able to bind the HJ substrate as efficiently as the wt protein (Fig. 1B, lanes 6, 7, 56, and 57). Despite its “normal” HJ-binding activity, mutant H87A displayed the lowest cleavage activity of all proteins in group B (5% of wt level). This activity is only just visible in Fig. 1C (lane 25). The cleavage activity of the other group B mutants (G7A, L10A, and V46A) was higher and therefore readily detectable (Fig. 1C, lanes 4, 6, 8, and 10). Clearly, the altered amino acids in the group B mutants are not indispensable. Nevertheless, the relatively low cleavage activity of these mutants indicates that the altered residues may at least have an indirect involvement in the catalysis of HJ resolution, e.g., by changing the local architecture of the amino acid chain. This is particularly relevant for two of the residues, i.e., L10 and H87, which both neighbor amino acid residues that have an essential function in catalysis (E11 and Q88, respectively; the roles of these residues are described below).
Group C mutants are unable to resolve HJ substrates.
The members of the third group of mutants (group C) showed the most remarkable phenotype of all the mutants in this study. While all 12 group C mutants displayed HJ-binding activity, they were unable to cleave the DNA substrate (Fig. 1B and C and Table 1). This phenomenon was most striking for five of the mutants (E11A, D57A, D68A, E70A, and Q88A), which were able to bind to the HJ substrate with an efficiency comparable to that of wt RecU (Fig. 1B). The other seven mutants (K31A, Y58A, Y66A, K72A, T74A, K76A, and L92A) displayed an HJ-binding activity that was somewhat lower than that of the wt protein. It is likely that the amino acid residues that are mutated in the group C proteins play a role in the catalysis of HJ resolution. This notion is founded not only on the phenotype of the group C mutants, but also on structural information that is available for two homologs of RecUMge, i.e., B. subtilis RecU (RecUBsu) (18) and B. stearothermophilus RecU (RecUBst) (17). Based on this information, we hypothesized that three of the targeted residues within the group C mutants, i.e., D57, E70, and Q88, form part of the catalytic site of RecUMge and are involved in the interaction with a divalent cation. The amino acid counterparts of these 3 residues were found to interact with Mg2+ in the three-dimensional structure of RecUBsu (18) (Table 2).
Table 2.
Group C amino acid residuesa from RecUMge and their corresponding residues in RecUBst and RecUBsu
| RecU species | Putative catalytic site residues | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RecUMge | E11 | K31 | D57 | Y58 | Y66 | D68 | E70 | K72 | T74 | K76 | Q88 | L92 |
| RecUBst | E34 | K54 | D86b | Y87 | Y95 | D97 | E99 | K101 | T103 | K106 | Q118b | M122 |
| RecUBsu | E36 | K56 | D88b | Y89 | Y97 | D99 | E101b | K103 | T105 | K108 | Q120b | M124 |
A group C residue is a (putative) catalytic site residue that, when mutated, gives rise to a RecUMge protein (a group C mutant [Table 1]) with an HJ-binding-positive, HJ resolution-negative phenotype. The residues in boldface were subjected to mutagenesis. The RecUMge mutants are from this study. The RecUBst mutant is described by Rigden and coworkers (21). The RecUBsu mutants are from McGregor et al. (E36, D88, and D99) (18) and Canas et al. (K56) (3).
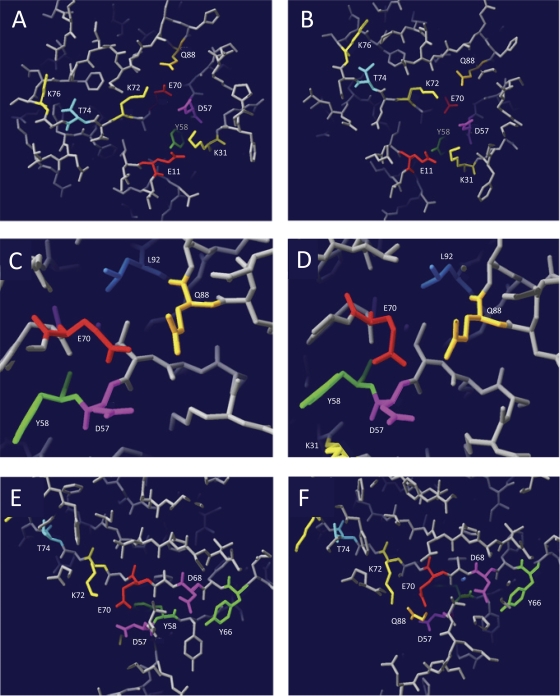
To relate the HJ resolution-negative phenotype of the group C mutants to the structure of RecUMge and the architecture of the enzyme's active site, a three-dimensional model of the protein was predicted using the protein homology/analogy recognition engine Phyre (16). Using the RecUBsu and RecUBst structures as templates, high-confidence 3D models of RecUMge, with an estimated precision of 100%, were generated. These models are remarkably similar, which emphasizes their reliability (Fig. 2; compare the models in panels A, C, and E, which are based on the RecUBst structure, to those in panels B, D, and F, based on the RecUBsu structure). As shown in Fig. 2A and B, 9 of the 12 group C residues cluster around the putative active site of RecUMge, the center of which is formed by the aforementioned triad of catalytic residues, i.e., D57, E70, and Q88. Four residues are located in the vicinity of this triad (K72, Y58, E11, and K31), whereas two other residues are somewhat further removed from the putative catalytic site (T74 and K76).
Fig. 2.
Three-dimensional models of the putative catalytic site of RecUMge. Three-dimensional models of RecUMge were constructed using the protein homology/analogy recognition engine Phyre (16). The lowest Phyre E values (3.4e−22 and 3e−21), with 100% estimated precision, were obtained with the structures of RecUBst and RecUBsu, respectively. The generated structure (PDB) files, which were based on either the RecUBst structure (A, C, and E) or the RecUBsu structure (B, D, and F), were analyzed using DeepView/Swiss-PdbViewer v4.0.1 (11). (A and B) Overview of the putative catalytic site of RecUMge. The group C amino acid residues, i.e., the amino acids that are crucial in the catalysis of HJ resolution by RecUMge but are not essential for HJ binding (Tables 1 and 2), are indicated by their names and residue-specific colors. In this slab view of the structure, the positions of 9 of the 12 group C residues can be observed. (C and D) Details of the three RecUMge residues, i.e., D57, E70, and Q88 (the putative catalytic triad residues), which are potentially involved in coordination of a divalent cation within the catalytic site. Residue L92, located nearby, which is also a group C residue, is clearly visible. (E and F) Overview of the putative catalytic site of RecUMge as viewed from a different angle from that shown in panels A and B. The positions of group C residues D68 and Y66, as opposed to the catalytic triad residues, is illustrated. In panel F, part of the side chain of residue Q88 can just be seen in the slab view of the model. In panel E, this residue cannot be discerned.
When zooming in to the putative catalytic triad from a different angle than that shown in Fig. 2A and B, the proximity of another group C residue to the catalytic site is also apparent (L92, the amino acid shown in blue in Fig. 2C and D). Two other residues, however, D68 and Y66, are more remote from the catalytic triad within the model, at a site opposite residue T74 with respect to the triad (Fig. 2E and F). Based on the characteristics of the group C mutants, as well as on the structural models of RecUMge, the group C residues may be divided into two groups, i.e., (i) residues with a direct function in site-specific hydrolysis of the HJ substrates and (ii) residues that function in the binding and positioning of the HJ DNA. The first group consists of residues E11, K31, D68, and L92, as well as the residues from the putative catalytic triad (D57, E70, and Q88); modification of these residues rendered RecUMge catalytically inactive but did not have a significant influence on the HJ-binding characteristics of the protein. The second group consists of residues Y58, Y66, K72, K74, and K76, the modification of which reduced the HJ-binding efficiency of RecUMge and also rendered the protein catalytically inactive (Table 1).
Four of the group C mutants, i.e., D57A, D68A, E70A, and Q88A, displayed a somewhat higher HJ-binding activity than the wt protein. This was also observed for group A mutant D112A and group B mutant H87A (Fig. 1B). As these six proteins do not have obvious common characteristics, it is difficult to provide a general explanation for their relatively high HJ-binding activities.
Group D mutants are inactive in HJ binding and resolution.
The proteins from the fourth group of mutants (group D) did not display any activity either in HJ binding or in HJ resolution (Fig. 1B and C and Table 1). These mutants include G60A, F69A, F79A, H91A, G100A, F103A, and F108A. The residues that are targeted in these mutants (the group D residues), are all located within, or very close to, putative secondary structures of RecUMge (Fig. 1A). One of the group D residues, F69, may be centrally oriented in a β-sheet that is predicted to constitute a crucial part of the catalytic site of RecUMge, as described above (Fig. 1A and 2E and F). Because F69 is flanked by two group C residues (D68 and E70) that exhibited normal levels of HJ-binding activity, it is tempting to speculate that F69 functions in the proper binding and positioning of the HJ DNA substrate in the protein's catalytic site. Like F69, residues G60, F79, F103, and F108 may also form part of predicted β-sheets. The other two group D residues, H91 and G100, are located within a putative α-helix and at the N-terminal side of a predicted β-sheet, respectively (Fig. 1A). Each of the group D residues may therefore be crucial to the integrity of putative secondary structures of RecUMge and thereby play a role in the proper folding of specific regions of the protein. This notion is supported by biochemical findings, as the expression of the group D mutants in E. coli led to a relatively high proportion of insoluble protein, which was not observed for any of the other proteins from this study (data not shown). Nevertheless, analysis of the proteins by far-UV circular dichroism (CD) spectroscopy did not reveal significant differences between the mutants in their secondary structures (data not shown).
Activities of N- and C-terminal deletion mutants of RecUMge.
To investigate the role of the N and C termini of RecUMge, a series of deletion mutants were generated and tested in HJ-binding and resolution assays (Table 1). Up to 3 amino acids could be deleted from the N terminus of RecUMge without having a significant effect on the HJ-binding and resolution activity of the protein. However, after deletion of five or more of the N-terminal amino acids, both HJ binding and cleavage activities of the mutants were significantly reduced. While mutant NΔ5 showed a strongly reduced HJ resolution activity as opposed to the wt protein, mutant NΔ8 was catalytically inactive and displayed only limited HJ-binding activity (Table 1). These data corroborate the finding that mutation of residue G7 (in mutant G7A), which is deleted in NΔ8, results in a protein that is severely restricted in its catalytic activity.
At the C terminus, deletion of only 3 amino acids from RecUMge already resulted in a protein that was significantly impaired in HJ binding and cleavage (Table 1). Deletion of six additional residues gave rise to a protein (CΔ9) that did not display any detectable activity. Because the 10 C-terminal amino acids of RecUMge may form part of an α-helical structure (Fig. 1A), it is probable that a small deletion of only 3 amino acid residues may have a dramatic effect on the protein's structure and thereby on its activities.
DISCUSSION
The RecU protein superfamily is a large group of (putative) HJ resolvases that are highly conserved in the bacterial phylum Firmicutes. Hitherto, RecU homologs have not been found in Gram-negative bacteria; in these species, the HJ resolvase function is performed by RuvC- and RusA-like enzymes. Despite the important role that resolvases generally play in homologous DNA recombination and DNA repair, only three members of the RecU superfamily have previously been studied, i.e., the RecU proteins from B. subtilis, B. stearothermophilus, and M. genitalium. Both RecUBsu and RecUMge were found to bind to branched DNA substrates, including HJs, in a substrate sequence-independent fashion and to cleave HJ substrates in a sequence-dependent manner (1, 26). In addition, high-resolution 3D structures of RecUBsu (18), as well as RecUBst (17), have been determined, which showed a remarkable similarity between these proteins and archaeal resolvases, as well as type II restriction endonucleases. While the 3D structures have proven to be an invaluable tool in the prediction of the functions of various amino acid residues within the RecU proteins, the importance of only a limited number of (conserved) residues has previously been assessed by mutational analysis (3, 4, 18, 21).
In this study, we performed an unbiased and comprehensive mutational analysis in which we targeted all amino acid residues that are completely conserved among RecU and RecU-like sequences. Thus, we were able to identify 7 amino acid residues that are important for both HJ binding and resolution by RecUMge (Table 1). These residues are likely to have a structural role within the polypeptide, as they form part of predicted secondary structures. In addition to these structurally important residues, we identified a set of 12 residues (termed group C residues) that are probably involved in the catalysis of endonucleolytic cleavage of HJ substrates by RecUMge. The most interesting residues from this set are E11, K31, D57, D68, E70, Q88, and L92; single point mutation of these residues resulted in proteins that displayed relatively high levels of HJ-binding activity while being inactive in HJ cleavage. Based on the phenotypes of the group C mutants, as well as on structural models of the RecUMge protein, it is likely that three of the group C residues (D57, E70, and Q88) play a direct role in the catalysis of HJ resolution by interacting with a divalent cation within the protein's catalytic center. Within the structure of RecUBsu, the counterparts of these three residues were found to coordinate an Mg2+ ion (18) (Table 2 shows a direct comparison of the amino acid numbering of the 12 group C residues from RecUMge, RecUBst, and RecUBsu). Counterparts of residues D57 and Q88 also appeared to be involved in the coordination of Mg2+ within the catalytic center of RecUBst (17). However, the third residue involved in Mg2+ binding in this protein appeared to be a nonconserved residue, i.e., T84 (17). While a threonine is present at the congruent position of T84 in RecUBsu (residue T86), a phenylalanine is present at this position (K55) in RecUMge (Fig. 1A). Residue K55, however, is not predicted to be in close proximity to the putative catalytic site within structural models of RecUMge (Fig. 2). Likewise, residue T86 was not found to be localized close to the putative catalytic center of RecUBsu (18). We therefore hypothesize that the organization of the catalytic center of RecUMge is similar to that of RecUBsu and that residues D57, E70, and Q88 together form a catalytic triad that is involved in the coordination of a divalent cation.
The importance of the counterparts of the catalytic triad residues D57, E70, and Q88 has previously been shown only for D57; mutation of residue D88, the RecUBsu counterpart of D57, resulted in a protein that was capable of binding to HJs but was unable to resolve these substrates (18). In this study, we found that single point mutation of each of the three catalytic triad residues resulted in proteins with an HJ binding-positive but HJ cleavage-negative phenotype. Other putative catalytic site residues that have previously been targeted for mutagenesis are the counterparts of RecUMge residues E11, D68, and K31. Mutation of these residues in RecUBsu (E36, D99, and K56) gave rise to mutants that bound to HJs but were either significantly impaired in their HJ cleavage activity (D99A and K56A) (3, 18) or catalytically inactive (mutant E36A) (18).
The corresponding mutants from RecUMge (E11A, D68A, and K31A) were found to have similar phenotypes, although none of these mutants displayed any detectable catalytic activity.
The RecUBsu counterparts of the critical lysine residues K31 and K72 of RecUMge, K56 and K103 (Table 2), have been proposed to play a direct role in the catalysis of HJ resolution by analogy with other endonucleases (18). Both lysines, which were seen in close proximity to D88 and E101 in the RecUBsu structure, could act as a general base to deprotonate the attacking nucleophile (water) during phosphodiester bond cleavage (18). Alternatively, these residues could be involved in either the stabilization of the catalytic acidic residues (D88 and E101) or the stabilization of a substrate transition state during endonucleolysis. A similar role in catalysis has been proposed for the lysine residue K101 from RecUBst, which was hypothesized to act as a Lewis acid and stabilize the putative pentacoordinated phosphate intermediate during cleavage of the phosphodiester bond (17). In line with this hypothesis, we found the RecUMge counterpart of this lysine residue, K72, to have a crucial function in HJ cleavage. Moreover, both K72 and K31 were found in close proximity to the catalytic triad residues D57, E70, and Q88 in the structural models of RecUMge, which is in line with a direct function of these residues in cleavage of the scissile phosphodiester bond of HJ substrates.
While the similarities in sequence and structure are obvious among the RecU proteins that have been studied, there is also a clear difference between the Mollicutes sequences and the “Bacillus-like” RecU sequences (Fig. 1A). In contrast to the first group of sequences, the Bacillus-like sequences carry a long, flexible N-terminal region of approximately 30 amino acid residues. It was reported that deletion of the 32 N-terminal amino acids of RecUBsu resulted in a protein (Δ1-32) that was able to bind to HJs but was catalytically inactive (4). This deletion would correspond to a deletion of the N-terminal 7 amino acids of RecUMge. In agreement with the characteristics of the RecUBsu Δ1-32 mutant, we found RecUMge mutant NΔ8 to be HJ binding positive but catalytically negative (Table 1). Because mutant NΔ5 did display cleavage activity, the catalytic inactivity of NΔ8 was caused by deletion of the RGM sequence at positions 6 to 8 of RecUMge. Indeed, substitution of either residue G7 or M8 did lead to significant reductions in HJ cleavage activity. Because residue R6 is not conserved among all known RecU sequences, it was not targeted for mutagenesis in this study. However, mutation of the residue corresponding to R6 in RecUBsu, i.e., R31, did not have an effect on the HJ-binding or cleavage activity of the protein (4). We therefore conclude that the inactivity of mutant NΔ8 is caused by the cumulative effects of the deletion of both residues G7 and M8 and that deletion of the counterparts of these residues from RecUBsu is the cause of the catalytic inactivity of the RecUBsu mutant Δ1-32.
Finally, regarding the divalent-cation dependence of RecUMge, it is interesting that this protein displays HJ resolution activity only in the presence of Mn2+ (26). While the HJ resolvases from E. coli, i.e., RuvCEco and RusAEco, were found to be catalytically active in the presence of either Mg2+ or Mn2+ (5, 22–24), the HJ resolution activity of RecUBsu has been reported only in the presence of Mg2+ (1). Thus, it is unknown whether RecUBsu is also active in the presence of Mn2+ instead of Mg2+. Nevertheless, an Mn2+ ion was shown to be coordinated by the catalytic amino acid residues D88 and E101 in the structure of RecUBsu after metal ion-free crystals of the protein were soaked with Mn2+ (18). It is clear, however, that a protein's catalytic site may adopt a different conformation upon interaction with either Mn2+ or Mg2+. The determination of the structure of the active site of RecUMge may help in understanding the strict Mn2+ dependence of this protein for its catalytic activity. In addition, this structure may shed light on the positions, orientations, and functions of the catalytically important residues that have been identified in this study.
ACKNOWLEDGMENTS
We thank G. van der Zwan, A. Wiskerke, and K. van den Boom from the Biomolecular Spectroscopy Group, Faculty of Sciences, Free University Amsterdam, for help in determination of the far-UV CD spectra of the proteins used in this study. We thank E. Spuesens for critically reading the manuscript.
A.M.C.V.R. is supported by grants from the European Society for Pediatric Infectious Diseases, ZonMW, and the Erasmus MC.
Footnotes
Published ahead of print on 3 June 2010.
REFERENCES
- 1. Ayora S., Carrasco B., Doncel-Perez E., Lurz R., Alonso J. C. 2004. Bacillus subtilis RecU protein cleaves Holliday junctions and anneals single-stranded DNA. Proc. Natl. Acad. Sci. U. S. A. 101:452–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burgos R., et al. 2006. Mycoplasma genitalium P140 and P110 cytadhesins are reciprocally stabilized and required for cell adhesion and terminal-organelle development. J. Bacteriol. 188:8627–8637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Canas C., Carrasco B., Ayora S., Alonso J. C. 2008. The RecU Holliday junction resolvase acts at early stages of homologous recombination. Nucleic Acids Res. 36:5242–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carrasco B., Canas C., Sharples G. J., Alonso J. C., Ayora S. 2009. The N-terminal region of the RecU Holliday junction resolvase is essential for homologous recombination. J. Mol. Biol. 390:1–9 [DOI] [PubMed] [Google Scholar]
- 5. Chan S. N., Harris L., Bolt E. L., Whitby M. C., Lloyd R. G. 1997. Sequence specificity and biochemical characterization of the RusA Holliday junction resolvase of Escherichia coli. J. Biol. Chem. 272:14873–14882 [DOI] [PubMed] [Google Scholar]
- 6. Clausen H. F., et al. 2001. Serological investigation of Mycoplasma genitalium in infertile women. Hum. Reprod. 16:1866–1874 [DOI] [PubMed] [Google Scholar]
- 7. Dallo S. F., Baseman J. B. 1991. Adhesin gene of Mycoplasma genitalium exists as multiple copies. Microb. Pathog. 10:475–480 [DOI] [PubMed] [Google Scholar]
- 8. Dhandayuthapani S., Rasmussen W. G., Baseman J. B. 1999. Disruption of gene mg218 of Mycoplasma genitalium through homologous recombination leads to an adherence-deficient phenotype. Proc. Natl. Acad. Sci. U. S. A. 96:5227–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dhandayuthapani S., Rasmussen W. G., Baseman J. B. 2002. Stability of cytadherence-related proteins P140/P110 in Mycoplasma genitalium requires MG218 and unidentified factors. Arch. Med. Res. 33:1–5 [DOI] [PubMed] [Google Scholar]
- 10. Fraser C. M., et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397–403 [DOI] [PubMed] [Google Scholar]
- 11. Guex N., Peitsch M. C. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 [DOI] [PubMed] [Google Scholar]
- 12. Hu P. C., et al. 1987. A Mycoplasma genitalium protein resembling the Mycoplasma pneumoniae attachment protein. Infect. Immun. 55:1126–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ingleston S. M., et al. 2002. Holliday junction binding and processing by the RuvA protein of Mycoplasma pneumoniae. Eur. J. Biochem. 269:1525–1533 [DOI] [PubMed] [Google Scholar]
- 14. Iverson-Cabral S. L., Astete S. G., Cohen C. R., Rocha E. P., Totten P. A. 2006. Intrastrain heterogeneity of the mgpB gene in Mycoplasma genitalium is extensive in vitro and in vivo and suggests that variation is generated via recombination with repetitive chromosomal sequences. Infect. Immun. 74:3715–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iverson-Cabral S. L., Astete S. G., Cohen C. R., Totten P. A. 2007. mgpB and mgpC sequence diversity in Mycoplasma genitalium is generated by segmental reciprocal recombination with repetitive chromosomal sequences. Mol. Microbiol. 66:55–73 [DOI] [PubMed] [Google Scholar]
- 16. Kelley L. A., Sternberg M. J. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 17. Kelly S. J., Li J., Setlow P., Jedrzejas M. J. 2007. Structure, flexibility, and mechanism of the Bacillus stearothermophilus RecU Holliday junction resolvase. Proteins 68:961–971 [DOI] [PubMed] [Google Scholar]
- 18. McGregor N., et al. 2005. The structure of Bacillus subtilis RecU Holliday junction resolvase and its role in substrate selection and sequence-specific cleavage. Structure 13:1341–1351 [DOI] [PubMed] [Google Scholar]
- 19. Peterson S. N., et al. 1995. Characterization of repetitive DNA in the Mycoplasma genitalium genome: possible role in the generation of antigenic variation. Proc. Natl. Acad. Sci. U. S. A. 92:11829–11833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peterson S. N., Hu P. C., Bott K. F., Hutchison C. A., III 1993. A survey of the Mycoplasma genitalium genome by using random sequencing. J. Bacteriol. 175:7918–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rigden D. J., et al. 2002. PrfA protein of Bacillus species: prediction and demonstration of endonuclease activity on DNA. Protein Sci. 11:2370–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shah R., Bennett R. J., West S. C. 1994. Activation of RuvC Holliday junction resolvase in vitro. Nucleic Acids Res. 22:2490–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shah R., Bennett R. J., West S. C. 1994. Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell 79:853–864 [DOI] [PubMed] [Google Scholar]
- 24. Sharples G. J., Chan S. N., Mahdi A. A., Whitby M. C., Lloyd R. G. 1994. Processing of intermediates in recombination and DNA repair: identification of a new endonuclease that specifically cleaves Holliday junctions. EMBO J. 13:6133–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sluijter M., Hoogenboezem T., Hartwig N. G., Vink C. 2008. The Mycoplasma pneumoniae MPN229 gene encodes a protein that selectively binds single-stranded DNA and stimulates Recombinase A-mediated DNA strand exchange. BMC Microbiol. 8:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sluijter M., et al. 2010. The Mycoplasma genitalium MG352-encoded protein is a Holliday junction resolvase that has a non-functional orthologue in Mycoplasma pneumoniae. Mol. Microbiol. 77:1261–1277 [DOI] [PubMed] [Google Scholar]
- 27. Sluijter M., Spuesens E. B., Hartwig N. G., van Rossum A. M., Vink C. 2009. The Mycoplasma pneumoniae MPN490 and Mycoplasma genitalium MG339 genes encode RecA homologs that promote homologous DNA strand exchange. Infect. Immun. 77:4905–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Svenstrup H. F., Jensen J. S., Gevaert K., Birkelund S., Christiansen G. 2006. Identification and characterization of immunogenic proteins of Mycoplasma genitalium. Clin. Vaccine Immunol. 13:913–922 [DOI] [PMC free article] [PubMed] [Google Scholar]