Abstract
The application of forward genetics can reveal new factors required for the virulence of intracellular pathogens. To facilitate such virulence screens, we developed macrophage cell lines with which the number of intact host cells following infection with intracellular pathogens can be rapidly and easily ascertained through the expression of a constitutive lacZ transgene. Using known virulence mutants of Francisella novicida and Histoplasma capsulatum, we confirmed the applicability of these host cells for the quantitative assessment of bacterial and fungal virulence, respectively. To identify new genes required for Histoplasma virulence, we employed these transgenic macrophage cells to screen a collection of individual transfer DNA (T-DNA) insertion mutants. Among the mutants showing decreased virulence in macrophages, we identified an insertion in the locus encoding the Histoplasma Hsp82 homolog. The lesion caused by the T-DNA insertion localizes to the promoter region, resulting in significantly decreased HSP82 expression. Reduced HSP82 expression markedly attenuates the virulence of Histoplasma yeast in vivo. While the HSP82 hypomorph grows normally in vitro at 37°C and under acid and salinity stresses, its ability to recover from high-temperature stress is impaired. These results provide genetic proof of the role of stress chaperones in the virulence of a thermally dimorphic fungal pathogen.
INTRODUCTION
Histoplasma capsulatum is a thermally dimorphic fungal pathogen endemic to the midwest of the United States and parts of Latin America. The acquisition of Histoplasma by inhalation causes respiratory histoplasmosis, the severity of which ranges from subclinical to acute pulmonary disease (e.g., pneumonitis), depending on dose and host immune status (15). In individuals with impaired adaptive immunity, the extrapulmonary spread of Histoplasma leads to life-threatening disseminated disease. The temperature change that characterizes the switch from the soil environment to the mammalian host triggers a morphological switch in Histoplasma growth from filamentous mycelia to the yeast form (21). This dimorphism also reflects a switch from a saprobic to a parasitic lifestyle that is required for Histoplasma to cause disease (23, 28). Pathogenic yeast cells infect host macrophages in which they establish an intracellular niche permissive for replication, ultimately leading to host cell lysis (40).
The switch to higher temperature growth during infection suggests that Histoplasma yeast cells need to adapt to increased temperature stresses, potentially through molecular chaperones such as heat shock proteins. Various heat shock response proteins have been identified in Histoplasma, including Hsp60, Hsp70, and Hsp90 family members. Temperature shift from ambient to 37°C transiently increases the expression of Histoplasma Hsp70 and Hsp82 homologs, suggesting a role for these stress proteins in adaptation to the higher temperature environment of the mammalian host (5, 25, 35). Histoplasma Hsp60 is a well-established immunoreactive antigen from yeast cell membrane/cell wall fractions (10). The vaccination of mice with Hsp60 confers protection from Histoplasma through both cellular (CD4+ T cells [7]) and humoral immune mechanisms (11). In addition, Hsp60 can function as an adhesin for yeast binding to complement receptors on host macrophages (12, 19). Indicating the importance of ameliorating heat stresses, virulence differences among Histoplasma strains are correlated with in vitro thermosensitivity and differences in heat shock response (5). Although it is widely assumed that Hsp factors contribute to the adaptation of thermally dimorphic fungal pathogens to mammalian body temperature during infection, genetic proof of the role(s) of heat shock proteins has been absent.
Limited forward-genetic capabilities for Histoplasma have hampered progress in understanding the molecular mechanisms of Histoplasma pathogenesis. Such approaches, which enable the discovery of unsuspected or novel virulence factors, require (i) an effective mutagen and (ii) an efficient screen for a phenotype of interest. The development of Agrobacterium-mediated transformation techniques for many fungi, including Histoplasma (24, 36), has solved the first task; the transfer and integration of transfer DNA (T-DNA) into the fungal genome provides the means of generating insertion mutants that can be quickly mapped with molecular techniques (44).
To identify genes important for Histoplasma pathogenesis, genetic screening strategies rely on the identification of mutants with attenuated virulence. The number of mutants that can be screened is limited by the efficiency of virulence assays that rely on the infection of host cells or animals with individual mutants. Outside of an animal host, the virulence of Histoplasma mutants has been determined by infecting macrophages with yeast cells and either measuring yeast viability and intracellular replication or by quantifying the Histoplasma-dependent killing of macrophages after infection. Methods for measuring the intracellular viability and replication of yeast include the microscopic enumeration of intracellular yeast (30), quantitation of CFU following dilution plating of host cell lysates (8, 14), or computation of viable yeast in host cell lysates by the incorporation of radioactive metabolites (29). All of these approaches are laborious and unsuited for mass screening. The alternative virulence detection method, macrophage killing by Histoplasma yeast, has been performed by quantifying the number of macrophages remaining through the tritiated uridine or bromodeoxyuridine labeling of cells (2, 9), measurement of macrophage DNA (33), or microscopy (13). Individual visual examination of each infection and/or the multiple steps required to perform these assays makes them unattractive for high-throughput application. Furthermore, since general DNA stains and metabolic assays are not specific for mammalian cells, steps must be taken to remove any yeast cells.
To facilitate the high-throughput application of Histoplasma virulence screens and better implement forward-genetic approaches to understanding Histoplasma virulence, we developed an assay for macrophage lethality that is simple, efficient, and quantitative, and it provides for specific measurement of intact host cells following infection. Transgenic macrophages were engineered with a colorimetric reporter gene, and its use as a surrogate marker to quantify surviving macrophages validated using both fungal and bacterial pathogen species. By screening a collection of Histoplasma insertion mutants with this technique, we identified Histoplasma Hsp82, an Hsp90 family member, as a factor necessary for full Histoplasma virulence. This provides genetic proof that heat shock proteins enable Histoplasma to manage stresses associated with infection of mammalian hosts.
MATERIALS AND METHODS
Microbial strains and culture.
Histoplasma capsulatum strains used in this study were derived from the North America class 2 wild-type strain G217B (ATCC 26032) and are listed in Table 1. Histoplasma yeast cells were grown in Histoplasma-macrophage medium (HMM [42]) at 37°C with shaking (200 rpm) until late log/early stationary phase unless otherwise noted. For the growth of uracil auxotrophs, medium was supplemented with uracil (100 μg/ml). For tests of salinity stress, Histoplasma was grown on HMM plates supplemented with NaCl. For tests of pH stress, Histoplasma was grown on solid HMM buffered with 4-morpholineethanesulfonic acid (MES). The growth rate and growth stage of strains were determined by the measurement of liquid culture turbidity at 595 nm. Yeast cells were precisely enumerated by hemacytometer counts. For growth on solid medium, HMM was solidified with 0.6% agarose and supplemented with 25 μM FeSO4. Histoplasma yeast cells were transformed with linear plasmids by electroporation (41) and plated on solid HMM to select Ura+ transformants. F. novicida strains used in this study were F. novicida U112 and its derivatives ΔmglA::Erm (17) and ΔpmrA::Kan (26). F. novicida strains were grown in tryptic soy medium containing 0.1% l-cysteine and quantified by using optical density at 600 nm to estimate the number of bacteria (optical density at 600 nm [OD600] of 1, which is equivalent to 3 × 109 bacteria per ml).
Table 1.
Microbial strains used
| Strain | Genotype |
|---|---|
| H. capsulatum | |
| G217B | Wild type (ATCC 26032), North American class 2 |
| WU15a | ura5-Δ42 |
| OSU8b | ura5-Δ42 cbp1-9::T-DNA [hph] |
| OSU10 | ura5-Δ42 hsp82-1::T-DNA [hph] |
| OSU25c | ura5-Δ42/pCR473 [URA5, gfp-RNAi] |
| OSU74 | ura5-Δ42 hsp82-1::T-DNA [hph]/pCR468 [URA5, gfp] |
| OSU86 | ura5-Δ42 hsp82-1::T-DNA [hph]/pCR545 [URA5, HSP82] |
| F. novicida | |
| U112d | Wild type |
| ΔmglAe | ΔmglA::Erm |
| JSG2845d | ΔpmrA::Kan |
Macrophage culture and infection.
The macrophage cell lines P388D1 and J774.1 and lacZ transgenic derivatives of each were cultured in Ham's F-12 medium or Dulbecco's modified Eagle medium (DMEM), respectively, with 10% fetal bovine serum (FBS) and 2 mM l-glutamine at 37°C under 5% CO2 and 95% air. Bone marrow-derived macrophages (BMDM) were obtained by differentiating bone marrow stem cells obtained from the femurs of C57BL/6 mice for 5 days in DMEM with 10% FBS, 2 mM l-glutamine, and 30% L929 conditioned medium as a source of macrophage colony-stimulating factor (M-CSF). For the infection of macrophages, Histoplasma yeast cells or Francisella bacteria were diluted to the desired concentration in cell culture medium (HMM buffered to pH 7.2 with 25 mM bicarbonate [HMM-M] for P388D1 infections, DMEM for J774 infections, and DMEM with 30% L929 conditioned medium for BMDM). Culture media were supplemented with 10% FBS and 2 mM l-glutamine. For infection, the macrophage culture medium was replaced with the yeast or bacterial suspension and the cells were cocultured at 37°C in 5% CO2 and 95% air. For Francisella infections, macrophages and bacteria were coincubated for 2 h at 37°C in 5% CO2 and 95% air to allow for bacterial uptake, after which extracellular bacteria were killed by incubating cells for 30 min in complete medium containing 50 μg/ml gentamicin (26). For both Histoplasma and Francisella infections, 96-well culture plates were shaken for 60 s at 1,000 rpm twice daily during the time course of the assay. At the desired time point, macrophages were quantified by staining DNA with PicoGreen as described previously (8) or by β-galactosidase activity as detailed below.
Retroviral transformation of macrophages.
Retroviral particles were generated by the calcium phosphate-mediated transient transfection (16) of 293T cells with pCR396 and plasmids encoding MMLV gag-pol and the VSVg protein. For transfection, 2 × 106 293T cells were seeded into collagen-coated 60-mm tissue culture plates and cultured in DMEM with 10% FBS. pUMVC (4.5 μg), 0.5 μg of pVSVg, and 5 μg of pCR396 were combined with 200 μl of 250 mM CaCl2. Two hundred μl of 2× HEPES-buffered saline (50 mM HEPES, 275 mM NaCl, pH 7.1) with 1.5 mM sodium phosphate was added dropwise to the DNA-CaCl2 solution, and the mixture was added to the 293T cells. The culture medium was replaced 24 h posttransfection. Forty-eight hours posttransfection, the culture supernatant was removed and filtered through a 0.45-μm syringe filter. The retrovirus-containing culture medium was added to 1 × 106 macrophages (either P388D1 or J774.1 cells) in a T-25 flask, and the medium was supplemented with 8 μg/ml protamine sulfate. Retrovirus infection was carried out for 6 h at 37°C, after which the culture medium was replaced and the cells cultured for an additional 40 h. Transformed macrophages were selected by subculturing in culture medium containing 500 μg/ml G418 (AG Scientific). Clonal cell lines were established by a limiting dilution of G418-resistant cells, and the β-galactosidase activity was determined for each line as described below.
LacZ-based assay of macrophage number.
Transgenic macrophages were seeded into wells of a 96-well tissue culture plate at 3 × 104 per well (P388D1 cells), 5 × 104 per well (J774 cells), or 3.4 × 104 per well (BMDM cells) in culture medium with 10% FBS and 2 mM l-glutamine. For infection experiments, Histoplasma yeast cells or Francisella bacteria were added after the overnight incubation of macrophages as described above. At the desired time point, the culture medium was removed and replaced with β-galactosidase assay buffer: phosphate-buffered saline (PBS) with 0.5% Triton X-100, 2 mM MgCl2, and 2 mg/ml o-nitrophenyl-β-d-galactopyranoside (ONPG). The absorbance of each well at 420 and 600 nm was determined for 60 min at 25°C using a Synergy2 plate reader (BioTek). Relative β-galactosidase activity was determined by analyzing the 420 nm absorbances during the linear range of the assay (typically 20 to 60 min), after subtracting the values of absorbance at 600 nm and normalization to results for control wells. Triplicate wells were analyzed, and the mean relative activities were compared by Student's t test for statistical significance.
Mutagenesis of Histoplasma and screening for attenuated mutants.
Agrobacterium tumefaciens was used to mutagenize Histoplasma yeast as described previously (44). Briefly, A. tumefaciens strain LBA1100 harboring plasmid pCM41 (22) was cocultured with WU15 Histoplasma yeast at 25°C on cocultivation medium (18 mM NH4Cl, 1.25 mM MgSO4, 2 mM KCl, 100 μM CaCl2, 10 μM FeSO4, 5 mM KH2PO4, 0.5% glucose, 40 mM MES, pH 5.3, 700 μM cystine, 100 μg/ml uracil, and 0.1 mM acetosyringone). Bacteria and yeast cells were combined in PBS and plated on Whatman number 5 filter paper on solid cocultivation medium at densities of 7.5 × 106 bacteria/cm2 and 1.8 × 106 yeast cells/cm2. After 48 h, filters were transferred to HMM solid medium containing 100 μg/ml uracil, 200 μM cefotaxime, and 100 μg/ml hygromycin to select for transformed yeast. Plates were incubated at 37°C for 8 to 12 days until transformant colonies appeared. Transformants were picked into wells of a 96-well plate containing 100 μl of HMM with 100 μg/ml uracil, and the plate was incubated for 3 days at 37°C. The density of yeast in each well was estimated by the measurement of the optical density at 595 nm and comparison to a standard curve of known numbers of Histoplasma yeast. Yeast cells from each well were diluted into liquid HMM to a density of approximately 3 × 106/ml, and 10 μl was transferred with a multichannel pipettor to a 96-well plate previously seeded with 3 × 104 P388D1-lacZ cells/well in HMM-M with 10% FBS and 2 mM l-glutamine. Plates were mixed by shaking for 60 s at 1,000 rpm and then incubated at 37°C in 5% CO2 and 95% air. When the majority of the wells had less than 10% confluence (typically 6 to 7 days), the remaining macrophages were quantified using the β-galactosidase activity as described above.
Determination of the site of T-DNA insertion.
One of the attenuated mutants recovered from the screen (16E1) was chosen for further characterization and was designated OSU10. Sequences flanking the T-DNA insertion site in OSU10 were determined using thermal asymmetric interlaced PCR (TAIL-PCR) (18). For the primary PCR, 200 ng of total nucleic acid was used in a PCR with the left border primer LB6 (TGTTGGACTGACGCAACGACCTTGTCAACC) and the semirandom primer LAD1 (ACGATGGACTCCAGAGCGGCCGCVNVNNNGGAA). For secondary PCR, a 1,000-fold dilution of the primary PCR was used as the template for PCR performed with the LB7 left border primer (CGGACAGACGGGGCAAAGCTGCCTACCA) and the AC1 primer (ACGATGGACTCCAGAG), which binds to the 5′ end of the LAD1 primer. The secondary PCR product was sequenced using primer LB8 (CAGGGACTGAGGGACCTCAGCAGGTCG). For sequence flanking the right border of the T-DNA element, sequences were amplified by PCR using the T-DNA-specific primer ORI-7 (CAAGAACTCTGTAGCACCGCC) and the HSP82-5 primer (GAACCTCCTTCTCAGTCTCCTTG). Flanking sequences were aligned to the Histoplasma genome sequence (http://genome.wustl.edu/genomes/view/Histoplasma_capsulatum/) to determine the location and nature of the T-DNA insertion.
Quantitative reverse transcription-PCR (qRT-PCR).
Histoplasma RNA was collected from exponentially growing yeast cells by beating with 0.5-μm diameter glass beads in TRIzol reagent (Invitrogen) and precipitation of the aqueous phase with isopropanol. Total RNA was resuspended in RNase-free H2O and subjected to two sequential treatments with DNase I (Ambion). Five μg of total RNA was reverse transcribed with Superscript III (Invitrogen) using 250 ng random 15-mers to prime reverse transcription. HSP82 transcripts were amplified by PCR using primers HSP82-11 (GACCAAGCCTATCTGGACTCGCA) and HSP82-12 (TCACGAGAGAGGTTGAGAGGAAGGTC) and 1.2 μl of a 1:10 dilution of reverse-transcribed RNA as the template. PCR was performed in a Mastercycler EP Realplex2 thermocycler (Eppendorf), and amplicons were quantified using the SYBR green-based reaction mix (Bio-Rad). Relative changes in HSP82 transcription were determined using the ΔΔCT method (32) after normalization to actin transcripts amplified with primer ACT1-5 (GGTTTCGCTGGCGATGATGCTC) and primer ACT1-9 (AAGGACGGCCTGGATGGAGACG).
In vivo virulence determination.
To enable murine infections, the OSU10 strain was transformed with a URA5 plasmid to restore uracil prototrophy (strain OSU74) (Table 1). The construction of the complemented strain OSU86 was achieved by the transformation of OSU10 with a URA5 plasmid that also carries the native HSP82 locus. The complementing HSP82 locus fragment was amplified from wild-type Histoplasma DNA and included 1,202 bp upstream and 939 bp downstream of the HSP82 coding sequence (CDS).
C57BL/6 mice were infected with Histoplasma yeast by the intranasal delivery of 1 × 104 yeast cells suspended in HMM. Histoplasma yeast cells were collected from exponentially growing liquid cultures and enumerated by hemacytometer. After 8 days, mice were euthanized and lungs and spleens collected. Lung and spleen tissues were homogenized in 5 and 3 ml HMM, respectively, and serial dilutions of the homogenates were plated on solid HMM to determine the fungal burden in each organ. Mean CFU counts were compared between infections by the Wilcoxon rank-sum test for statistical significance.
Oxidative and nitrosative stress challenge.
Suspensions of Histoplasma yeast cells were incubated for 4 h either in PBS with hydrogen peroxide (Sigma) or in HMM with DETA NONOate (Cayman Chemical). A stock solution of 100 mM DETA NONOate was prepared fresh by solubilization in 10 mM NaOH. To liberate nitric oxide, the DETA NONOate was diluted into liquid HMM at pH 7.0 to the desired concentration. Dilution into neutral pH liberates two molecules of NO per molecule of DETA NONOate. After oxidative or nitrosative challenge, dilutions of each suspension were spotted onto solid HMM to assess yeast viability. Plates were incubated at 35 to 37°C until colonies appeared.
RESULTS
Development of a LacZ-based assay for enumeration of macrophages.
To facilitate the rapid quantitation of macrophages necessary for high-throughput virulence screens, we created transgenic macrophage cell lines that constitutively express the lacZ gene encoding β-galactosidase. By using a constitutive promoter, the expression of this reporter gene can be reliably used to determine the number of cells. The well-established β-galactosidase assay is specific and quantitative using both colorimetric and fluorimetric substrates. Furthermore, the assay can be performed directly in a detergent-containing cell lysis solution, thereby reducing manipulation steps and greatly simplifying the assay. The Escherichia coli lacZ gene and the gene encoding resistance to G418 were separated by an IRES sequence and placed downstream of the human EF1α promoter to provide for constitutive expression (Fig. 1A). To enable the creation of transgenic cells by retroviral transformation, the expression construct was inserted between the Moloney murine leukemia virus (MMLV) 5′ and 3′ repeats, and retroviral particles were produced by the transfection of epithelial cells with the MMLV vector. The retroviral particles were used to infect P388D1 and J774.1 macrophage-like cells and transformed cells selected with G418. Numerous attempts to create transgenic P388D1 cells by direct calcium phosphate- or liposome-mediated transfection or by the electroporation of cells failed to produce G418-resistant cells; transfection procedures and electroporation resulted in a high degree of cell death despite ensuring that the DNA was free of endotoxin (data not shown). Following retrovirus transformation, P388D1 and J774.1 clonal lines were established by a limiting dilution of G418-resistant cells.
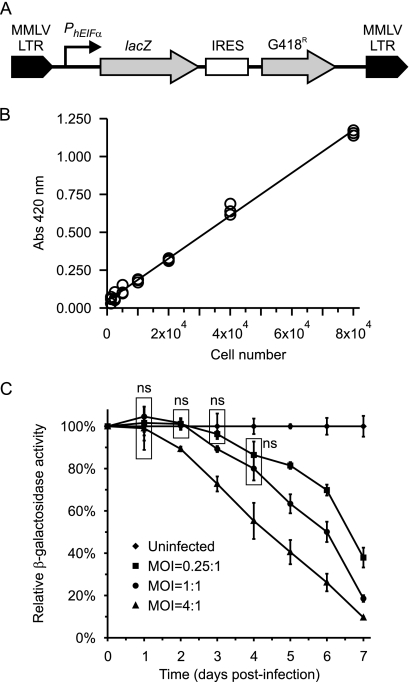
Fig. 1.
Quantitation of macrophages engineered to express lacZ through β-galactosidase activity. (A) Schematic representation of the MMLV retroviral vector used to generate retroviral particles to transform P388D1 and J774.1 macrophages. The human EIFα promoter provides for the constitutive expression of lacZ in transformed macrophages, which were selected by resistance to G418. IRES, internal ribosome entry site. (B) β-Galactosidase hydrolysis of ONPG is proportional to macrophage numbers. Increasing numbers of P388D1-lacZ macrophages were lysed, and β-galactosidase activity was measured as absorbance at 420 nm. Data points represent individual measurements from triplicate wells, and a line of best fit shows the linearity of the assay for the quantitation of macrophages. (C) The dose-dependent kinetics of macrophage killing by Histoplasma yeast. P388D1-lacZ macrophages were infected with wild-type G217B Histoplasma yeast at MOIs of 0.25:1 (squares), 1:1 (circles), or 4:1 (triangles) yeast to macrophages. At days 1 through 7 postinfection, remaining macrophages were assayed for residual β-galactosidase activity and normalized to that of uninfected wells (diamonds). Values are presented as averages ± standard deviations (SD) from three replicate infections. Data points were statistically compared to each other at each time point. Nonsignificant differences (ns) are indicated by boxes around the relative data points. All other points were statistically different (P < 0.05) from uninfected samples and samples infected at other MOIs.
We validated that β-galactosidase activity was directly proportional to the number of lacZ-expressing macrophages. Eight to 12 individual lines were examined for β-galactosidase activity, and a line with the greatest level of β-galactosidase activity was selected for further testing. A twofold dilution series of P388D1-lacZ cells was plated to establish the linearity and dynamic range of the assay. Following the adherence of cells, the culture medium was exchanged with the lysis solution containing the β-galactosidase substrate o-nitrophenyl-β-d-galactopyranoside (ONPG), and the hydrolysis of ONPG was monitored by absorbance at 420 nm over time. No β-galactosidase activity is detected from the parental P388D1 cell line (data not shown). The β-galactosidase activity of P388D1-lacZ cells is linear between 1 × 103 and 8 × 104 cells (R2 = 0.995) (Fig. 1B) and highly correlated with the number of macrophages present.
Using the P388D1-lacZ cells, we established that the kinetics of host cell destruction by Histoplasma show dose dependence. With immortalized cell lines, the degree of host cell killing results from a net balance between host cell replication and host cell lysis by the pathogen. To monitor this process, we infected P388D1-lacZ cells with wild-type G217B Histoplasma yeast and quantified the number of host cells remaining for 7 days using the β-galactosidase reporter for host cell number. With increasing time, fewer macrophages remain after infection, and as predicted, higher multiplicities of infection (MOI) result in a higher rate of destruction of the P388D1-lacZ cells (Fig. 1C). With P388D1 cells, low MOIs of 1:1 and 4:1 can be used with maximal host cell killing occurring in 6 to 7 days.
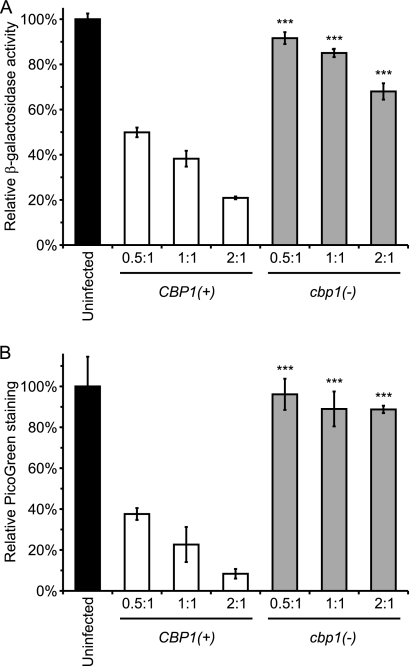
To determine whether the infection of the P388D1-lacZ cell line could be used to differentiate virulent and attenuated Histoplasma yeast, cells were infected with CBP1(+) and cbp1 mutant yeast. Yeast cells that do not produce the secreted Cbp1 virulence factor are attenuated in the ability to kill host macrophages in vitro (8, 33). Consistent with previous findings, infection of P388D1-lacZ cells and enumeration of the β-galactosidase activity remaining after 7 days show that the cbp1 mutant yeast cells are diminished in virulence (Fig. 2A). Similar results were seen with a second phylogenetic strain of Histoplasma yeast (G186A) and an isogenic ags1Δ attenuated mutant (data not shown). The results using β-galactosidase activity to indicate the number of remaining macrophages closely parallels that obtained using the nucleic acid-binding dye PicoGreen to enumerate macrophages by their DNA (Fig. 2B). These findings validate the use of P388D1-lacZ cells and their β-galactosidase activity to assess the virulence of Histoplasma yeast by virtue of their ability to kill host macrophages.
Fig. 2.
Macrophage enumeration by β-galactosidase activity parallels macrophage DNA quantification. P388D1-lacZ macrophages were infected with virulent (CBP1(+); WU15) or attenuated [cbp1(−); OSU8] Histoplasma yeast at MOIs of 0.5:1, 1:1, or 2:1 yeast per macrophage. At 6 days postinfection, the relative number of macrophages remaining was determined. (A) The remaining β-galactosidase activity was quantified by absorbance at 420 nm after the addition of ONPG in cell lysis buffer. (B) The remaining macrophage DNA was quantified by the removal of yeast following host cell lysis and staining of the host cell lysate with the fluorescent nucleic acid dye PicoGreen. Values were normalized to those from wells of uninfected cells and are presented as the averages ± SD from three wells. Data are representative of three independent experiments. Significant differences from CBP1(+) infections as determined by Student's t test are indicated by asterisks (***, P < 0.001).
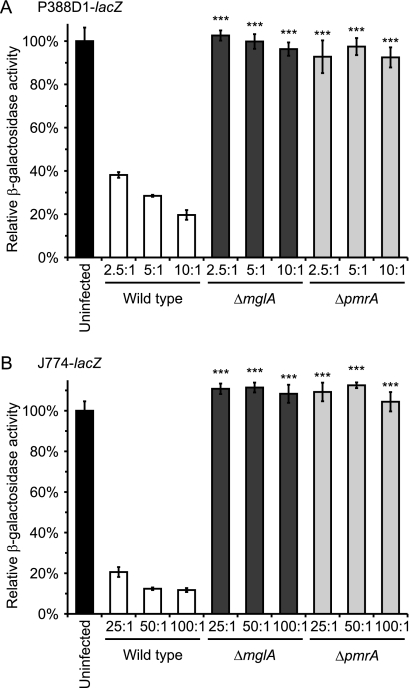
As destruction of host cells is not unique to the pathogenesis of Histoplasma, we established the utility of these β-galactosidase-expressing cell lines for assessing bacterial pathogen virulence. F. novicida infects macrophages and escapes the phagosome to replicate in the cytosol of the host cell. Like Histoplasma yeast, Francisella replication within macrophages is followed by host cell death. mglA and pmrA mutants of Francisella are defective in intramacrophage replication (1, 26). The infection of P388D1-lacZ cells with mglA and pmrA mutants does not significantly reduce the number of viable macrophages, whereas fully virulent Francisella efficiently destroys the macrophages (Fig. 3A). The infection of lacZ transgenic cells derived from the more commonly used J774 line with Francisella shows a similar readout of decreased virulence for the mglA and pmrA mutants (Fig. 3B). These results, which were determined by the assay of the macrophage-expressed β-galactosidase activity, are comparable to results obtained with lactate dehydrogenase release assays testing other Francisella virulence mutants (38). Taken together with the data using Histoplasma yeast, the results with Francisella show the general validity and applicability of the lacZ-expressing macrophage lines to monitor the microbe-dependent destruction of host cells as an assay for intracellular pathogen virulence.
Fig. 3.
LacZ-expressing macrophages accurately quantify the virulence of F. novicida. (A) P388D1-lacZ macrophages were infected with wild-type F. novicida U112, F. novicida ΔmglA, or F. novicida ΔpmrA at MOIs of 2.5:1, 5:1, or 10:1 bacteria per macrophage. After 2 days, remaining macrophages were quantified by assaying the macrophage-specific β-galactosidase activity and the activity normalized to that of uninfected wells. (B) J774-lacZ macrophages were infected with wild-type F. novicida U112, F. novicida ΔmglA, or F. novicida ΔpmrA at MOIs of 25:1, 50:1, or 100:1 bacteria per macrophage. After 2 days, remaining macrophages were quantified by assaying the macrophage-specific β-galactosidase activity, and the activity normalized to that of uninfected wells. Values are presented as the averages ± SD from three wells. Data are representative of two independent experiments. Significant differences from wild-type infections as determined by Student's t test are indicated by asterisks (***, P < 0.001).
Isolation of a Histoplasma mutant with reduced virulence.
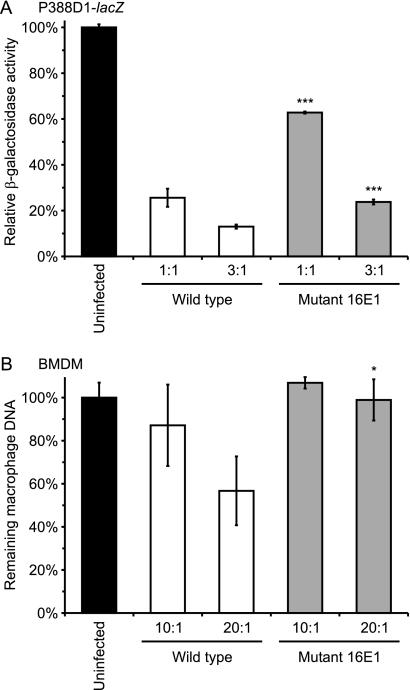
To discover new genes important for Histoplasma virulence, we utilized the efficiency of the P388D1-lacZ assay to screen insertion mutants for decreased ability to destroy host macrophages. Insertional mutagenesis was accomplished through A. tumefaciens-mediated transformation of Histoplasma yeast (22, 36). Individual transformants of strain WU15 were isolated from transformation plates and used to infect P388D1-lacZ monolayers at an MOI of approximately 1:1 (yeast to macrophage) based on optical density measurements. A total of 2,090 mutants were screened in 96-well plates, and the quantitation of the remaining β-galactosidase activity after 7 days revealed 30 candidate mutants less efficient in killing host cells (at least 33% of macrophages remaining compared to less than 10% remaining when infected with virulent Histoplasma yeast). Secondary tests of these candidate mutants in P388D1-lacZ cells using multiple MOIs and more rigorous inoculum determination (i.e., hemacytometer counts of yeast cells instead of optical density) confirmed that three are significantly attenuated (greater than 25% reduction in macrophage killing compared to that of the wild type). One of these mutants has an insertion at the CBP1 locus, which presumably accounts for its attenuated virulence and provides validation of the genetic screen for the discovery of genes important for pathogenesis. For mutant 16E1, the ability to kill P388D1 cells is reduced up to 50% compared to that of the parental strain (37% killing versus 74% killing by the wild type) (Fig. 4A). We confirmed that the mutant also was attenuated in primary cells by infecting murine bone marrow-derived macrophages. In these primary cells, the majority of macrophage killing by mutant 16E1 is lost (1% versus 43% killed by the wild type) (Fig. 4B), which is consistent with results using the P388D1-lacZ cell line.
Fig. 4.
Application of the P388D1-lacZ screen identifies a candidate virulence mutant. Macrophages infected with wild-type Histoplasma yeast or the 16E1 insertion mutant demonstrate that the 16E1 mutant is attenuated in virulence in macrophages. (A) P388D1-lacZ macrophages were infected with wild-type (WU15) or mutant yeast (OSU10) at MOIs of 1:1 or 3:1 yeast per macrophage. At 7 days postinfection, remaining macrophages were quantified by β-galactosidase activity, and results are plotted following normalization to results for uninfected wells. (B) Murine bone marrow-derived macrophages (BMDM) were infected with wild-type (OSU25) or mutant (OSU74) yeast at MOIs of 10:1 and 20:1 for 48 h. The number of intact macrophages was determined by measurement of the remaining macrophage DNA using PicoGreen fluorescence. Values are presented as averages ± SD from three wells and are representative of three independent experiments. Statistical differences from wild-type results (*, P < 0.05; ***, P < 0.001) were determined by Student's t test.
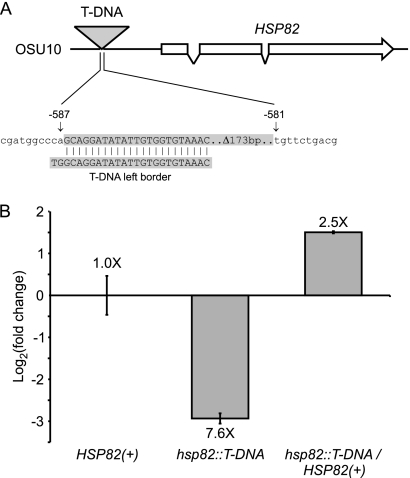
We designated the 16E1 mutant OSU10 (Table 1) and determined the sequence flanking the T-DNA insertion to ascertain the gene that was required for full yeast virulence. By comparison to the Histoplasma genome, the sequence of a TAIL-PCR fragment extending from the left border of the T-DNA element localizes the lesion to the region immediately upstream of the gene encoding the Histoplasma Hsp82 homolog (Fig. 5A). The complete delineation of the mutant locus showed that the right border of the T-DNA element was truncated by 173 bp, and the T-DNA inserts into the 5′ region of the gene 581 bp upstream of the HSP82 start codon. This localization suggests that the insertional mutation disrupts HSP82 promoter activity. We compared the expression of HSP82 in OSU10 to that in parental HSP82(+) yeast by quantitative RT-PCR. The insertion of the T-DNA element in OSU10 correlates with a 7-fold reduction in HSP82 transcript levels, which is consistent with the impairment of HSP82 promoter activity (Fig. 5B).
Fig. 5.
Mutant 16E1 contains a T-DNA insertion that decreases HSP82 expression. (A) Schematic depiction of the molecular lesion caused by T-DNA insertion at the HSP82 locus in mutant 16E1 (designated strain OSU10). The T-DNA element was localized by the TAIL-PCR amplification of the sequence extending from the T-DNA left border. The insertion element is located 581 bp upstream of the HSP82 CDS, which is encoded by three exons. (B) T-DNA insertion upstream of the HSP82 gene decreases HSP82 transcription. HSP82 transcript levels were determined by quantitative RT-PCR of RNA isolated from HSP82(+) (WU15), the hsp82::T-DNA mutant (OSU10), or yeast in which the hsp82 mutation was complemented with the wild-type locus (hsp82::T-DNA/HSP82(+); OSU86). Yeast cells were grown in liquid culture at 37°C and RNA isolated during exponential growth. HSP82 transcript levels were normalized to those for actin mRNA, and the relative fold change compared to results for the wild type was determined by the ΔΔCT method.
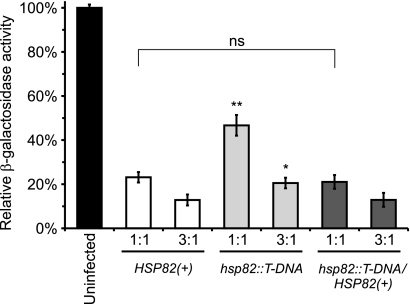
We constructed an HSP82-complemented strain to verify that the hsp82::T-DNA insertion is the source of the virulence attenuation. A linear plasmid containing the HSP82 locus was constructed that contained the native HSP82 promoter, HSP82 coding sequence, and putative terminator regions. The transformation of OSU10 yeast with the HSP82 plasmid, pCR545, restores expression of HSP82 to at least wild-type levels (Fig. 5B). HSP82 complementation also restores the ability of the hsp82 mutant to kill host macrophages in vitro (Fig. 6). These results establish the link between virulence attenuation in vitro and the mutation of the HSP82 locus.
Fig. 6.
Complementation of HSP82 restores yeast virulence in P388D1-lacZ macrophages. P388D1-lacZ macrophages were infected with HSP82(+) (OSU25), the hsp82::T-DNA insertion mutant (OSU74), or the HSP82-complemented strain (OSU86) at an MOI of 1:1 or 3:1 yeast per macrophage. At 6 days postinfection, remaining macrophages were quantified by β-galactosidase activity. Relative β-galactosidase activities are plotted after normalization to those for uninfected wells. Values represent the averages ± SD from three replicates. Significant (*, P < 0.05; **, P < 0.01) or nonsignificant (ns; P > 0.05) differences from infections with HSP82(+) yeast were determined by Student's t test. Results are representative of three independent experiments.
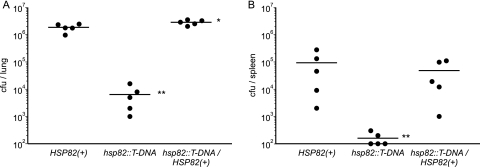
To determine the in vivo requirement for Hsp82 in Histoplasma virulence, we tested hsp82 mutant yeast in a murine model of histoplasmosis. Following intranasal inoculation of wild-type mice, the hsp82 mutant has a dramatically impaired ability to replicate within the lung (Fig. 7A), and dissemination to the spleen is nearly absent (Fig. 7B). The restoration of HSP82 expression rescues the mutant phenotype, restoring most of the pathogenicity as measured by lung and spleen fungal burdens. Thus, the Hsp82 factor significantly contributes to Histoplasma virulence in vivo.
Fig. 7.
HSP82 is required for full Histoplasma virulence in vivo. C57BL/6 mice were intranasally infected with HSP82(+) (strain OSU25), the hsp82 insertion mutant (hsp82::T-DNA; strain OSU74), or HSP82-complemented [hsp82::T-DNA/HSP82(+); strain OSU86] Histoplasma yeast. At 8 days postinfection, the fungal burdens in the lungs (A) and spleens (B) were determined. Data points represent CFU counts per organ for each mouse (n = 5). Data points on the x axis represent fungal burdens lower than the assay's limit of detection (100 CFU). Actual yeast cells delivered were determined by the enumeration of CFU counts in the inocula: 1.5 × 104 for HSP82(+), 1.2 × 104 for the hsp82 mutant, and 1.5 × 104 for the HSP82-complemented strain. Asterisks indicate statistically significant decreases (*, P < 0.05; **, P < 0.01) in CFU/organ compared to results of HSP82(+) infections as determined by the Wilcoxon rank-sum test.
Involvement of Hsp82 in Histoplasma stress tolerance.
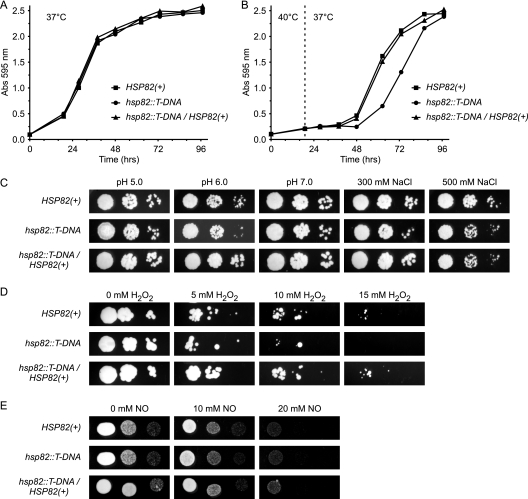
As Hsp82 is a member of the Hsp90 family of heat shock proteins that enable cells to withstand cellular stresses, we examined the tolerance of hsp82 mutant yeast to in vitro stresses. Hsp90 family members are expressed even in unstressed cells, but their function helps activate the heat shock response that leads to the induction of chaperone-like proteins to cope with thermal stress. Consistently with this, the liquid culture growth of wild-type and hsp82 mutant yeast is identical at the physiological temperature of 37°C (Fig. 8A). At febrile temperatures, Histoplasma yeast cells have reduced growth rates which are exacerbated for the hsp82::T-DNA mutant (data not shown). The transient incubation of yeast at 40°C shows that Hsp82 is required for efficient recovery from thermal stress (Fig. 8B). Normal levels of HSP82 expression facilitate the resumption of normal growth kinetics when heat stress is removed. However, yeast cells with decreased HSP82 show a prolonged delay before returning to normal growth rates. Compared to yeast with normal HSP82 levels, diminished HSP82 does not increase sensitivity to stresses such as salinity and pH (Fig. 8C). The role of Hsp82 in resistance to oxidative and nitrosative challenge, two stresses relevant to in vivo infection, also were examined. Normal HSP82 expression improves the survival of Histoplasma yeast treated with peroxide (Fig. 8D) but not with nitric oxide (Fig. 8E).
Fig. 8.
Hsp82 improves Histoplasma survival of heat and oxidative stresses. Growth of HSP82(+) (OSU25; squares), hsp82 mutant (OSU74; circles), and hsp82/HSP82-complemented (OSU86; triangles) Histoplasma yeast when challenged with the following in vitro stresses: growth at 37°C (A) or after transient incubation at 40°C (B), growth under salinity and pH stress (C), and survival of oxidative (D) or nitrosative (E) stress. Liquid HMM cultures were inoculated at identical densities from a suspension of yeast cells and incubated at 37°C (A) or held at 40°C for 19 h (the time of temperature shift is denoted by the dashed line) and then returned to 37°C (B). The growth of yeast was measured by the optical density at 595 nm. Data are representative of three independent experiments. (C) Dilutions of Histoplasma yeast from cultures of HSP82(+), hsp82::T-DNA, or the complemented hsp82 mutant were diluted and spotted onto solid medium (HMM) at different pH values (5.0, 6.0, and 7.0) or increasing salt concentration (pH 7.0 medium supplemented to 300 mM NaCl or 500 mM NaCl). (D and E) Suspensions of Histoplasma yeast were incubated with increasing amounts of H2O2 in PBS (D) or the NO donor DETA NONOate in HMM (E) for 4 h, after which aliquots were removed and dilutions spotted onto solid HMM plates. Concentrations of H2O2 and NO are indicated, with two molecules of NO liberated for each molecule of DETA NONOate.
DISCUSSION
A number of microbes infect and replicate within mammalian phagocytes, and the survival of these host cells following infection is one measure of the virulence of intracellular pathogens. With this in mind, we developed an efficient assay for measuring host cell survival based on the macrophage-specific expression of the lacZ gene. As demonstrated using attenuated strains of F. novicida and H. capsulatum, this assay is broadly applicable for the analysis of intracellular pathogens, although the subtraction of any endogenous β-galactosidase activity may be required for some bacterial species. The assay's strength is its simplicity; multiple steps are not required, and the assay is performed with the addition of a single solution containing a colorimetric substrate. This makes it easily adapted to high-throughput applications. The quantitative and sensitive nature of the enzymatic assay also allows for the detection of subtle decreases in pathogen virulence in vitro.
Our intent for developing the transgenic lacZ-expressing macrophage lines was to facilitate the more efficient screening of insertional mutants of Histoplasma to identify new factors required for intracellular pathogenesis. The application of the assay to a small-scale mutant collection identified three isolates with significant attenuation of virulence in macrophages. This frequency (0.14%) underscores the need for an efficient assay to screen large numbers of mutants. Some attenuated mutants may have been missed, because the assay with Histoplasma yeast is highly sensitive to the multiplicity of infection (Fig. 1C). For example, inoculation with an attenuated mutant at slightly higher MOIs than expected could resemble the virulence of wild-type yeast. Similarly, false virulence mutants would be incorrectly isolated if inoculated at lower-than-expected MOIs. The maintenance of the efficiency required for initial screening necessitated that we use optical density measurements as an estimate of yeast cell numbers despite the potential false positives the inaccuracy could produce. False mutants were pruned from the initial candidate collection through subsequent validation tests that used the precise enumeration of yeast by hemacytometer counts and multiple MOIs.
Admittedly, not all virulence-contributing genes will be isolated using this approach. Since the assay measures the survival of macrophages and not Histoplasma, mutations that attenuate yeast intracellular replication but not their ability to lyse host cells would be missed. While the infection of macrophages is a core facet of Histoplasma pathogenesis, it represents only a part of the overall virulence phenotype. Thus, the degree of attenuation in vitro does not necessarily equate to the magnitude of attenuation in vivo when Histoplasma is confronted with multiple components of the mammalian immune system. In addition, the macrophage assay will not identify mutations in genes required only for virulence in the animal host. Despite these limitations, the macrophage screen provides a practical means of discovering some of the factors important for Histoplasma virulence.
Screening for mutations attenuating Histoplasma virulence in macrophages isolated a hypomorphic allele of HSP82. Hsp82 is an essential factor for normal growth that also participates in the response to cellular stresses (3). The thermal dimorphism of Histoplasma makes the role of Hsp82 complex, since temperature change serves as both an inducer of stress and a signal for alternate-phase growth. The transcription of HSP82 is equivalent in wild-type Histoplasma mycelia and yeast maintained at 25 and 37°C, respectively (data not shown), which is consistent with Hsp82's function in general cellular homeostasis. Following the shift of mycelia from 25°C to temperatures above 34°C, Hsp82 production dramatically increases for 1 to 3 h but returns to baseline levels after 6 h and during continuous growth as yeast (25).
Data presented here focus on the role of Hsp82 during yeast-phase growth. T-DNA insertion in the HSP82 promoter region significantly diminishes but does not eliminate HSP82 expression. Despite no effects on growth at 37°C in vitro, decreased Hsp82 reduces Histoplasma virulence in macrophages and severely impairs its ability to infect murine lungs. This suggests that a low basal level of Hsp82 expression is sufficient to preserve cellular functions at mammalian body temperature but is unable to ameliorate other stresses encountered during infection. We hypothesize that febrile temperatures in the host is one stress that requires Hsp82 function, since the hsp82 mutant is less able to recover from the transient incubation of Histoplasma yeast at 40°C in vitro. This establishes a causal link between Hsp82 and Histoplasma's ability to manage temperature stress. However, hsp82 mutant yeast cells also have attenuated virulence in macrophages at 37°C despite identical yeast growth in broth culture at this same temperature. This suggests that the infection of host cells imposes additional, nonthermal stresses that require Hsp82 function. Consistently with this, challenge of Histoplasma with peroxide demonstrates that HSP82 expression can improve the ability of yeast to survive oxidative stress. Our in vitro data suggest that Histoplasma adaptation to decreasing pH of the phagosome does not require Hsp82 function.
The mechanisms by which stresses are perceived and the downstream targets of Hsp82 function in Histoplasma yeast currently are unknown. The alteration of the saturated/unsaturated fatty acid ratio of lipids affects the stress response to temperature change, linking the Histoplasma heat shock response to the physical state of the membrane (4, 20). The stress-induced exposure of normally unexposed hydrophobic regions of cellular proteins is thought to trigger the activation of the stress response (27). Heat shock proteins generally function as molecular chaperones, and Hsp82 can stabilize genetic variants of proteins, permitting the evolution of new phenotypes ranging from cancer to antifungal drug resistance (6, 39). Hsp82 binds to heat shock factor (Hsf), a client protein that orchestrates the transcription of stress response proteins and maintains Hsf in an inactive state (31). Other Hsp82 clients are linked to signal transduction, transcription factors, and chromatin remodeling, which can dramatically influence cellular phenotypes (37). In Candida albicans, Hsp82 has been linked to morphological changes via a Ras-PKA pathway (34). Further studies will be necessary to define the molecular players downstream of Hsp82 in Histoplasma's response to stress and whether temperature-induced dimorphic transition operates through an Hsp-dependent signaling pathway. Nonetheless, the isolation of an hsp82 mutant provides an important foothold in deciphering how Histoplasma manages stresses, particularly those associated with infection of the mammalian host.
ACKNOWLEDGMENTS
We thank Nrusingh Mohapatra and John Gunn for providing F. novicida strains and William E. Goldman for the T-DNA vector used in Agrobacterium-mediated insertional mutagenesis.
This research was supported, in part, by grant 0865450D from the American Heart Association and by research grant AI083335 from the National Institutes of Health.
Footnotes
Published ahead of print on 23 May 2011.
REFERENCES
- 1. Baron G. S., Nano F. E. 1998. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol. Microbiol. 29:247–259 [DOI] [PubMed] [Google Scholar]
- 2. Bohse M. L., Woods J. P. 2007. RNA interference-mediated silencing of the YPS3 gene of Histoplasma capsulatum reveals virulence defects. Infect. Immun. 75:2811–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borkovich K. A., Farrelly F. W., Finkelstein D. B., Taulien J., Lindquist S. 1989. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9:3919–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carratù L., et al. 1996. Membrane lipid perturbation modifies the set point of the temperature of heat shock response in yeast. Proc. Natl. Acad. Sci. U. S. A. 93:3870–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caruso M., Sacco M., Medoff G., Maresca B. 1987. Heat shock 70 gene is differentially expressed in Histoplasma capsulatum strains with different levels of thermotolerance and pathogenicity. Mol. Microbiol. 1:151–158 [DOI] [PubMed] [Google Scholar]
- 6. Cowen L. E., Lindquist S. 2005. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309:2185–2189 [DOI] [PubMed] [Google Scholar]
- 7. Deepe G. S., Jr., Gibbons R. S. 2002. Cellular and molecular regulation of vaccination with heat shock protein 60 from Histoplasma capsulatum. Infect. Immun. 70:3759–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edwards J. A., Alore E. A., Rappleye C. A. 2011. The yeast-phase virulence requirement for alpha-glucan synthase differs among Histoplasma capsulatum chemotypes. Eukaryot. Cell 10:87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eissenberg L. G., West J. L., Woods J. P., Goldman W. E. 1991. Infection of P388D1 macrophages and respiratory epithelial cells by Histoplasma capsulatum: selection of avirulent variants and their potential role in persistent histoplasmosis. Infect. Immun. 59:1639–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gomez F. J., Gomez A. M., Deepe G. S., Jr 1991. Protective efficacy of a 62-kilodalton antigen, HIS-62, from the cell wall and cell membrane of Histoplasma capsulatum yeast cells. Infect. Immun. 59:4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guimarães A. J., Frases S., Gomez F. J., Zancope-Oliveira R. M., Nosanchuk J. D. 2009. Monoclonal antibodies to heat shock protein 60 alter the pathogenesis of Histoplasma capsulatum. Infect. Immun. 77:1357–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Habich C., et al. 2006. Heat shock protein 60: identification of specific epitopes for binding to primary macrophages. FEBS Lett. 580:115–120 [DOI] [PubMed] [Google Scholar]
- 13. Hilty J., Smulian A. G., Newman S. L. 2008. The Histoplasma capsulatum vacuolar ATPase is required for iron homeostasis, intracellular replication in macrophages and virulence in a murine model of histoplasmosis. Mol. Microbiol. 70:127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hwang L. H., Mayfield J. A., Rine J., Sil A. 2008. Histoplasma requires SID1, a member of an iron-regulated siderophore gene cluster, for host colonization. PLoS Pathog. 4:e1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kauffman C. A. 2009. Histoplasmosis. Clin. Chest Med. 30:217–225 [DOI] [PubMed] [Google Scholar]
- 16. Kingston R. E., Chen C. A., Rose J. K. Calcium phosphate transfection. Curr. Protoc. Mol. Biol. 2003 doi: 10.1002/0471142727.mb0901s63. Chapter 9:Unit 9.1. [DOI] [PubMed] [Google Scholar]
- 17. Lauriano C. M., Barker J. R., Nano F. E., Arulanandam B. P., Klose K. E. 2003. Allelic exchange in Francisella tularensis using PCR products. FEMS Microbiol. Lett. 229:195–202 [DOI] [PubMed] [Google Scholar]
- 18. Liu Y. G., Chen Y. 2007. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43:649–654 [DOI] [PubMed] [Google Scholar]
- 19. Long K. H., Gomez F. J., Morris R. E., Newman S. L. 2003. Identification of heat shock protein 60 as the ligand on Histoplasma capsulatum that mediates binding to CD18 receptors on human macrophages. J. Immunol. 170:487–494 [DOI] [PubMed] [Google Scholar]
- 20. Maresca B., Kobayashi G. 1993. Changes in membrane fluidity modulate heat shock gene expression and produced attenuated strains in the dimorphic fungus Histoplasma capsulatum. Arch. Med. Res. 24:247–249 [PubMed] [Google Scholar]
- 21. Maresca B., Kobayashi G. S. 1989. Dimorphism in Histoplasma capsulatum: a model for the study of cell differentiation in pathogenic fungi. Microbiol. Rev. 53:186–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marion C. L., Rappleye C. A., Engle J. T., Goldman W. E. 2006. An alpha-(1,4)-amylase is essential for alpha-(1,3)-glucan production and virulence in Histoplasma capsulatum. Mol. Microbiol. 62:970–983 [DOI] [PubMed] [Google Scholar]
- 23. Medoff G., et al. 1986. Irreversible block of the mycelial-to-yeast phase transition of Histoplasma capsulatum. Science 231:476–479 [DOI] [PubMed] [Google Scholar]
- 24. Michielse C. B., Hooykaas P. J., van den Hondel C. A., Ram A. F. 2005. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 48:1–17 [DOI] [PubMed] [Google Scholar]
- 25. Minchiotti G., Gargano S., Maresca B. 1992. Molecular cloning and expression of hsp82 gene of the dimorphic pathogenic fungus Histoplasma capsulatum. Biochim. Biophys. Acta 1131:103–107 [DOI] [PubMed] [Google Scholar]
- 26. Mohapatra N. P., et al. 2007. Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect. Immun. 75:3305–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Neckers L., Mimnaugh E., Schulte T. W. 1999. The Hsp90 chaperone family, p. 9–42.In Latchman D. S. (ed.), Stress proteins, vol.136 Springer, New York, NY. [Google Scholar]
- 28. Nemecek J. C., Wuthrich M., Klein B. S. 2006. Global control of dimorphism and virulence in fungi. Science 312:583–588 [DOI] [PubMed] [Google Scholar]
- 29. Newman S. L., Gootee L. 1992. Colony-stimulating factors activate human macrophages to inhibit intracellular growth of Histoplasma capsulatum yeasts. Infect. Immun. 60:4593–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Porta A., et al. 2010. Changes in membrane fluid state and heat shock response cause attenuation of virulence. J. Bacteriol. 192:1999–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sakurai H., Enoki Y. 2010. Novel aspects of heat shock factors: DNA recognition, chromatin modulation and gene expression. FEBS J. 277:4140–4149 [DOI] [PubMed] [Google Scholar]
- 32. Schmittgen T. D., Livak K. J. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 33. Sebghati T. S., Engle J. T., Goldman W. E. 2000. Intracellular parasitism by Histoplasma capsulatum: fungal virulence and calcium dependence. Science 290:1368–1372 [DOI] [PubMed] [Google Scholar]
- 34. Shapiro R. S., et al. 2009. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr. Biol. 19:621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shearer G., Jr, Birge C. H., Yuckenberg P. D., Kobayashi G. S., Medoff G. 1987. Heat-shock proteins induced during the mycelial-to-yeast transitions of strains of Histoplasma capsulatum. J. Gen. Microbiol. 133:3375–3382 [DOI] [PubMed] [Google Scholar]
- 36. Sullivan T. D., Rooney P. J., Klein B. S. 2002. Agrobacterium tumefaciens integrates transfer DNA into single chromosomal sites of dimorphic fungi and yields homokaryotic progeny from multinucleate yeast. Eukaryot. Cell 1:895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Taipale M., Jarosz D. F., Lindquist S. 2010. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11:515–528 [DOI] [PubMed] [Google Scholar]
- 38. Tempel R., Lai X. H., Crosa L., Kozlowicz B., Heffron F. 2006. Attenuated Francisella novicida transposon mutants protect mice against wild-type challenge. Infect. Immun. 74:5095–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Whitesell L., Lindquist S. L. 2005. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5:761–772 [DOI] [PubMed] [Google Scholar]
- 40. Woods J. P. 2003. Knocking on the right door and making a comfortable home: Histoplasma capsulatum intracellular pathogenesis. Curr. Opin. Microbiol. 6:327–331 [DOI] [PubMed] [Google Scholar]
- 41. Woods J. P., Heinecke E. L., Goldman W. E. 1998. Electrotransformation and expression of bacterial genes encoding hygromycin phosphotransferase and beta-galactosidase in the pathogenic fungus Histoplasma capsulatum. Infect. Immun. 66:1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Worsham P. L., Goldman W. E. 1988. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J. Med. Vet. Mycol. 26:137–143 [PubMed] [Google Scholar]
- 43. Youseff B. H., Dougherty J. A., Rappleye C. A. 2009. Reverse genetics through random mutagenesis in Histoplasma capsulatum. BMC Microbiol. 9:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zemska O., Rappleye C. A. 2011. Agrobacterium-mediated insertional mutagenesis in Histoplasma capsulatum. In Brand A., MacCallum D. Host-Fungal interactions: manipulation of fungal gene expression. Humana Press, New York, NY. [Google Scholar]