Abstract
Cell wall-associated (CWA) proteins made by Gram-positive pathogens play a fundamental role in pathogenesis. Staphylococcus pseudintermedius is a major animal pathogen responsible for the canine skin disease bacterial pyoderma. Here, we describe the bioinformatic analysis of the family of 18 predicted CWA proteins encoded in the genome of S. pseudintermedius strain ED99 and determine their distribution among a phylogenetically diverse panel of S. pseudintermedius clinical isolates and closely related species of the Staphylococcus intermedius group. In parallel, we employed a proteomic approach to identify proteins presented on the surface of strain ED99 in vitro, revealing a total of 60 surface-localized proteins in one or more phases of growth, including 6 of the 18 genome-predicted CWA proteins. Based on these analyses, we selected two CWA proteins (SpsD and SpsL) encoded by all strains examined and investigated their capacity to mediate adherence to extracellular matrix proteins. We discovered that SpsD and SpsL mediated binding of a heterologous host, Lactococcus lactis, to fibrinogen and fibronectin and that SpsD mediated binding to cytokeratin 10, a major constituent of mammalian skin. Of note, the interaction with fibrinogen was host-species dependent, suggestive of a role for SpsD and SpsL in the host tropism of S. pseudintermedius. Finally, we identified IgG specific for SpsD and SpsL in sera from dogs with bacterial pyoderma, implying that both proteins are expressed during infection. The combined genomic and proteomic approach employed in the current study has revealed novel host-pathogen interactions which represent candidate therapeutic targets for the control of bacterial pyoderma.
INTRODUCTION
Skin diseases are a major cause of morbidity in dogs and an important animal welfare issue (33, 42). In particular, bacterial pyoderma caused by Staphylococcus pseudintermedius (formerly regarded as Staphylococcus intermedius [2]) is one of the most common diseases seen in small-animal veterinary practice worldwide (21). Although this organism is now regarded as a resident of the nares and perianal areas (33), disruption of the normal skin flora (e.g., by inappropriate antibacterial therapy), damage to the cutaneous barrier by a pruritic condition (e.g., hypersensitivities), immunosuppression by a systemic disease (e.g., hypothyroidism and hyperadrenocorticism), or primary immunodeficiencies (e.g., selective IgA deficiency) can lead to surface, superficial, or deep pyodermas caused by this nomadic organism (42). Treatment of canine pyoderma is often difficult without resorting to aggressive, medium-term administration of systemic antibacterial agents to prevent relapse of infection, and such therapy predisposes to the development of bacterial resistance that may be transferred to bacteria infecting humans (18). Of note, methicillin-resistant S. pseudintermedius has recently emerged as a major problem in veterinary clinics worldwide (2, 34). Although rare, several episodes of life-threatening infections of humans by S. pseudintermedius have been reported, with the typical route of transmission being through dog bite wounds (17, 36). Previously, crude vaccine preparations based on Staphylococcus aureus phage lysate or S. pseudintermedius autogenous bacterin have shown promise as adjunctive therapies for treatment of pyoderma (7, 8), and a rationally designed effective vaccine would be a highly desirable means to reducing the suffering associated with the disease.
Recent studies of cell wall-associated (CWA) proteins of the human and animal pathogen S. aureus indicate that some CWA proteins represent promising targets for immunotherapeutic and vaccine design (14, 49). However, our knowledge of the diversity and function of surface-presented proteins of S. pseudintermedius is very limited. Recent reports demonstrated that S. pseudintermedius has several host protein-binding properties in common with S. aureus, including the ability to bind to the extracellular matrix (ECM) proteins fibrinogen, fibronectin, and cytokeratin (15, 40). In addition, we along with others have demonstrated that S. pseudintermedius attaches to epidermal cells of healthy and atopic dogs (12, 29, 44, 45). However, the bacterial determinants responsible for these interactions have not been identified.
In order to provide a framework for investigating the pathogenesis of S. pseudintermedius and to identify new targets for the design of disease intervention strategies, we recently sequenced the whole genome of a clinical isolate of S. pseudintermedius (strain ED99) from a case of canine pyoderma, revealing the complement of encoded putative virulence factors (3). In the current study, we describe the family of CWA proteins encoded by S. pseudintermedius and their distribution among clinical isolates and closely related species of the S. intermedius group. In parallel, we identify the complement of proteins presented on the S. pseudintermedius ED99 cell surface during growth in vitro. Finally, we identify specific CWA proteins expressed by S. pseudintermedius during canine infection which mediate binding to the ECM proteins fibrinogen, fibronectin, and cytokeratin 10, suggesting an important role in disease pathogenesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. pseudintermedius strain ED99 was isolated from a dog with bacterial pyoderma presented to the Dermatology Service of The Hospital for Small Animals, The Royal (Dick) School of Veterinary Studies [R(D)SVS], The University of Edinburgh (2, 45). Phylogenetically diverse clinical isolates of S. pseudintermedius, Staphylococcus delphini, and S. intermedius used for CWA gene screening were selected to represent the diversity identified in a previous population genetic study of the S. intermedius group (SIG) (2). The study had employed concatenated sequences specific for five core genome loci, including the 16S rRNA, cpn60 (hsp60), tuf, pta, and agrD genes, to examine the genetic diversity among strains phenotypically identified as SIG members (2). S. aureus strain SH1000 (22), S. aureus strain Newman, a Newman ΔclfA strain (deficient in ClfA expression) (11), and Lactococcus lactis expressing FnbpA, encoded by plasmid pKS80 (27), were used as controls for binding to fibrinogen, fibronectin, and cytokeratin 10. L. lactis constructs expressing CWA proteins of S. pseudintermedius ED99 were generated using L. lactis subspecies cremoris strain MG1363 and expression plasmid pOri23 (37). S. aureus strains were grown to stationary phase overnight or to exponential phase (optical density at 600 nm [OD600] of 0.5 to 0.8) at 37°C in tryptic soy broth (TSB) with shaking. L. lactis bacterial cultures were grown statically at 30°C overnight in GM17 broth.
In silico structural analysis of cell wall-anchored proteins.
The complete genome sequence of S. pseudintermedius strain ED99 was deposited in the GenBank database under the accession number CP002478. The predicted CWA proteins were examined for functional domains using EMBL-EBI InterPro Scan (http://www.ebi.ac.uk/interpro). Structural analysis was carried out with the PHYRE (Protein Homology/analogY Recognition Engine) fold recognition server, available from the Structural Bioinformatics Group, Imperial College London, London, United Kingdom (http://www.sbg.bio.ic.ac.uk/phyre/). Repeat sequences were predicted by generating nucleic acid dot plots, using software available from Colorado State University (http://www.vivo.colostate.edu/molkit/dnadot/), applying tandem repeat finder software from Boston University (http://tandem.bu.edu/trf/trf.html) and variable sequence tandem repeat extraction and architecture modeling (XSTREAM), available from the University of California (http://jimcooperlab.mcdb.ucsb.edu/xstream/). Sequence alignments and pairwise sequence comparisons were generated with Clustal W2 (http://www.ebi.ac.uk/Tools/clustalw2). Amino acid composition and molecular weight predictions were generated using ProtParam on the ExPASy Proteomics Server (http://www.expasy.ch/tools/protparam.html).
Southern blot analysis.
Genomic DNA was extracted using a PerElute genomic DNA isolation kit (Edge Biosystems) as previously described (2), restriction digested with HindIII restriction endonuclease (New England BioLabs, United Kingdom), resolved by agarose electrophoresis, and transferred to nylon membranes as per the method of Southern (47). Probes were designed using oligonucleotides designed to amplify ∼500-bp regions specific for each gene encoding putative CWA proteins (Table 1) and were labeled using a Direct Nucleic Acid Labeling and Detection System (Amersham BioSciences, GE Healthcare, United Kingdom).
Table 1.
Oligonucleotides employed during the study
| Primer namea | Sequence (5′–3′)b | Purpose |
|---|---|---|
| spsD-F | CATGGGATCCAGAGAAGAAGAGGGA | L. lactis cloning |
| spsD-R | GCACCTGCAGGGGTTTTTTAATGTG | L. lactis cloning |
| spsL-F | GTTATGAGTTAGTCGACAGGGAGTT | L. lactis cloning |
| spsL-R | GAAACTGCAGATATATTGACCAC | L. lactis cloning |
| spsD-region A-Fc | TCAGAAGTGAGCGCAACAACAC | A domain cloning |
| spsD-region A-Rc | ATTCCAGTTTTCTAACGGGAAGAAACCAGC | A domain cloning |
| spsL-region A-Fc | AATGAAGATGTCACTGAAACAACTGGGAG | A domain cloning |
| spsL-region A-Rc | CCATCCTATATCAATCACATGGC | A domain cloning |
| spsA-F | TTGGTAGTGGACTGCAAATGCC | Southern probe |
| spsA-R | GGTCTTTCAGTGCGCTCACATC | Southern probe |
| spsB-F | GCCTTGATTTCGGTGTCAGACA | Southern probe |
| spsB-R | GCTGCCACCTAGAACGATTGC | Southern probe |
| spsC-F | CACGCCACTCGGTTGGTTCA | Southern probe |
| spsC-R | TGGAATGCGCCATAGTACCAAG | Southern probe |
| spsD-F | TGGTCAAGGAGAATTGCCACA | Southern probe |
| spsD-R | TTTTCACCACGATCCTCACCA | Southern probe |
| spsE-F | AACGCGTCAAACAACGTGCAA | Southern probe |
| spsE-R | CGCCATCAGTCGAAACGTAGGA | Southern probe |
| spsF-F | AGTGGAAGCAACAGTTGAACGC | Southern probe |
| spsF-R | TGGACCTACTTGGCTACCACCA | Southern probe |
| spsG-F | AACACAGCTGACAAAGCAACCG | Southern probe |
| spsG-R | CGATGTTTGCAGTTTGGCATTT | Southern probe |
| spsH-F | GATCAAAGGGCACACATGGATG | Southern probe |
| spsH-R | CCGTTATCAGTGGCAGGTGGT | Southern probe |
| spsI-F | GCAAACAAAAAAGCACCTGCAC | Southern probe |
| spsI-R | AAGATTCGCCTGCAATGTCGTA | Southern probe |
| spsJ-F | GTACGTCGGTTTCATTGAGCCA | Southern probe |
| spsJ-R | TTGACACTGATGCCGAAGCAC | Southern probe |
| spsK-F | ATTTCACAAGGGAACGCACATG | Southern probe |
| spsK-R | TGAGCCGCACGTCTATTCTGAA | Southern probe |
| spsL-F | ATGGCAAGCAAAATTTGACTGC | Southern probe |
| spsL-R | GTGTCAGCTTGCGCATCATATG | Southern probe |
| spsM-F | GGTGCTAAAGCCTACGATGCG | Southern probe |
| spsM-R | CCTCAACAATACTGCCCGATGA | Southern probe |
| spsN-F | GTGGCAAGGTCGTTATGACGGG | Southern probe |
| spsN-R | TGCCCTGATCCTTGATGTTTTGT | Southern probe |
| spsO-F | GGTAGTGTATCAGTGCTAATAGGAGCC | Southern probe |
| spsO-R | TTGACAAATCAGTAGCTGATGCATC | Southern probe |
| spsP-F | CAGGAGGACTAGGGTAATGTTCC | PCR detection |
| spsP-R | GCAAAACTTGGCGTGTTTACAAG | PCR detection |
| spsQ-F | CCGCTCTATTTTTAGGTTAATC | PCR detection |
| spsQ-R | GCGCTTCATCGAAACTTGGCGCAGG | PCR detection |
| spsR-F | TGGTCTAGGGCTATTAGCAACA | PCR detection |
| spsR-R | TCAGATTCACCTGTGTTTGG | PCR detection |
F, forward primer; R, reverse primer.
Inserted restriction sites are underlined.
Gateway specific nucleotides were added on each side of the DNA fragments (forward extension, 5′-AAAAAGCAGGCTCCGCCATG-3′; reverse extension, 5′-AGAAAGCTGGGTC-3′).
Tryptic digestion and processing of bacterial surface proteins.
An overnight culture of S. pseudintermedius ED99 was inoculated into 50 ml of brain heart infusion (BHI) broth to an OD600 of 0.04 and was grown to an early exponential (OD600 of 0.2), mid-exponential (OD600 of 0.5), or late exponential (OD600 of 0.65) phase of growth. Cells were harvested by centrifugation at 4,500 × g for 10 min and washed twice in ice-cold phosphate-buffered saline (PBS), followed by resuspension in 0.5 ml of PBS containing 40% (wt/vol) sucrose. Digestion was carried out with 20 μg of trypsin (Promega, United Kingdom) in the presence of 5 mM dithiothreitol (DTT; Millipore) for 2 h at 37°C with gentle shaking. The digestion mixture was centrifuged at 4,500 × g for 10 min, and the supernatant was incubated overnight at 37°C, followed by passage through a 4-mm by 0.45-μm Millex-HV polyvinylidene difluoride (PVDF) membrane filtration unit (Millipore) to remove microparticulates. Filtered digests (0.5 ml) were injected directly onto a 250-mm by 4.6-mm Jupiter 5U, C5, 300-Å reversed-phase column (Phenomenex) equilibrated with 2% acetonitrile and 0.1% (wt/vol) trifluoroacetic acid (TFA) at a flow rate of 0.5 ml/min. UV absorbance at 214 nm was monitored until the reading stabilized at baseline level. Bound material was eluted for 5 min by applying a linear gradient of 2% to 80% acetonitrile–0.1% (wt/vol) TFA, and fractions of 1 ml were collected every 2 min for 30 min.
LC-ESI-MS/MS.
High-performance liquid chromatography (HPLC) fractions corresponding to peaks of UV-absorbent material were frozen at −70°C before lyophilization overnight in a ModlyoD freeze dryer (Thermo Electron Corporation) in accordance with the manufacturer's recommendations. Lyophilized residues were each reconstituted in a total of 100 μl per digest of HPLC-grade water (Rathburn Chemicals, Ltd). Each reconstituted sample was passed through a second 4-mm by 0.45-μm Millex-HV PVDF membrane filtration unit to remove any insoluble material and microparticulates. Filtered samples were transferred to HPLC sample vials and stored at 4°C until required for liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) analysis. Liquid chromatography was performed using an Ultimate 3000 nano-HPLC system (Dionex) consisting of a WPS-3000 well plate micro autosampler, an FLM-3000 flow manager and column compartment, a UVD-3000 UV detector, an LPG-3600 dual-gradient micropump, and an SRD-3600 solvent rack controlled by Chromeleon chromatography software (Dionex). A micropump flow rate of 246 μl/min was used in combination with a cap-flow splitter cartridge, affording a 1/82 flow split and a final flow rate of 3 μl/min through a 5-cm by 200-μm (internal diameter) monolithic reversed-phase column (Dionex-LC Packings) maintained at 50°C. Samples of 1 to 4 μl were applied to the column by direct injection, and peptides were eluted by the application of a 15-min linear gradient from 8% to 45% solvent B (80% acetonitrile, 0.1% [vol/vol] formic acid), directed through a 3-nl UV detector flow cell. LC was interfaced directly with a three dimensional (3D) high-capacity ion trap mass spectrometer (Esquire HCTplus; Bruker Daltonics) via a low-volume (50 μl/min maximum) stainless steel nebulizer (catalog number G1946-20260; Agilent) and ESI. Parameters for tandem MS analysis were set as previously described (2a).
Database mining.
Deconvoluted MS/MS data were submitted to an in-house MASCOT server and searched against a specific and fully annotated S. pseudintermedius strain ED99 genome sequence using the MASCOT search algorithm (Matrix Science). The presentation and interpretation of MS/MS data were performed in accordance with published guidelines (51). To this end, fixed and variable modifications selected were carbamidomethyl (C) and oxidation (M), respectively, and mass tolerance values for MS and MS/MS were set at 1.5 Da and 0.5 Da, respectively. Molecular weight search (MOWSE) scores attained for individual protein identifications were inspected manually and considered significant only if (i) two peptides were matched for each protein and (ii) each peptide contained an unbroken b or y ion series of a minimum of 4 amino acid residues.
Expression of S. pseudintermedius CWA proteins in L. lactis MG1363.
PCR amplification of the full-length spsD and spsL genes was carried out with oligonucleotides which included PstI, SalI, or BamHI restriction sites to facilitate cloning (Table 1). PCR mixtures of 50 μl contained 0.25 μM forward and reverse primers, 1× Pfu Ultra II reaction buffer (Stratagene), 0.25 mM (each) deoxynucleoside triphosphate (dNTP; Promega), 5 U of Pfu Ultra II Fusion HS DNA polymerase (Stratagene), and approximately 100 ng of genomic DNA template. The thermocycler program included an initial denaturation step at 95°C for 2 min, followed by 30 cycles of 95°C for 20 s, 50°C for 20 s, and 72°C for 2 min, with a final incubation at 72°C for 3 min. PCR products were visualized on 0.8% (wt/vol) agarose gels and purified using a QIAquick Gel Extraction Kit (Qiagen, United Kingdom) according to the manufacturer's instructions. Purified DNA fragments were cloned into the StrataClone Blunt PCR cloning vector pSC-B (Stratagene) according to the manufacturer's instructions. Vector inserts were then subcloned into the Escherichia coli-L. lactis shuttle vector pOri23 using PstI, SalI, or BamHI restriction enzyme (37) and transformed into electrocompetent E. coli DH5α cells. CWA protein-encoding pOri23 constructs were purified using a Qiagen MiniPrep Kit (Qiagen, United Kingdom) and transformed into electrocompetent L. lactis cells, as described previously (37). L. lactis transformants were screened by plasmid isolation using a Qiagen MiniPrep Kit (Qiagen, United Kingdom) with addition of 100 U/ml mutanolysin (Sigma-Aldrich, United Kingdom) and 100 μg/ml lysozyme (Sigma-Aldrich, United Kingdom) to buffer P1, followed by diagnostic restriction digestion with appropriate restriction enzymes. DNA sequencing of each gene insert was carried out to confirm the absence of spurious mutations.
Cloning, expression, and purification of CWA A-domain peptides.
To generate recombinant proteins of the putative ligand-binding A domains of the predicted CWA proteins SpsD and SpsL, Gateway recombination cloning technology was employed (Invitrogen, United Kingdom). Oligonucleotides were designed for PCR amplification of the region encoding the A domain of spsD and spsL which included Gateway-compatible upstream and downstream nucleotide extensions (Table 1), and cloning was carried out according to the manufacturer's instructions. The destination vector (petG-K-NHis) based on a pET expression vector conferring kanamycin resistance and encoding a His6 tag at the N terminus was employed, and the expression construct was transformed into electrocompetent E. coli BL21 cells.
For the purification of recombinant protein, 4 liters of E. coli BL21 exponential phase liquid cultures was induced by adding 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma-Aldrich) for 4 h at 37°C in a shaking incubator at 225 rpm. Bacterial cultures were centrifuged at 1,912 × g for 10 min, pellets were resuspended in 20 ml of nickel column buffer A (20 mM NaPO4, 0.5 M NaCl), and bacterial cells were lysed using a French press (SLM Aminco; SLM Instruments, Inc.). Samples were ultracentrifuged at 180,000 × g at 4°C for 20 min and filtered using a 0.45-μm-pore-size filter device (Millipore). Proteins were purified by applying the samples to nickel columns (Nickel HisTrap HP; GE Healthcare) attached to a fast protein liquid chromatography (FPLC) machine (ÄKTA, GE Healthcare) using nickel column buffer A and buffer B (20 mM NaPO4, 0.5 M NaCl, 0.5 M imidazole). Samples were further purified using a Q Sepharose column (HiTrap 5-ml QHP; GE Healthcare) attached to an FPLC machine (ÄKTA, GE Healthcare) with Q Sepharose column buffer A (1 mM EDTA in PBS, pH 7.4) and Q Sepharose column buffer B (1 mM EDTA, 1 M NaCl in PBS, pH 7.4). Samples were extensively dialyzed (Fisherbrand dialysis tubing with a molecular weight cutoff of 12,000 to 14,000; Fisherbrand) against PBS at 4°C after each column.
Western blot analysis of recombinant CWA-associated proteins.
Protein samples were resolved by SDS-PAGE in 10% polyacrylamide gels, and Western blot transfer onto Amersham Hybond ECL Nitrocellulose Membrane (GE Healthcare, United Kingdom) was carried out under standard conditions; the resulting blot was blocked overnight in 5% (wt/vol) nonfat dried milk powder (Fluka) in PBS at 4°C. One liter of Western transfer buffer contained 0.15 M glycine, 8.4 mM SDS, and 20% (vol/vol) methanol. The blocked membranes were incubated with a 1:1,000 dilution of pooled serum samples from three canine bacterial pyoderma cases obtained from patients of the Dermatology Service of The Hospital for Small Animals, Division of Veterinary Clinical Sciences, The Royal (Dick) School of Veterinary Studies, The University of Edinburgh) in 1% (wt/vol) milk for 1 h at room temperature, followed by three washes with TBST buffer (0.14 M NaCl, 2.7 mM KCl, 25 mM Tris base, 0.05% Tween 20, pH 7.5). Secondary antibody incubation was carried out with a 1:3,000 dilution of horseradish peroxidase (HRP)-conjugated sheep anti-canine IgG (Bethyl Laboratories, Inc.) for 1 h at room temperature, and bound antibodies were detected after exposure to Amersham High Performance chemiluminescent film (GE Healthcare, United Kingdom).
Bacterial adherence assays.
Bacterial adherence assays were carried out using commercially available fibrinogen of canine, human, feline, and bovine origin (Sigma-Aldrich, United Kingdom) and bovine fibronectin (Calbiochem, EMD Biosciences). In addition, fibrinogen was purified from canine, equine, and avian blood (Lampire) by modified cold ethanol fractionation according to previously published protocols (6, 9). Recombinant cytokeratin 10 was expressed and purified from a pQE30 expression clone which encoded the C terminus of mouse cytokeratin 10 (rMCK10 294-570) (54). Recombinant protein expression in E. coli XL1-Blue cells (Stratagene, United Kingdom) was induced with IPTG (0.5 mM) and protein purified under hybrid conditions according to the manufacturer's instructions using a Ni-nitrilotriacetic acid (NTA) benchtop purification system (Invitrogen, United Kingdom). Solid-phase adherence assays with S. pseudintermedius or L. lactis cells binding to fibrinogen, fibronectin, or cytokeratin 10 were carried out as described previously (11). Each assay was performed in triplicate, and each experiment was repeated at least once, independently. Statistical significance of adherence values at the 20 μg/ml concentration, relative to controls, was determined using a Student's t test.
RESULTS AND DISCUSSION
S. pseudintermedius strain ED99 adheres to extracellular matrix proteins.
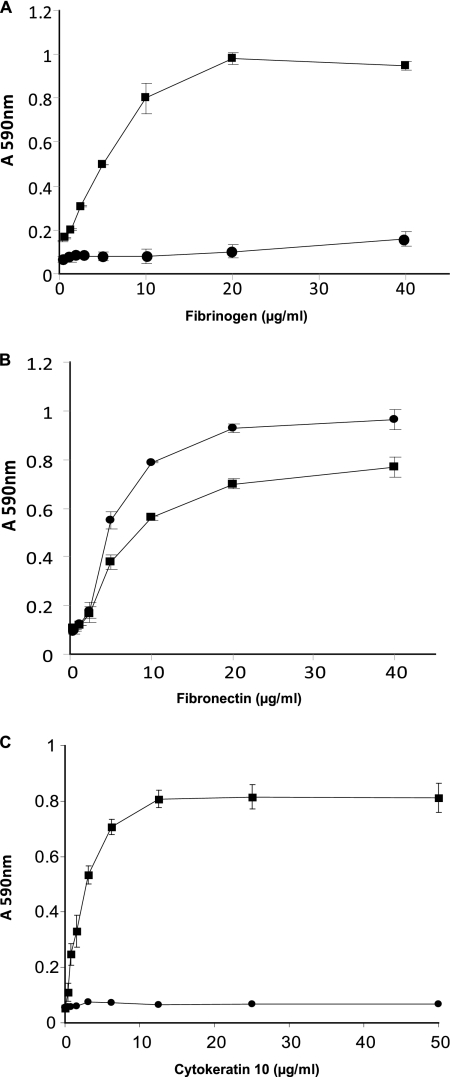
In order to investigate the capacity of S. pseudintermedius strain ED99 to bind to the extracellular matrix (ECM), we examined the adherence of exponential (OD600 of 0.5) and stationary phase cultures to doubling dilutions of fibrinogen, fibronectin, and cytokeratin 10 in solid-phase assays (Fig. 1). In the exponential phase of growth, S. pseudintermedius ED99 demonstrated adherence to canine fibrinogen, bovine fibronectin, and mouse cytokeratin 10 (P < 0.01) (Fig. 1). When grown to stationary phase of growth, S. pseudintermedius ED99 adhered also to fibronectin (P < 0.01) but did not adhere to fibrinogen or cytokeratin 10 (Fig. 1). The variation in adherence of S. pseudintermedius ED99 in exponential and stationary phases of growth implies that some surface factors mediating adherence to the extracellular matrix are expressed in a growth phase-dependent manner. Of note, the previously characterized S. pseudintermedius strain 326 exhibited similar trends for the binding of fibronectin and cytokeratin 10 but, in contrast to strain ED99, could bind to fibrinogen in the stationary phase of growth (15). These findings highlight strain-dependent differences in the surface presentation of extracellular matrix-binding proteins by S. pseudintermedius, as observed previously (15). Overall, the data suggest that S. pseudintermedius ED99 expresses surface components which mediate binding to fibrinogen, fibronectin, and cytokeratin 10 and that these components are expressed in a growth phase-dependent manner.
Fig. 1.
Adherence of S. pseudintermedius strain ED99 to immobilized ligands. Plates were coated with doubling dilutions of fibrinogen (A), fibronectin (B), and cytokeratin 10 (C) and incubated with S. pseudintermedius ED99 grown to exponential (▪) or stationary (•) phase of growth. Absorbance was measured at 590 nm, and results are expressed as mean values of triplicate samples; error bars indicate standard deviations. S. aureus strain SH1000, strain Newman, and an isogenic derivative of strain Newman deficient in ClfA production were used as controls (data not shown).
In silico analysis of the family of CWA proteins encoded by S. pseudintermedius ED99.
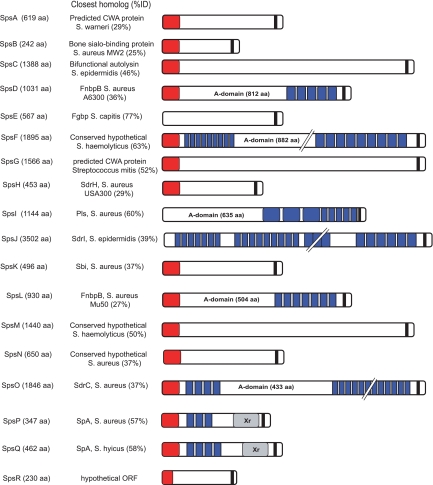
Analysis of the genome of strain ED99 revealed 18 genes encoding putative CWA proteins with a predicted minimum length of approximately 250 amino acids (aa) which contained one or more sequence motifs typical of CWA proteins, including an N terminus signal sequence, characteristic repeat regions, and an LPXTG cell wall anchor motif (named S. pseudintermedius surface protein; SpsA to SpsR) (Fig. 2). BLASTx analysis of the NCBI database resulted in sequence identities with known staphylococcal proteins ranging from 25% to 77% (Fig. 3). Signal sequences necessary for Sec-dependent protein secretion (13) were predicted for 15 putative Sps proteins, but no obvious signal sequence was predicted for SpsE, SpsI, and SpsJ (Fig. 3).
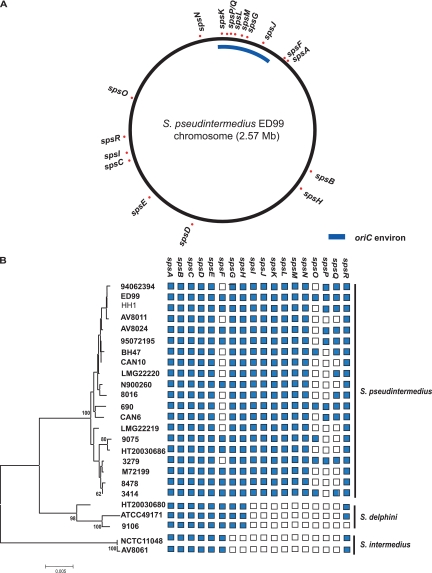
Fig. 2.
Genome location and distribution of genes encoding CWA proteins. (A) Genome map of S. pseudintermedius ED99 indicating the genomic location of each gene. (B) Distribution of the genes encoding putative CWA proteins among 20 S. pseudintermedius strains, representatives of the closely related S. delphini and S. intermedius, and other staphylococcal species associated with animal skin disease. The diversity of strains is indicated by a phylogenetic tree constructed using the neighbor-joining method, with bootstrap values over 60 indicated; a blue square denotes presence of a gene, and an empty square indicates its absence, based on Southern blot analysis (for spsA to spsO) or PCR amplification (for spsP, spsQ, and spsR).
Fig. 3.
Schematic representation of predicted CWA proteins encoded in the S. pseudintermedius ED99 genome. The predicted length in amino acids of each protein is indicated; typical MSCRAMM features are shown as appropriate with signal sequence (red), the A domain with IgG-like folds (labeled), repeat regions (blue), and a cell wall-anchoring LPX(TSA)/(GANS) motif (black). For SpsP and SpsQ (S. aureus SpA orthologues), the repeated IgG-binding domains (blue) and X repeat regions (Xr; gray) are also indicated. All organisms belong to Staphylococcus species unless otherwise noted. ORF, open reading frame.
Repeat regions are a common feature of staphylococcal CWA proteins, such as the Sdr-family, members of which contain a serine-aspartate dipeptide region (13). Accordingly, the 18 putative CWA proteins were examined for the presence of tandem repeat regions, which were detected for SpsD, SpsF, SpsI, SpsJ, SpsL, SpsO, SpsP, and SpsQ (Table 2and Fig. 3). Of note, SpsI contains repeats which belong to the B-repeat superfamily, sharing 52% overall identity with the B repeats of SdrD of S. aureus strain COL. None of the predicted repeats are serine-aspartate, characteristic of proteins in the Sdr family (13), but the repeat motifs of SpsO are serine rich, containing 578 serine residues (49.9% of the amino acid composition in the repeat region). In addition, SpsJ contains mainly serine (447 residues, or 49.9%), threonine (163 residues, or 18.2%), and aspartate (113 residues, or 12.6%) residues, resulting in conserved motifs SDSD and STS (Table 2). Repeat regions of previously characterized staphylococcal CWA proteins such as the fibronectin-binding protein A (FnbpA) have been shown to be involved in ligand binding (41) but may also function as molecular stalks which project the ligand-binding domain away from the bacterial cell surface (19).
Table 2.
Repeat regions of S. pseudintermedius CWA proteins
| CWA protein | No. of repeats | Repeat length (aa) | Pairwise identity (%) | Characteristic(s) |
|---|---|---|---|---|
| SpsD | 5 | 21 | 90–100 | |
| SpsF | 9 | 86 | 72–100 | |
| 9 | 15 | 73–100 | ||
| SpsI | 3 | 112 | 77–96 | B-repeat family |
| 5 | 14 | 100 | ||
| SpsJ | 6 | 80 | 65–92 | |
| 3 | 18–20 | 85–100 | ||
| 9 | 46 | 80–100 | ||
| 61 | 12–20 | 42–100 | SDSD and STS rich | |
| SpsL | 7 | 37 | 91–100 | |
| SpsO | 96 | 12 | 91–100 | Serine rich |
| 4 | 26 | 76–96 | ||
| SpsP | 3 | 55 | 78–89 | IgG binding |
| SpsQ | 4 | 52–59 | 59–86 | IgG binding |
In staphylococcal CWA proteins, β-sheet-rich IgG-like folds of the A domain have been shown to be important for ligand binding (35). Therefore, the putative CWA proteins identified in the S. pseudintermedius ED99 genome were examined for predicted IgG-like folds. Based on InterPro Scan and PHYRE predictions, two IgG-like folds each were identified within the nonrepeated A domains of SpsD, SpsI, SpsL, and SpsO (Fig. 3), dividing the A domains into N1, N2, and N3 subdomains. The predicted locations of the IgG-like folds are residues 167 to 320 (N2) and 322 to 519 (N3) for SpsD, residues 220 to 363 (N2) and 364 to 531 (N3) for SpsL, residues 202 to 351 (N2) and 347 to 512 (N3) for SpsI, and residues 339 to 492 (N2) and 487 to 659 (N3) for SpsO.
Of the 18 putative CWA proteins of S. pseudintermedius ED99, SpsD, SpsL, and SpsO contained each of the MSCRAMM (microbial surface components recognizing adhesive matrix molecules) features screened for, including a signal sequence at the N terminus, followed by a nonrepeated A domain with two IgG-like folds dividing the A domain into N1, N2, and N3 subdomains, a tandemly repeated domain at the C terminus (and at the N terminus for SpsO), and a C-terminal LPXTG anchor motif (Table 3). Of note, a TYTFTDYVD motif or variant, important for the “dock, lock, and latch” ligand-binding mechanism (35), was found in SpsD, SpsL, and SpsO, and putative latching sequences were also identified (Table 3). Further, putative Fn-binding motifs with weak homology to FnbpA-10 of FnbpA in S. aureus (41) were detected in the repeat region of SpsL (24% identity in pairwise alignments for SpsL1 to SpsL6 and 21% for SpsL7). No homology to Fn-binding motifs of FnbpA was detected in the repeat regions of SpsD and SpsO. The genes encoding SpsD, SpsL, and SpsO have distinct locations in the S. pseudintermedius ED99 genome. While spsD is located in a conserved region of the genome, spsL is in the oriC environ (50) (Fig. 2), and spsO is located adjacent to two putative transposases, suggesting that the locus may have been acquired by horizontal gene transfer. Consistent with this observation, the spsO gene was detected in only 7 of 20 S. pseudintermedius strains examined, which represented several distinct clonal lineages (Fig. 2).
Table 3.
Characteristics of the predicted CWA proteins SpsD, SpsL, and SpsO of S. pseudintermedius ED99
| Protein | Length (aa) | Mass (kDa)a | Signal peptide length (aa) | LPXTG motif | Position of IgG-like fold(s)b | TYTFTDYVD-like motif (position)b | Putative latching sequence (position)b | Position of repeat region(s) (aa) | Copy no. repeats |
|---|---|---|---|---|---|---|---|---|---|
| SpsD | 1,031 | 115 | 36 | LPDTG | 167–320 | RYRFMDYVN (267–275) | NNASGEG (491–497) | 867–959 | 5 |
| 322–519 | |||||||||
| SpsL | 930 | 103 | 38 | LPKTG | 220–363 | VYTFKDYVN (298–306) | NSASGSG (502–508) | 543–818 | 7 |
| 364–531 | |||||||||
| SpsO | 1,846 | 198 | 44 | LPNTG | 339–492 | TYTFTDYVD (424–432) | DKSTALG (635–641) | 661–1800 | 96 |
| 487–659 | 97–216c | 4c |
Predicted approximate molecular mass.
Motifs are within the A domain. Positions are amino acid numbering.
N-terminal repeat.
S. pseudintermedius ED99 encodes two putative S. aureus staphylococcal protein A (SpA) orthologues.
S. pseudintermedius ED99 has two predicted orthologues of S. aureus staphylococcal protein A (SpA) (SpsP and SpsQ) which are encoded adjacent to each other in the oriC environ of the genome (Fig. 2). The genes encode proteins with amino acid identity of 73%, each with a predicted N-terminal signal sequence of 33 aa, followed by a repeat region consisting of three and four predicted IgG-binding domains in SpsP and SpsQ, respectively (Fig. 3). Each IgG-binding domain of SpsP comprises 55 aa, with 78% to 89% identity between domains, whereas the four IgG-binding domains of SpsQ vary from 59 aa for the first and second, to 55 aa for the third, and 52 aa for the fourth domain and have between 59% to 86% protein sequence identity in any pairwise alignment. The repeated IgG-binding domains are well conserved between SpsP and SpsQ, with 67% to 90% sequence identity in pairwise alignments. On the C-terminal side of the IgG-binding domains, SpsP and SpsQ contain a predicted X region (Fig. 3). For SpsP, this region has 63% overall sequence similarity to region X of S. aureus Newman SpA, and for SpsQ the region consists of a 77-aa-long repeat sequence (Xr) and a constant region (Xc) with 70% similarity to the X region of S. aureus SpA. The Xr repeat region of SpsQ was recently used to develop a typing method analogous to the spa typing technique widely employed for epidemiological analysis of S. aureus (30, 43).
Distribution of genes encoding CWA proteins among the S. intermedius group.
In order to investigate the distribution of the putative CWA proteins identified in the S. pseudintermedius ED99 genome among other S. pseudintermedius clinical isolates, closely related species of the S. intermedius group (SIG), and S. aureus, Southern blot analysis and PCR amplification were performed. A total of 20 S. pseudintermedius strains representing the genetic diversity within the species, members of the closely related S. delphini and S. intermedius species, and a representative S. aureus isolate were screened for the presence of the putative CWA protein-encoding genes by Southern blot analysis (spsA to spsO) (Fig. 2). For the S. aureus spa orthologues spsP and spsQ, PCR amplification was employed as the genes share 70% nucleotide identity, which precluded design of gene-specific probes for Southern blot analysis. For similar reasons, the primers designed for PCR amplification were located upstream of spsP (primer spsP-F), in the nonrepeated region of spsP (primer spsP-R), in the unique region between spsP and spsQ (primer spsQ-F), and in a region unique for spsQ (primer spsQ-R) (Fig. 2). In addition, PCR was used to screen for spsR. Of the 18 genes examined, 14 were found in all S. pseudintermedius strains investigated. The remaining four genes (spsF, spsO, and the S. aureus spa orthologues spsP and spsQ) were present in 6 to 11 of the 20 S. pseudintermedius strains, depending on the gene. Furthermore, homologs of 9 of the 18 genes were detected in S. delphini, 7 were found in S. intermedius, and 10 genes were exclusive to S. pseudintermedius (Fig. 2). None of the genes encoding putative CWA proteins was detected by Southern blotting or PCR analysis of S. aureus (data not shown). It cannot be excluded that DNA sequence variation in PCR primer annealing sites for spsP, spsQ, and spsR could contribute to the lack of gene detection for some strains. Of note, five of seven genes conserved in all S. pseudintermedius strains but not found in S. delphini or S. intermedius were located in the oriC environ (Fig. 2). The oriC environ represents ∼250 kb around the origin of replication in staphylococcal genomes and is typically the only region of the genome which does not demonstrate synteny between different staphylococcal species (50). Our data are consistent with a role for the oriC environ in the speciation of S. pseudintermedius and imply that the oriC-encoded CWA proteins may have a function which is specific for the niche occupied by S. pseudintermedius. An understanding of the distribution of genes encoding CWA proteins among S. pseudintermedius strains is important because CWA proteins made by all strains could represent novel targets for broadly effective therapeutics.
Determination of the surface proteome of S. pseudintermedius ED99.
Genome sequence interrogation for motifs characteristic of genes encoding cell envelope-associated proteins often results in identification of only a proportion of surface-presented proteins. In order to identify the complement of proteins presented on the surface of S. pseudintermedius in vitro, we carried out surface proteome identification during early, mid-, and late exponential phases of growth. Manual inspection of the MS/MS spectrum was carried out for a total of 917 protein hits, representing 140 unique proteins. To be considered a positive result, protein identification required at least one protein hit above stringent cutoff values for two of the three technical replicates in at least one biological replicate. As a result of the stringent filtering conditions applied to protein identification, all proteins positively identified, except one, had two or more peptides detected with a MOWSE score superior or equal to 50. In total, 60 surface-localized proteins were identified by proteomic analysis of S. pseudintermedius ED99 in one or several growth phases tested (Table 4). According to the functional annotation and additional tests performed to predict subcellular localization, 10 of the proteins were predicted to be associated with the cell surface, and 7 were predicted to be secreted into the extracellular milieu (Table 4). Of the 18 putative CWA proteins predicted in the genome, 6 were identified in one or more phases, namely SpsD, SpsK, SpsL, SpsN, SpsO, and SpsQ, suggesting that the other 12 predicted CWA proteins were either not expressed under the conditions tested, were present in quantities below the threshold of detection of the LC-ESI-MS/MS technique, or were masked by their location in the cell envelope.
Table 4.
The surface proteome of S. pseudintermedius strain ED99
| Gene no. | Functional annotationa | Molecular mass (Da) | Proteomic analysis by growth phase |
Predicted subcellular localization | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Early exponential |
Mid-exponential |
Late exponential |
||||||||||
| MOWSE score | Coverage (%) | No. of peptides | MOWSE score | Coverage (%) | No. of peptides | MOWSE score | Coverage (%) | No. of peptides | ||||
| 40 | CWA protein, SpsQ | 50,892 | 207 | 14 | 7 | 252 | 14 | 8 | 467 | 22 | 14 | Cell wall anchored |
| 43 | CWA protein, SpsL | 102,621 | 452 | 16 | 11 | 372 | 16 | 9 | 152 | 6 | 3 | Cell wall anchored |
| 2230 | CWA protein, SpsN | 72,139 | 246 | 17 | 7 | 786 | 34 | 16 | 377 | 22 | 14 | Cell wall anchored |
| 1279 | CWA protein, SpsD | 114,479 | 439 | 15 | 13 | 267 | 15 | 10 | Cell wall anchored | |||
| 1860 | CWA protein, SpsO | 165,911 | 56 | 2 | 3 | 167 | 5 | 6 | Cell wall anchored | |||
| 2283 | CWA protein, SpsK | 55,381 | 235 | 25 | 9 | 829 | 47 | 20 | Cell wall anchored | |||
| 2089 | LSU ribosomal protein L7/L12 (L23e) | 12,543 | 171 | 59 | 5 | 525 | 87 | 13 | 408 | 69 | 7 | Cytoplasmic |
| 1811 | NAD-dependent glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12) | 36,477 | 119 | 17 | 3 | 503 | 34 | 12 | 622 | 39 | 11 | Cytoplasmic |
| 2081 | Translation elongation factor Tu | 43,243 | 199 | 24 | 5 | 360 | 20 | 5 | 212 | 14 | 5 | Cytoplasmic |
| 1018 | Cell division trigger factor (EC 5.2.1.8) | 49,186 | 193 | 20 | 4 | Cytoplasmic | ||||||
| 2090 | LSU ribosomal protein L10p (P0) | 17,856 | 141 | 33 | 4 | Cytoplasmic | ||||||
| 2092 | LSU ribosomal protein L11p (L12e) | 14,965 | 112 | 17 | 2 | Cytoplasmic | ||||||
| 2091 | LSU ribosomal protein L1p (L10Ae) | 24,985 | 124 | 19 | 4 | Cytoplasmic | ||||||
| 553 | LSU ribosomal protein L5p (L11e) | 20,195 | 296 | 45 | 9 | Cytoplasmic | ||||||
| 997 | Pyruvate kinase (EC 2.7.1.40) | 63,114 | 75 | 15 | 3 | Cytoplasmic | ||||||
| 555 | SSU ribosomal protein S8p (S15Ae) | 14,784 | 157 | 25 | 4 | Cytoplasmic | ||||||
| 195 | Fructose-bisphosphate aldolase class I (EC 4.1.2.13) | 33,045 | 107 | 14 | 3 | Cytoplasmic | ||||||
| 743 | Heat shock protein 60 family cochaperone GroES | 10,555 | 77 | 23 | 2 | Cytoplasmic | ||||||
| 1096 | Chaperone protein DnaK | 66,086 | 143 | 11 | 5 | Cytoplasmic | ||||||
| 1397 | Translation elongation factor Ts | 32,533 | 109 | 11 | 4 | Cytoplasmic | ||||||
| 1434 | Acyl carrier protein/HmrB protein involved in methicillin resistance | 8,546 | 114 | 31 | 2 | Cytoplasmic | ||||||
| 2270 | Alkyl hydroperoxide reductase protein C (EC 1.6.4.-) | 21,086 | 193 | 21 | 5 | Cytoplasmic | ||||||
| 2282 | SSU ribosomal protein S6p | 11,687 | 111 | 24 | 2 | Cytoplasmic | ||||||
| 58 | Arginine deiminase (EC 3.5.3.6) | 47,385 | Cytoplasmic | |||||||||
| 1807 | Enolase (EC 4.2.1.11) | 47,061 | 368 | 24 | 8 | 206 | 16 | 4 | Cytoplasmic | |||
| 650 | Fructose-bisphosphate aldolase class II (EC 4.1.2.13) | 31,285 | 131 | 29 | 5 | 211 | 33 | 8 | Cytoplasmic | |||
| 908 | Transaldolase (EC 2.2.1.2) | 25,774 | 108 | 18 | 3 | 97 | 12 | 2 | Cytoplasmic | |||
| 2082 | Translation elongation factor G | 76,914 | 381 | 24 | 14 | 159 | 13 | 5 | Cytoplasmic | |||
| 672 | ATP synthase beta chain (EC 3.6.3.14) | 51,230 | 186 | 13 | 4 | 100 | 11 | 3 | Cytoplasmic | |||
| 1559 | Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex (EC 2.3.1.12) | 46,732 | 113 | 7 | 2 | 148 | 12 | 3 | Cytoplasmic | |||
| 1560 | Pyruvate dehydrogenase E1 component beta subunit (EC 1.2.4.1) | 35,417 | 191 | 25 | 6 | 217 | 20 | 5 | Cytoplasmic | |||
| 1558 | Dihydrolipoamide dehydrogenase of pyruvate dehydrogenase complex (EC 1.8.1.4) | 49,655 | 150 | 11 | 4 | Cytoplasmic | ||||||
| 343 | Pyruvate formate-lyase (EC 2.3.1.54) | 85,061 | 133 | 8 | 4 | Cytoplasmic | ||||||
| 1035 | LSU ribosomal protein L21p | 11,382 | 139 | 39 | 4 | 90 | 39 | 2 | Cytoplasmic | |||
| 549 | LSU ribosomal protein L29p (L35e) | 8,027 | 165 | 42 | 3 | 129 | 34 | 2 | Cytoplasmic | |||
| 556 | LSU ribosomal protein L6p (L9e) | 19,623 | 237 | 30 | 5 | 50 | 8 | 2 | Cytoplasmic | |||
| 863 | Putative transcriptional regulator | 13,325 | 168 | 43 | 4 | 122 | 19 | 2 | Cytoplasmic | |||
| 540 | SSU ribosomal protein S10p (S20e) | 11,553 | 130 | 43 | 4 | 105 | 24 | 2 | Cytoplasmic | |||
| 566 | SSU ribosomal protein S11p (S14e) | 13,872 | 100 | 18 | 10 | 85 | 10 | 2 | Cytoplasmic | |||
| 2083 | SSU ribosomal protein S7p (S5e) | 17,801 | 123 | 25 | 4 | 107 | 17 | 3 | Cytoplasmic | |||
| 551 | LSU ribosomal protein L14p (L23e) | 13,152 | 107 | 12 | 2 | Cytoplasmic | ||||||
| 568 | LSU ribosomal protein L17p | 13,748 | 177 | 28 | 3 | Cytoplasmic | ||||||
| 552 | LSU ribosomal protein L24p (L26e) | 11,359 | 98 | 20 | 2 | Cytoplasmic | ||||||
| 1037 | LSU ribosomal protein L27p | 10,293 | 91 | 25 | 3 | Cytoplasmic | ||||||
| 544 | LSU ribosomal protein L2p (L8e) | 30,342 | 208 | 29 | 6 | Cytoplasmic | ||||||
| 559 | LSU ribosomal protein L30p (L7e) | 6,369 | 101 | 52 | 3 | Cytoplasmic | ||||||
| 565 | SSU ribosomal protein S13p (S18e) | 13,712 | 140 | 39 | 5 | Cytoplasmic | ||||||
| 1421 | SSU ribosomal protein S16p | 10,208 | 213 | 51 | 5 | Cytoplasmic | ||||||
| 547 | SSU ribosomal protein S3p (S3e) | 24,097 | 95 | 24 | 5 | Cytoplasmic | ||||||
| 415 | hypothetical protein similar to TpgX | 25,769 | 146 | 23 | 3 | Lipoprotein | ||||||
| 956 | Serine protease, DegP/HtrA, do-like (EC 3.4.21.-) | 45,084 | 219 | 15 | 6 | 211 | 15 | 4 | Membrane associated | |||
| 670 | ATP synthase alpha chain (EC 3.6.3.14) | 54,803 | 180 | 8 | 4 | 115 | 13 | 5 | Membrane associated | |||
| 1809 | Triosephosphate isomerase (EC 5.3.1.1) | 27,504 | 65 | 5 | 1 | Membrane associated | ||||||
| 495 | Secretory antigen precursor SsaA | 25,482 | 104 | 19 | 2 | 132 | 13 | 2 | 129 | 18 | 2 | Secreted |
| 964 | Extracellular adherence protein of broad specificity Eap/Map | 27,424 | 97 | 12 | 4 | Secreted | ||||||
| 302 | Immunodominant staphylococcal antigen A precursor | 27,639 | 95 | 18 | 3 | Secreted | ||||||
| 1057 | Hypothetical protein | 6,856 | 144 | 34 | 4 | Secreted | ||||||
| 151 | Malate:quinone oxidoreductase (EC 1.1.99.16) | 56,325 | 142 | 8 | 3 | 130 | 8 | 3 | Secreted | |||
| 946 | General stress protein-like protein | 22,079 | 92 | 18 | 3 | Secreted | ||||||
| 477 | Transcriptional regulator, LytR family | 35,263 | 260 | 35 | 9 | Secreted | ||||||
SSU, small subunit; LSU, large subunit.
Of the 60 proteins, 11 were detected in early exponential phase, including all six surface proteome-detected CWA proteins with the exception of SpsK, two putative secreted antigens, a protein with homology to the extracellular adherence protein (Eap) of S. aureus, and three cytoplasmic proteins (large subunit [LSU] ribosomal protein L7/L12, NAD-dependent glyceraldehyde-3-phosphate dehydrogenase [GAPDH], and transcription elongation factor Tu). Most of the proteins identified in early exponential phase were also found in both mid- and late exponential phases, with the exception of SpsD, Eap, and one of the two secretory antigens.
The majority of proteins identified (n = 50) were detected in mid-exponential phase, of which 25 were specific for this phase. Of note, 40 of the proteins recognized on the S. pseudintermedius surface in mid-exponential phase were predicted to be intracellular proteins. In addition, five of the six predicted CWA proteins (SpsD, SpsK, SpsL, SpsN, and SpsQ), the ATP synthase alpha-chain, a triosephosphate isomerase, and a putative serine protease (HtrA) were identified on the cell surface during mid-exponential phase.
During late exponential phase, a smaller number of proteins was identified, with 33 proteins identified in total, of which only 7 were specific for this phase: arginine deiminase, dihydrolipoamide dehydrogenase, pyruvate formate-lyase, a putative lipoprotein, two putative general stress proteins, and a transcriptional regulator of the LytR family (Table 4). The identification of intracellular proteins on the cell surface is not unexpected, and numerous studies of S. aureus have reported a similar phenomenon (10, 16, 46, 53, 58). In some cases, alternative functions associated with the cell surface have been described for cytoplasmic proteins, including GAPDH (26, 52), enolase (4, 23), transcription elongation factors (1, 24), and chaperone proteins GroES and DnaK (20, 57). In addition, ribosomal proteins are commonly identified on the bacterial cell surface and can exhibit a high degree of immunogenicity for humans (25, 48).
Overall, these data represent the first identification of the complete surface proteome for a strain of S. pseudintermedius. Although some of the proteins were in common with other studies of surface proteomes of Gram-positive bacteria, 27 S. pseudintermedius proteins were identified for which homologs had not been previously identified on the surface of other bacterial species. Ribosomal proteins represented 16 proteins, and 9 of the 11 other proteins identified were predicted to be involved in host-pathogen interactions, including putative cell wall-anchored proteins, secreted antigens, and a putative lipoprotein (Table 4). These data highlight the complementary role that proteomics can play in identifying surface-presented proteins which are not predicted from genome sequence analysis. In particular, our understanding of the function of intracellular proteins when presented on the cell surface is very limited, and the possibility that such proteins may represent valid therapeutic targets has not been well examined to date.
S. pseudintermedius CWA proteins mediate binding to extracellular matrix proteins.
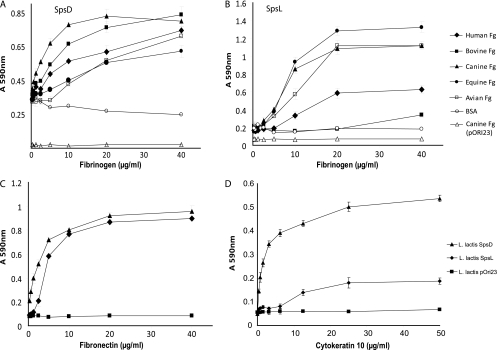
In order to investigate the molecular basis of S. pseudintermedius adherence to the extracellular matrix, we selected the only two CWA proteins (SpsD and SpsL) which were detected in the surface proteome and encoded by all strains and which had each of the CWA characteristics searched for, including a signal sequence at the N terminus, a nonrepeated A domain with two IgG-like folds, a tandemly repeated domain at the C terminus, and a C-terminal LPXTG anchor motif, and investigated their capacity to mediate adherence to ECM proteins. L. lactis expressing SpsD and SpsL and L. lactis carrying the vector pOri23 alone were tested for their ability to adhere to human fibronectin. L. lactis expressing SpsD and SpsL demonstrated adherence to human fibronectin in a dose-dependent manner (Fig. 4C). Of note, SpsD and SpsL contain 32% and 27% amino acid identity, respectively, with fibronectin-binding protein B (FnbpB) encoded by S. aureus strain Mu50. Further, SpsL contains seven repeats with weak homology to repeats of FnbpA involved in adherence to fibronectin (41), suggesting that a related mechanism of binding to fibronectin may exist for SpsL. In contrast, SpsD did not contain sequence homology with the fibronectin-binding region of FnbpA, suggesting a distinct mechanism of interaction with fibronectin for SpsD.
Fig. 4.
Adherence of L. lactis expressing specified CWA proteins to immobilized ligands. (A and B) Plates were coated with doubling dilutions of fibrinogen (Fg) of human, bovine, canine, equine, or avian origin or with bovine serum albumin (BSA), as indicated in the legend of panel B, and incubated with L. lactis expressing SpsD or SpsL. L. lactis with empty expression vector pOri23 incubated with immobilized canine fibrinogen was included as a negative control. (C and D) For panel C plates were coated with doubling dilutions of bovine fibronectin and incubated with L. lactis expressing SpsD or SpsL, as indicated on panel D. L. lactis with empty expression vector pOri23 incubated with immobilized bovine fibronectin was included as a negative control. For panel D, plates were coated with doubling dilutions of recombinant mouse cytokeratin 10 and incubated with L. lactis expressing SpsD or SpsL. L. lactis with empty expression vector pOri23 incubated with immobilized recombinant mouse cytokeratin 10 was included as a negative control. Absorbance was measured at 590 nm, and results are expressed as mean values of triplicate samples. Error bars indicate standard deviation. S. aureus strain SH1000, strain Newman, and an isogenic derivative of strain Newman deficient in ClfA production were used as controls (data not shown).
Previous studies have reported considerable variation in the sequence of fibrinogen from different vertebrate species, implying a requirement for host specialization of fibrinogen-binding proteins made by bacteria colonizing different host species (28). We tested the capacity of CWA proteins to mediate binding of L. lactis to fibrinogen of canine, human, feline, avian, equine, and bovine origin. Both SpsD and SpsL mediated adherence of L. lactis to canine fibrinogen with high affinity (P < 0.01) (Fig. 4A and B). In addition, L. lactis expressing SpsD adhered to bovine fibrinogen with similar affinity but adhered less well to human, equine, and avian fibrinogens (P < 0.01) (Fig. 4A). We speculate that the region of fibrinogen that is involved in binding to SpsD is conserved in canine and bovine hosts, leading to a similar high-affinity interaction. In contrast, in addition to canine fibrinogen, SpsL mediated strong adherence to equine and avian fibrinogens (P < 0.01) but demonstrated weak or no adherence to fibrinogens of human and bovine origins. These data imply that SpsD and SpsL have different affinities for fibrinogens from different host species which could involve different regions of the host ligand. The data also suggest that S. pseudintermedius has evolved host-specialized CWA proteins which mediate efficient binding to canine fibrinogen. Further molecular analysis is required to identify the bacterial protein and host ligand domains involved in the interaction.
Finally, SpsD mediated robust adherence of L. lactis to recombinant mouse cytokeratin 10 in solid-phase assays (P < 0.001), but SpsL demonstrated only weak adherence (Fig. 4D). The interaction of S. aureus with cytokeratin 10, a major substituent of human skin, is mediated by at least two CWA-associated proteins, including ClfB and IsdA (5, 54), and is considered to be important for S. aureus nasal colonization (56). The identification of a cytokeratin 10-binding surface protein of S. pseudintermedius suggests that such an interaction may also be important for canine host colonization. SpsD appears to be a promiscuous binding protein with the capacity to bind to at least three host ligands, including fibrinogen, fibronectin, and cytokeratin 10, in a dose-dependent and saturable manner. Of note, the marked elevation of binding of S. pseudintermedius strain ED99 to fibrinogen and cytokeratin 10 in the exponential phase of growth correlates with the identification of SpsD in the surface proteome of ED99 during the early and mid-exponential phases of growth and its absence at the late exponential time point (Fig. 1and Table 4).
Several CWA proteins made by S. aureus have been shown to interact with multiple protein ligands. For example, FnbpA mediates binding to fibronectin, fibrinogen, and elastin (27, 39, 55), and ClfB mediates binding to fibrinogen and cytokeratin 10 (31, 54). The existence of tandem IgG-related sequences in SpsD and SpsL suggests that a ligand-binding mechanism analogous to that of other microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) may exist (14, 35).
S. pseudintermedius CWA proteins SpsD and SpsL are produced during colonization or infection of dogs.
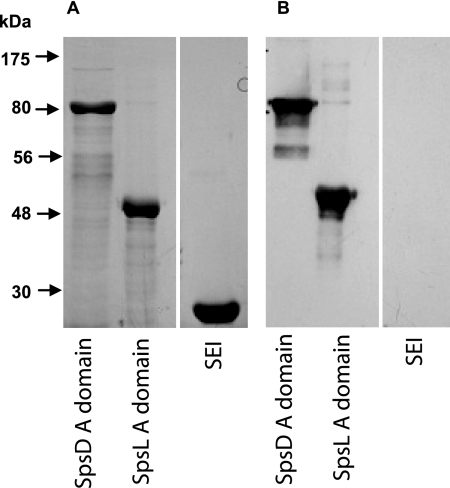
The surface proteome of S. pseudintermedius during growth under nutrient-replete conditions in vitro may differ considerably from that during infection. In vivo expression and host immune recognition are important characteristics of candidate protective antigens. Reactivity of recombinant A domains from SpsD and SpsL with canine serum obtained from bacterial pyoderma cases was examined by Western blot analysis. Both recombinant SpsD (rSpsD) and rSpsL, but not the negative-control recombinant staphylococcal enterotoxin I (rSEI) of S. aureus, bound to IgG present in the canine serum (Fig. 5). These data indicate that IgG specific for SpsD and SpsL is present in sera from dogs with bacterial pyoderma, indicating that both proteins are produced during either S. pseudintermedius colonization or infection, are recognized by the host, and induce a humoral immune response.
Fig. 5.
SpsD and SpsL are expressed during canine infection. SDS-PAGE (A) and Western immunoblot analysis (B) of recombinant A domains of SpsD and SpsL with sera obtained from dogs with S. pseudintermedius pyoderma. Recombinant SEI encoded by the human pathogen S. aureus was included as a negative control.
Concluding comments.
Overall, these results provide important new information relating to the adhesive proteins presented on the surface of S. pseudintermedius. Stranger-Jones et al. screened the genome of S. aureus for all genes predicted to encode CWA proteins and immunized mice with each protein to determine their capacity to protect against lethal or invasive infection (49). Several of the proteins were combined into a multiple protein vaccine which induced high levels of protection against S. aureus invasive disease of mice (49), stimulating renewed optimism for a vaccine for the prevention of human S. aureus infections. In addition to their potential use as vaccine components, CWA proteins are being examined as passive immunotherapeutic targets (32, 38). In particular, the major fibrinogen-binding protein, ClfA, has been the focus of antibody-based therapeutics which have reached the phase III trial stage, albeit unsuccessfully (32). Recently, we have published the structure for ClfA binding to its major ligand fibrinogen, resulting in the design of peptides which block ClfA function but do not interfere with fibrinogen binding to platelets, an essential step in platelet aggregation (14). These peptides are currently under investigation as potential therapeutic agents for treating staphylococcal disease (14). Overall, we report the genome-scale prediction and proteomic identification of the complement of S. pseudintermedius surface proteins and describe novel host-pathogen interactions likely to be important in pathogenesis. The data presented here represent an important step toward the rational design of preventive or therapeutic approaches for controlling bacterial canine pyoderma.
ACKNOWLEDGMENTS
This work was funded by the Petplan Charitable Trust (United Kingdom) and by Pfizer Animal Health. Moredun Research Institute receives support from RERAD, the Rural and Environment Research and Analysis Directorate of the Scottish government.
L. lactis strain MG1363 and pOri23 were kindly provided by P. Moreillon, University of Lausanne, Switzerland. We are grateful to R. Doolittle for helpful advice regarding fibrinogen purification and to T. Foster for provision of the recombinant mouse cytokeratin 10 construct and helpful discussions.
Footnotes
Published ahead of print on 16 May 2011.
REFERENCES
- 1. Balasubramanian S., Kannan T. R., Baseman J. B. 2008. The surface-exposed carboxyl region of Mycoplasma pneumoniae elongation factor Tu interacts with fibronectin. Infect. Immun. 76:3116–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bannoehr J., et al. 2007. Population genetic structure of the Staphylococcus intermedius group: insights into agr diversification and the emergence of methicillin-resistant strains. J. Bacteriol. 189:8685–8692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a. Batycka M., et al. 2006. Ultra-fast tandem mass spectrometry scanning combined with monolithic column liquid chromatography increase throughput in proteomic analysis. [DOI] [PubMed] [Google Scholar]
- 3. Ben Zakour N. L., Bannoehr J., van den Broek A. H. M., Thoday K. L., Fitzgerald J. R. 2011. Complete genome sequence of the canine pathogen Staphylococcus pseudintermedius. J. Bacteriol. 193:2363–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carneiro C. R., Postol E., Nomizo R., Reis L. F., Brentani R. R. 2004. Identification of enolase as a laminin-binding protein on the surface of Staphylococcus aureus. Microbes Infect. 6:604–608 [DOI] [PubMed] [Google Scholar]
- 5. Clarke S. R., et al. 2009. Iron-regulated surface determinant protein A mediates adhesion of Staphylococcus aureus to human corneocyte envelope proteins. Infect. Immun. 77:2408–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cohn E. J., et al. 1946. Preparation and properties of serum and plasma proteins; a system for the separation into fractions of the protein and lipoprotein components of biological tissues and fluids. J. Am. Chem. Soc. 68:459–475 [DOI] [PubMed] [Google Scholar]
- 7. Curtis C. F., Lamport A. I., Lloyd D. H. 2006. Masked, controlled study to investigate the efficacy of a Staphylococcus intermedius autogenous bacterin for the control of canine idiopathic recurrent superficial pyoderma. Vet. Dermatol. 17:163–168 [DOI] [PubMed] [Google Scholar]
- 8. DeBoer D. J., Moriello K. A., Thomas C. B., Schultz K. T. 1990. Evaluation of a commercial staphylococcal bacterin for management of idiopathic recurrent superficial pyoderma in dogs. Am. J. Vet. Res. 51:636–639 [PubMed] [Google Scholar]
- 9. Doolittle R. F., Fuller G. M. 1967. Quantitative determination of amino-terminal amino acids in crosslinked and non-crosslinked fibrin. Biochem. Biophys. Res. Commun. 26:327–333 [DOI] [PubMed] [Google Scholar]
- 10. Dreisbach A., et al. 2010. Profiling the surfacome of Staphylococcus aureus. Proteomics 10:3082–3096 [DOI] [PubMed] [Google Scholar]
- 11. Fitzgerald J. R., et al. 2006. Fibronectin-binding proteins of Staphylococcus aureus mediate activation of human platelets via fibrinogen and fibronectin bridges to integrin GPIIb/IIIa and IgG binding to the FcγRIIa receptor. Mol. Microbiol. 59:212–230 [DOI] [PubMed] [Google Scholar]
- 12. Forsythe P. J., Hill P. B., Thoday K. L., Brown J. 2002. Use of computerized image analysis to quantify staphylococcal adhesion to canine corneocytes: does breed and body site have any relevance to the pathogenesis of pyoderma? Vet. Dermatol. 13:29–36 [DOI] [PubMed] [Google Scholar]
- 13. Foster T. J., Hook M. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484–488 [DOI] [PubMed] [Google Scholar]
- 14. Ganesh V. K., et al. 2008. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog. 4:e1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Geoghegan J. A., Smith E. J., Speziale P., Foster T. J. 2009. Staphylococcus pseudintermedius expresses surface proteins that closely resemble those from Staphylococcus aureus. Vet. Microbiol. 138:345–352 [DOI] [PubMed] [Google Scholar]
- 16. Glowalla E., Tosetti B., Kronke M., Krut O. 2009. Proteomics-based identification of anchorless cell wall proteins as vaccine candidates against Staphylococcus aureus. Infect. Immun. 77:2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guardabassi L., Loeber M. E., Jacobson A. 2004. Transmission of multiple antimicrobial-resistant Staphylococcus intermedius between dogs affected by deep pyoderma and their owners. Vet. Microbiol. 98:23–27 [DOI] [PubMed] [Google Scholar]
- 18. Guardabassi L., Schwarz S., Lloyd D. H. 2004. Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 54:321–332 [DOI] [PubMed] [Google Scholar]
- 19. Hartford O., Francois P., Vaudaux P., Foster T. J. 1997. The dipeptide repeat region of the fibrinogen-binding protein (clumping factor) is required for functional expression of the fibrinogen-binding domain on the Staphylococcus aureus cell surface. Mol. Microbiol. 25:1065–1076 [DOI] [PubMed] [Google Scholar]
- 20. Hickey T. B., Ziltener H. J., Speert D. P., Stokes R. W. 2010. Mycobacterium tuberculosis employs Cpn60.2 as an adhesin that binds CD43 on the macrophage surface. Cell Microbiol. 12:1634–1647 [DOI] [PubMed] [Google Scholar]
- 21. Hill P. B., et al. 2006. Survey of the prevalence, diagnosis and treatment of dermatological conditions in small animals in general practice. Vet. Rec. 158:533–539 [DOI] [PubMed] [Google Scholar]
- 22. Horsburgh M. J., et al. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kolberg J., et al. 2006. Streptococcus pneumoniae enolase is important for plasminogen binding despite low abundance of enolase protein on the bacterial cell surface. Microbiology 152:1307–1317 [DOI] [PubMed] [Google Scholar]
- 24. Kunert A., et al. 2007. Immune evasion of the human pathogen Pseudomonas aeruginosa: elongation factor Tuf is a factor H and plasminogen binding protein. J. Immunol. 179:2979–2988 [DOI] [PubMed] [Google Scholar]
- 25. Kurar E., Splitter G. A. 1997. Nucleic acid vaccination of Brucella abortus ribosomal L7/L12 gene elicits immune response. Vaccine 15:1851–1857 [DOI] [PubMed] [Google Scholar]
- 26. Madureira P., et al. 2007. Streptococcus agalactiae GAPDH is a virulence-associated immunomodulatory protein. J. Immunol. 178:1379–1387 [DOI] [PubMed] [Google Scholar]
- 27. Massey R. C., et al. 2001. Fibronectin-binding protein A of Staphylococcus aureus has multiple, substituting, binding regions that mediate adherence to fibronectin and invasion of endothelial cells. Cell Microbiol. 3:839–851 [DOI] [PubMed] [Google Scholar]
- 28. McCarthy A. J., Lindsay J. A. 2010. Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: implications for vaccine design and host-pathogen interactions. BMC Microbiol. 10:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McEwan N. A., Mellor D., Kalna G. 2006. Adherence by Staphylococcus intermedius to canine corneocytes: a preliminary study comparing noninflamed and inflamed atopic canine skin. Vet. Dermatol. 17:151–154 [DOI] [PubMed] [Google Scholar]
- 30. Moodley A., Stegger M., Ben Zakour N. L., Fitzgerald J. R., Guardabassi L. 2009. Tandem repeat sequence analysis of staphylococcal protein A (Spa) gene in methicillin-resistant Staphylococcus pseudintermedius. Vet. Microbiol. 135:320–326 [DOI] [PubMed] [Google Scholar]
- 31. Ni Eidhin D., et al. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245–257 [DOI] [PubMed] [Google Scholar]
- 32. Otto M. 2008. Targeted immunotherapy for staphylococcal infections: focus on anti-MSCRAMM antibodies. BioDrugs 22:27–36 [DOI] [PubMed] [Google Scholar]
- 33. Patel A., Forsythe P. J. 2008. Small animal dermatology. Saunders/Elsevier, St. Louis, MO [Google Scholar]
- 34. Perreten V. 2010. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J. Antimicrob. Chemother. 65:1145–1154 [DOI] [PubMed] [Google Scholar]
- 35. Ponnuraj K., et al. 2003. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115:217–228 [DOI] [PubMed] [Google Scholar]
- 36. Pottumarthy S., et al. 2004. Clinical isolates of Staphylococcus intermedius masquerading as methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 42:5881–5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Que Y. A., Haefliger J. A., Francioli P., Moreillon P. 2000. Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infect. Immun. 68:3516–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rivas J. M., Speziale P., Patti J. M., Hook M. 2004. MSCRAMM-targeted vaccines and immunotherapy for staphylococcal infection. Curr. Opin. Drug Discov. Devel. 7:223–227 [PubMed] [Google Scholar]
- 39. Roche F. M., et al. 2004. The N-terminal A domain of fibronectin-binding proteins A and B promotes adhesion of Staphylococcus aureus to elastin. J. Biol. Chem. 279:38433–38440 [DOI] [PubMed] [Google Scholar]
- 40. Schmidt V., Nuttall T., Fazakerley J., McEwan N. 2009. Staphylococcus intermedius binding to immobilized fibrinogen, fibronectin and cytokeratin in vitro. Vet. Dermatol. 20:502–508 [DOI] [PubMed] [Google Scholar]
- 41. Schwarz-Linek U., et al. 2003. Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature 423:177–181 [DOI] [PubMed] [Google Scholar]
- 42. Scott D. W., Miller W. H., Jr., Griffin C. E. 2001. Bacterial skin diseases, p. 275–335 In Scott D. W., Miller W. H., Jr., Griffin C. E. (ed.), Small animal dermatology, 6th ed. W. B. Saunders, Philadelphia, PA [Google Scholar]
- 43. Shopsin B., et al. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simou C., Hill P. B., Forsythe P. J., Thoday K. L. 2005. Species specificity in the adherence of staphylococci to canine and human corneocytes: a preliminary study. Vet. Dermatol. 16:156–161 [DOI] [PubMed] [Google Scholar]
- 45. Simou C., Thoday K. L., Forsythe P. J., Hill P. B. 2005. Adherence of Staphylococcus intermedius to corneocytes of healthy and atopic dogs: effect of pyoderma, pruritus score, treatment and gender. Vet. Dermatol. 16:385–391 [DOI] [PubMed] [Google Scholar]
- 46. Solis N., Larsen M. R., Cordwell S. J. 2010. Improved accuracy of cell surface shaving proteomics in Staphylococcus aureus using a false-positive control. Proteomics 10:2037–2049 [DOI] [PubMed] [Google Scholar]
- 47. Southern E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503–517 [DOI] [PubMed] [Google Scholar]
- 48. Spence J. M., Clark V. L. 2000. Role of ribosomal protein L12 in gonococcal invasion of Hec1B cells. Infect. Immun. 68:5002–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stranger-Jones Y. K., Bae T., Schneewind O. 2006. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 103:16942–16947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Takeuchi F., et al. 2005. Whole-genome sequencing of staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292–7308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Taylor G. K., Goodlett D. R. 2005. Rules governing protein identification by mass spectrometry. Rapid Commun. Mass Spectrom. 19:3420. [DOI] [PubMed] [Google Scholar]
- 52. Terao Y., Yamaguchi M., Hamada S., Kawabata S. 2006. Multifunctional glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pyogenes is essential for evasion from neutrophils. J. Biol. Chem. 281:14215–14223 [DOI] [PubMed] [Google Scholar]
- 53. Ventura C. L., et al. 2010. Identification of a novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PLoS One 5:e11634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Walsh E. J., O'Brien L. M., Liang X., Hook M., Foster T. J. 2004. Clumping factor B, a fibrinogen-binding MSCRAMM (microbial surface components recognizing adhesive matrix molecules) adhesin of Staphylococcus aureus, also binds to the tail region of type I cytokeratin 10. J. Biol. Chem. 279:50691–50699 [DOI] [PubMed] [Google Scholar]
- 55. Wann E. R., Gurusiddappa S., Hook M. 2000. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem. 275:13863–13871 [DOI] [PubMed] [Google Scholar]
- 56. Wertheim H. F., et al. 2008. Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans. PLoS Med. 5:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xolalpa W., et al. 2007. Identification of novel bacterial plasminogen-binding proteins in the human pathogen Mycobacterium tuberculosis. Proteomics 7:3332–3341 [DOI] [PubMed] [Google Scholar]
- 58. Ziebandt A. K. 2010. Proteomics uncovers extreme heterogeneity in the Staphylococcus aureus exoproteome due to genomic plasticity and variant gene regulation. Proteomics 10:1634–1644 [DOI] [PubMed] [Google Scholar]