Abstract
Chlamydia spp. are among the many pathogenic Gram-negative bacteria that employ a type III secretion system (T3SS) to overcome host defenses and exploit available resources. Significant progress has been made in elucidating contributions of T3S to the pathogenesis of these medically important, obligate intracellular parasites, yet important questions remain. Chief among these is how secreted effector proteins traverse eukaryotic membranes to gain access to the host cytosol. Due to a complex developmental cycle, it is possible that chlamydiae utilize a different complement of proteins to accomplish translocation at different stages of development. We investigated this possibility by extending the characterization of C. trachomatis CopB and CopB2. CopB is detected early during infection but is depleted and not detected again until about 20 h postinfection. In contrast, CopB2 was detectible throughout development. CopB is associated with the inclusion membrane. Biochemical and ectopic expression analyses were consistent with peripheral association of CopB2 with inclusion membranes. This interaction correlated with development and required both chlamydial de novo protein synthesis and T3SS activity. Collectively, our data indicate that it is unlikely that CopB serves as the sole chlamydial translocation pore and that CopB2 is capable of association with the inclusion membrane.
INTRODUCTION
Chlamydia trachomatis is a prevalent and medically significant agent of oculogenital disease in humans. Infection of the conjunctiva with serovars A to C leads to corneal scarring and eventually blinding trachoma (11), whereas genital infections with serovars D to K or lymphogranuloma venereum ([LGV] serovars L1 to L3) manifest as sexually transmitted disease. Chlamydia pneumoniae represents an additional human pathogen associated with respiratory infection while infections with species such as Chlamydia caviae and Chlamydia muridarum are confined to animals. Regardless of host and tissue range, all species preferentially infect epithelial cells, where they develop entirely within a membrane-bound parasitophorous vesicle termed an inclusion.
All Chlamydia spp. possess a biphasic developmental cycle that is initiated when infectious yet metabolically inert particles termed elementary bodies (EBs) invade a host cell (36). Chlamydial gene expression occurs in at least three temporally regulated classes, early, mid-, and late cycle (5). During the early cycle, internalized EBs convert to noninfectious, metabolically active forms termed reticulate bodies (RBs). Midcycle is dominated by vegetative growth of RBs, and development is concluded during the late cycle by the asynchronous differentiation of a subset of RBs back to EBs. The environmental stimuli triggering these events remain unknown, yet physical contact with undefined ligands in the host plasma membrane and parasitophorous vesicle is thought to play a predominant role (55). Several clues exist that suggest how chlamydial development might be orchestrated. Perhaps the most prominent of these is that temporally regulated gene expression ensures de novo synthesis of stage-specific proteins at appropriate developmental phases (reviewed in reference 1).
In the apparent absence of overt exotoxins (50), the success of chlamydial infection, and therefore pathogenesis, is doubtless governed largely by the ability to efficiently create and maintain the inclusion as a privileged intracellular niche. Although a diverse array of chlamydial factors likely contributes to infection, secretion of antihost proteins by a nonflagellar type III secretion system (T3SS) represents an essential mechanism for virulence. T3SSs are complex machines that are expressed by an array of Gram-negative bacteria, in which they enable vectorial secretion and translocation of antihost proteins (termed effectors) into the cytosol of an infected eukaryotic cell (19, 22, 51). These effector proteins vary depending on the pathogen but generally act by manipulating host cellular processes to promote survival of the respective bacterium. All chlamydial genomes encode the capacity for T3S (4, 28, 42, 43, 50, 52), and effector proteins are secreted throughout development (reviewed in reference 53). For example, effectors such as Tarp (14) and CT694 (25) that affect actin rearrangement are first secreted during invasion. Modification of the inclusion membrane with chlamydial Inc proteins (reviewed in reference 45) begins during early-cycle development while later-acting effectors such as CT847 (13) are secreted beginning during midcycle. The continual accumulation of these proteins suggests that T3S remains active through later development. Hence, it is likely that T3S contributes to chlamydial survival on multiple levels and that secretion activity is present throughout the developmental cycle.
Cumulative evidence from genetically tractable systems indicates that a multiprotein complex of T3S apparatus proteins comprising the basal body resides in the bacterial envelope and mediates secretion of effector proteins across the inner and outer bacterial membranes (34, 35). However, translocation of effectors requires a set of T3S proteins, collectively referred to as the needle complex and translocon. Translocation across the eukaryotic membrane requires a hydrophilic protein associated with the distal terminus of the needle filament and two hydrophobic domain-containing proteins that associate with the eukaryotic membrane (37). The needle, tip, and hydrophobic translocator proteins are exemplified by Yersinia YscF, LcrV, YopB, and YopD, respectively. YopB is presumed to represent the primary component of a translocation pore that traverses the eukaryotic membrane and allows passage of effectors into the host cytosol. Osmoprotection (21) and dye permeabilization (39) studies indirectly indicate formation of YopB-dependent pores. The Shigella homolog IpaB is capable of oligomerization and depends on hydrophobic domains to interact with lipid bilayers (26).
Sequence conservation among T3SS components has facilitated the identification of many chlamydial T3SS proteins (reviewed in reference 9). However, the identity and function of some essential components are less obvious. We have previously provided direct evidence that CdsF represents the chlamydial needle protein (8) while biophysical (33) and protein interaction (49) studies suggest that CT584 may be the tip protein. The composition of translocation pores remains incompletely explored. C. trachomatis CopB (CT578) has been proposed as a logical possibility since it is secreted by a type III mechanism, is detected in the inclusion membrane, and exhibits homology to YopB family translocators (15). Since de novo synthesis of CopB occurs during late-cycle development, the question arises as to what forms a competent translocon during earlier development. CopB2 (CT861) represents one attractive candidate for an alternative translocator protein. CopB2 is also secreted via a T3S mechanism when ectopically expressed in Yersinia (14), and expression studies have shown that copB2 is a midcycle gene (5). These observations have led to the hypothesis that CopB and CopB2 serve as alternative translocator proteins that could mediate translocation of effector proteins across the chlamydial inclusion membrane at different stages of development. Indeed, labeling of cytosolic proteins in selectively permeabilized infected cells detected a pool of CopB2 exposed to the host cytosol (15). Unfortunately, this secreted CopB2 is below detection in immunolocalization studies, making it unclear whether CopB2 could be a mediator of translocation. In the absence of a tractable genetic system, we have begun to test whether CopB2 characteristics are consistent with this protein serving a translocator function. We report here the finding that CopB2 is capable of associating with inclusions but that this interaction is distinct from mechanisms typical of YopB-like proteins.
MATERIALS AND METHODS
Cell culture and organisms.
C. trachomatis serovar L2 (LGV 434) and C. pneumoniae AR-39 were cultivated in HeLa 229 epithelial cells (CCL 1.2; American Type Culture Collection, Manassas, VA) routinely maintained at 37°C in the presence of 5% CO2–95% humidified air in RPMI 1640 medium (Invitrogen Corp., Carlsbad, CA) supplemented with 10% (vol/vol) fetal bovine serum (Sigma, St. Louis, MO) and 10 μg/ml gentamicin (Teknova, Hollister, CA). Purified EBs, prepared as described previously (12) by centrifugation through MD-76R (Mallinckrodt, St. Louis, MO) density gradients, were used as infection sources for all experiments. C. trachomatis (12) and C. pneumoniae (57) infections of HeLa cells were carried out for 1 to 2 h at 37°C as described previously. When indicated in the figure legends, chloramphenicol (Cm; 200 μg/ml) was included to inhibit chlamydial de novo protein synthesis. Where appropriate, INP007 was included at 50 μM to inhibit T3S as described previously (56). Escherichia coli NovaBlue (EMD Biosciences, Gibbstown, NJ) was used for routine cloning procedures and was grown at 37°C in either Luria-Bertani (LB) broth or on LB agar plates containing 50 μg/ml carbenicillin (Cb) or kanamycin (Km), where appropriate.
Bioinformatic analyses.
Detection of primary sequence similarities between CopB or CopB2 and potential homologs were assessed using PSI-BLAST using default parameters (3). Predicted structural features within CopB and CopB2 were analyzed using COILS (32) to identify putative coiled-coil domains, and the method of Kyte and Doolittle (29) was employed to detect hydrophobic domains. Phylogenetic comparisons of CopB and CopB2 chlamydial orthologs were carried out using profile alignment with predicted local structure (PROMALS) alignment based on primary sequence and predicted secondary structure (41). Graphic representations of the output comparisons were generated using EMBOSS-Plotcon (44).
Membrane fractionation.
Potential membrane association of CopB and CopB2 was tested by examining partitioning during fractionation of endogenous chlamydial proteins in preparations of whole-culture membranes. Semiconfluent HeLa monolayers were mock treated or infected as described above with C. trachomatis L2 at a multiplicity of infection (MOI) of 10 or 1 where indicated in the figure legends. Separation of membranes through sucrose density gradients was accomplished using modifications of previously reported methods (18, 27). Briefly, HeLa cells cultured in T75 flasks were mock treated or infected with C. trachomatis L2 (MOI of 1) for 24 h, and material was scraped into 8 ml of ice-cold sucrose buffer (10 mM Tris, pH 7.5, 1 mM EDTA, 0.25 M sucrose,) supplemented with protease inhibitors (complete cocktail; Roche). Whole-culture material was Dounce homogenized to disrupt host cells, and lysates were cleared of large particles via sequential centrifugation at 900 × g and 7,500 × g. Cleared input was layered onto sucrose gradients containing 4-ml layers of 24%, 28%, 32%, 36%, 40%, and 44% (wt/vol) sucrose, and membranes were separated by centrifugation in a JS-24 rotor at 100,000 × g for 1.5 h. Material entering the gradient was collected in 12 2-ml fractions, and proteins were concentrated by addition of trichloroacetate (TCA) to 10% (vol/vol). For Triton X-114 extractions, soluble and integral membrane proteins were separated based on solubility in Triton X-114 (Sigma) to examine the partitioning of selected proteins during chlamydial infection. Cultures were harvested at 2 or 20 h postinfection in the presence of protease inhibitors (Roche), and material was extracted with 1% Triton X-114 essentially as described previously (10). Proteins were precipitated by addition of ice-cold acetone (Sigma) added to a concentration of 10% (vol/vol), followed by washes with cold 70% (vol/vol) ethanol. For immunoblot analyses, all proteins were solubilized in electrophoresis sample buffer (2.3% [wt/vol] sodium dodecyl sulfate [SDS], 5% [vol/vol] β-mercaptoethanol, 25% [vol/vol] glycerol, and 60 mM Tris pH 6.8), and equal volumes of material were resolved in 12% (vol/vol) polyacrylamide gels by SDS-PAGE (30). Resolved material was then transferred to Immobilon-P (Millipore Corp., Bedford, MA) in Tris-glycine buffer (25 mM Tris, 192 mM glycine, 10% methanol), and detection of specific chlamydial proteins was accomplished using anti-major outer membrane protein (MOMP) (25), anti-HSP60 (59), anti-CopN (16), anti-IncG (47), anti-CopB (15), anti-CopB2 (15), anti-CdsD (17), or anti-CdsJ (17). After incubation with specific primary antibodies, immunoblots were probed with the appropriate alkaline phosphatase or horseradish peroxidase-conjugated secondary antibodies (Sigma) and developed with One Step nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP; Pierce Biotechnology, Inc., Rockford, IL) or ECL Plus reagent (GE Healthcare), respectively.
DNA methods and transfections.
Full-length coding sequences for CopB (CT578) or CopB2 (CT861) were PCR amplified from C. trachomatis L2 genomic DNA using KOD HiFi DNA polymerase (EMD Biosciences). For green fluorescent protein (GFP)-containing constructs, genes were amplified using primers (for CopB, 5′-CCGCTCGAGTCCCTTTCATCTTCTTCGTCTTCCG-3′ and 5′-CGGAATTCTTAAGCTGCGGCGGCTAAGGCGCCACTTATTGCGG-3′; for CopB2, 5′-AAACCGCTCGAGTCATCCTGGTTTGCTCAAGCGACAGACGTCGC-3′ and 5′-CGGAATTCTTACGTAAGGCGTCTGTAAACGTTTGGTC-3′) containing engineered XhoI and EcoRI restriction sites (Integrated DNA Technologies, Coralville, IA). For Flag-tagged CopB2, primers contained EcoRI and KpnI sites (5′-GCGAATTCATCATCCTGGTTTGCTCAAGCGACAGACGTCGC-3′ and 5′-ACTGGTACCTTATCGTAAGGCGTCTGTAAACGTTTGGTCAATTTC-3′). The appropriately digested PCR products were subsequently ligated into similarly digested pEGFP-C3 (Clontech, Mountain View, CA) to create GFP-CopB and GFP-CopB2 or into p3XFlag-CMV-10 (Sigma) to create 3XFT-CopB2 (CopB2 carrying three copies of the Flag tag). In-frame deletion mutants of CopB2 were generated using 3XFT-CopB2 as a template and QuikChange (Agilent Technologies, Santa Clara, CA) technology according to the manufacturer's instructions. Custom primers were used to generate 3XFT-CopB2Δcc1 (deletion of codons 170 to 196; 5′-CCTATCATGGAAGAACAAAAACATCCTC-3′ and 5′-GAGGATGTTTTTGTTCTTCCATGATAGG-3′) and 3XFT-CopB2Δcc2 (deletion of codons 276 to 311; 5′-GCAGATTTTTCTGCCCGATTTTGGGATTC-3′ and 5′-GAATCCCAAAATCGGGCAGAAAAATCTGC-3′). In addition, a 3′ truncation mutant was generated (deletion of codons 470 to 506) via amplification of CopB2 with primers containing engineered EcoRI and KpnI sites (5′-GCGAATTCATCATCCTGGTTTGCTCAAGCGACAGACGTCGC-3′ and 5′-ACTGGTACCTTAATATTCATCCCGTTCCCAATTTATTAATTCGATGCG-3′). The amplification product was digested and mobilized into similarly digested p3XFlag-CMV-10 to create 3XFT-CopB2Δcc3. All construct sequences were verified by DNA sequencing (Genewiz, South Plainfield, NJ). Transfections were accomplished using EndoFree plasmid DNA preparations (Qiagen, Valencia, CA) and Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. When coupled with infection, HeLa cells were generally first infected with C. trachomatis L2 and then transfected at ca. 6 h postinfection. Alternatively, HeLa cells were transfected 48 h postinfection for C. pneumoniae or 12 h prior to infection for experiments examining development, as presented in Fig. 7.
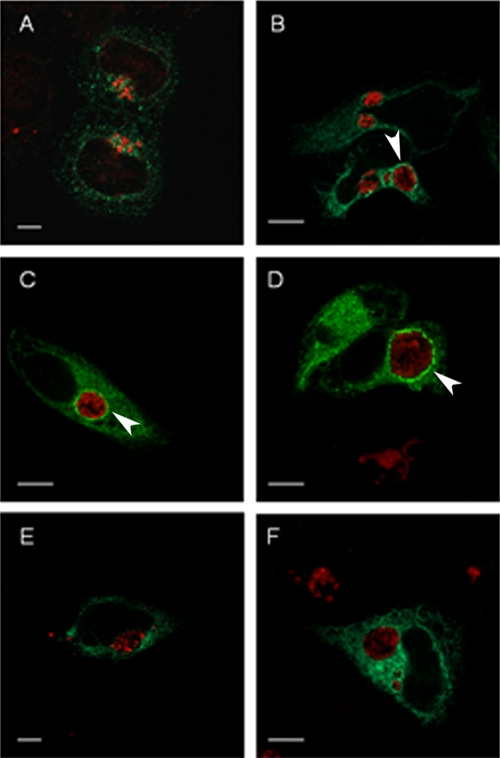
Fig. 7.
3XFT-CopB2 association with inclusions requires chlamydial development. HeLa cells were transfected with p3XFT-CopB2 12 h prior to infection with C. trachomatis L2. Chloramphenicol (E) or INP007 (F) was added to selected cultures at 6 h or 8 h postinfection, respectively. Cultures were processed for microscopy at 8 h (A), 14 h (B), 18 h (C), or 22 h (D to F) postinfection. 3XFT-CopB2 was detected using anti-FLAG (green), and chlamydiae were detected using MOMP-specific antibodies (red). Proteins were visualized by probing with appropriate secondary antibodies and viewed using confocal laser scanning microscopy. Arrows designate colocalization of 3XFT-CopB2 with chlamydial inclusions. Scale bar, 5 μm.
Microscopy.
HeLa monolayers were grown on 12-mm-diameter glass coverslips for immunofluorescence analyses. Monolayers were directly infected with Chlamydia followed by transfection with appropriate plasmid DNAs. Unless otherwise indicated, samples were routinely fixed with methanol at ca. 24 h postinfection for immunofluorescence analyses. Samples were blocked for at least 1 h in Tris-buffered saline plus Tween 20 (TBST; 137 mM NaCl, 2.68 mM KCl, 2.48 mM Tris base [pH 7.4], 0.5% [vol/vol] Tween 20). Samples requiring only visualization of GFP by direct fluorescence were immediately mounted on slides for microscopy. For indirect immunofluorescence analyses, monoclonal Flag-M2- (Sigma) or MOMP-specific antibody (25, 58) was diluted in 5% bovine serum albumin (BSA) in TBST and used to detect ectopically expressed fusion proteins or chlamydiae, respectively. Proteins were visualized with secondary antibodies conjugated to Alexa-488, Alexa-594, or Alexa-546 (Invitrogen). Images were acquired using a Zeiss Axiovert Zoom LSM 510 confocal microscope and Ziess LSM Image Software (Carl Zeiss Microimaging GmbH, Bernried, Germany) or by epifluorescence microscopy using a 60× apochromat objective plus ×1.5 intermediate magnification on a TE2000U inverted photomicroscope (Nikon Inc., Melville, NY) equipped with a Retiga EXi 1394, 12-bit monochrome charge-coupled device (CCD) camera (QImaging, Surrey, BC, Canada) and MetaMorph imaging software, version 6.3r2 (Molecular Devices, Downingtown, PA). All images were equivalently processed using Adobe Photoshop CS2, version 9.0 (Adobe Systems, San Jose, CA).
RESULTS
Bioinformatic analysis of CopB and CopB2.
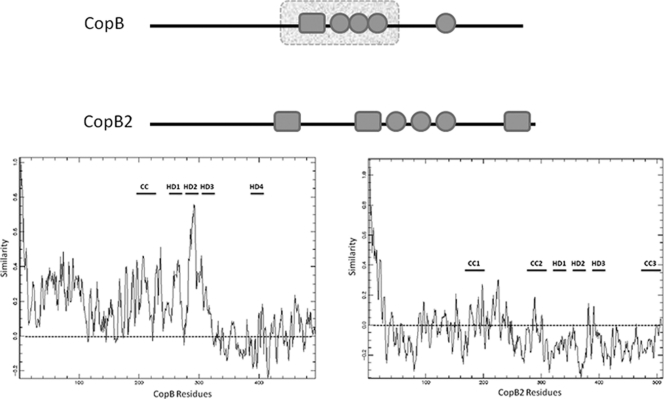
Similarities between CopB and CopB2 have been noted that include similar gene positioning in apparently duplicated T3S-related operons and conserved secondary structures reminiscent of the YopB family of T3S translocator proteins (14). Indeed, PSI-BLAST searches of primary sequence similarities reveal 49% similarity of CopB residues 176 to 335 with translocon protein SipB of Salmonella pathogenicity island 1. CopB2 fails to demonstrate this sequence similarity. However, residues 320 to 400 are predicted to contain hydrophobic residues (Fig. 1, top diagram) that could represent up to three transmembrane (TM) helices. This feature raises the possibility of CopB2 functioning as an integral membrane pore-forming protein. Interestingly, CopB2 primary sequence is more divergent across chlamydial species than CopB (Fig. 1, bottom tracing). PROMALS comparison of CopB from all sequenced chlamydial genomes reveals conserved N-terminal 320 residues that maintain at least 68% similarity between the most divergent orthologs. The hydrophobic region of CopB containing similarity to SipB represents the most conserved portion of the protein. CopB2 sequences are comparatively more divergent, as indicated by a significant portion of the primary sequence falling below the 0.0 PROMALS threshold. In fact the most divergent ortholog, C. pneumoniae Cpn1020, displays 55% overall similarity with C. trachomatis and contains only two predicted TM domains (not shown). Despite this primary sequence diversity, all CopB2 sequences contain three separate domains predicted to form coiled-coil structural motifs. Overall, these analyses are consistent with CopB2 functioning as a translocator protein, yet noted differences raise the possibility of somewhat divergent functions.
Fig. 1.
Schematic representation of predicted secondary structures and areas of species conservation for C. trachomatis CopB and CopB2. (Top) A linear representation of respective proteins is shown including the positions of predicted coiled-coil (solid-line rectangle) and hydrophobic (circles) domains as well as areas containing primary sequences similar to nonchlamydial proteins (broken-lined rectangle). (Bottom) PROMALS output of primary peptide sequence comparisons of CopB or CopB2 from C. trachomatis serovars A, D, and L2, C. muridarum, C. felis, C. abortus, C. caviae, and C. pneumoniae. Tracings indicate overall similarity (y axis; 1.0 = 100%) of residues at a given position (x axis) within respective proteins. A dashed line indicates no similarity threshold, and the positions of coiled-coil and hydrophobic domains are indicated by CC and HD, respectively.
Temporal analysis of CopB and CopB2.
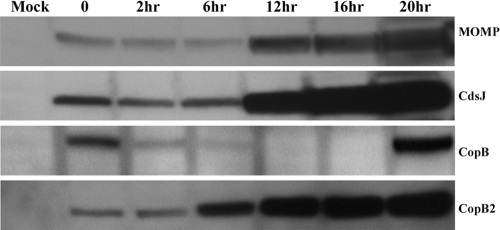
Comparative temporal expression analyses have indicated that CopB and CopB2 are differentially synthesized during chlamydial development (6, 23). De novo expression of copB occurs during late-cycle development whereas copB2 message is detected earlier during midcycle development. We have previously shown that CopB is present in purified EBs (17), raising the possibility that this protein may function during invasion or early-cycle development. Whether this pool of CopB is secreted and how long the protein persists, however, have not been investigated. It is also unclear when CopB2 may be present during chlamydial development. To address these open questions, we infected HeLa cell monolayers with C. trachomatis L2 and monitored protein levels throughout development via immunoblotting (Fig. 2). As expected, full-length CopB was readily detected at time points corresponding to the presence of EBs (0 h) and de novo expressed copB (20 h). CopB abundance was greatly decreased after 6 h of infection and was not detectible at 12 or 16 h postinfection. CopB2 also migrated as an apparent full-length protein (ca. 56 kDa) but was detected throughout development. CopB2 was detectible in EB material, and levels persisted at the early (2 h) time point. An increase in CopB2 was detected by 6 h, and this accumulation continued through remaining samples similarly to the control midcycle gene products CdsJ and MOMP. These data reinforce the concept that CopB cannot represent the sole YopB-class chlamydial translocator protein and are consistent with independent CopB2 function during midcycle development.
Fig. 2.
Temporal analysis of CopB and CopB2 levels during chlamydial infection. HeLa cells were mock infected or infected with C. trachomatis L2 at an MOI of 10 for 1 h at 37°C. Cultures were thoroughly washed, supplemented with fresh medium, and subsequently harvested at the indicated times (in hours) after the medium addition. TCA-concentrated, whole-cell culture lysates were probed via immunoblotting with rabbit anti-CopB or anti-CopB2. Antibodies specific for the T3S structural protein CdsJ and chlamydial envelope protein MOMP were used as controls. Proteins were visualized by probing with secondary antibodies conjugated to horseradish peroxidase and development with ECL Plus chemiluminescence reagent.
Biochemical evidence of membrane association.
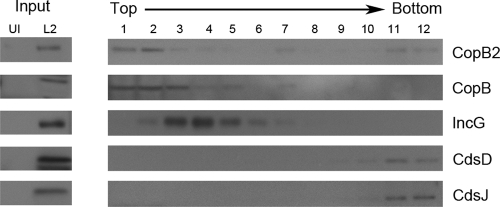
The ability to associate with host membranes is one feature typical of YopB family T3S translocator proteins (37). Indeed, immunolocalization analyses indicated an expected colocalization of CopB with inclusion membranes. However, protein localization remained unclear for CopB2 since levels of this protein were below detection in immunolocalization assays (15). We therefore employed fractionation and biochemical separation of membranes to address localization of endogenous CopB and CopB2 in cultures infected with C. trachomatis. We first examined whether CopB or CopB2 generally associated with membranes by examining protein partitioning in membrane flotation gradients. Material from mock- or Chlamydia-infected cells was harvested 24 h postinfection and gently disrupted via Dounce homogenization to leave chlamydiae intact. Debris was pelleted via low-speed centrifugation, and membrane-containing lysates were separated on sucrose density gradients (Fig. 3). Immunoblot analysis of column input indicated detectible levels of CopB and CopB2 as well as controls for inclusion membrane protein (IncG) or intact bacteria (CdsD and CdsJ) in infected but not mock-infected material. As expected, CdsD and CdsJ were detected solely in the bottom column fractions and likely corresponded to sedimentation of intact chlamydiae as seen previously (18, 27). Detection of IncG in fractions 2 to 7 was also as expected and was consistent with membrane association. Neither CopB nor CopB2 distribution completely overlapped with IncG since both were detected most prominently in fractions 1 to 3 and 7. Importantly, CopB2 fractionation in the upper column fractions was consistent with membrane association and mirrored that of the established inclusion membrane protein CopB. CopB2 was also detectable with intact bacteria in the bottom two gradient fractions, whereas CopB and IncG were not. These data are consistent with detection of CopB2 within intra-inclusion chlamydiae via indirect immunofluorescence (15). In contrast, both CopB and IncG are efficiently secreted and are not detected accumulating within bacteria at this stage of development (15, 47). While these data do not indicate precise localization for CopB or CopB2, they indicate that both proteins are capable of associating with membranes.
Fig. 3.
CopB and CopB2 fractionation in whole-culture membrane flotation gradients. C. trachomatis-infected (L2) or mock-infected (UI) HeLa cultures were disrupted 24 h postinfection, and membrane-containing, clarified lysates (input) were generated. Lysates were subjected to density gradient centrifugation through sucrose. Twelve individual column fractions were collected, and proteins were concentrated via TCA precipitation. Equivalent volumes of material corresponding to the top (1) to the bottom (12) fractions of the column were resolved by SDS-PAGE in 12% polyacrylamide gels. Immunoblots of input and column fractions were probed with anti-CopB and anti-CopB2 or with anti-IncG, anti-CdsD, and anti-CdsJ as fractionation controls. Proteins were visualized by probing with secondary antibodies conjugated to horseradish peroxidase and development with ECL Plus chemiluminescence reagent.
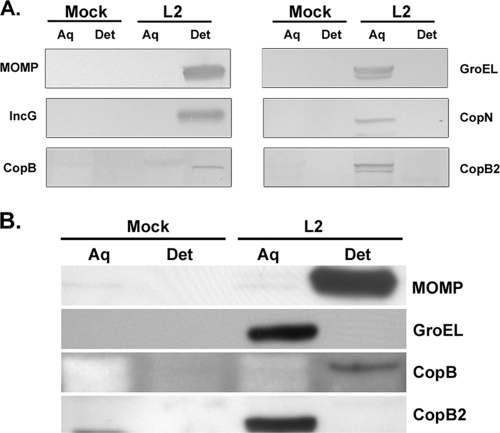
Since YopB family translocator proteins are integral membrane proteins (37), we further examined the mode of membrane association for both CopB and CopB2. Membrane and soluble proteins were separated via Triton X-114 extraction (5), and material was probed by immunoblotting with specific antibodies (Fig. 4A). Material from mock-infected cultures was processed in parallel as an antibody specificity control for immunoblotting. To ensure complete separation of protein pools, C. trachomatis MOMP and GroEL partitioning were examined as controls for bacteria-associated membrane and soluble proteins, respectively. In addition, the T3SS proteins IncG and CopN were tested as controls for integral and peripheral inclusion membrane association, respectively. As expected, both MOMP and IncG were detected solely in the membrane-containing detergent fractions, whereas GroEL and CopN partitioned into the nonmembrane, aqueous fractions. The majority of CopB was detected in the detergent fraction, a localization consistent with a role as an integral membrane translocator protein. CopB2 partitioned similarly to GroEL, and CopN and was detected in only the aqueous protein fraction. These data indicate that CopB2 is not an integral membrane protein and are seemingly inconsistent with CopB2 function as a prototypical translocator protein. In contrast to these infected-culture data, CopB from purified EBs partitions as a soluble protein (17). Since both CopB and CopB2 were detected during early hours of infection, we extended the partitioning analysis to include 2 h postinfection (Fig. 4B). As described previously, CopB was detected primarily in the aqueous fraction of purified EBs (data not shown). After 2 h of infection, CopB2 remained associated with the aqueous fraction. Similar to 20-h cultures, CopB was predominantly detected in the detergent fraction. These data are consistent with CopB secretion and localization in the inclusion membrane during early infection.
Fig. 4.
CopB and CopB2 partition differently in Triton X-114 extracts. Integral membrane association was tested by extracting C. trachomatis L2-infected HeLa cultures with Triton X-114 and separating proteins into aqueous and detergent soluble fractions. (A) Cultures were harvested at 20 h postinfection, and a parallel, mock-treated HeLa culture was harvested as a specificity control. Equivalent quantities of total protein from aqueous (Aq)- and detergent (Det)-soluble samples were resolved by SDS-PAGE in 12% polyacrylamide gels. Immunoblots were probed with anti-MOMP, anti-GroEL, anti-CopN, or anti-IncG as a fractionation control and with antibody specific for CopB or CopB2. Proteins were visualized by probing with secondary antibodies conjugated to alkaline phosphatase and development with NBT-BCIP. (B) HeLa cultures were mock treated or C. trachomatis L2 infected at an MOI of 10. Cultures were harvested at 2 h postinfection, extracted with Triton X-114, and probed by immunoblotting with the indicated antibodies. Proteins were visualized by probing with secondary antibodies conjugated to horseradish peroxidase and development with ECL Plus chemiluminescence reagent.
GFP-CopB and GFP-CopB2 in HeLa cells.
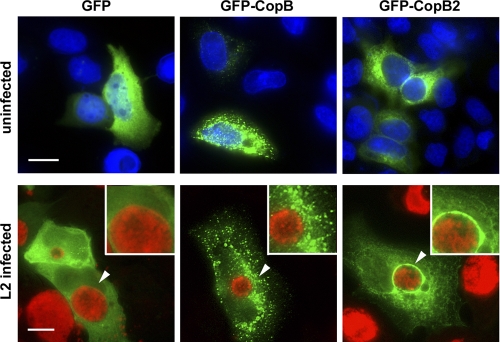
Despite the use of multiple antibodies reactive to all or various portions of CopB2 and raised in either rabbits or mice, immunolocalization studies have been unable to detect secreted CopB2 during infection (data not shown). Since detection limits continually confounded efforts to examine endogenous CopB2 via indirect immunofluorescence, we employed ectopic expression of recombinant protein as a means to address subcellular localization. HeLa cells were transiently transfected with plasmids expressing GFP alone or fused to either CopB or CopB2 coding sequences (Fig. 5). Cultures were visualized by direct fluorescence at 18 h posttransfection. As expected, GFP-only signal was apparent diffusely throughout cells. GFP-CopB2-specific signal was also diffuse in character in the cytosol of HeLa cells but was typically excluded from 4′,6′-diamidino-2-phenylindole (DAPI)-stained nuclear areas. Efficiency of GFP-CopB transfections was much lower than transfection of GFP-CopB2 or GFP, and significant numbers of dead cells were observed. When viable transfected cells were detected, GFP-CopB was routinely observed in a punctate pattern distributed throughout respective cells. We next examined the localization of proteins in the context of infection by transfecting C. trachomatis-infected HeLa cultures. Chlamydiae were stained to indicate localization of inclusions. Expression of neither CopB nor CopB2 had an apparent impact on chlamydial infection since similar numbers of inclusions were detected in all transfected cells (data not shown). However, we did detect striking differences in protein localization. Changes in the distribution of GFP and GFP-CopB were not apparent in the presence of infection except that both proteins were efficiently excluded from inclusions. In striking contrast, concentration of GFP-CopB2 around the periphery of inclusions was prominently detected. These data raise the possibility that, like endogenous CopB (15), CopB2 is capable of association with inclusion membranes.
Fig. 5.
GFP-CopB2 colocalizes with inclusions when ectopically expressed in C. trachomatis-infected HeLa cells. pGFP-CopB and pGFP-CopB2 as well as the negative control, pEGFP, were transfected into uninfected or C. trachomatis L2-infected HeLa cells. Cultures were fixed at 18 h posttransfection, and localization of recombinant proteins (EGFP, GFP-CopB, and GFP-CopB2; green) was observed. HeLa cell nuclei of uninfected cultures were visualized by staining with DAPI (blue), whereas chlamydiae (red) in infected cultures were visualized with anti-GroEL and Alexa-594-conjugated secondary antibodies. Images were acquired via epifluorescence microscopy, and arrows indicate areas presented in insets. Scale bar, 10 μm.
Species-specific localization of 3XFT-CopB2.
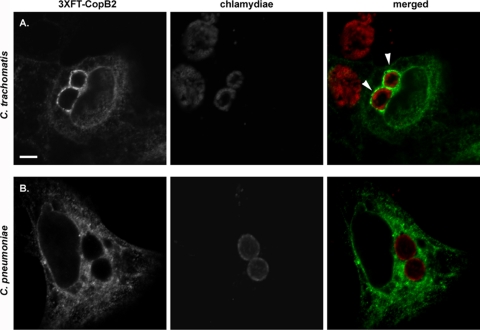
Given the interesting and potentially revealing localization of GFP-CopB2, we next sought to address the physiological relevance of GFP-CopB2 association with inclusion membranes. As noted above, the primary sequence of CopB2 orthologs in other chlamydial species is somewhat divergent. We therefore reasoned that species-specific association of CopB2 could be indicative of whether colocalization with inclusions represents function. To address this question and control for potential spurious effects arising from fusion partners, we examined the localization of Flag-tagged CopB2 during infections with either C. trachomatis or C. pneumoniae (Fig. 6). Similarly to GFP-CopB2, Flag-tagged CopB2 was prominently detected in apparent association with C. trachomatis inclusions. In contrast C. pneumoniae inclusions failed to colocalize with 3XFT-CopB2. This apparent species specificity greatly reduces the possibility that recombinant CopB2 spuriously associates with membranes. Instead, association with inclusions requires a defined environment particular to each species.
Fig. 6.
3XFT-CopB2 colocalization with C. trachomatis, but not C. pneumoniae, inclusions. HeLa cells were transfected with p3XFT-CopB2 after infection with C. trachomatis L2 (A) or C. pneumoniae AR-39 (B) and processed for microscopy at 18 h posttransfection. These times corresponded to ca. 24 h postinfection for C. trachomatis and 64 h for C. pneumoniae. Recombinant protein was detected using anti-FLAG (green), and chlamydiae were detected using MOMP-specific antibodies (red). Proteins were visualized by probing with appropriate secondary antibodies and viewed using confocal laser scanning microscopy. Arrows designate colocalization of 3XFT-CopB2 with chlamydial inclusions. Scale bar, 5 μm.
Requirement for chlamydial development in CopB2 localization.
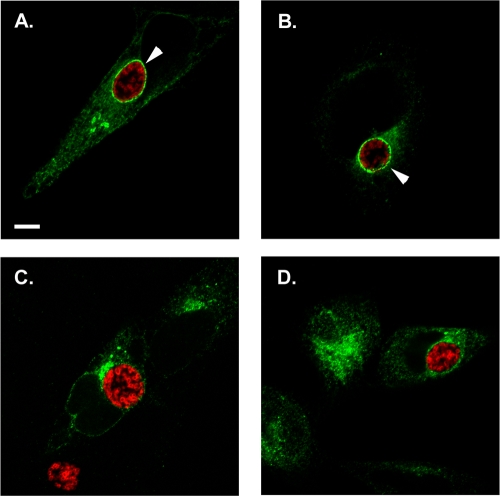
Given that endogenous CopB2 is also detectible during early development, we wanted to test the temporal manifestation of 3XFT-CopB2 association with inclusions. We therefore transfected HeLa cells with 3XFT-CopB2 and examined cultures after 8, 14, 18, and 22 h postinfection with C. trachomatis L2 (Fig. 7). At 8 h postinfection we were unable to detect significant association of 3XFT-CopB2 with chlamydiae. However, colocalization was evident at 14 h as concentrations of 3XFT-CopB2 surrounding chlamydial inclusions. Consistent with previous results, 3XFT-CopB2 was clearly visible in a rim-like pattern surrounding chlamydial inclusions at 18 and 22 h postinfection. To test the requirement for de novo chlamydial protein synthesis and T3S activity, we treated 3XFT-CopB2-transfected cells at 6 to 8 h postinfection with either chloramphenicol or INP007. Cultures were then fixed, and 3XFT-CopB2 was visualized at 22 h postinfection. Association of 3XFT-CopB2 with inclusions was not detected under either of these conditions. Collectively, these data indicate that the ability of CopB2 to associate with inclusions is developmentally dependent and suggest an interaction with protein(s) in the chlamydial inclusion membrane.
Deletion analysis of CopB2.
The observations that endogenous CopB2 did not behave biochemically like an integral membrane protein and association of recombinant CopB2 with inclusions seemed to require interaction with another protein(s) led us to consider which regions of CopB2 might be required for peri-inclusion localization. The primary sequence of CopB2 in all chlamydial species contains three domains predicted to form coiled-coil regions. Since this structural motif is often involved in mediating protein-protein interactions, we generated deletion mutants of CopB2 by removing these domains. Three constructs were generated containing in-frame deletions within 3XFT-CopB2 of coiled-coil domain 1 (residues 170 to 196), 2 (residues 276 to 311), or 3 (residues 470 to 506). These plasmids, along with full-length CopB2 as a control, were transfected into HeLa cells that were subsequently infected with C. trachomatis L2. Cultures were fixed at 18 h posttransfection (24 h postinfection), stained for chlamydiae and the Flag-tag of respective versions of CopB2, and analyzed by confocal microscopy (Fig. 8). Both full-length CopB2 and CopB2 lacking coiled-coil domain 1 were readily detected in association with chlamydial inclusions, whereas CopB2 lacking either coiled-coil domain 2 or 3 failed to localize to the peri-inclusion region. These data provide further evidence of specific interaction of CopB2 with inclusions and are consistent with protein-protein interactions being essential for this localization.
Fig. 8.
C-terminal domains of CopB2 are required for association with inclusion membranes. Full-length CopB2 (A) or constructs carrying in-frame deletions of residues 170 to 196 (B), 276 to 311 (C), or 470 to 506 (D) were expressed in C. trachomatis-infected HeLa cells for 18 h posttransfection. Flag-tagged CopB2 chimeras were detected using anti-FLAG (green), and chlamydiae were detected using MOMP-specific antibodies (red). Proteins were visualized by probing with appropriate secondary antibodies and viewed using confocal laser scanning microscopy. Arrows designate colocalization of CopB2 with chlamydial inclusions. Scale bar, 5 μm.
DISCUSSION
The process of T3S effector translocation remains an incompletely understood process. Basic characteristics have emerged, and it is clear from studies in other T3SSs that at least three proteins are secreted via T3S and directly participate in translocation (37). In Yersinia, a reasonable working model depicts a situation where the tip complex protein LcrV senses contact with a eukaryotic cell. Subsequent to this interaction, secreted YopD forms a scaffold for insertion and assembly of the integral membrane, YopB-containing translocation pore (reviewed in reference 37). Prior to secretion, a single chaperone interacts with both YopB and YopD to prevent their premature association in the bacterial cytoplasm (40). How chlamydiae may engage and deliver effector proteins into eukaryotic cells represents a largely unexplored question. Due to the complex developmental cycle exhibited by Chlamydia spp., the identity and role(s) of translocator proteins have important implications for infection biology. Both CopB and CopB2 have been intuitively implicated as potential pore proteins, but evidence corroborating this hypothesis has not been forthcoming. Indeed the difficulty of working with this class of proteins has confounded significant advances even in genetically tractable systems. We therefore tested whether the biochemical and spatial parameters of CopB and CopB2 are consistent with translocator function.
A comparatively strong case can be made for a role of CopB as a T3SS translocator protein. Importantly, CopB primary sequence contains areas of homology with YopB family proteins, and this area includes a conserved triad of hydrophobic domains capable of forming transmembrane helices. The C-terminal domain of CopB is more divergent, yet PSI-BLAST searches indicated ca. 43% similarity of this region with the predicted SseC-like translocator protein from Chromobacterium violaceum (7). Previous studies have revealed that CopB is secreted via a T3S mechanism and is detectible by immunolocalization in a pattern consistent with inclusion membrane localization (15). Similarly to Yersinia YopB, CopB interacts with a T3S chaperone (Scc2) that is encoded immediately upstream from its own gene (15, 49). Scc2 also interacts with CopD, the putative YopD-like translocator component (49).
Our data extend these observations and further corroborate a role of CopB in translocation. CopB is present in EBs, yet partitions as a nonmembrane protein after Triton X-114 extraction of purified EBs (17; also data not shown). This would be consistent with an interaction with Scc2 while associated with chlamydiae. However, CopB fractionated at the top of membrane flotation gradients and partitioned as an integral membrane protein after Triton X-114 extraction of infected cultures. Membrane association was detected at 20 to 24 h postinfection and is consistent with immunolocalization studies indicating an accumulation of CopB in inclusion membranes (15). Interestingly, Triton X-114 solubility was also apparent as early as 2 h postinfection. These data are consistent with early secretion of the EB pool of CopB and insertion into the nascent inclusion membrane. We were unable to recapitulate CopB membrane localization in ectopic expression studies. Epitope-tagged CopB was most often detected in punctate patterns when expressed in HeLa cells. These did not colocalize with fluorescent dextran added as a fluid-phase endocytic marker. CopB-containing punctate areas also did not colocalize with a panel of endocytic and Golgi markers including Lamp-1, GM-130, Rab4, Rab5, Rab6, or Rab9 (data not shown) and could represent multimeric aggregates of the protein.
If CopB truly represents a chlamydial translocation pore, it is unclear how T3S could function throughout development since our immunoblot data failed to detect CopB during a window roughly corresponding to midcycle development. One possibility is that T3S does not occur in the interim when CopB is not detectible. We regard this as unlikely, given that midcycle T3S effectors such as IncA and CT847 are secreted during this time. Both IncA-mediated homotypic fusion of chlamydial inclusions (20) and degradation of the CT847 target protein GCIP (13) are detectible by 12 h postinfection. Alternatively, a low-abundance pool of CopB could be present in the inclusion membrane during this time. It is possible that CopB is diffusible in the inclusion membrane and could enable translocation in trans instead of mediating flow of effectors in a contiguous process from bacterium to host. A contiguous, rim-like staining pattern is apparent for CopB by 18 to 20 h postinfection (15), and this is in contrast to a more punctate distribution typical of the needle protein CdsF (8). Moreover, Akopyan, et al., recently demonstrated that Yersinia and Salmonella effector proteins could be translocated via an intermediate step from the bacterial exterior (2). A final possibility is that an alternative translocator pore exists that could function in the absence of CopB. CopB2 represents a possible candidate for this role. CopB2 also contains hydrophobic domains, is encoded downstream of a T3SS chaperone and upstream of a putative YopD-like translocator protein, is secreted via the T3SS, and gains access to the host cytosol (15). Our immunoblot analysis indicated that a pool of CopB2 is present throughout the developmental cycle, and ectopic expression of CopB2 revealed an association with inclusions.
The data, however, are not entirely consistent with CopB2 functioning as a prototypical translocation pore. For example, the upstream-encoded T3S chaperone Scc3 does not associate with CopB2 (15) but instead appears to bind another secreted protein, CopN (48, 49). CopB2 does not exhibit apparent homology with any YopB family proteins. The apparent association of CopB2 with membranes is also unusual. While membrane flotation experiments did indicate that endogenous CopB2 is capable of associating with membranes, Triton X-114 extractions were inconsistent with CopB2 being an integral membrane protein. Detectible levels of tagged CopB2 association with inclusions required progression of the chlamydial developmental cycle. We did not see colocalization at 8 h postinfection, and arrest of the developmental cycle by addition of chloramphenicol or INP007 prevented inclusion association. Finally, although we cannot rule out aberrant protein folding in truncation mutants, colocalization required two coiled-coil domains. Although we did not rule out a role of the hydrophobic domain, the data taken together strongly support a peripheral association of CopB2 with inclusion membranes.
In the absence of a tractable genetic system, ectopic expression of recombinant chlamydial proteins has afforded one efficacious mechanism to investigate function. We took advantage of this approach to examine CopB2 but acknowledge the inherent risk in these artificial settings. Unfortunately, recombinant C. trachomatis CopB2 appears to be poorly antigenic (data not shown). This may also be the case with endogenous CopB2 during infection since antibody profiling studies of Chlamydia-infected humans fail to detect significant reactivity (54). After generating multiple CopB2-specific antibodies, we were unable to confirm inclusion membrane localization of endogenous CopB2. Our interpretation of the ectopic expression data is that the inclusion association is not a spurious artifact of protein overexpression. In the absence of infection, neither GFP- nor Flag-tagged versions of CopB2 exhibited a propensity to concentrate in eukaryotic membranes. Colocalization with inclusions was species specific since C. trachomatis CopB2 did not colocalize with C. pneumoniae inclusions. Indeed, the fact that CopB2 is less well conserved across species is consistent with this species specificity. Furthermore, epitope-tagged CopB2 did colocalize with the closely related C. muridarum inclusions but not with those of the more divergent C. caviae, and ectopically expressed C. pneumoniae Cpn1020 did not colocalize with C. trachomatis inclusions. It should be noted that recombinant Cpn1020 has also been ectopically expressed in C. pneumoniae-infected cells, but an inclusion membrane localization was not detected (38). It is therefore possible that peri-inclusion localization represents a divergent property of C. trachomatis CopB2.
In aggregate, the data support a working model (Fig. 9) in which CopB is an integral membrane translocator protein while CopB2 associates with inclusion membranes peripherally. The requirement of inclusion colocalization of CopB2 on developmental progression raises the possibility that CopB2 interacts with another chlamydial protein in the inclusion membrane. Although host-specific, spurious INP007 effects cannot be ruled out, the addition of iron to cultures did not restore 3XFT-CopB2 association with inclusions. These data therefore suggest that a type III substrate interacts with CopB2. Likely candidates include CopB2 itself or an Inc protein. However, we failed to detect a CopB2-CopB2 interaction during copurification of recombinant protein tests or in yeast two-hybrid or coimmunoprecipitation assays (data not shown). We have also not excluded the possibility that CopB2 interacts with a host protein interacting with the inclusion. Unfortunately, technical issues have confounded our strenuous efforts to identify an interaction with other proteins. It is currently unclear whether CopB2 could fulfill a role as a translocator protein. Inclusion association was not detected during the early cycle even though immunoblotting clearly indicates that endogenous protein is present. Also, endogenous C. pneumoniae CopB2 (Cpn1020) was detected in the cytosol of infected cells (31). It is possible that CopB2 has diverged to accomplish other functions. In fact, there is precedent for alternative functions of translocator class proteins. Shigella IpaB may indirectly affect host cell cytoskeletal rearrangements during invasion through interactions with the integrin CD44 (46) while Salmonella SipB has been detected in macrophage mitochondria, where it may lead to cell death by autophagy (24). Until systems to genetically modify chlamydiae are available, it will be difficult to provide definitive evidence for how CopB2 contributes to chlamydial pathogenesis.
Fig. 9.
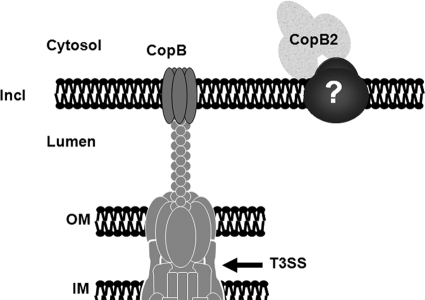
A working model for association of CopB and CopB2 with inclusion membranes. As T3S substrates, both CopB and CopB2 are secreted across the chlamydial inner and outer membranes (IM and OM, respectively) via the T3S apparatus to gain access to the inclusion membrane (Incl). CopB is predicted to form a prototypical translocation pore based on homology to YopB class translocator proteins and detection of endogenous CopB colocalization with the inclusion membrane (14). Association of CopB2 with the cytosolic face of inclusion membranes, however, is likely peripheral. Both the C-terminal and internal coiled-coil domains are involved in interactions with a chlamydial protein that is likely integral to the inclusion membrane.
ACKNOWLEDGMENTS
We thank H. Betts-Hampikian, H. Maslowski, and M. McKuen for critical reading of the manuscript. We also thank C. Linton for excellent technical assistance and G. Plano for fruitful discussions.
This work was supported by Public Health Service grants from the National Institutes of Health, NIAID (AI065530), to K. Fields. T. Hackstadt was supported by the Intramural Research Program of the NIAID/NIH.
Footnotes
Published ahead of print on 23 May 2011.
REFERENCES
- 1. Abdelrahman Y. M., Belland R. J. 2005. The chlamydial developmental cycle. FEMS Microbiol. Rev. 29:949–959 [DOI] [PubMed] [Google Scholar]
- 2. Akopyan K., et al. 2011. Translocation of surface-localized effectors in type III secretion. Proc. Natl. Acad. Sci. U. S. A. 108:1639–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Azuma Y., et al. 2006. Genome sequence of the cat pathogen, Chlamydophila felis. DNA Res. 13:15–23 [DOI] [PubMed] [Google Scholar]
- 5. Baehr W., et al. 1988. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc. Natl. Acad. Sci. U. S. A. 85:4000–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belland R. J., et al. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U. S. A. 100:8478–8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Betts H. J., Chaudhuri R. R., Pallen M. J. 2004. An analysis of type III secretion gene clusters in Chromobacterium violaceum. Trends Microbiol. 12:476–482 [DOI] [PubMed] [Google Scholar]
- 8. Betts H. J., Twiggs L. E., Sal M. S., Wyrick P. B., Fields K. A. 2008. Bioinformatic and biochemical evidence for the identification of the type III secretion system needle protein of Chlamydia trachomatis. J. Bacteriol. 190:1680–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Betts-Hampikian H. J., Fields K. A. 2010. The chlamydial type III secretion mechanism: revealing cracks in a tough nut. Front. Microbiol. 1:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bordier C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604–1607 [PubMed] [Google Scholar]
- 11. Burton M. J., Mabey D. C. 2009. The global burden of trachoma: a review. PLoS Negl. Trop. Dis. 3:e460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caldwell H. D., Kromhout J., Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chellas-Gery B., Linton C. N., Fields K. A. 2007. Human GCIP interacts with CT847, a novel Chlamydia trachomatis type III secretion substrate, and is degraded in a tissue-culture infection model. Cell Microbiol. 9:2417–2430 [DOI] [PubMed] [Google Scholar]
- 14. Clifton D. R., et al. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U. S. A. 101:10166–10171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fields K. A., Fischer E. R., Mead D. J., Hackstadt T. 2005. Analysis of putative Chlamydia trachomatis chaperones Scc2 and Scc3 and their use in the identification of type III secretion substrates. J. Bacteriol. 187:6466–6478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fields K. A., Hackstadt T. 2000. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol. Microbiol. 38:1048–1060 [DOI] [PubMed] [Google Scholar]
- 17. Fields K. A., Mead D. J., Dooley C. A., Hackstadt T. 2003. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol. Microbiol. 48:671–683 [DOI] [PubMed] [Google Scholar]
- 18. Gabel B. R., Elwell C. A., van Ijzendoorn S. C., Engel J. N. 2004. Lipid raft-mediated entry is not required for Chlamydia trachomatis infection of cultured epithelial cells. Infect. Immun. 72:7367–7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghosh P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hackstadt T., Scidmore-Carlson M., Shaw E., Fischer E. 1999. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell. Microbiol. 1:119–130 [DOI] [PubMed] [Google Scholar]
- 21. Hakansson S., et al. 1996. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 15:5812–5823 [PMC free article] [PubMed] [Google Scholar]
- 22. He S. Y., Nomura K., Whittam T. S. 2004. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta 1694:181–206 [DOI] [PubMed] [Google Scholar]
- 23. Hefty P. S., Stephens R. S. 2007. Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements. J. Bacteriol. 189:198–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hernandez L. D., Pypaert M., Flavell R., Galan J. E. 2003. A Salmonella protein causes macrophage cell death by inducing autophagy. J. Cell Biol. 163:1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hower S., Wolf K., Fields K. A. 2009. Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol. Microbiol. 72:1423–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hume P. J., McGhie E. J., Hayward R. D., Koronakis V. 2003. The purified Shigella IpaB and Salmonella SipB translocators share biochemical properties and membrane topology. Mol. Microbiol. 49:425–439 [DOI] [PubMed] [Google Scholar]
- 27. Jorgensen I., Valdivia R. H. 2008. Pmp-Like Proteins Pls1 and Pls2 are secreted into the lumen of the Chlamydia trachomatis inclusion. Infect. Immun. 76:3940–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kalman S., et al. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385–389 [DOI] [PubMed] [Google Scholar]
- 29. Kyte J., Doolittle R. F. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132 [DOI] [PubMed] [Google Scholar]
- 30. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 31. Lugert R., Kuhns M., Polch T., Gross U. 2004. Expression and localization of type III secretion-related proteins of Chlamydia pneumoniae. Med. Microbiol. Immunol. 193:163–171 [DOI] [PubMed] [Google Scholar]
- 32. Lupas A., Van Dyke M., Stock J. 1991. Predicting coiled coils from protein sequences. Science 252:1162–1164 [DOI] [PubMed] [Google Scholar]
- 33. Markham A. P., Jaafar Z. A., Kemege K. E., Middaugh C. R., Hefty P. S. 2009. Biophysical characterization of Chlamydia trachomatis CT584 supports its potential role as a type III secretion needle tip protein. Biochemistry 48:10353–10361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marlovits T. C., Stebbins C. E. 2010. Type III secretion systems shape up as they ship out. Curr. Opin. Microbiol. 13:47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moraes T. F., Spreter T., Strynadka N. C. 2008. Piecing together the type III injectisome of bacterial pathogens. Curr. Opin. Struct. Biol. 18:258–266 [DOI] [PubMed] [Google Scholar]
- 36. Moulder J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mueller C. A., Broz P., Cornelis G. 2008. The type III secretion system tip complex and translocon. Mol. Microbiol. 68:1085–1095 [DOI] [PubMed] [Google Scholar]
- 38. Muller N., Sattelmacher F., Lugert R., Gross U. 2008. Characterization and intracellular localization of putative Chlamydia pneumoniae effector proteins. Med. Microbiol. Immunol. 197:387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neyt C., Cornelis G. R. 1999. Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: requirement for translocators YopB and YopD. Mol. Microbiol. 33:971–981 [DOI] [PubMed] [Google Scholar]
- 40. Neyt C., Cornelis G. R. 1999. Role of SycD, the chaperone of the Yersinia Yop translocators YopB and YopD. Mol. Microbiol. 31:143–156 [DOI] [PubMed] [Google Scholar]
- 41. Pei J., Grishin N. 2007. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics 23:802–808 [DOI] [PubMed] [Google Scholar]
- 42. Read T., et al. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31:2134–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Read T. D., et al. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rice P., Longden I., Bleasby A. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 45. Rockey D. D., Scidmore M. A., Bannantine J. P., Brown W. J. 2002. Proteins in the chlamydial inclusion membrane. Microbes Infect. 4:333–340 [DOI] [PubMed] [Google Scholar]
- 46. Schroeder G. N., Hilhi H. 2008. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin. Microbiol. Rev. 21:134–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scidmore-Carlson M. A., Shaw E. I., Dooley C. A., Fischer E. R., Hackstadt T. 1999. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol. Microbiol. 33:753–765 [DOI] [PubMed] [Google Scholar]
- 48. Slepenkin A., M. de la Maza L., Peterson E. M. 2005. Interaction between components of the type III secretion system of Chlamydiaceae. J. Bacteriol. 187:473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Spaeth K. E., Chen Y. S., Valdivia R. H. 2009. The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog. 5:e1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stephens R. S., et al. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759 [DOI] [PubMed] [Google Scholar]
- 51. Tampakaki A. P. 2004. Conserved features of type III secretion. Cell Microbiol. 6:805–816 [DOI] [PubMed] [Google Scholar]
- 52. Thomson N. R., et al. 2005. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 15:629–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Valdivia R. H. 2008. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr. Opin. Microbiol. 11:53–59 [DOI] [PubMed] [Google Scholar]
- 54. Wang J., et al. 2010. A genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J. Immunol. 185:1670–1680 [DOI] [PubMed] [Google Scholar]
- 55. Wilson D. P., Whittum-Hudson J. A., Timms P., Bavoil P. 2009. Kinematics of intracellular Chlamydiae provide evidence for contact-dependent development. J. Bacteriol. 191:5734–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wolf K., et al. 2006. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol. Microbiol. 61:1543–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wolf K., Fischer E., Hackstadt T. 2000. Ultrastructural analysis of developmental events in Chlamydia pneumoniae-infected cells. Infect. Immun. 68:2379–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wolf K., et al. 2001. Chlamydia pneumoniae major outer membrane protein is a surface-exposed antigen that elicits antibodies primarily directed against conformation-dependent determinants. Infect. Immun. 69:3082–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yuan Y., Lyng K., Zhang Y. X., Rockey D. D., Morrison R. P. 1992. Monoclonal antibodies define genus-specific, species-specific, and cross-reactive epitopes of the chlamydial 60-kilodalton heat shock protein (hsp60): specific immunodetection and purification of chlamydial hsp60. Infect. Immun. 60:2288–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]