Abstract
As an etiological agent of bacterial sepsis and wound infections, Vibrio vulnificus is unique among the Vibrionaceae. The most intensely studied of its virulence factors is the capsular polysaccharide (CPS). Over 100 CPS types have been identified, yet little is known about the genetic mechanisms that drive such diversity. Chitin, the second-most-abundant polysaccharide in nature, is known to induce competence in Vibrio species. Here, we show that the frequency of chitin-induced transformation in V. vulnificus varies by strain and that (GlcNAc)2 is the shortest chitin-derived polymer capable of inducing competence. Transformation frequencies (TFs) increased 8-fold when mixed-culture biofilms were exposed to a strain-specific lytic phage, suggesting that the lysis of dead cells during lytic infection increased the amount of extracellular DNA within the biofilm that was available for transfer. Furthermore, we show that V. vulnificus can undergo chitin-dependent carbotype conversion following the uptake and recombination of complete cps loci from exogenous genomic DNA (gDNA). The acquisition of a partial locus was also demonstrated when internal regions of homology between the endogenous and exogenous loci existed. This suggested that the same mechanism governing the transfer of complete cps loci also contributed to their evolution by generating novel combinations of CPS biosynthesis genes. Since no evidence that cps loci were preferentially acquired during natural transformation (random transposon-tagged DNA was readily taken up in chitin transformation assays) exists, the phenomenon of chitin-induced transformation likely plays an important but general role in the evolution of this genetically promiscuous genus.
INTRODUCTION
After 3.8 billion years of evolution (39), bacteria, whether commensal or pathogenic, have evolved numerous ways to cope with the sophisticated defensive strategies of the host's immune system. A simple but very effective strategy is the production and variation of surface polysaccharides that can alter proper host immune function (13, 21). This type of antigenic variation is an important mechanism used by pathogenic microorganisms for escaping the neutralizing activities of antibodies and phagocytes. Antigenic variation may occur during the course of an infection, or an organism can exist in nature as multiple antigenic types (antigenic variety). The colonized host often provides the ideal selective environment for the emergence of new antigenic variants of bacteria, providing that the organism's other virulence determinants remain intact.
Vibrio vulnificus is an estuarine bacterium that is pathogenic to humans and animals (35). Its continued environmental persistence and transmission is bolstered by its ability to colonize shellfish and form biofilms on various marine biotic surfaces, such as plankton, algae, fish, and eels (4, 40, 42). Primary septicemia and wound infections can arise following the consumption or handling of contaminated food or water, and the fatality rate of septicemic patients is greater than 50% (for a review, see reference 35). It alone is responsible for 95% of all seafood-related deaths in the United States, and it carries the highest death rate of any food-borne disease agent (68). The most intensely studied of its virulence factors is the capsular polysaccharide (CPS), which mediates resistance to complement-mediated bacteriolysis and phagocytosis (31, 36, 66, 76). All virulent strains produce copious amounts of CPS; acapsulated strains produce little or no CPS and are attenuated (46, 73). CPS is composed of monosaccharides joined by glycosidic linkages, and an incredibly diverse range of branched and modified CPS molecules is possible. More than 100 capsular types (carbotypes) have been identified among natural V. vulnificus isolates (8, 29), and completely different CPS biosynthesis loci are found in the same chromosomal region in different V. vulnificus strains (44). This suggests that the elaboration of CPS structures is an important survival strategy for V. vulnificus and likely allows the organism to stave off elimination by the host's immune system, yet little is known about the genetic mechanisms that elaborate such a degree of CPS diversity.
Chitin, the second-most-abundant polysaccharide in nature after cellulose, is an insoluble polymer of N-acetylglucosamine (GlcNAc). It is found in the exoskeletons of marine invertebrates, such as crustaceans, cephalopods, diatoms, and sponges (20, 37). The epidemic V. cholerae O139 serovar emerged from the pandemic El Tor O1 serovar through the replacement of a 22-kb O1 lipopolysaccharide (lps) locus with a 40-kb O139 lps locus (3, 12, 14). It has been proposed that this may have occurred in a process of natural transformation that was induced by chitin (6, 41). Chitin-induced competence was also reported to occur in V. vulnificus (27); however, the uptake of large contiguous DNA fragments was not investigated. Hence, we determined the role of chitin-induced transformation in carbotype conversion in V. vulnificus. We show that the frequency of chitin-induced transformation in V. vulnificus varies by strain and that (GlcNAc)2 was the shortest chitin-derived polymer capable of inducing competence. Transformation frequencies (TFs) increased when mixed-culture biofilms were exposed to a strain-specific lytic phage, which effectively augmented the amount of DNA within the biofilm that was available for transfer. We also show that V. vulnificus can undergo chitin-dependent carbotype conversion following the uptake and recombination of complete cps loci from exogenous genomic DNA (gDNA). The incorporation of partial cps loci led to the generation of novel combinations of cps biosynthesis genes.
MATERIALS AND METHODS
Bacterial strains and media.
All strains are listed in Table 1. Rifampin-resistant (Rfr) derivatives of V. vulnificus strains 27562 (Collection de l'Institut Pasteur), YJ016 and CMCP6 (Anita Wright, University of Florida), and MO-6/24 (Paul Gulig, University of Florida) were used as recipients in chitin transformation assays. Strains were grown in Luria-Bertani (LB) broth (Sigma), HEPES minimal medium (HMM) (56), or Instant Ocean at 25 ppt (IO-25) as indicated below. Antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg/ml; Rf, 100 μg/ml; kanamycin (Km), 160 μg/ml for V. vulnificus and 25 μg/ml for Escherichia coli; and chloramphenicol (Cm), 3 μg/ml for V. vulnificus and 10 μg/ml for E. coli. IPTG (isopropyl-β-d-thiogalactopyranoside) and l-arabinose were added to 1 mM and 0.2%, respectively.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| V. vulnificus | ||
| 27562 | Type strain; clinical isolate from Florida; Rfr | Collection de l'Institut Pasteur |
| 27562wzc::pSW23T | Acapsular mutant of 27562 with a disruption of the wzc tyrosine autokinase required for CPS transport; Rfr Cmr | 46 |
| 27562-Tn | Mutant transposon library; Rfr Kmr | This study |
| YJ016 | Clinical isolate from Taiwan; Rfr | 10 |
| YJ016-Tn | Mutant transposon library; Rfr Kmr | This study |
| YJ016vv0343::pSW23T | YJ016 variant tagged at vv0343 with pSW23T; Rfr Cmr | This study |
| YJ016wbfY::pSW23T | YJ016 variant tagged at wbfY with pSW23T; Rfr Cmr | This study |
| MO-6/24 | Clinical isolate from United States; Rfr | 73 |
| MO-6/24-Tn | Mutant transposon library; Rfr Kmr | This study |
| MO-6/24wbfT::pSW23T | MO-6/24 variant tagged at wbfT with pSW23T; Rfr Cmr | This study |
| CMCP6 | Clinical isolate from Korea; Rfr | 10 |
| CMCP6-wzc::pSW23T | Acapsular mutant of CMCP6 with a disruption of the wzc tyrosine autokinase required for CPS transport; Rfr Cmr | This study |
| CMCP6-cps27562 | Acapsular mutant of CMCP6 that acquired the cps locus of 27562wzcpSW23T; Rfr Cmr | This study |
| CMCP6-cps27562/pBAD24T::wzc | Acapsular mutant of CMCP6 that acquired the cps locus of 27562wzc::psw23T and was complemented with wild-type wzc; Rfr Cmr | This study |
| CMCP6-wcvCYJ016 | CMCP6 that acquired the 3′ end of the YJ016wbfY::pSW23T cps locus; Rfr Cmr | This study |
| CMCP6-cpsMO-6/24 | CMCP6 that acquired the entire cps locus of MO-6/24wbfT::pSW23T; Rfr Cmr | This study |
| E. coli | ||
| S17.1λpir | Donor strain for conjugation | 58 |
| Plasmids | ||
| pNKTXI-SceI | Mini-Tn10 mutagenesis plasmid; Kmr Apr | 46 |
| pBAD24T | Mobilizable derivative of pBAD24; Apr | 46 |
| pBAD24T::wzc | pBAD24T containing wzc under the control of PBAD; Apr | 46 |
| pSW23T | Suicide vector; Cmr | 17 |
| pSW23T::vv0343 | pSW23T containing vv0343; Cmr | This study |
| pSW23T::wbfY | pSW23T containing wbfY; Cmr | This study |
| pSW23T::wbfT | pSW23T containing wbfT; Cmr | This study |
| pGFPTAT | GFP-producing plasmid containing the RP4 origin of conjugative transfer; Apr | 32 |
DNA manipulation.
gDNA was extracted with DNAZOL (Invitrogen). Plasmid DNA was purified using the Sigma plasmid miniprep kit. PCR was performed in 50-μl volumes using Taq (BioTools) or Platinum Pfx (Invitrogen) DNA polymerase. When necessary, PCR products were cloned into pCR2.1 (Invitrogen) and sequenced at the Hospital for Sick Children (Toronto, Canada) using the M13F and -R primers. All primers are listed in Table S1 in the supplemental material.
Constructing pGFPTAT.
The RP4 origin of transfer was amplified from pSW23T (17) with the primers oriT1ERI and oriT2ERI, digested with EcoRI, and cloned into the EcoRI site of pGFPTA (32) to create pGFPTAT.
Constructing YJ016vv0343::pSW23T, YJ016wbfY::pSW23T, and MO-6/24wbfT::pSW23T.
The vv0343, wbfY, and wbfT genes were amplified from the respective strains with primers vv0343-1/vv0343-2, vv0364-1/vv0364-2, and wbfT-1/wbfT-2. Each PCR product was digested with BamHI and ECORI and separately cloned into the same sites of pSW23T. The resulting plasmids were conjugated from S17.1λpir to the appropriate V. vulnificus strain. Correct integration was verified by PCR using a primer targeting the cat gene of pSW23T and a second primer that annealed to chromosomal DNA flanking the target gene (vv0341For, vv0363For, or wbuBendF).
Generation of Tn libraries.
The pNKTXI-SceI plasmid (46) was conjugated from S17.1λpir to V. vulnificus Rfr derivatives of strains 27562, YJ016, CMCP6, and MO-6/24. Kmr transposon (Tn) mutants were selected on LB-Rf-Km plates. Mutants from each library (designated 27562-Tn, YJ016-Tn, CMCP6-Tn, and MO-6/24-Tn) were pooled and used in mixed-culture transformation assays. Where indicated below, gDNA was extracted and used as the donor in transformation experiments.
Chitin transformation assays.
Chitin transformation assays were performed as described in references 6, 27, and 41, with some minor modifications. Overnight cultures were diluted 1:20 into 5 ml of fresh LB and grown with shaking at 30°C to an optical density at 600 nm (OD600) of 0.5 to 0.6. The cells were washed with 1 ml of IO-25 and resuspended in 10 ml of IO-25. Two milliliters of this was added to sterile crab or shrimp shell pieces in 12-well plates and incubated for 24 h at 30°C. The supernatant was removed and replaced by 2 ml of fresh IO-25. Two micrograms of donor gDNA was added to each well, and the plates were incubated for an additional 24 h. The crab shells were removed and vortexed vigorously in 4 ml of LB for 30 s to detach the bacteria. Dilutions were plated on LB with and without antibiotic. Transformation frequency was calculated as the number of resistant CFU divided by the total number of viable CFU. gDNA was extracted from selected transformants and analyzed by PCR with primers that targeted genes specific for the donor, recipient, and transformant in each experiment. The CMCP6-specific target was vv12532 (53). The 27562- and YJ016-specific targets were the respective integron-integrase (intIA) genes (54). The mini-Tn10-specific target was its Km resistance marker. Carbotype-specific genes served as targets for tracking the movement of cps loci. In mixed-culture assays, only one of the strains was Rfr. To test the competence-inducing effects of GlcNAc or (GlcNAc)2, the procedure described above was followed except that the IO-25 media contained a 5 mM concentration of the mono- or disaccharide and the bacterial suspension (2 ml) was added to sterile glass coverslips.
For transformation assays using colloidal chitin, colloidal chitin was prepared essentially as described previously (57). Briefly, 3 g of chitin flakes (Sigma) was mixed with 50 ml of concentrated HCl and the mixture was incubated with shaking at 4°C overnight. To precipitate the colloidal chitin, 250 ml of 50% ethanol was added to the HCl-chitin flake mixture and the solution was incubated with shaking at room temperature overnight. The mixture was centrifuged at 10,000 rpm for 20 min to pellet the colloidal chitin. The pellet was washed with distilled water until pH 7 was reached and was then resuspended in 20 ml of distilled water. For transformation assays, CMCP6 was grown, washed, and resuspended in IO-25 as described earlier. Two milliliters of bacteria was added to 200 μl of the colloidal chitin suspension, and the culture was incubated with shaking at 30°C for 3 h. Two micrograms of donor gDNA was added, and growth was continued overnight. The samples were collected and vortexed to detach bacteria from the colloidal chitin, and dilutions were plated on LB with and without antibiotic. Transformation frequency was calculated as the number of resistant CFU divided by total number of viable CFU. Statistical analyses (P values) were calculated using the Student t test.
Slide agglutination assays.
Assays with rabbit antisera raised against formalin-killed V. vulnificus 27562 were done as previously described (46). Samples were tested in triplicate.
Phage infection.
V. vulnificus-specific phages (Paul Gulig, University of Florida) were propagated on strain MO-6/24. Phage supernatants were sterilized by filtration through 0.22-μm filters (Millipore), and aliquots were plated on LB agar to confirm that the filtrates did not contain bacteria. To test phage sensitivity, overnight cultures of the V. vulnificus strains were diluted 1:100 in fresh LB-Rf and grown to an OD600 of 0.4 at 30°C. Aliquots of the filtrates were added to 100 μl of each strain at a multiplicity of infection (MOI) of 10 in glass culture tubes, and the sample was incubated at 30°C for 20 min. Three milliliters of prewarmed top agar was added to the tube, and the mixture was gently swirled before being laid over an LB-Rf plate. Plates were incubated at 30°C overnight, and plaques were counted the next day. Phage 152-A10 caused nearly complete lysis of MO-6/24/O, while 27562, YJ016, and CMCP6 were resistant to infection.
CLSM.
pGFPTA was transferred to V. vulnificus by conjugation from S17.1λpir. Strains were grown in HMM with the appropriate antibiotics at 30°C. Expression of gfp was induced with IPTG when the OD600 reached 0.4, and growth was continued overnight. The next day, cultures were adjusted to an OD600 of 1.0 and diluted 1:3 in fresh medium containing IPTG. Three milliliters was then added to sterile crab shells or glass coverslips in 12-well plates and incubated statically at 30°C for 24 or 72 h. Where indicated, phage 152-A10 (1 × 109 PFU) was then added and the incubation was continued for an additional 16 or 24 h. Biofilms were rinsed in Vibrio-specific phosphate-buffered saline (PBS) (VPBS; NaCl [130 mM], Na2HPO4 [5 mM], KH2PO4 [1.5 mM, pH 7.4]) (9) and stained with 10 μM propidium iodide (PI) for 15 min at room temperature. Strains lacking pGFPTA were also stained with a 1:100 dilution of NanoOrange (NO) (Molecular Probes) for 1 h at room temperature. Biofilms were washed three times with 3 ml of VPBS, and image stacks were captured with a Zeiss LSM 510 Axioplan 2 confocal laser scanning microscope (CLSM) fitted with a W-plan Apochromat 40× objective. Three-dimensional (3-D) biofilm images were visualized with Volocity (PerkinElmer). For green fluorescent protein (GFP)-PI-stained images, green areas indicate live intact cells and red areas indicate dead cells. For NO-PI-stained images, green areas indicate live cells, red areas indicate lysed cells, and yellow areas (signal overlap) indicate intact cells that are dead or dying. The 3-D axis indicator in Fig. 1 and 2 (x axis, green arrow; y axis, red arrow; z axis, blue arrow) is shown for each image. COMSTAT was used to calculate biomass volume (total number of biomass pixels multiplied by voxel size and divided by the substratum area). The percentage of dead cells was calculated as the number of PI-stained pixels divided by the total number of biomass pixels. Stacked 3-D surface plots were generated with ImageJ 10.2 (National Institutes of Health). Where indicated, biofilms formed by pGFPTA-carrying cells were treated with 2 U RNase-free DNase (NEB) at 0, 24, and 48 h postinoculation. Control samples were not treated. The biofilms were rinsed 3 times for 5 min in VPBS and imaged as described above.
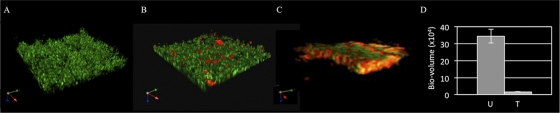
Fig. 1.
Composition of V. vulnificus biofilms grown on crab shells. The 27562 and CMCP6 strains were grown in mixed culture on crab shells in 12-well plates for 24 h. The structure and composition of the biofilms were examined by CSLM. (A) GFP-tagged cells; (B) GFP-tagged cells stained with PI; (C) staining with NanoOrange and PI; (D) plot of the bio-volume for DNase-treated (T) and untreated (U) biofilms of GFP-tagged cells. Results are representative of at least three independent experiments. Error bars represent standard deviations (P < 0.005).
Fig. 2.
Changes in V. vulnificus biofilm composition in response to a lytic phage. The MO-6/24-Tn and CMCP6 strains were grown in mixed culture on crab shells in 12-well plates for 24 h. The biofilms were then exposed to the MO-6/24-specific lytic phage 152-A10 for 0 (top row), 16 (middle row), and 24 (bottom row) h. The structures and compositions of the biofilms were examined by CSLM. The image on the left of each row is GFP-tagged cells stained with PI. To the right are representative plots showing the relative signal intensities and distribution in the separate GFP and PI channels for the same volumetric horizontal section near the surface of the corresponding biofilm.
RESULTS
Chitin induces competence in V. vulnificus.
To verify that chitin induces competence in V. vulnificus, we monitored the uptake by CMCP6 of gDNA isolated from kanamycin-resistant (Kmr) transposon (Tn) libraries of strains 27562 and YJ016. The transformation frequency (TF) in the absence of donor gDNA was used as a control. Crab shells were used as the source of chitin in transformation assays. The donor, recipient, and transformants were analyzed by PCR with gene-specific primers. Kmr transformants were readily obtained (TF = 1.2 × 10−6 ± 4.7 × 10−7) when the recipient was incubated in the presence of crab shells (Table 2). No transformants were detected in the absence of donor gDNA. As expected, the TFs were slightly higher when the recipient strain also served as the source of the donor gDNA. CMCP6 was the most competent of the strains tested (P < 0.005), and similar TFs were obtained when it was grown in the presence of shrimp shells (1.3 × 10−6 ± 3.8 × 10−7) or colloidal chitin (3.7 × 10−6 ± 6.8 × 10−7). The TFs for MO-6/24, 27562, and YJ016 were similar to one another (P > 0.3) but at least 25-fold lower than that for CMCP6 (P < 0.001). When glass coverslips, mussel shells, or oyster shells were used as the colonization surface, no transformants were recovered for any of the strains (TF < 10−9). These results confirm that chitin induces competence in V. vulnificus. To determine the shortest oligosaccharide capable of inducing competence, Rfr CMCP6 was grown on glass coverslips in media containing GlcNAc or (GlcNAc)2 and the uptake of gDNA isolated from the MO-6/24-Tn library was monitored. Rfr Kmr transformants were obtained at a frequency of 1.4 × 10−6 ± 5.9 × 10−7 when (GlcNAc)2 was added to the media. No transformants were recovered with GlcNAc. This suggested that (GlcNAc)2 was the smallest chitin-derived polymer capable of inducing competence.
Table 2.
Transformation frequencies of V. vulnificus strains grown on crab shells
| Recipient strain | TF with indicated source of donor gDNAa |
|
|---|---|---|
| 27562-Tn | YJ016-Tn | |
| 27562 | 8.0 × 10−7 ± 2.1 × 10−7 | 5.6 × 10−8 ± 2.7 × 10−8 |
| YJ016 | <DL | 4.8 × 10−8 ± 1.4 × 10−8 |
| CMCP6 | 1.2 × 10−6 ± 4.7 × 10−7 | 1.5 × 10−6 ± 5.6 × 10−7 |
| MO-6/24 | ND | 5.2 × 10−8 ± 3.1 × 10−8 |
<DL, below the detection limit of 10−9; ND, not determined.
Kmr Tn libraries of strains 27562, YJ016, and MO-6/24 were also grown in mixed culture with an Rfr derivative of CMCP6 on crab shells. Transformants were selected on plates containing Km and Rf. PCR analysis with strain-specific primers indicated that CMCP6 transformants were obtained with TFs of 1.6 × 10−5 ± 3.7 × 10−6, 6.9 × 10−5 ± 6.7 × 10−6, and 2.9 × 10−5 ± 5.6 × 10−6, respectively. This suggested that DNA was available within the biofilm for uptake and recombination and supported previous reports of V. cholerae and V. vulnificus (5, 27, 41, 75). This was verified by CLSM. Cells expressing gfp were grown in the presence of crab shells for 24 h. A substantial biofilm formed on the crab shell surface (Fig. 1A). The biofilms were then stained with propidium iodide (PI), which stains nucleic acid of dead or dying cells but is excluded from live bacteria. GFP-PI staining revealed pockets of dead/dying cells (red staining) within the biofilm (Fig. 1B). To ascertain if extracellular nucleic acid was present, biofilms were stained with NanoOrange (NO) and PI. NO is a general protein stain that is virtually nonfluorescent in aqueous solution but undergoes a dramatic fluorescence enhancement upon interaction with proteins (34). As such, it has proven extremely useful in the staining of bacterial cell bodies and flagella (2, 25, 27, 69). Thus, NO-PI staining was anticipated to reveal a mixture of live (green staining due to NO), intact dead or dying (yellow staining due to signal overlap of NO and PI), and dead lysed (red staining due to PI) bacteria within the biofilm. We observed areas of live, intact dead, and dead lysed bacteria at the surface of the biofilm, with the last two dominating the biofilm's interior (Fig. 1C). The presence of lysed dead bacteria (red staining) throughout the biofilm suggested that extracellular nucleic acid was present in the biofilm and available for transfer. In support of this notion, treatment of the biofilm with DNase resulted in a 20-fold decrease in biomass volume relative to that of the control sample (Fig. 1D). These results supported previous studies suggesting that DNA is freely available within biofilms for uptake by the biofilm consortium (18, 38, 43, 65).
Lytic phage facilitate natural transformation in mixed-culture biofilms.
To better mimic environmental conditions in which competence develops, mixed-culture biofilms of gfp-tagged MO-6/24-Tn and CMCP6 were grown on crab shells and exposed for various lengths of time to a lytic phage (152-A10) that specifically infects MO-6/24. The biofilms were stained with PI and analyzed by CLSM (Fig. 2). The relative amount of PI staining within the biofilm increased with increasing phage exposure times (from 22.7% ± 1% of the total signal initially to 57.7% ± 5.6% of the total signal after 16 h). Interestingly, this level decreased thereafter (19% ± 1.2% of the total signal at 24 h), hinting at the lysis of dead cells during lytic infection and a potential increase in the amount of extracellular DNA. At the 16-h time point, CMCP6 transformants were obtained at a frequency of 2.3 × 10−4 ± 4.1 × 10−5. This was an increase of 8 times over that in samples in which the phage was omitted. These results support the notion that lytic phages may facilitate the exchange of DNA within biofilms during chitin-induced transformation (70).
Chitin-induced competence mediates carbotype conversion in V. vulnificus.
To determine if V. vulnificus could exchange complete or partial cps loci by natural transformation, gDNA from a chloramphenicol-resistant (Cmr) derivative of strain 27562 that contained a lesion in a gene (27562wzc::pSW23T) that is part of its 24-kb cps locus (Fig. 3) (45) was used as the donor in transformation experiments with strain CMCP6, which had been grown in the presence of crab shells. Cmr CMCP6 transformants were obtained at a frequency of 2.2 × 10−7 ± 3.7 × 10−8. The clones had a translucent phenotype, which suggested that they had acquired the wzc::pSW23T mutation. To confirm this, pBAD24T::wzc (45) was conjugated to 10 random transformants, and the strains were grown in the absence or presence of arabinose. The same transformants carrying the empty vector served as controls. Control strains remained translucent whether arabinose was present in the media or not. All of the transformants carrying pBAD24T::wzc remained translucent on media lacking arabinose but had an opaque phenotype when arabinose was added to the media. gDNA from each transformant was analyzed by PCR with 15 primer pairs that spanned the 24-kb 27562 cps locus (Fig. 3). The results indicated that 9 of the transformants had acquired only the wzc::pSW23T mutation. However, one transformant acquired the entire cps locus in a single transformation event (Fig. 4). This was confirmed in slide agglutination assays with antisera to wild-type 27562 cells. Antisera reacted strongly with the 27562 parental strain and with the CMCP6 transformant when it was grown under inducing conditions, while no agglutination was observed with the CMCP6 parental strain or with the CMCP6 transformant that was grown under noninducing conditions. Thus, the CMCP6 transformant that acquired the complete cps locus of strain 27562 (CMCP6-cps27562) began producing the 27562 CPS (carbotype conversion) when the wild-type wzc gene was supplied in trans. Since there was no internal region of homology between the cps loci of the wzc::pSW23T donor and CMCP6 recipient strains (46) and wcvA was also acquired, this suggested that the exchange was mediated by recombination between the homologous wza, wzb, and wzc regions at the 5′ end of the 27562 and CMCP6 cps loci and homologous regions at the 3′ end beyond wcvC in 27562 and vv1_0771 in CMCP6.
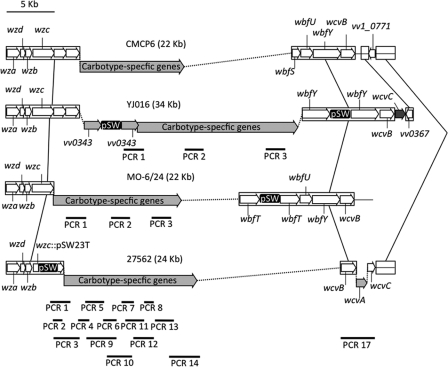
Fig. 3.
Tracking the transfer of complete and partial cps loci. The loci are aligned via the conserved wza-wzc genes at the 5′ end and wcvB at the 3′ end of each locus (white arrows). These regions flank carbotype-specific genes (light-gray arrows). Dotted lines join regions that are otherwise contiguous on the chromosomes of the respective strains. The cps locus of donor strains was tagged via targeted disruption of wzc in 27562, duplication of vv0343 or wbfY in YJ016, and duplication of wbfT in MO-6/24 using the pSW23T suicide vector (indicated as pSW), which includes a cat gene that confers chloramphenicol resistance. The black lines below the cps loci represent PCR products from primer pairs that target the region (see Table S2 in the supplemental material). The boxed areas that are joined by connecting lines denote regions where homologous recombination could occur in chitin-induced transformation experiments with CMCP6 as the recipient. The dark-gray arrow denotes the wcvC gene within the 3′ conserved region of the YJ016 cps locus, which is absent from the same region of the CMCP6 cps locus. A scale bar (in kb) is shown at the top left.
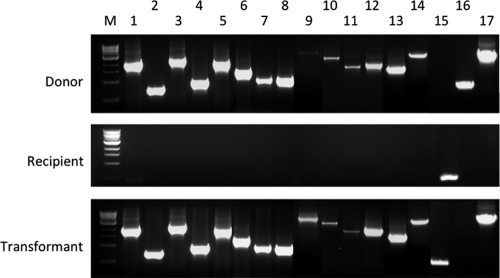
Fig. 4.
Chitin-induced transfer of the 27562 cps locus to CMCP6. gDNAs from the donor wzc::pSW23T strain, the recipient CMCP6 strain, and a representative CMCP6 transformant were used as templates in PCRs. Lanes 1 to 14 and 17 contain the corresponding PCR fragments that span the CPS region of strain 27562, depicted in Fig. 2. Lane M, molecular size markers; lane 15, PCR using primers targeting vv12532 of CMCP6; lane 16, PCR using primers targeting intIA of 27562.
We also tagged the 24-kb cps locus of strain MO-6/24 and the 34-kb cps locus of strain YJ016 with the cat gene of pSW23T by targeted duplication of the wbfT and vv0343 genes, respectively (Fig. 3). This allowed us to tag the loci with a Cmr marker without disrupting cps expression. gDNA from the MO-6/24wbfT::pSW23T or YJ016vv0343::pSW23T strain was used as the donor in chitin transformation experiments, with strain CMCP6 as the recipient. PCR with primers targeting genes at the beginning, middle, and end of the respective cps loci was used to determine how much of the locus was transferred. Cmr CMCP6 transformants were obtained at a frequency of 1.3 × 10−8 ± 0.7 × 10−8 when gDNA from the MO-6/24wbfT::pSW23T strain was used as the donor. This TF was similar to that for the 27562-tagged cps locus, and PCR analysis indicated that the entire MO-6/24 cps locus was incorporated in this strain (CMCP6-cpsMO-6/24). No transformants were recovered using the vv0343-tagged gDNA of strain YJ016 as the donor (TF < 10−9). However, when the cps locus was similarly tagged at wbfY (YJ016wbfY::pSW23T), CMCP6 transformants were obtained at a TF of 6.7 × 10−7 ± 4.1 × 10−8. PCR analysis revealed that the CMCP6 transformants had acquired the wbfY, wcvB, and wcvC genes near the 3′ end of the donor YJ016 cps locus, but the genes upstream of wbfY were not transferred (Fig. 5). The net affect of this recombination event was the incorporation of an additional polysaccharide biosynthesis gene (wcvC) into the parental CMCP6 locus (CMCP6-wcvCYJ016). Thus, novel combinations of CPS biosynthesis genes can be generated via chitin-induced natural transformation. Although these results suggested that there might be a limitation with respect to the length of gDNA that could be successfully transferred via chitin-induced transformation (i.e., the 24-kb cps loci of strains 276562 and MO-6/24 were acquired by CMCP6, but the 34-kb cps locus of YJ016 was not), our inability to detect the transfer of the larger YJ016 cps locus may have been due to limitations associated with obtaining sufficient quantities of high-molecular-weight gDNA that would encompass the entire 34-kb cps locus. Even under the gentlest conditions, the bulk of the gDNA isolated from the various V. vulnificus strains was typically <35 kb (data not shown).
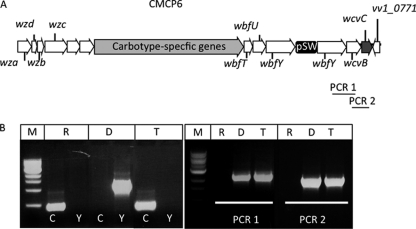
Fig. 5.
Creation of hybrid cps loci by chitin-induced natural transformation. (A) Illustration of the hybrid cps locus of a CMCP6 transformant obtained following transformation with gDNA from YJ016wbfT::pSW23T. (B) PCR analysis of gDNAs from the CMCP6 recipient (lanes R), YJ016wbfT::pSW23T donor (lanes D), and a representative CMCP6 transformant (lanes T). (Left) PCRs with recipient- and donor-specific primers. Lanes M, molecular size markers; lanes C, PCR for the CMCP6-specific vv12532 gene; lanes Y, PCR for the YJ016-specific intIA gene. (Right) PCR of the same samples used in panel A to detect products PCR 1 and PCR 2.
DISCUSSION
CPS is one of the few known virulence factors of V. vulnificus that is recognized to be absolutely required for pathogenicity (35). Strains that produce a CPS are cleared from the bloodstream more slowly and are more invasive in subcutaneous tissue than acapsular cells (76). Our genomic comparison of the CPS regions from V. vulnificus strains with different CPS carbotypes revealed that the genetically varied cps loci were flanked by conserved chromosomal regions, which suggested that they were acquired by horizontal gene transfer (44). Here, we demonstrate that V. vulnificus can undergo carbotype conversion following the uptake and expression of complete cps loci from exogenous DNA and that the conserved regions flanking the carbotype-specific cps loci serve as targets for homologous recombination. The acquisition of a partial locus was also demonstrated when internal regions of homology between the endogenous and exogenous loci existed. This suggested that the same mechanism governing the transfer of complete cps loci also contributed to their evolution by generating novel combinations of CPS biosynthesis genes. Furthermore, phase variation, in which the bacteria switch from the opaque (CPS+) to translucent (CPS−) phenotype, has been widely observed among V. vulnificus isolates and even within cells of a single colony (24, 30, 52, 74). In some cases, this switch is due to the site-specific deletion of the wzb gene (10, 52), which encodes a phosphatase required for CPS expression (72). These wzb deletion mutants were “locked” in the translucent phase (10). Chitin-induced transformation could provide a mechanism for these strains to restore CPS production and revert to an opaque phenotype. Finally, the putative lps locus is adjacent to the cps locus in the genomes of CMCP6, YJ016, and 27562. The separate or simultaneous exchange of these cps and lps loci in the Vibrionaceae may provide a mechanistic basis for promoting the emergence of new pathogenic strains.
Carbotype conversion in V. vulnificus was induced by chitin, and surfaces lacking chitin or with low chitin bioavailability were unable to support natural transformation in wild-type cells. Since the majority of chitin in marine environments is incorporated into the exoskeletons of invertebrates, the induction of competence in V. vulnificus necessitates sustained contact with a chitin-containing surface. Biofilm formation promotes the establishment of stable surface-associated communities. The stability of bacterial biofilms and the physical proximity of bacteria within them provide optimal environments for gene transfer to occur. The cells are sheathed in a hydrated matrix composed of polysaccharides, nucleic acids, and proteins that are produced by the resident microorganisms (61). The nucleic acid component is derived from DNA that is released by the biofilm consortia and functions as a cell-to-cell interconnecting component that provides stability to the structure (1, 18, 38, 43, 65). It also provides genetic material for exchange by natural transformation. DNA release may occur naturally with cell aging or through the actions of bacteriocins (51) or phages (63). Biofilms can concentrate phage-sized particles over 100-fold relative to the concentration in the surrounding water column (22), and lysis of susceptible cells can lead to the accumulation of extensive amounts of DNA within the biofilm, as we have shown. This DNA is available for uptake by species that may be or may become naturally competent. When mixed-culture biofilms of strains MO-6/24 and CMCP6 were exposed to a MO-6/24-specific lytic phage, there was an increase in the number of CMCP6 transformants obtained. This was in agreement with previous reports demonstrating the role of lytic phages and chitin-induced competence in the emergence of new variants of pathogenic V. cholerae (70) and suggests that phage-induced lysis of susceptible strains can augment natural transformation in marine biofilm environments.
In nature, V. vulnificus exists as hundreds of different carbotypes (8, 29). The diversity of genes involved in capsule production underscores the complexity and importance of this surface structure for V. vulnificus. It also implies that carbotype conversion provides a selective advantage; otherwise, a single or small number of carbotypes would be expected to be dominant among clinical and environmental isolates. The exchange of cps loci by chitin-induced transformation is unlikely to occur during the course of human infection, since there is no chitin in the human body. Carbotype conversion most likely occurs in marine environments; however, the rationale for this remains nebulous. Despite competition studies demonstrating that encapsulated strains of V. vulnificus were better adapted than acapsular strains for survival in oysters and in oyster hemocytes (28, 60), there is no indication that the type of capsule produced is important, and although many marine invertebrates are known to produce antibacterial compounds (7, 19, 26, 48, 59, 62, 67, 71), it is generally accepted that they possess only an innate immune response (11, 33). This, in theory, should not drive the need for carbotype conversion, since this would better benefit bacteria confronting an acquired immune response. However, recent studies suggest the existence of specific or “primed” immunity in invertebrates (for a review, see reference 55). If this proves to be the case, then the colonization of these hosts by V. vulnificus could drive carbotype conversion and promote bacterial survival. In this scenario, the use of chitin, which is abundant in the exoskeletons of crustaceans that V. vulnificus is known to colonize (15, 16, 49, 64), as an inducer of natural competence is an ingenious evolutionary development. Since no evidence exists to suggest that polysaccharide loci are preferentially acquired during natural transformation, the phenomenon of chitin-induced transformation likely plays an important but general role in the evolution of this genetically promiscuous genus (23, 27, 41, 47, 50).
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Steven Doyle and Battista Calvieri (University of Toronto) for their expertise with CLSM analysis.
Y.G. is an Ontario Graduate Scholarship (OGS) award recipient. This work was supported by a Canadian Institutes of Health Research (CIHR) grant to D.A.R.-M.
Footnotes
Supplemental material for this article may be found at http://iai.asmusa.org/.
Published ahead of print on 13 June 2011.
REFERENCES
- 1. Allesen-Holm M., et al. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114–1128 [DOI] [PubMed] [Google Scholar]
- 2. Alonso J. L., Mascellaro S., Moreno Y., Ferrus M. A., Hernandez J. 2002. Double-staining method for differentiation of morphological changes and membrane integrity of Campylobacter coli cells. Appl. Environ. Microbiol. 68:5151–5154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bik E. M., Bunschoten A. E., Gouw R. D., Mooi F. R. 1995. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 14:209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bisharat N., et al. 1999. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354:1421–1424 [DOI] [PubMed] [Google Scholar]
- 5. Blokesch M., Schoolnik G. K. 2008. The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae. J. Bacteriol. 190:7232–7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blokesch M., Schoolnik G. K. 2007. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 3:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bragadeeswaran S., Thangaraj S. 2011. Hemolytic and antibacterial studies on skin mucus of eel fish, Anguilla anguilla Linnaues, 1758. Asian J. Biol. Sci. 4:272–276 [Google Scholar]
- 8. Bush C. A., et al. 1997. Classification of Vibrio vulnificus strains by the carbohydrate composition of their capsular polysaccharides. Anal. Biochem. 250:186–195 [DOI] [PubMed] [Google Scholar]
- 9. CFSAN 1998. Bacteriological analytical manual, 8th ed. Centre for Food Safety and Applied Nutrition, U.S. Food and Drug Administration, Silver Spring, MD [Google Scholar]
- 10. Chatzidaki-Livanis M., Jones M. K., Wright A. C. 2006. Genetic variation in the Vibrio vulnificus group 1 capsular polysaccharide operon. J. Bacteriol. 188:1987–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen F. Y., Liu H. P., Bo J., Ren H. L., Wang K. J. 2010. Identification of genes differentially expressed in hemocytes of Scylla paramamosain in response to lipopolysaccharide. Fish Shellfish Immunol. 28:167–177 [DOI] [PubMed] [Google Scholar]
- 12. Comstock L. E., Johnson J. A., Michalski J. M., Morris J. G., Jr., Kaper J. B. 1996. Cloning and sequence of a region encoding a surface polysaccharide of Vibrio cholerae O139 and characterization of the insertion site in the chromosome of Vibrio cholerae O1. Mol. Microbiol. 19:815–826 [DOI] [PubMed] [Google Scholar]
- 13. Comstock L. E., Kasper D. L. 2006. Bacterial glycans: key mediators of diverse host immune responses. Cell 126:847–850 [DOI] [PubMed] [Google Scholar]
- 14. Comstock L. E., et al. 1995. The capsule and O antigen in Vibrio cholerae O139 Bengal are associated with a genetic region not present in Vibrio cholerae O1. Infect. Immun. 63:317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davis J. W., Sizemore R. K. 1982. Incidence of Vibrio species associated with blue crabs (Callinectes sapidus) collected from Galveston Bay, Texas. Appl. Environ. Microbiol. 43:1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. d'Ayala G. G., Malinconico M., Laurienzo P. 2008. Marine derived polysaccharides for biomedical applications: chemical modification approaches. Molecules 13:2069–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Demarre G., et al. 2005. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 156:245–255 [DOI] [PubMed] [Google Scholar]
- 18. Dominiak D. M., Nielsen J. L., Nielsen P. H. 2011. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ. Microbiol. 13:710–721 [DOI] [PubMed] [Google Scholar]
- 19. Dorrington T., Villamil L., Gomez-chiarri M. 2011. Upregulation in response to infection and antibacterial activity of oyster histone H4. Fish Shellfish Immunol. 30:94–101 [DOI] [PubMed] [Google Scholar]
- 20. Falini G., Fermani S. 2004. Chitin mineralization. Tissue Eng. 10:1–6 [DOI] [PubMed] [Google Scholar]
- 21. Finlay B. B., Falkow S. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flood J. A., Ashbolt N. J. 2000. Virus-sized particles can be entrapped and concentrated one hundred fold within wetland biofilms. Adv. Environ. Res. 3:403–411 [Google Scholar]
- 23. Frischer M. E., Thurmond J. M., Paul J. H. 1990. Natural plasmid transformation in a high-frequency-of-transformation marine Vibrio strain. Appl. Environ. Microbiol. 56:3439–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grau B. L., Henk M. C., Pettis G. S. 2005. High-frequency phase variation of Vibrio vulnificus 1003: isolation and characterization of a rugose phenotypic variant. J. Bacteriol. 187:2519–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grossart H. P., Steward G. F., Martinez J., Azam F. 2000. A simple, rapid method for demonstrating bacterial flagella. Appl. Environ. Microbiol. 66:3632–3636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gueguen Y., et al. 2009. Oyster hemocytes express a proline-rich peptide displaying synergistic antimicrobial activity with a defensin. Mol. Immunol. 46:516–522 [DOI] [PubMed] [Google Scholar]
- 27. Gulig P. A., Tucker M. S., Thiaville P. C., Joseph J. L., Brown R. N. 2009. USER friendly cloning coupled with chitin-based natural transformation enables rapid mutagenesis of Vibrio vulnificus. Appl. Environ. Microbiol. 75:4936–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harris-Young L., Tamplin M. L., Mason J. W., Aldrich H. C., Jackson J. K. 1995. Viability of Vibrio vulnificus in association with hemocytes of the American oyster (Crassostrea virginica). Appl. Environ. Microbiol. 61:52–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hayat U., et al. 1993. Capsular types of Vibrio vulnificus: an analysis of strains from clinical and environmental sources. J. Infect. Dis. 168:758–762 [DOI] [PubMed] [Google Scholar]
- 30. Hilton T., Rosche T., Froelich B., Smith B., Oliver J. 2006. Capsular polysaccharide phase variation in Vibrio vulnificus. Appl. Environ. Microbiol. 72:6986–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hor L. I., Chang T. T., Wang S. T. 1999. Survival of Vibrio vulnificus in whole blood from patients with chronic liver diseases: association with phagocytosis by neutrophils and serum ferritin levels. J. Infect. Dis. 179:275–278 [DOI] [PubMed] [Google Scholar]
- 32. Ito Y., Suzuki M., Husimi Y. 2000. A T-extended vector using a green fluorescent protein as an indicator. Gene 245:59–63 [DOI] [PubMed] [Google Scholar]
- 33. Iwanaga S., Lee B. L. 2005. Recent advances in the innate immunity of invertebrate animals. J. Biochem. Mol. Biol. 38:128–150 [DOI] [PubMed] [Google Scholar]
- 34. Jones L. J., Haugland R. P., Singer V. L. 2003. Development and characterization of the NanoOrange protein quantitation assay: a fluorescence-based assay of proteins in solution. Biotechniques 34:850–858,passim [DOI] [PubMed] [Google Scholar]
- 35. Jones M. K., Oliver J. D. 2009. Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77:1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kashimoto T., et al. 2003. Vibrio vulnificus induces macrophage apoptosis in vitro and in vivo. Infect. Immun. 71:533–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kurita K. 2006. Chitin and chitosan: functional biopolymers from marine crustaceans. Mar. Biotechnol. (NY) 8:203–226 [DOI] [PubMed] [Google Scholar]
- 38. Mann E. E., et al. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4:e5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marais D. J. 1983. Earth's early biosphere. Princeton University Press, Princeton, NJ [Google Scholar]
- 40. Marco-Noales E., Milan M., Fouz B., Sanjuan E., Amaro C. 2001. Transmission to eels, portals of entry, and putative reservoirs of Vibrio vulnificus serovar E (biotype 2). Appl. Environ. Microbiol. 67:4717–4725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meibom K. L., Blokesch M., Dolganov N. A., Wu C. Y., Schoolnik G. K. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827 [DOI] [PubMed] [Google Scholar]
- 42. Morris J. G., Jr. 2003. Cholera and other types of vibriosis: a story of human pandemics and oysters on the half shell. Clin. Infect. Dis. 37:272–280 [DOI] [PubMed] [Google Scholar]
- 43. Mulcahy H., Charron-Mazenod L., Lewenza S. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 4:e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakhamchik A., Wilde C., Chong H., Rowe-Magnus D. A. 2010. Evidence for the horizontal transfer of an unusual capsular polysaccharide biosynthesis locus in marine bacteria. Infect. Immun. 78:5214–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakhamchik A., Wilde C., Rowe-Magnus D. A. 2008. Cyclic-di-GMP regulates extracellular polysaccharide production, biofilm formation, and rugose colony development by Vibrio vulnificus. Appl. Environ. Microbiol. 74:4199–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakhamchik A., Wilde C., Rowe-Magnus D. A. 2007. Identification of a Wzy polymerase required for group IV capsular polysaccharide and lipopolysaccharide biosynthesis in Vibrio vulnificus. Infect. Immun. 75:5550–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Paul J. H., Thurmond J. M., Frischer M. E., Cannon J. P. 1992. Intergeneric natural plasmid transformation between E. coli and a marine Vibrio species. Mol. Ecol. 1:37–46 [DOI] [PubMed] [Google Scholar]
- 48. Pelon W., Luftig R. B., Johnston K. H. 2005. Vibrio vulnificus load reduction in oysters after combined exposure to Vibrio vulnificus-specific bacteriophage to an oyster extract component. J. Food Prot. 68:1188–1191 [DOI] [PubMed] [Google Scholar]
- 49. Peters W. 1972. Occurrence of chitin in mollusca. Comp. Biochem. Physiol. B Comp. Biochem. 41:541–544 [Google Scholar]
- 50. Pollack-Berti A., Wollenberg M. S., Ruby E. G. 2010. Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ. Microbiol. [Epub ahead of print.] doi:10.1111/j.1462-2920.2010.02250.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Riley M. A., Wertz J. E. 2002. Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol. 56:117–137 [DOI] [PubMed] [Google Scholar]
- 52. Rosche T. M., Smith B., Oliver J. D. 2006. Evidence for an intermediate colony morphology of Vibrio vulnificus. Appl. Environ. Microbiol. 72:4356–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rowe-Magnus D. A. 2009. Integrase-directed recovery of functional genes from genomic libraries. Nucleic Acids Res. 37:e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rowe-Magnus D. A., Guerout A. M., Biskri L., Bouige P., Mazel D. 2003. Comparative analysis of superintegrons: engineering extensive genetic diversity in the vibrionaceae. Genome Res. 13:428–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rowley A. F., Powell A. 2007. Invertebrate immune systems—specific, quasi-specific, or nonspecific? J. Immunol. 179:7209–7214 [DOI] [PubMed] [Google Scholar]
- 56. Ruby E. G., Nealson K. H. 1977. Pyruvate production and excretion by the luminous marine bacteria. Appl. Environ. Microbiol. 34:164–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shimahara K., Takiguchi Y. 1988. Preparation of crustacean chitin. Methods Enzymol. 161:417–423 [Google Scholar]
- 58. Simon R., Priefer U. B., Puhler A. 1983. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 59. Smith V. J., Desbois A. P., Dyrynda E. A. 2010. Conventional and unconventional antimicrobials from fish, marine invertebrates and micro-algae. Mar. Drugs 8:1213–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Srivastava M., Tucker M. S., Gulig P. A., Wright A. C. 2009. Phase variation, capsular polysaccharide, pilus and flagella contribute to uptake of Vibrio vulnificus by the Eastern oyster (Crassostrea virginica). Environ. Microbiol. 11:1934–1944 [DOI] [PubMed] [Google Scholar]
- 61. Stoodley P., Sauer K., Davies D. G., Costerton J. W. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187–209 [DOI] [PubMed] [Google Scholar]
- 62. Subramanian S., Ross N. W., MacKinnon S. L. 2008. Comparison of antimicrobial activity in the epidermal mucus extracts of fish. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 150:85–92 [DOI] [PubMed] [Google Scholar]
- 63. Sutherland I. W., Hughes K. A., Skillman L. C., Tait K. 2004. The interaction of phage and biofilms. FEMS Microbiol. Lett. 232:1–6 [DOI] [PubMed] [Google Scholar]
- 64. Suzuki M., Sakuda S., Nagasawa H. 2007. Identification of chitin in the prismatic layer of the shell and a chitin synthase gene from the Japanese pearl oyster, Pinctada fucata. Biosci. Biotechnol. Biochem. 71:1735–1744 [DOI] [PubMed] [Google Scholar]
- 65. Svensson S. L., et al. 2009. The CprS sensor kinase of the zoonotic pathogen Campylobacter jejuni influences biofilm formation and is required for optimal chick colonization. Mol. Microbiol. 71:253–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tamplin M. L., Specter S., Rodrick G. E., Friedman H. 1985. Vibrio vulnificus resists phagocytosis in the absence of serum opsonins. Infect. Immun. 49:715–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tasumi S., et al. 2002. Primary structure and characteristics of a lectin from skin mucus of the Japanese eel Anguilla japonica. J. Biol. Chem. 277:27305–27311 [DOI] [PubMed] [Google Scholar]
- 68. Todd E. C. 1989. Costs of acute bacterial foodborne disease in Canada and the United States. Int. J. Food Microbiol. 9:313–326 [DOI] [PubMed] [Google Scholar]
- 69. Tsang P. H., Li G., Brun Y. V., Freund L. B., Tang J. X. 2006. Adhesion of single bacterial cells in the micronewton range. Proc. Natl. Acad. Sci. U. S. A. 103:5764–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Udden S. M., et al. 2008. Acquisition of classical CTX prophage from Vibrio cholerae O141 by El Tor strains aided by lytic phages and chitin-induced competence. Proc. Natl. Acad. Sci. U. S. A. 105:11951–11956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wei O. Y., Xavier R., Marimuthu K. 2010. Screening of antibacterial activity of mucus extract of snakehead fish, Channa striatus (Bloch). Eur. Rev. Med. Pharmacol. Sci. 14:675–681 [PubMed] [Google Scholar]
- 72. Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75:39–68 [DOI] [PubMed] [Google Scholar]
- 73. Wright A. C., Powell J. L., Kaper J. B., Morris J. G., Jr 2001. Identification of a group 1-like capsular polysaccharide operon for Vibrio vulnificus. Infect. Immun. 69:6893–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wright A. C., Simpson L. M., Oliver J. D., Morris J. G., Jr 1990. Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect. Immun. 58:1769–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yamamoto S., Morita M., Izumiya H., Watanabe H. 2010. Chitin disaccharide (GlcNAc)2 induces natural competence in Vibrio cholerae through transcriptional and translational activation of a positive regulatory gene, tfoXVC. Gene 457:42–49 [DOI] [PubMed] [Google Scholar]
- 76. Yoshida S., Ogawa M., Mizuguchi Y. 1985. Relation of capsular materials and colony opacity to virulence of Vibrio vulnificus. Infect. Immun. 47:446–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.