Abstract
Haemophilus ducreyi, the etiologic agent of chancroid, has an obligate requirement for heme. Heme is acquired by H. ducreyi from its human host via TonB-dependent transporters expressed at its bacterial surface. Of 3 TonB-dependent transporters encoded in the genome of H. ducreyi, only the hemoglobin receptor, HgbA, is required to establish infection during the early stages of the experimental human model of chancroid. Active immunization with a native preparation of HgbA (nHgbA) confers complete protection in the experimental swine model of chancroid, using either Freund's or monophosphoryl lipid A as adjuvants. To determine if transfer of anti-nHgbA serum is sufficient to confer protection, a passive immunization experiment using pooled nHgbA antiserum was conducted in the experimental swine model of chancroid. Pigs receiving this pooled nHgbA antiserum were protected from a homologous, but not a heterologous, challenge. Passively transferred polyclonal antibodies elicited to nHgbA bound the surface of H. ducreyi and partially blocked hemoglobin binding by nHgbA, but were not bactericidal. Taken together, these data suggest that the humoral immune response to the HgbA vaccine is protective against an H. ducreyi infection, possibly by preventing acquisition of the essential nutrient heme.
INTRODUCTION
The sexually transmitted genital ulcer disease (GUD) chancroid is caused by the Gram-negative bacterium Haemophilus ducreyi (reviewed in references 9, 22, and 44). Although chancroid is currently considered rare in the United States (http://www.cdc.gov/std/stats08/other.htm), outbreaks of H. ducreyi infection occurred in large American cities throughout the 1980s and 1990s (10, 25). During these times, chancroid was endemic in sub-Saharan Africa, Asia, and the Caribbean (6, 44). It is difficult to assess the current epidemiology of chancroid because of syndromic management of GUDs and a lack of reporting and diagnostic tools. Some publications regarding the epidemiology of GUDs have described declining numbers of chancroid cases worldwide (8, 29, 40), while others have shown that H. ducreyi infections are still found in pockets throughout the world (2, 5, 15, 31). H. ducreyi has recently been shown to be the cause of lower limb cutaneous ulcers in patients from the South Pacific (24, 30, 46). Chancroid is also an important cofactor in the heterosexual transmission of the human immunodeficiency virus (HIV) (18, 32) and may have been particularly critical early in the HIV epidemic (38).
H. ducreyi is an obligate human pathogen. Unable to synthesize heme, H. ducreyi is thought to acquire this essential compound from its host by binding hemoglobin (Hb) or free heme using the TonB-dependent transporters (TBDTs) HgbA and TdhA, respectively (11, 21, 27, 41). Only 3 TBDTs are expressed by H. ducreyi: HgbA, an Hb receptor; TdhA, a heme receptor; TdX, which has not been assigned a function and is not expressed by all H. ducreyi strains (19). An isogenic hgbA mutant of prototypical strain 35000HP is avirulent in the human and rabbit experimental models of chancroid (3, 39), proving that HgbA is a virulence factor for H. ducreyi. Conversely, a double tdhA/tdX mutant was fully virulent in the human experimental model (19), which suggests the following conclusions: (i) TdX and TdhA are not necessary for virulence in early steps of H. ducreyi infection in the experimental human model of chancroid; (ii) Hb is the most important source of heme for H. ducreyi; (iii) HgbA is the most important TBDT for acquisition of heme/iron by H. ducreyi.
By homology to other TBDTs, HgbA is thought to assume a pore-like structure in the outer membrane of H. ducreyi, with 22 β-strands, 11 putative surface-exposed loops, and a plug region present in the periplasm (B. Temple, unpublished data) (26). Using antisera from swine immunized with HgbA, our laboratory showed that loops 4, 5, and 7 of HgbA are immunogenic and that loop 4- and loop 5-specific antisera block Hb binding to HgbA (26). By generating single-loop deletion mutants of hgbA, we demonstrated that only deletion of loops 5 and 7 of HgbA substantially reduced Hb binding by HgbA. However, deletion of any loop of the HgbA protein prevented the use of Hb as a source of heme/iron by H. ducreyi (26). Taken together, these data indicate that a central domain of the primary amino acid sequence of HgbA is important for binding Hb by H. ducreyi.
Previous studies have shown that active immunization with a native preparation of HgbA purified from H. ducreyi prototypical class I strain 35000HP (nHgbAI) protects against a homologous challenge in the experimental swine model of chancroid (1, 13). Protection was observed when using either Freund's adjuvant or an adjuvant approved for use in humans, monophosphoryl lipid A (MPL). Anti-nHgbAI antisera from both vaccine trials bound HgbA at the surface of H. ducreyi and partially blocked binding of Hb to nHgbAI. These in vitro correlates of protection suggest that the humoral immune response elicited to the HgbA vaccine may be protective. To obtain evidence that antisera developed to the HgbA vaccine may protect against an infectious challenge, we performed classic passive immunization studies with antisera elicited against nHgbAI in the experimental swine model of chancroid.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
H. ducreyi strain 35000HP is the human-passaged variant (4) of wild-type isolate 35000 (14) and the prototypical strain for H. ducreyi class I strains (48). Strain FX547 is an isogenic hgbA deletion mutant of strain 35000HP (26), and FX548, an isogenic 35000HP strain in which the hgbA gene was replaced with hgbA of strain DMC111, a class II H. ducreyi strain (13). In this report, H. ducreyi strains 35000HP, FX547, and FX548 are designated 35000HPhgbAI, 35000HPΔhgbA, and 35000HPhgbAII, respectively. Other H. ducreyi strains used in this report include the 35000HP isogenic dsrA mutant FX517 (12) and the isogenic momp (43) and ompA2 mutants (20), as well as the gmhA mutant, termed 35000.252 (7).
H. ducreyi strains were routinely grown on chocolate agar plates (CAPs) containing gonococcal (GC) medium base (Difco, Detroit, MI) and 1% bovine Hb (Becton Dickinson, Sparks, MD) supplemented with 5% FetalPlex (Gemini Bio-Products, West Sacramento, CA) and 1% GGC (0.1% glucose, 0.001% glutamine, 0.026% cysteine) at 34.5°C in 5% CO2. For the purpose of nHgbAI purification and whole-cell binding enzyme-linked immunosorbent assays (ELISAs), H. ducreyi strains were cultured in low-heme GC broth (GCB; 1% GGC, 5% FetalPlex, and no addition of heme [1]).
Animals.
A total of eight Yorkshire Cross (York) pigs (four pigs in each of two separate passive immunization experiments) were obtained at 3 weeks of age and housed at ambient temperature (20 to 25°C) in individual pens at the North Carolina State University (NCSU) School of Veterinary Medicine. Animals were given water and antibiotic-free high-protein feed ad libitum beginning 3 weeks prior to the start of and throughout the study. During inoculation and biopsy procedures, pigs were sedated with 2 mg of ketamine-HCl (Fort Dodge Labs, Fort Dodge, IA) and 2 mg of xylazine (Miles Laboratories, Shawnee Mission, KS) per kg of body weight, injected intramuscularly. At the time of biopsy, pigs generally weighed between 15 and 25 kg. The Institutional Animal Care and Use Committees (IACUC) at NCSU approved the methods and use of animals for these experiments.
Preparation and passive immunization of the anti-nHgbAI polyclonal swine antisera.
A native preparation of the HgbA protein from class I H. ducreyi strain 35000HP (nHgbAI) was prepared as previously described from 12 liters of strain 35000HP grown in low-heme GCB (1). To ensure homogeneity of the nHgbAI preparation used for active immunization, the preparation was monitored by SDS-PAGE followed by Coomassie and silver staining (45), as well as Western blotting with monoclonal antibody 2C7 to assess for the presence of the major outer membrane proteins MOMP and OmpA2 (37). Based on these assays, the nHgbAI protein preparation was over 95% pure, with very little contamination with lipooligosaccharide (LOS) or MOMP/OmpA2 proteins (data not shown).
To generate the nHgbAI antisera, nHgbAI protein was sent to Covance (Custom Immunology Services, Denver, PA) for immunization of four York pigs. Since all published pig studies, including the passive challenge described herein, were done at NCSU, it is likely that the pigs used to develop anti-nHgbAI at Covance came from a family line unrelated to the animals used for passive immunization. This may explain the cross-reactivity seen in the antisera from these animals (see Fig. 3 and 4 below). At Covance, each pig received three immunizations of 250 μg of nHgbAI in complete (first immunization) and incomplete (second and third immunizations) Freund's adjuvant (Sigma-Aldrich, St. Louis, MO) at 3-week intervals, exactly as previously described (1). Three weeks after the last immunization, animals were exsanguinated, and serum was extracted from blood, aliquoted, and frozen at −20°C before being sent to our laboratory.
Fig. 3.
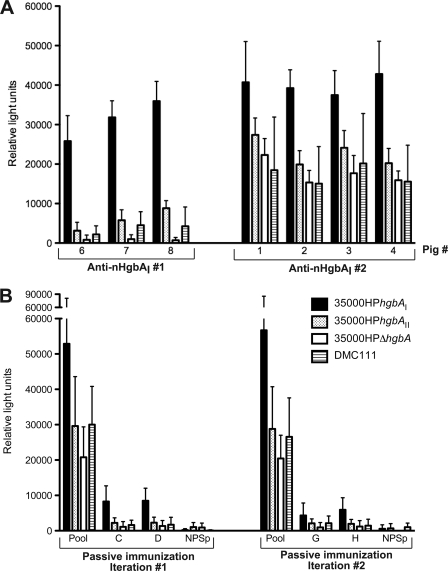
Whole-cell binding of anti-nHgbAI Abs. (A) The reactivities of the Abs from individual pigs actively immunized with nHgbAI (pigs 1 to 4; anti-nHgbAI 2) to 4 different H. ducreyi strains (see the legend for Fig. 3B) were tested in a whole-cell binding ELISA. Antisera from the first nHgbAI/Freund's vaccine trial (anti-nHgbAI 1, pigs 6 to 8) were used positive controls. (B) Pooled nHgbAI antisera used for infusion in each passive immunization experiment and antisera from passively immunized animals (animals C, D, G, and H) were tested in a whole-cell binding assay using the same H. ducreyi strains as shown in Fig. 3A. Pool, anti-nHgbAI 2 pool; NPSp, normal pig serum pool.
Fig. 4.
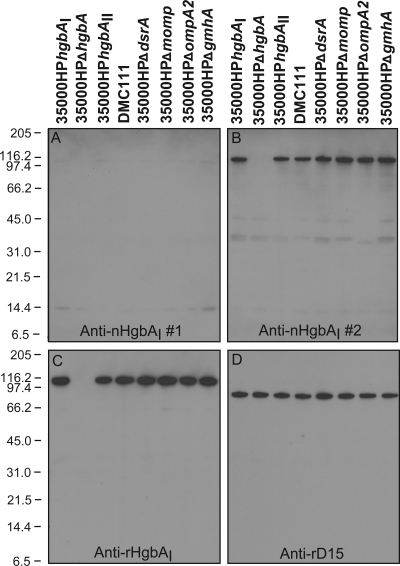
Reactivities of anti-HgbAI Abs to total cellular proteins from a panel of H. ducreyi strains. The reactivities of pooled anti-nHgbAI 2 to total cellular proteins from H. ducreyi wild-type strains 35000HPhgbAI and DMC111 and several mutants in the 35000HP background were determined in Western blot assays. Anti-nHgbAI 1, pooled swine polyclonal nHgbAI antiserum from a previous nHgbA vaccine trial (pool of antisera from pigs 6, 7, and 8) (1); anti-nHgbAI 2, pooled swine polyclonal nHgbAI antiserum from the current nHgbA vaccine trial (pool of antisera from pigs 1, 2, 3, and 4); anti-rHgbAI, rabbit polyclonal antiserum to denatured rHgbA (28); anti-rD15, rabbit polyclonal antiserum to denatured rD15 (42). The Western blot with anti-rD15 shows equal loading.
Two days prior to infusion, 50 ml of antiserum from each of the 4 nHgbAI-immunized pigs was pooled into one preparation, filter sterilized, and divided into 50-ml aliquots. A pool of normal pig serum (NPS) was prepared in the same manner from sera obtained from three pigs from the NCSU Veterinary School which had previously been used for purposes other than infection (surgery) and were already scheduled for euthanasia. The aliquots were kept at 4°C until the day of the infusion. On the day of passive immunization, animals were sedated as described above. Fifty milliliters of blood was removed from each animal, and 50 ml of pooled nHgbAI antiserum or NPS was passively administered through the brachiocephalic vein at the trunk by using a 60-ml syringe and a 1.50-in. 20-gauge needle. Depending on the size of the pigs (between 15 and 25 kg), and assuming a mean blood volume of 62.5 ml/kg (http://www.iacuc.ucsf.edu/Proc/awSwineNorm.asp), the infused nHgbAI antisera accounted for 3 to 5% of the blood volume of passively immunized animals. Preparations of anti-nHgbAI and NPS were tested for the presence of endotoxin by using the end point chromogenic Limulus amebocyte assay from Lonza (catalog number 50-647U) following the manufacturer's instructions. All infused sera had endotoxin concentrations below 1 endotoxin unit/ml (data not shown).
For experiments described here, sera generated in the previously published active immunization HgbA vaccine trial with Freund's adjuvant (1) were used as positive controls in many assays (see Fig. 3 to 6 below). These sera were purified from blood taken 3 weeks after the third immunization (prior to infection) from nHgbAI/Freund's-immunized animals (pigs number 6, 7, and 8; serum from pig 5 was unavailable).
Fig. 6.
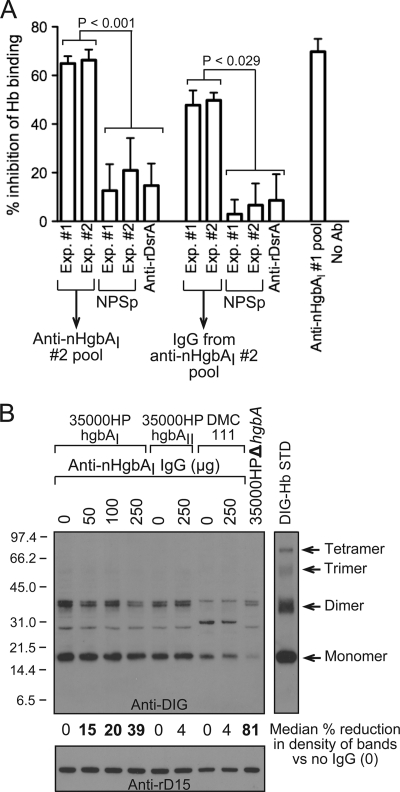
Anti-nHgbAI Abs partially block Hb binding by HgbA. (A) The abilities of pooled nHgbAI antisera or purified IgG to block binding of DIG-Hb to nHgbAI were measured using an Hb blocking ELISA. The anti-nHgbAI 1 pool from a previous study was used as a positive control (1), while the irrelevant polyclonal antisera to the outer membrane trimeric autotransporter DsrA (anti-rDsrA) was used as a negative control (48). Results are expressed as the percentage of a no-antibody control, arbitrarily defined as 0% inhibition of DIG-Hb binding to nHgbA. Data were compared using a Mann-Whitney rank sum test. NPSp, normal pig serum pool. (B) The abilities of anti-nHgbAI 2 IgG to block binding of DIG-Hb to H. ducreyi strains 35000HPhgbAI, 35000HPhgbAII, and DMC111 were analyzed using a whole-cell Hb blocking assay. Bands on the Western blot were analyzed with NIH Image (version 1.62) and are arbitrarily expressed as the percent reduction in band density compared to the strain plus DIG-Hb without the addition of IgG (indicated as 0). Bold numbers indicate a statistically significant reduction in band density (Mann-Whitney rank sum test). The DIG-Hb lane (10 ng) served as a standard (STD). Shown is a representative Western blot from 4 different experiments with similar results.
ELISA studies.
A direct ELISA (see Fig. 2, below) was used to evaluate the reactivities of individual and pooled nHgbAI antisera to purified nHgbA, using changes to the protocol previously described (1, 13). An indirect ELISA, based on a kit from Bethyl Laboratories (catalog number E100-104; Montgomery, TX), was used to measure the quantity of nHgbAI-specific IgGs in the passively transferred antisera and the antisera from passively immunized animals. In this assay, wells coated either with nHgbAI or goat anti-pig antibodies (Abs) from the Bethyl kit were incubated with dilutions of the passively transferred antisera or antisera from animals passively immunized with anti-nHgbAI. Pig sera with known amounts of IgG were added to wells with anti-pig IgG to generate a standard curve. Wells were thereafter washed, incubated with anti-pig IgG conjugated to horseradish peroxidase (HRP), and developed as previously described (1, 13). The amount of nHgbAI-specific IgG in the pig antisera was determined by comparing the optical density obtained with the antisera to that of the standard curve (49). A whole-cell binding ELISA was also used to measure the reactivity of the antisera to HgbA expressed on the surface of intact H. ducreyi strains 35000HPhgbAI, 35000HPhgbAII, 35000HPΔhgbA, and DMC111. A vacuum manifold was used to remove unbound components of the antisera, and Ab reactivity was determined using HRP-conjugated anti-pig IgGs, as previously described (1, 13).
Fig. 2.
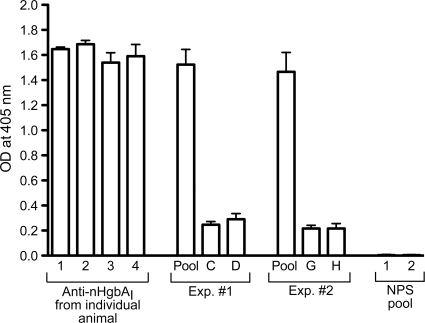
Reactivity of anti-nHgbAI Abs to purified nHgbAI. The activity of the nHgbAI/Freund's antisera was measured using an ELISA with nHgbAI as coating antigen. Data are expressed as OD405 readings, shown as means ± standard deviations obtained from at least 3 separate experiments. The 4 left bars indicate the reactivity of the antiserum from each donor pig actively immunized with nHgbAI. The middle section of the graph (Exp. 1 and 2) shows the reactivity of the pooled antisera for each passive immunization experiment (pool) and that of the antiserum from each animal after infusion with the pooled nHgbAI antiserum (C, D, G, and H). The right side of the graph shows the reactivity of NPS to purified nHgbAI. Antisera were diluted 1:5,000.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
Total cellular proteins (from 2.5 × 107 CFU of H. ducreyi strains grown in low-heme GCB) were separated on a 4-to-12% gradient denaturing/reducing bis-Tris NuPAGE gel (Invitrogen, Carlsbad, CA) (150 constant volts) and transferred onto nitrocellulose for 2 h at 200 mA. The nitrocellulose was stained with Ponceau S for 10 min to monitor loading of the wells. After an overnight incubation in blocking solution (0.5% Tween 20 in phosphate-buffered saline [PBS]), four nitrocellulose membranes with the same bacterial antigens were concurrently processed and developed with the following antisera for 1 h at room temperature: anti-nHgbAI 1 (1), anti-nHgbAI 2 (current study), anti-recombinant HgbAI (rHgbAI) (28), all at 1:25,000, and anti-rD15 at 1:10,000 (42). After three 10-min washes with 0.05% Tween 20-PBS, blots were incubated with an alkaline-phosphatase (AP)-conjugated anti-pig or anti-rabbit secondary Ab for 1 h at room temperature. Blots were washed 3 more times before development with the AP chemiluminescence substrate Lumi-Phos WB (Thermo Scientific, Rockford, IL).
Immunoprecipitation.
H. ducreyi strains were grown overnight under heme-limiting conditions to induce maximal expression of HgbA (11). Cultures were centrifuged and pellets resuspended in GCB to an optical density at 600 nm (OD600) of 1.0 (approximately 5 × 108 CFU/ml). Ten microliters of serum was added to 1 ml of the bacterial suspension in a microcentrifuge tube and rocked at room temperature for 20 min. To remove unbound antibody and serum components, the suspension was centrifuged for 1 min at 14,000 rpm, the supernatant discarded, and the cell pellet washed with 1 ml GCB. The cell pellet was resuspended in 100 μl PBS, and 1 ml of 2% Zwittergent 3,14 (ZW 3,14) in TEN buffer (50 mM Tris-HCl, 5 mM EDTA, 150 mM NaCl, pH 8.0) was added to solubilize proteins. After incubation at 37°C with agitation for 1 h, the tube was centrifuged for 10 min at 14,000 rpm to remove insoluble debris. The supernatant (1.0 ml), containing ZW 3,14-soluble proteins and Ab complexes, was moved to a new tube containing 25 μl of a 50% slurry of protein A/G-agarose beads (ExAlpha Biologicals, Shirley, MA). The tubes were incubated for 2 h to allow binding of Ab (and their respective bound cognate antigens) to protein A/G, centrifuged, and then washed thrice using 0.5% ZW 3,14 in TEN. The agarose pellet was resuspended in 1.0 ml TEN and moved to a fresh tube and centrifuged, and the supernatant was discarded. Forty microliters of 1× Laemmli sample buffer lacking any reducing agents was added to the washed agarose, the tubes were boiled for 5 min at 95°C, and 15 μl was subjected to a 4-to-12% gradient SDS-PAGE gel and rapid Coomassie blue staining (20-min soak in 37 ml of 0.114% [wt/vol] Coomassie blue R-250 plus 20 ml of 0.0214% [wt/vol] Bismark Brown in 40% methanol, 7% acetic acid).
Hb blocking assays.
The ability of anti-nHgbAI to block binding of digoxigenin-labeled Hb (DIG-Hb) to nHgbAI was measured using two methods: an Hb-blocking ELISA, as previously described (13), and a whole-cell blocking ELISA. For both methods, 1 mg of bovine Hb (Sigma-Aldrich, St. Louis, MO) was labeled with DIG following the manufacturer's instructions (Roche Diagnostics GmbH, Germany) and frozen at −80°C. In the Hb blocking ELISA, wells of an ELISA plate (Costar flat-bottom, high binding plate; catalog number 3590; Cambridge, MA) were incubated overnight with 100 ng of nHgbAI before blocking with 2% bovine serum albumin (BSA) in PBS. Purified anti-nHgbAI IgG (20 μg of IgG purified using a protein A/G resin) was then added to each well and incubated for 30 min before addition of 400 ng of DIG-Hb in 1% BSA-PBS, allowing the incubation to continue for an additional hour. Wells were washed 3 times with 0.05% Tween 20-PBS, and then AP-conjugated anti-DIG (1:5,000; Roche Diagnostics, Indianapolis, IN) was added to each well and incubated for 1 h. After 3 more washes, the One-Step PNPP substrate (Pierce, Rockford, IL) was added to the wells, the plate incubated for 45 min, and optical density was measured at 405 nm by using the 1420 Victor2 multilabel reader (Perkin-Elmer, MA).
For the whole-cell Hb blocking assay, 1-ml aliquots of suspensions at an OD600 of 0.5 of H. ducreyi strains 35000HPhgbAI and 35000HPΔhgbA (negative control), grown in low-heme GCB, were mixed with 50, 100, or 250 μg of anti-nHgbAI IgG (purified using protein A/G) for 30 min at room temperature. DIG-Hb (200 ng) was then added to the bacteria/IgG suspensions and incubated for another 30 min at room temperature. The cells were subsequently washed 3 times with GCB, moved to a new microcentrifuge tube, and washed one more time in GCB. The bacterial cell pellets were suspended in Laemmli sample buffer containing β-mercaptoethanol and subjected to SDS-PAGE (4-to-12% gradient gel) and Western blotting (1 h at 200 mA) with an AP-conjugated anti-DIG Ab (1:500; 2 h at room temperature; Roche Diagnostics, Germany). Blots were developed using an AP chemiluminescent substrate as described above.
Infection and processing of biopsy specimens.
Twenty-four hours after infusion of the pooled nHgbAI antiserum or the NPS, ears of passively immunized animals were infected with H. ducreyi strains 35000HPhgbAI and 35000HPhgbAII by using multitest skin test applicators (Lincoln Diagnostics, Decatur, IL) as previously described (1, 13). Inocula were prepared by scraping H. ducreyi grown on CAPs (grown for 15 h at 34.5°C in 5% CO2) in GCB (OD600, 1.5; approximately 1 × 109 CFU/ml). Prior to inoculation, the ears of the animals were thoroughly washed with isopropyl alcohol-soaked wipes. The left ear of each animal was inoculated in separate sites with strain 35000HPhgbAI at 104 CFU (10 μl of a 109-CFU/ml inoculum; [1.06 ± 0.19] × 104 CFU [mean ± standard deviation]) or 103 CFU (10 μl of a 108-CFU/ml inoculum; [0.924 ± 0.17] × 103 CFU). The right ear was similarly infected with approximately 104 or 103 CFU of 35000HPhgbAII ([1.11 ± 0.2] × 104 CFU and [1.03 ± 0.34] × 103 CFU, respectively). Seven days after infection, six 6-mm biopsy punches of the lesion sites were removed from each ear and processed for culture and histology. Four biopsy specimens for each strain and inoculum size were minced with a sterile scalpel and incubated for up to 72 h on CAPs supplemented with 1% GGC, 5% FetalPlex, and 3 μg/ml vancomycin at 34.5°C in 5% CO2. Cultures of H. ducreyi were confirmed by colony characteristics (color, morphology, and cohesiveness) and PCR using primers specific for hgbAI and hgbAII (13). The remaining 2 biopsy specimens were incubated in 4% paraformaldehyde prior to hematoxylin and eosin (H&E) staining and processing at the Histology Laboratory of the College of Veterinary Medicine at NCSU. A Leica DM IRB inverted microscope (Leica Microsystems, Bannockburn, IL) was used to view the slides, and images were saved using Q capture software (Q-Imaging, Surrey, BC, Canada). Slides were graded independently by two persons using the previously described histologic scale (1, 13, 35). Cohen's kappa statistic for the two raters (κ = 0.544) indicated moderate agreement.
Statistics.
Statistical analyses were performed using Sigma Stat (version 3.5; Systat Software, Chicago, IL).
RESULTS
Passive immunization with anti-nHgbAI protected pigs from a homologous challenge.
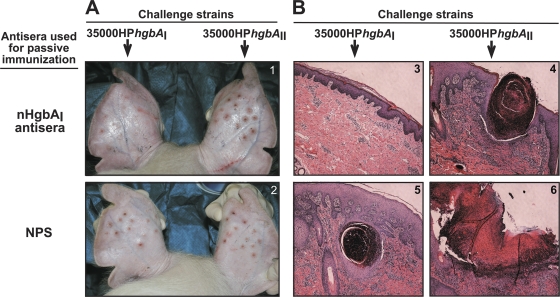
Two parameters were used to determine if passive immunization with anti-nHgbAI protected against a challenge: tissue damage and bacterial recovery. Tissue damage was measured by determining the severity of the lesions at the macroscopic and microscopic levels. Macroscopic examination of the sites infected with the homologous strain 35000HPhgbAI in animals passively immunized with anti-nHgbAI revealed a few small, pink lesions, and in most cases, no lesion was visible, except for markings left by the applicator device (Fig. 1A, panel 1, left ear). Conversely, lesions resulting from infection with the heterologous strain 35000HPhgbAII, an isogenic strain of H. ducreyi 35000HP that differs only in the expression of the heterologous HgbAII instead of HgbAI, were larger, raised, red, and inflamed (Fig. 1A, panel 1, right ear), similar to sites that developed in animals passively immunized with NPS after infection with either 35000HPhgbAI or 35000HPhgbAII (Fig. 1A, panel 2).
Fig. 1.
Macroscopic (A) and microscopic (B) examination of experimental H. ducreyi lesions from passively immunized pigs. Anti-nHgbAI (top) or NPS (bottom) was passively transferred to naïve pigs 24 h prior to a challenge with homologous (35000HPhgbAI) or heterologous (35000HPhgbAII) H. ducreyi strains. (A) Representative photographs of pig ears challenged with either H. ducreyi strain 35000HPhgbAI (left ear) or 35000HPhgbAII (right ear). Photos were taken 7 days after an infectious challenge with the designated strains and immediately before biopsy specimens were harvested. (B) Representative H&E-stained biopsy specimens of lesions from passively immunized animals challenged with either H. ducreyi strain 35000HPhgbAI (panels 3 and 5) or 35000HPhgbAII (panels 4 and 6). Magnification, ×50.
Microscopic analysis of H&E-stained biopsy specimens was consistent with macroscopic observations. Sites infected with strain 35000HPhgbAI in animals passively immunized with anti-nHgbAI showed a low-level inflammatory infiltrate, and the dermis, epidermis, and basement membranes were intact (Fig. 1B, panel 3). Conversely, biopsy specimens from NPS-immunized animals infected with the homologous strain showed destruction of the dermis and epidermis and a massive inflammatory infiltrate (Fig. 1B, panel 5). A large influx of inflammatory cells and tissue destruction was also the hallmark of sites infected with the heterologous strain, regardless of the antiserum used to passively immunize the animals (Fig. 1B, panels 4 and 6). H&E-stained biopsy sections were graded using the 1 to 5 grading system previously described (1, 13). Briefly, a score of 1 was assigned to healthy skin, while 5 characterized a fully developed ulcerative lesion. Animals passively immunized with anti-nHgbAI and infected with the homologous strain had a mean lesion grade of 1.81 ± 1.1, compared to 4.42 ± 0.74 in animals infused with NPS (P < 0.001, t test). Conversely, there was not a statistically significant difference between the mean lesion grades of sites infected with the heterologous strain in nHgbAI antisera-infused animals (4.1 ± 1.1) and animals that received NPS (3.75 ± 1; P = 0.567, t test).
Bacterial recovery was determined by culturing lesions taken from animals immunized with either anti-nHgbAI or NPS. At the lower inoculum dose (103 CFU), animals that received the nHgbAI antiserum were completely protected from a homologous challenge; we were unable to recover viable H. ducreyi from any of the 16 sites biopsied from 4 pigs (Table 1), compared to recovery of viable bacteria from all 12 sites in 3 pigs that received NPS (P < 0.001, Fisher's exact test). At the higher inoculum dose (104 CFU), viable homologous H. ducreyi organisms were recovered from 3 out of 16 sites biopsied from the 4 animals immunized with the nHgbAI antiserum (Table 1), compared to culture from all 12 lesions recovered in the 3 NPS-immunized animals (P < 0.001). H. ducreyi 35000HPhgbAII expressing the heterologous HgbA protein was recovered from all sites on all animals, regardless of their immunization or the inoculum size (Table 1). Thus, there was complete protection from homologous infection at the lower challenge dose but no protection from a heterologous challenge was observed.
Table 1.
Recovery of H. ducreyi from immunized pigsa
| Challenge strain | Expt no. | NPS immunization |
Anti-nHgbAI immunization |
||||
|---|---|---|---|---|---|---|---|
| Pig |
H. ducreyi-positive biopsies (% of 4 total biopsies) after inoculation with: |
Pig |
H. ducreyi-positive biopsies (% of 4 total biopsies) after inoculation with: |
||||
| 103 CFU | 104 CFU | 103 CFU | 104 CFU | ||||
| Class I strain, | 1 | C | 0 | 0 | |||
| 35000HPhgbAI | B | 4 (100) | 4 (100) | D | 0 | 0 | |
| 2 | E | 4 (100) | 4 (100) | G | 0 | 2 (50) | |
| F | 4 (100) | 4 (100) | H | 0 | 1 (25) | ||
| Total | 12 (100) (A) | 12 (100) (B) | 0 (0) (A) | 3 (19) (B) | |||
| Class II strain, | 1 | C | 4 (100) | 1 (25) | |||
| 35000HPhgbAII | B | 4 (100) | 4 (100) | D | 4 (100) | 4 (100) | |
| 2 | E | 4 (100) | 4 (100) | G | 4 (100) | 4 (100) | |
| F | 4 (100) | 4 (100) | H | 4 (100) | 4 (100) | ||
| Total | 12 (100) | 12 (100) | 16 (100) | 13 (81) | |||
Results followed by the same uppercase letter were significantly different: A and B, P < 0.001.
Potential mechanisms of protection of anti-nHgbAI.
There are several mechanisms that may account for the protection observed in this passive immunization trial with anti-nHgbAI. From active immunization trials (1, 13), correlates of protection of the nHgbAI vaccine included cell surface binding as well as bactericidal activity and blocking of Hb binding; there was no indication that opsonophagocytosis was involved in the mechanism of protection of the HgbA vaccine (1). Sera from individual animals actively immunized with nHgbAI, the antisera used for passive transfer, as well as sera from passively immunized animals were therefore tested for reactivity to nHgbAI, binding to viable H. ducreyi, and bactericidal and Hb blocking activities.
Anti-nHgbAI binds purified nHgbAI.
The reactivities of individual and pooled antisera to nHgbAI were first tested in a direct ELISA with purified nHgbAI. As shown in Fig. 2, sera from each of the 4 animals actively immunized with nHgbAI (Fig. 2, left, numbers 1 through 4), as well as the pooled sera delivered in both passive immunization experiments (Fig. 2, pool), were highly reactive to purified nHgbAI. Sera obtained from swine 24 h after passive immunization with the nHgbAI antisera (Fig. 2, bars C, D, G, and H) also showed reactivity to nHgbAI, albeit at lower levels than sera from animals actively immunized with nHgbAI. Based on the relative size of the animals and the assumption that pigs have a mean blood volume of 62.5 ml per kg of weight (see Materials and Methods for more details), the dilution of the antisera was about 1/20 to 1/30 (3 to 5% of blood volume). Pooled antisera from naïve pigs (NPS pool) did not react to purified nHgbAI (Fig. 2).
The amount of nHgbAI-specific Abs present in the antisera of passively immunized animals was also measured using an indirect ELISA. The concentrations of nHgbAI-specific Abs in antisera from pigs C and D were 0.233 ± 0.05 and 0.277 ± 0.055 mg/ml, respectively. In sera from pigs G and H, the concentration of anti-nHgbAI-specific Abs was 0.177 ± 0.025 and 0.183 ± 0.045 mg/ml, respectively.
Anti-nHgbAI 2 exhibited cross-reactivity to H. ducreyi strain 35000HPΔhgbA in a whole-cell binding ELISA.
To avoid confusion between antisera produced using Freund's adjuvant, antisera from the study published in 2006 (1) were designated anti-nHgbAI 1 and the antisera from the present study were designated anti-nHgbAI 2. nHgbAI antisera 1 was used in the various in vitro assays as a positive control.
In both active immunization trials with the HgbA vaccine, using either Freund's or MPL adjuvants, reactivities of the antisera to HgbA at the surface of H. ducreyi correlated with protection (1, 13). To determine if nHgbAI antisera 2 bound HgbA at the surface of intact, viable H. ducreyi cells, individual and pooled nHgbAI antisera were subjected to a whole-cell binding ELISA. Reactivities of these different antisera to H. ducreyi strains 35000HPhgbAI, 35000HPhgbAII, and DMC111 were compared to the reactivity of the antisera to the isogenic hgbA mutant 35000HPΔhgbA. There was high reactivity of all individual and pooled anti-nHgbAI 2 to H. ducreyi strain 35000HPhgbAI (Fig. 3A, right, and B, pool, black bars). However, anti-nHgbAI 2 also exhibited high reactivities to strains 35000HPΔhgbA, 35000HPhgbAII, and DMC111 (Fig. 3A, right), which was not present in anti-nHgbAI 1 (Fig. 3A, left).
To identify the antigen(s) targeted by the cross-reactive anti-nHgbAI 2, Western blot assays were performed using total cellular protein from H. ducreyi strains 35000HPhgbAI, 35000HPΔhgbA, 35000HPhgbAII, DMC111, and a panel of isogenic H. ducreyi mutants grown in low-heme GCB. Anti-nHgbAI 1 showed little reactivity to any denatured bacterial component, including HgbA, in a Western blotting format (Fig. 4A), as previously described (1); however, anti-nHgbAI 2 reacted with denatured HgbA (Fig. 4B). Anti-nHgbAI 2 also showed minor reactivity to the major outer membrane proteins MOMP and OmpA2 (range, 31 to 45 kDa) (16). We were able to identify these bands as MOMP and OmpA2, as they were absent in the lanes containing isogenic mutants 35000HPΔmomp and 35000HPΔompA (Fig. 4B).
Anti-nHgbAI 2 binds HgbA at the surface of H. ducreyi as measured in an immunoprecipitation assay.
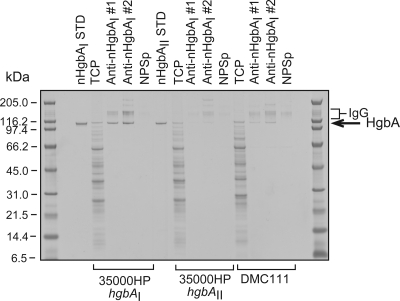
To ensure that anti-nHgbAI 2 bound HgbA in a native conformation in the context of whole H. ducreyi cells and to identify putative targets of the cross-reactivity displayed in the whole-cell binding ELISA with these pooled antisera, anti-nHgbAI 1 and 2 pools as well as control antisera were subjected to an immunoprecipitation assay (Fig. 5). Anti-nHgbAI 2 precipitated only HgbA from H. ducreyi strain 35000HPhgbAI grown in a low-heme culture. To confirm that this band was HgbA, these same immunoprecipitation samples were subjected to a Western blot assay using a rabbit polyclonal anti-rHgbAI antibody (28). The anti-rHgbAI Ab recognized a band only in those lanes where an HgbA band appeared in the Coomassie-stained gel (data not shown). Conversely, control lanes loaded with material from an immunoprecipitation assay using only protein A-agarose or agarose and IgG yielded either no band or bands that comigrated with the bands labeled IgG, respectively, in a Coomassie-stained SDS-PAGE gel (data not shown). Neither antiserum precipitated nHgbAII from strain 35000HPhgbAII, but both did so at a low level in strain DMC111. Pooled antiserum from naïve pigs (NPS) did not precipitate any protein from any of the tested strains.
Fig. 5.
Anti-nHgbAI Abs bind to the surface of H. ducreyi. The abilities of pooled nHgbAI antisera to bind HgbA at the surface of H. ducreyi strains 35000HPhgbAI, 35000HPhgbAI, and DMC111 were measured in an immunoprecipitation assay. Material obtained from the immunoprecipitation assay was subjected to a 4 to 12% SDS-PAGE and rapid Coomassie blue staining. Pooled antisera used in this assay were obtained from animals immunized 3 times with nHgbAI in Freund's adjuvant, either from a previous study (anti-nHgbAI 1) (1) or obtained in the course of the current study (anti-nHgbAI 2). nHgbAI STD and nHgbAII STD, 0.5 μg of purified nHgbAI or nHgbAII, respectively; TCP, total cellular protein from 1 × 107 CFU; NPSp, normal pig serum pool. Shown is a representative gel from at least 4 different experiments with similar results.
Anti-nHgbAI 2 partially blocks binding of DIG-Hb to HgbA.
Based on the data from the whole-cell ELISA and immunoprecipitation assays (Fig. 3 and 5, respectively) and the fact that anti-nHgbAI 2 protected against a homologous challenge (Fig. 1), this second preparation of nHgbAI antiserum appears to bind HgbA in its native conformation. We therefore studied the biological activities of this antiserum, including bactericidal activity and its capacity to block Hb binding to HgbA. No bactericidal activity was detected in any nHgbAI antisera (data not shown). However, both pools of anti-nHgbAI 2, as well as IgG purified from these antisera, partially blocked binding of DIG-Hb to purified nHgbAI in an ELISA-type assay (Fig. 6A). This activity was observed in ranges similar to that obtained with anti-nHgbAI 1 (69% inhibition compared to 64 to 66% inhibition for anti-nHgbAI 2). Anti-nHgbAI 2 and IgG blocked DIG-Hb binding to HgbA significantly better than NPS or irrelevant anti-rDsrA antisera and IgG (Fig. 6A).
To determine whether anti-nHgbAI 2 could block Hb binding in the context of viable bacteria, we developed a whole-cell Hb blocking assay. As shown in Fig. 6B, addition of 50, 100, or 250 μg of anti-nHgbAI IgG significantly reduced the density of the Hb band on the Western blot by 15, 20, and 39%, respectively (P = 0.026 for 50 μg, P = 0.004 for 100 μg, and P < 0.001 for 250 μg of anti-nHgbAI 2 IgG; Mann-Whitney rank sum test), consistent with the ability of the antisera to block Hb binding to purified nHgbAI in the ELISA (Fig. 6A, 47 and 49% reduction for anti-nHgbAI 2 IgG experiments 1 and 2, respectively). In contrast, 250 μg of anti-nHgbAI 2 IgG did not significantly reduce binding of DIG-Hb to H. ducreyi strains 35000HPhgbAII and DMC111 (4% reduction in band density, compared to results with no addition of anti-nHgbAI IgG), which express a class II HgbA protein on the bacterial surface.
DISCUSSION
Passive immunization with antisera elicited by the HgbAI vaccine protects against a homologous H. ducreyi challenge in the experimental swine model of chancroid.
Previous reports from our laboratory showed that a native preparation of HgbA, the Hb receptor of H. ducreyi, is a successful vaccine in the experimental swine model of chancroid (1, 13). Whether nHgbA is administered with Freund's adjuvant or MPL, an adjuvant currently used in humans, the antisera elicited to the HgbA vaccine bound the surface of H. ducreyi and partially blocked Hb binding to HgbA. These data suggested that protection by the HgbA vaccine is Ab mediated. To test this hypothesis, we sought to determine if passive immunization of naïve swine with nHgbAI antiserum could protect against an infectious challenge with homologous and heterologous H. ducreyi strains. Infusion of pigs with anti-nHgbAI prevented infection with the homologous H. ducreyi strain 35000HPhgbAI at an inoculum of 103 CFU. Thus, the humoral immune response elicited to the nHgbA vaccine protects against an infectious H. ducreyi challenge.
There was breakthrough infection at the higher inoculum dose (104 CFU) in 2 of 4 passively immunized animals. This may have been related to the concentration of nHgbAI-specific Abs present in the sera of infused pigs. Although the small sample size precluded a statistical analysis of correlations between antibody levels and passive protection, pigs with higher levels of nHgbAI-specific Abs were completely protected from challenge with either inoculum, while those with lower concentrations experienced breakthrough infections. These data suggest that the amount of nHgbAI-specific Abs is important for clearance of H. ducreyi in this animal model.
Anti-nHgbAI 2 displays the same in vitro correlates of protection as the ones identified in antisera from animals protected against a homologous H. ducreyi challenge.
In 2 previous active immunization trials using nHgbA as a vaccine, antisera from nHgbA-vaccinated animals were tested for the ability to bind purified nHgbA and HgbA in the context of whole H. ducreyi, the ability to inhibit Hb binding to HgbA, and bactericidal and opsonophagocytic activities. In the first vaccination trial in which the nHgbA vaccine was administered with Freund's adjuvant (anti-nHgbAI 1), the antisera from vaccinated animals bound purified nHgbA and HgbA at the surface of H. ducreyi, had modest bactericidal activity, and partially blocked DIG-Hb binding to nHgbAI, but lacked opsonophagocytic activity (1). In the second active immunization trial using MPL as the adjuvant, the antisera from animals immunized with nHgbAI/MPL bound nHgbAI purified from H. ducreyi and HgbA in its native conformation, but with much less reactivity than anti-nHgbAI 1. Nevertheless, anti-nHgbAI/MPL blocked Hb binding to nHgbA in an ELISA to levels similar to that of the nHgbAI/Freund's 1 antisera, but lacked bactericidal activity. Furthermore, the nHgbA vaccine administered with MPL was protective (13). Therefore, analysis of the humoral immune response developed to the HgbA vaccine from previous studies demonstrates that binding of anti-HgbA to HgbA in its native conformation as well as blocking of Hb binding appear to correlate with protection (1, 13). Because there was only modest in vitro bactericidal activity demonstrated by the nHgbAI antisera in the first trial, and none in the second, bactericidal activity does not appear to be necessary for protection.
The same in vitro assays described above were therefore used to assess the biological properties of individual and pooled anti-nHgbAI 2 used for the passive immunization experiments described here. These antisera bound purified nHgbAI and HgbAI on the surface of H. ducreyi and partially blocked Hb binding to HgbA, again at levels similar to those observed for anti-nHgbAI 1 (Fig. 2, 5, and 6, respectively). However, there were some differences between the activities of anti-nHgbAI 2 and 1. First, there was higher cross-reactivity to the surface of the isogenic hgbA mutant strain 35000HPΔhgbA by anti-nHgbAI 2. From Western blot assays, the anti-nHgbAI 2 bound denatured HgbA more than anti-nHgbAI 1 (Fig. 4A and B). Potential explanations for these differences include modification of the protein structure during preparation of the individual vaccines and genetic differences between swine herds; the animals used to generate anti-nHgbAI 2 came from a different farm and are likely distantly related to the animals that were previously used to conduct the HgbA vaccination trials. Anti-nHgbAI 2 also contained Abs that recognized the two major outer membrane proteins of H. ducreyi, MOMP and OmpA2 (Fig. 4B). There are a number of proteins from pig pathogens that have high homology to the major outer membrane proteins of H. ducreyi (23). Because these pathogens are early colonizers and infection by these pathogens is often endemic and mostly asymptomatic (P. Routh, personal communication), it is possible that one or more of the animals immunized with nHgbAI/Freund's vaccine may have been colonized with such cross-reacting bacteria. This would have contributed to the reactivity of the pooled anti-nHgbAI 2 to H. ducreyi MOMP and OmpA2. Western blot analysis of the nHgbAI preparation used to generate anti-nHgbAI 2 with monoclonal antibody 2C7, which recognizes both MOMP and OmpA2 (37), revealed that MOMP and/or OmpA2 was present in the preparations; however, the amount was undetectable by Coomassie blue staining or silver staining (data not shown).
Antisera elicited to nHgbAI only protect against a homologous challenge.
H. ducreyi strains are grouped into classes, termed class I and class II, according to the expression of variant outer membrane determinants and the structure of LOS (33, 34, 36, 48). Although the amino acid sequences of some H. ducreyi surface determinants, such as DsrA and NcaA, differ widely between H. ducreyi strains belonging to different classes, the HgbA protein is highly conserved, with more than 95% identity between HgbA proteins in the two strain classes (26, 48). Most of these differences reside in the large immunodominant loop 4 of HgbA, which contains 17 different putatively surface-exposed amino acids out of a total of only 27 different residues between full-length HgbA proteins of different classes (950 total amino acids) (26). Because of this high identity between HgbA proteins of different groups, we were surprised to discover that the nHgbAI vaccine did not protect against infection with H. ducreyi strain 35000HP expressing class II HgbA (13). The current data from passive immunization are consistent with these previous results. Pooled antisera elicited to the HgbAI vaccine were only protective against infection with an H. ducreyi strain expressing HgbAI. Taken together, these data suggest that differences in a small number of immunogenic, variable residues in HgbAI and HgbAII contribute to protection by an HgbA-based vaccine. Therefore, a bivalent HgbA vaccine may be necessary to prevent chancroid caused by both classes of H. ducreyi strains.
Another potential explanation for the lack of heterologous protection lies in the different exposure of the class II HgbA protein on the surface of H. ducreyi strains 35000HP and DMC111. The LOS of the class II strain DMC111 is truncated (48), and this smaller LOS structure may result in greater exposure of HgbA at the surface of class II H. ducreyi. However, because H. ducreyi class II strain DMC111 is noninfectious in the experimental swine model of infection, we were unable to examine the protective capacity of a class I antibody response against infection with a naturally occurring class II strain. It is therefore possible that natural class II strain infections may be protected by the class I vaccine. Further studies are needed to clarify this issue.
A possible mechanism of protection of the HgbA vaccine is nutritional immunity.
Iron is required for the growth of most bacteria. However, iron in the host is sequestered from invading pathogens by several different proteins (47). Kochan used the term “nutritional immunity” to describe this process of “depletion by the host of iron essential for bacterial growth.” He associated this term with acquired immunity to relate it to the limitation of an essential nutrient by the host iron/heme-scavenging proteins (17). The idea of preventing a pathogen from acquiring an essential nutrient is decades old; however, this report, along with others from our laboratory (1, 13), are the first to suggest that vaccine-induced nutritional immunity can actually occur in the host. Our studies with the HgbA vaccine show that antisera elicited to an Hb receptor can partially prevent Hb binding to the Hb receptor itself, suggesting that nutritional immunity is possible (Fig. 6). Further experiments are under way to determine if anti-nHgbAI IgG can prevent bacterial growth.
In conclusion, we have shown in this report that passive immunization with pooled antisera from swine immunized with the Hb receptor of H. ducreyi protected naïve pigs against a homologous challenge in the experimental swine model of chancroid. Our results also suggest that the mechanism of protection of the HgbA vaccine may be nutritional immunity, since Abs elicited to the H. ducreyi Hb receptor were not bactericidal but partially prevented HgbA from binding its ligand, Hb.
ACKNOWLEDGMENT
This work was supported by 5-R01-AI 05393 from the NIH to C.E.
Footnotes
Published ahead of print on 6 June 2011.
REFERENCES
- 1. Afonina G., et al. 2006. Immunization with the Haemophilus ducreyi hemoglobin receptor HgbA protects against infection in the swine model of chancroid. Infect. Immun. 74:2224–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Al-Mutairi N., et al. 2007. Clinical patterns of sexually transmitted diseases, associated sociodemographic characteristics, and sexual practices in the Farwaniya region of Kuwait. Int. J. Dermatol. 46:594–599 [DOI] [PubMed] [Google Scholar]
- 3. Al-Tawfiq J. A., et al. 2000. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis. 181:1049–1054 [DOI] [PubMed] [Google Scholar]
- 4. Al-Tawfiq J. A., et al. 1998. Standardization of the experimental model of Haemophilus ducreyi infection in human subjects. J. Infect. Dis. 178:1684–1687 [DOI] [PubMed] [Google Scholar]
- 5. Anand Kumar B. H., Vijaya D., Ravi R., Reddy R. R. 2003. Study of genital lesions. Indian J. Dermatol. Venereol. Leprol. 69:126–128 [PubMed] [Google Scholar]
- 6. Annan N. T., Lewis D. A. 2005. Treatment of chancroid in resource-poor countries. Expert Rev. Anti Infect. Ther. 3:295–306 [DOI] [PubMed] [Google Scholar]
- 7. Bauer B. A., Stevens M. K., Hansen E. J. 1998. Involvement of the Haemophilus ducreyi gmhA gene product in lipooligosaccharide expression and virulence. Infect. Immun. 66:4290–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Becker M., et al. 2010. Etiology and determinants of sexually transmitted infections in Karnataka State, south India. Sex. Transm. Dis. 37:159–164 [DOI] [PubMed] [Google Scholar]
- 9. Bong C. T., Bauer M. E., Spinola S. M. 2002. Haemophilus ducreyi: clinical features, epidemiology, and prospects for disease control. Microbes Infect. 4:1141–1148 [DOI] [PubMed] [Google Scholar]
- 10. Chen C. Y., Mertz K. J., Spinola S. M., Morse S. A. 1997. Comparison of enzyme immunoassays for antibodies to Haemophilus ducreyi in a community outbreak of chancroid in the United States. J. Infect. Dis. 175:1390–1395 [DOI] [PubMed] [Google Scholar]
- 11. Elkins C. 1995. Identification and purification of a conserved heme-regulated hemoglobin-binding outer membrane protein from Haemophilus ducreyi. Infect. Immun. 63:1241–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elkins C., Morrow K. J., Jr., Olsen B. 2000. Serum resistance in Haemophilus ducreyi requires outer membrane protein DsrA. Infect. Immun. 68:1608–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fusco W. G., et al. 2010. Immunization with the Haemophilus ducreyi hemoglobin receptor HgbA with adjuvant monophosphoryl lipid A protects swine from a homologous but not a heterologous challenge. Infect. Immun. 78:3763–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hammond G. W., Lian C. J., Wilt J. C., Ronald A. R. 1978. Antimicrobial susceptibility of Haemophilus ducreyi. Antimicrob. Agents Chemother. 13:608–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hope-Rapp E., et al. 2009. Etiology of genital ulcer disease. A prospective study of 278 cases seen in an STD clinic in Paris. Sex. Transm. Dis. 37:153–158 [DOI] [PubMed] [Google Scholar]
- 16. Klesney-Tait J., et al. 1997. The major outer membrane protein of Haemophilus ducreyi consists of two OmpA homologs. J. Bacteriol. 179:1764–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kochan I. 1973. The role of iron in bacterial infections, with special consideration of host-tubercle bacillus interaction. Curr. Top. Microbiol. Immunol. 60:1–30 [DOI] [PubMed] [Google Scholar]
- 18. Kreiss J. K., et al. 1989. Isolation of the human immunodeficiency virus from genital ulcers in Nairobi prostitutes. J. Infect. Dis. 160:380–384 [DOI] [PubMed] [Google Scholar]
- 19. Leduc I., et al. 2008. Evaluation of the repertoire of the TonB-dependent receptors of Haemophilus ducreyi for their role in virulence in humans. J. Infect. Dis. 197:1103–1109 [DOI] [PubMed] [Google Scholar]
- 20. Leduc I., et al. 2008. Outer membrane protein DsrA is the major fibronectin-binding determinant of Haemophilus ducreyi. Infect. Immun. 76:1608–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee B. C. 1991. Iron sources for Haemophilus ducreyi. J. Med. Microbiol. 34:317–322 [DOI] [PubMed] [Google Scholar]
- 22. Lewis D. A. 2003. Chancroid: clinical manifestations, diagnosis, and management. Sex. Transm. Infect. 79:68–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. MacInnes J. I., Desrosiers R. 1999. Agents of the “suis-ide diseases” of swine: Actinobacillus suis, Haemophilus parasuis, and Streptococcus suis. Can. J. Vet. Res. 63:83–89 [PMC free article] [PubMed] [Google Scholar]
- 24. McBride W. J., Hannah R. C., Le Cornec G. M., Bletchly C. 2008. Cutaneous chancroid in a visitor from Vanuatu. Australas. J. Dermatol. 49:98–99 [DOI] [PubMed] [Google Scholar]
- 25. Mertz K. J., et al. 1998. An investigation of genital ulcers in Jackson, Mississippi, with use of a multiplex PCR assay: high prevalence of chancroid and human immunodeficiency virus infection. J. Infect. Dis. 178:1060–1066 [DOI] [PubMed] [Google Scholar]
- 26. Nepluev I., et al. 2009. An immunogenic, surface-exposed domain of Haemophilus ducreyi outer membrane protein HgbA is involved in hemoglobin binding. Infect. Immun. 77:3065–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Noinaj N., Guillier M., Barnard T. J., Buchanan S. K. 2010. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64:43–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patterson K., et al. 2002. Development of a rapid immunodiagnostic test for Haemophilus ducreyi. J. Clin. Microbiol. 40:3694–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paz-Bailey G., et al. 2005. Changes in the etiology of sexually transmitted diseases in Botswana between 1993 and 2002: implications for the clinical management of genital ulcer disease. Clin. Infect. Dis. 41:1304–1312 [DOI] [PubMed] [Google Scholar]
- 30. Peel T. N., Bhatti D., De Boer J. C., Stratov I., Spelman D. W. 2010. Chronic cutaneous ulcers secondary to Haemophilus ducreyi infection. Med. J. Aust. 192:348–350 [DOI] [PubMed] [Google Scholar]
- 31. Phiri S., et al. 2010. Impact of acyclovir on ulcer healing, lesional, genital and plasma HIV-1 RNA among patients with genital ulcer disease in Malawi. Sex. Transm. Infect. 86:345–352 [DOI] [PubMed] [Google Scholar]
- 32. Plummer F. A., et al. 1991. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J. Infect. Dis. 163:233–239 [DOI] [PubMed] [Google Scholar]
- 33. Post D. M., Gibson B. W. 2007. Proposed second class of Haemophilus ducreyi strains show altered protein and lipooligosaccharide profiles. Proteomics 7:3131–3142 [DOI] [PubMed] [Google Scholar]
- 34. Post D. M., et al. 2007. Identification of genes involved in the expression of atypical lipooligosaccharide structures from a second class of Haemophilus ducreyi. Infect. Immun. 75:113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. San Mateo L. R., Toffer K. L., Orndorff P. E., Kawula T. H. 1999. Neutropenia restores virulence to an attenuated Cu,Zn superoxide dismutase-deficient Haemophilus ducreyi strain in the swine model of chancroid. Infect. Immun. 67:5345–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scheffler N. K., et al. 2003. Proteome of Haemophilus ducreyi by 2-D SDS-PAGE and mass spectrometry: strain variation, virulence, and carbohydrate expression. J. Proteome Res. 2:523–533 [DOI] [PubMed] [Google Scholar]
- 37. Spinola S. M., Griffiths G. E., Shanks K. L., Blake M. S. 1993. The major outer membrane protein of Haemophilus ducreyi is a member of the OmpA family of proteins. Infect. Immun. 61:1346–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Steen R. 2001. Eradicating chancroid. Bull. World Health Organ. 79:818–826 [PMC free article] [PubMed] [Google Scholar]
- 39. Stevens M. K., et al. 1996. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect. Immun. 64:1724–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suntoke T. R., et al. 2009. Evaluation of multiplex real-time PCR for detection of Haemophilus ducreyi, Treponema pallidum, herpes simplex virus type 1 and 2 in the diagnosis of genital ulcer disease in the Rakai District, Uganda. Sex. Transm. Infect. 85:97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thomas C. E., Olsen B., Elkins C. 1998. Cloning and characterization of tdhA, a locus encoding a TonB-dependent heme receptor from Haemophilus ducreyi. Infect. Immun. 66:4254–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thomas K. L., et al. 2001. Cloning, overexpression, purification, and immunobiology of an 85-kilodalton outer membrane protein from Haemophilus ducreyi. Infect. Immun. 69:4438–4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Throm R. E., et al. 2000. Evaluation of an isogenic major outer membrane protein-deficient mutant in the human model of Haemophilus ducreyi infection. Infect. Immun. 68:2602–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trees D. L., Morse S. A. 1995. Chancroid and Haemophilus ducreyi: an update. Clin. Microbiol. Rev. 8:357–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tsai C.-M., Frasch C. E. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 155:115–119 [DOI] [PubMed] [Google Scholar]
- 46. Ussher J. E., Wilson E., Campanella S., Taylor S. L., Roberts S. A. 2007. Haemophilus ducreyi causing chronic skin ulceration in children visiting Samoa. Clin. Infect. Dis. 44:e85–e87 [DOI] [PubMed] [Google Scholar]
- 47. Weinberg E. D. 2009. Iron availability and infection. Biochim. Biophys. Acta 1790:600–605 [DOI] [PubMed] [Google Scholar]
- 48. White C. D., et al. 2005. Haemophilus ducreyi outer membrane determinants, including DsrA, define two clonal populations. Infect. Immun. 73:2387–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhu W., et al. 2005. Comparison of immune responses to gonococcal PorB delivered as outer membrane vesicles, recombinant protein, or Venezuelan equine encephalitis virus replicon particles. Infect. Immun. 73:7558–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]