Abstract
Borrelia burgdorferi, an agent of Lyme disease, establishes persistent infection in immunocompetent animals and humans. Although the infection in humans can be cleared by antibiotic therapy, persistence in reservoir animals is necessary for the maintenance of the bacterium in the natural reservoir host⇔tick vector infectious cycle. B. burgdorferi binds to β1- and β3-chain integrins, and the P66 outer membrane protein is responsible for at least some of the integrin binding activity of the spirochete. Because integrins are transmembrane, bidirectional signaling molecules, integrin binding may alter the nature of the host response to the bacteria. We used isogenic B. burgdorferi p66+ and Δp66 strains to analyze the responses of cultured human cells to P66-integrin interaction during infection. Microarray results suggest that the response differs according to the cell type, infection time, and experimental conditions. Clusters of genes in functionally related categories that showed significant changes included proteins involved in cell-extracellular matrix interactions, actin dynamics, stress response, and immune responses. Integrin binding by P66 may therefore help B. burgdorferi establish infection by facilitating tissue invasion and modulating the activation of the immune system to other components of the bacteria, e.g., lipoproteins. These results provide insight into how B. burgdorferi is able to establish infection in immunocompetent hosts.
INTRODUCTION
Lyme disease is the most prevalent arthropod-borne illness in the United States and is also common throughout Europe and parts of Asia. It is caused by the spirochetes Borrelia burgdorferi, Borrelia garinii, and Borrelia afzelii, which are maintained in a zoonotic cycle involving small mammals and birds, as well as specific Ixodes species ticks. The Lyme disease Borrelia bacteria cause a multisystemic illness as a result of dissemination from the tick bite in the skin to distal sites in the body, including the heart, knee joints, and nervous system. These bacteria are able to establish persistent infection in some tissues in immunocompetent animals. Since B. burgdorferi is an extracellular pathogen, it must employ various strategies to avoid clearance by the host immune system, to disseminate, and to colonize distant tissues.
B. burgdorferi is known to bind glycosaminoglycans, decorin, fibronectin, native collagen, laminin, complement factor H, and integrins in vitro. Bacterial ligands that bind to each of these mammalian receptors, with the exception of collagen, have been identified (3, 5, 6, 10, 20-23, 25, 27, 29, 32, 34, 36). The B. burgdorferi outer membrane protein P66 was identified as a β3-chain integrin ligand (10) and is used as a diagnostic antigen in human Lyme disease serologic testing (8, 16). Although P66 does not have an RGD sequence, which is a common integrin-binding motif, recombinant P66 binds to both β3-chain integrins and can compete with whole B. burgdorferi for binding to purified β3-chain integrins and cultured cells (10). Replacement of the integrin-binding domain of P66 by a kanamycin resistance marker significantly decreases the bacterium's ability to bind both purified integrins and epithelial cells transfected to overexpress the integrin αvβ3 (11). P66 also binds integrin α3β1, and it may bind additional integrins.
Integrins are divalent, cation-dependent, heterodimeric transmembrane proteins involved in cell-to-cell contact and communication, inflammatory responses, changes in actin dynamics, and many other processes. Integrin-mediated signaling is complex, as the signaling intermediates and the end result depend on the particular integrin heterodimer present, the cell type in which it is expressed, and the ligand to which it binds. Since integrin signaling is also bidirectional (outside the cell to inside the cell and vice versa) (26), cells may respond in a variety of ways, including modulation of integrin signaling and availability in the active conformation, to integrin ligation. Expression of integrins varies between cell types and in response to other environmental stimuli; αIIbβ3 is present only on platelets megakaryocytes, and mast cells; αvβ3 is more widely distributed; and one or more β1 integrins are produced by all cell types except erythrocytes in mammals.
It has been hypothesized that to establish a persistent infection, B. burgdorferi evades the host immune system through generation of antigenic variants of the Vls surface protein (40-42) and through binding of the complement cascade inhibitor factor H (6, 23, 27, 29). It is also apparent that B. burgdorferi elicits a robust humoral and cellular immune response, in part mediated by Toll-like receptors (TLRs), particularly TLR2, in response to bacterial lipoproteins (2, 24, 37, 38). However, it is important to note that the lipoprotein acyl moiety recognized by TLR2 is not exposed on the surfaces of healthy, intact bacteria. Manipulation of host cell biology and the host response through binding of a bacterial surface protein to an integrin is therefore an important mechanism by which the bacteria may manipulate their environment in the host while still viable, intact, and able to replicate and disseminate.
Microarray analysis has made it possible to survey the whole transcriptome for an overview of changes in response to time, infection stresses, or a combination of factors. Here, we examined the mammalian host responses to infection with B. burgdorferi, focusing specifically on the role of P66 binding to integrins.
MATERIALS AND METHODS
Mammalian cell culture.
The human kidney epithelial cell line HEK 293 (abbreviated 293 here) and a derivative transfected to express the genes for the integrin αv and β3 subunits (hereafter referred to as 293+αvβ3 cells) were grown in monolayers at 37°C under 5% CO2 in Dulbecco modified Eagle medium (DMEM) and Ham's F-12 medium supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. The medium for the 293+αvβ3 cells was also supplemented with 400 ng/ml G418 (Sigma). The human macrovascular endothelial cell line EA.hy926 (17, 18) was grown in monolayers at 37°C under 5% CO2 in DMEM with 4.5 g/liter glucose, supplemented with 10% FBS and hypoxanthine-aminopterin-thymidine (HAT) (Sigma) to final concentrations of 100 μM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymidine, and with 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Each of these cell lines has previously been shown to bind Borrelia burgdorferi (13, 28), with different host cell substrates contributing to B. burgdorferi attachment. In particular, the pair of transfected 293+αvβ3 cells and parental HEK 293 cells were chosen in the hope of revealing β3-integrin-specific responses. The human microvascular endothelial cell line HMEC-1 (1) was used as a second endothelial line and was cultured in endothelial basal medium (Clonetics, San Diego, CA) supplemented with 15% heat-inactivated fetal bovine serum (HyClone, Logan, UT), 1 μg/ml hydrocortisone (Sigma-Aldrich, St. Louis, MO), and 10 ng/ml epidermal growth factor (Sigma-Aldrich).
For infections, two protocols were used. In one protocol, designed to allow longer-term infections of cells in association with the matrix, the cells were infected while adherent to the culture flask after washing the layers with phosphate-buffered saline (PBS) to remove the antibiotics in the culture medium. In the second approach, designed to maximize the availability of integrins, the cell monolayers were washed with PBS and lifted with PBS containing 5 mM EDTA, pelleted at 400 × g for 10 min, washed in medium without antibiotics, and resuspended in a total of 5 ml of medium without antibiotics per cell line. The cells were counted with a hemocytometer, and 6 × 106 to 1 × 107 cells were dispensed into 1.5-ml microcentrifuge tubes.
Culture of endothelial cells on Matrigel.
For some experiments, Ea.hy926 cells were cultured on Matrigel, which allows assessment of certain endothelial cell activities thought to model certain responses to vascular endothelial growth factor (VEGF) by endothelial cells in vivo. The cells were plated, and 2 days later, they were treated with purified VEGF (obtained from the NCI Repository) or infected with B. burgdorferi at a multiplicity of infection (MOI) of 10:1. Growth of the endothelial cells was monitored, and structure formation was quantified visually for 7 to 10 days, depending on the lot of Matrigel. Data were analyzed for statistical significance using one-way repeated-measure analysis of variance (ANOVA) and Dunnett's posttest as well as Tukey's multiple-comparison test.
Bacterial culture and infections of mammalian cells.
The high-passage-number, noninfectious Borrelia burgdorferi strain HB19/KO1 (p66+), which contains an intact p66 (bb0603) locus marked with an adjacent kanamycin resistance cassette, and HB19/KO4 (Δp66), in which the portion of p66 encoding the integrin-binding domain is replaced by the kanamycin resistance cassette, were grown in MKP medium supplemented with human serum and 200 μg/ml kanamycin as previously described (11). Mid-log-phase cultures supplemented with glycerol to 20% final volume were stored at −80°C until use (12, 13). For individual experiments, frozen aliquots of bacteria were thawed, washed twice with PBS containing 0.2% (wt/vol) bovine serum albumin (BSA), and then resuspended in 200 μl of cell culture medium without antibiotics. The bacteria were enumerated and assessed for viability (which for these experiments was assessed by morphology and motility) using a Petroff-Hausser counting chamber and dark-field microscopy; for all experiments, >90% of the bacteria were intact and motile. The bacteria were added to either tubes containing lifted cells or to adherent cell layers to achieve a multiplicity of infection of 10 bacteria per mammalian cell. Lifted cells were incubated with the bacteria by gently rocking at ambient temperature (for consistency with our previously published conditions) for 1 or 3 h. The mammalian cells were subsequently harvested by centrifugation at 400 × g for 10 min to remove unbound bacteria and washed with PBS, and total RNA was extracted. The adherent cells were incubated with the bacteria at 37°C under 5% CO2 (to allow survival under prolonged infection conditions) for 3, 6, or 24 h. After incubation, the monolayers were washed once with PBS, lifted with trypsin-EDTA (TE), centrifuged for 10 min at 2,000 rpm, and washed with PBS. In both protocols, the cells were then resuspended in lysis buffer for RNA extraction using the RNeasy kit (Qiagen, Valencia, CA).
RNA purification and cDNA synthesis.
RNA was extracted from uninfected and infected human cells using the Qiagen RNeasy kit with on-column DNase digestion using the Qiagen RNase-free DNase kit. The quality of RNA was checked using an Agilent bioanalyzer (Agilent, Santa Clara, CA). Labeled cDNA was prepared from 10 μg of RNA per HEEBO chip or 15 μg of RNA per Qiagen chip using an oligo(dT) primer with Superscript II reverse transcriptase in the presence of aminoallyl-dUTP. Each cDNA sample was then coupled to either a Cy3 or Cy5 dye in the presence of 0.1 M NaHCO3, and the reaction was ended by adding 4 M hydroxylamine. Labeled cDNAs were purified over a Cyscribe GFX column (Amersham Biosciences) separately and concentrated, and the two solutions were then mixed and hybridized to the microarray slide. Earlier hybridizations were performed using in-house-generated arrays made from Operon's human v2.1.2 plus v2.1.4 cDNA library, representing 27,175 human open reading frames (ORFs) (Operon Technologies, Inc., Alameda, CA). Later hybridizations employed HEEBO chips (Stanford Functional Genomics Facility, Stanford CA), consisting of 44,544 70-mer probes representing 30,718 known genes. A trial of both chips in parallel with a single set of samples yielded similar results, with the exception of genes that were not represented on the lower-complexity chips.
Microarray hybridization and analysis of data.
Hybridizations were generally performed in three biological replicate samples with two of the replicate slides in one dye configuration and the third as a dye swap. Hybridization was performed overnight using the Pronto microarray hybridization kit (Corning, Corning, NY). Images were obtained using a Packard Scanarray 4000 two-color microarray scanner (GMI, Ramsey, MN), and image analysis was done with ImaGene software (BioDiscovery, Inc., El Segundo, CA) (30). Statistical significance and fold changes were generated by using both GeneSifter (VizX Labs [Seattle, WA], which has since been acquired by Geospiza [Seattle, WA]) and GeneSpring (Agilent, Santa Clara, CA).
Statistical analysis.
The different chips used in these experiments required slightly different approaches to the analyses. For the chips from Operon Technologies, the data were first filtered based on flag values from the ImaGene analysis to eliminate bad spots. Raw intensity values were used in GeneSifter for data analysis. The median background of each spot was subtracted from the median signal and uploaded into the program. The spot intensity values were normalized to the median intensity of all spots over the entire chip, and projects were set up in GeneSifter placing all conditions side by side. Comparisons were made between each infection condition and the uninfected control within a time point, and each infection condition was also compared to the same condition at different time points.
The minimum fold change considered for further study in any comparison was 1.5-fold up or down. The genes that had a passing flag quality value as defined by ImaGene for a majority of the biological replicates were the only ones considered. A two-way ANOVA test was then performed to judge statistical significance. P values were calculated in relation to infection condition and time point. A P value of less than 0.05 for either infection or time is considered significant for the data obtained from the Operon Technologies chips, which were probed with cDNAs derived from the lifted Ea.hy926, 293, and 293+αvβ3 cells.
For the HEEBO chips, which were probed with cDNAs derived from adherent cells and lifted HMEC cells, background values were subtracted from each spot, and the median intensity expression ratios were uploaded into GeneSifter and normalized using LOWESS. A two-way ANOVA was used to filter the data using a P value of <0.01 and by a fold change of 1.5 or greater compared to uninfected cells.
For both sets of chips, genes that were significantly different between the infection conditions were sorted into functional groups using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (http://www.genome.jp/kegg) and gene ontologies, WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt) (39), and the Cluster and Treeview programs (http://rana.lbl.gov/eisen/?page_id=42). Gene ontologies and KEGG pathway information were applied for all genes by the GeneSifter program and used to group genes by function. Z-scores were calculated by GeneSifter to gauge over- or underrepresentation of certain functional groups. A z-score of over 2 or less than −2 was considered significant.
qRT-PCR.
Select genes representing the different functional groups identified by microarray analysis were chosen for validation by quantitative reverse transcriptase PCR (qRT-PCR). RNA preparations were those used for microarray analyses, and no PCR products were detected in control reactions from which reverse transcriptase was omitted. The primer sequences are shown and described in Table 1. qRT-PCR was performed in a Stratagene Mx3000P system (La Jolla, CA) and using Qiagen Quantitect SYBR green PCR kit (Qiagen, Valencia, CA). Each 20-μl reaction mixture contained 10 μl of 2× SYBR green master mix, 5 pmol of each primer, and either 150 ng of template cDNA (for 18S rRNA and actin) or 75 μg (for all other genes). These differences were due to the abundance of 18S rRNA and actin mRNA in the samples and the higher efficiency of these primer sets.
Table 1.
Primers used for quantitative reverse transcriptase PCR amplification
| mRNA target | 5′ primer sequence | 3′ primer sequence | Product length (bp) |
|---|---|---|---|
| β-Actin | CAGGCACCAGGGCGTGATG | GTACATGGCTGGGGTGTTGA | 286 |
| MAPK8 (JNK) | CAGTCAGGCAAGGGATTTGT | TGATGATGGATGCTGAGAGC | 270 |
| ROCK1 | GGTTAGGGCGAAATGGTGTA | AGTTGATTGCCAACGAAAGC | 207 |
| COL5A1 | GGCTGTGCTACCAAGAAAGG | GAGGTCACGAGGTTGCTCTC | 197 |
| 18S rRNA (normalization control) | CGGCTACCACATCCAAGGAA | GCTGGAATTACCGCGGCT | 186 |
To calculate fold changes, the ΔΔCt method was used. The cycle threshold (Ct) of the gene of interest was subtracted from the Ct of the 18S rRNA gene for each condition. The difference of the normalized Ct values of the two conditions was used to calculate fold change as follows: for infection condition 1, Ct1 of the gene of interest − Ct1 of the 18S rRNA gene=ΔCt1 and for infection condition 2, Ct2 of the gene of interest − Ct2 of the 18S rRNA gene=ΔCt2, where Ct1 is the cycle threshold for infection condition 1 and Ct2 is the cycle threshold for infection condition 2. The fold change was calculated as follows: fold change=2ΔCt1 − ΔCt2.
Fluorescence microscopy.
Mammalian cells were plated in tissue culture-treated chamber slides and incubated for 1 or 2 days at 37°C with 5% CO2. The cell layers were then washed and infected essentially as described above. After incubation for various periods of time, the layers were washed and then fixed with 3% paraformaldehyde in PBS. The samples were then permeabilized with Triton X-100. After washing and blocking in HEPES-buffered saline supplemented with 1% BSA, the slides were probed with anti-Lyme spirochete antiserum (gift from Allen Steere), washed, and then probed with phalloidin plus secondary anti-rabbit IgG, each conjugated to a different fluorescent dye. All images in each figure were processed the same way.
Assessment of GTPase activity.
Rho, Rac1, and CDC42 activation states in adherent cells, infected as described above for the microarray analyses, were determined using a pulldown plus immunoblot kit from Cell Biolabs (San Diego, CA), according to the manufacturer's instructions.
Assessment of iNOS production.
The levels of inducible nitric oxide synthase (iNOS) at the protein level were measured using a enzyme-linked immunosorbent assay (ELISA) kit from R&D Systems (Minneapolis, MN), according to the manufacturer's instructions. The means, standard deviations, and statistical significance were calculated using the unpaired t test in the GraphPad Prism software package.
Purified recombinant P66 proteins.
Recombinant P66 integrin-binding domain fused to maltose-binding protein (MBP-P66M) was prepared as described previously (15). MBP-βgal (MBP fused to β-galactosidase), the fusion protein encoded by genes on the pMalC2 vector, was purified in parallel and used as a control. Site-directed amino acid changes were made in the P66 portion of the MBP-P66 fusion protein using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The changes are designated by the single-letter amino acid code corresponding to the full-length protein encoded by the genes on the prototype sequenced B. burgdorferi strain B31M1 (7, 19). All constructs were sequenced to ensure that no additional mutations had been introduced. The purified proteins were compared to wild-type P66 and MBP alone for binding to purified integrin as previously described (10, 15).
Microarray data accession number.
All microarray data have been deposited at GEO under accession number GSE27996.
RESULTS
To assess the effect of the Borrelia burgdorferi integrin ligand P66 on human cell gene expression, we took two distinct approaches. In the first, we lifted adherent cells with EDTA, then restored the divalent cations required for integrin function, and incubated the cells with the bacteria in suspension. This approach was used to maximize the number of integrin heterodimers per cell available for interaction with B. burgdorferi. With prolonged time in suspension, however, adherent cells may undergo anoikis (apoptosis in response to loss of adhesion to substrate), so we also performed infections of cells that were adherent in the culture flasks, although this limits the availability of integrins. Three cell lines were used in both conditions: the human macrovascular endothelial cell line EA.hy926, the kidney epithelial cell line HEK 293 (293), which does not express the integrin β3 subunit, and HEK 293 cells transfected to overexpress integrin αvβ3 (293+αvβ3 cells). All experiments used a multiplicity of infection (MOI) of 10 bacteria per human cell, with no centrifugation step to facilitate bacterial and human cell interactions. In addition, all experiments included infections with bacteria that express P66 (p66+) and that do not express P66 (Δp66), as well as uninfected controls. Results for endothelial cells versus epithelial cells will be presented in separate sections. In all cases, analyses were focused on functional pathways, as opposed to individual genes, that showed statistically significant differences in expression between infection conditions.
P66 affects expression of genes in several functional pathways in EA.hy926 endothelial cells.
On-chip comparisons were done between cells that were not infected with B. burgdorferi, cells that were infected with p66+ bacteria, and cells infected with Δp66 bacteria. Using the designations assigned from the z-scores generated by GeneSifter and processed through WebGestalt, several pathways were found to be differentially regulated in the infections of both lifted and adherent cells with the p66+ bacteria versus the Δp66 bacteria (Table 2). Several pathways were also differentially expressed in either the adherent cells or the lifted cells, but we have focused on those common to both infection conditions, as they are most likely to be informative specifically regarding the cellular response to integrin binding by P66. In support of this choice, one pathway, apoptosis, showed significant differences between infection with p66+ bacteria versus Δp66 bacteria only in the lifted cells, as would be expected if the cells are “rescued” by P66-integrin interaction. While supportive of our other data demonstrating that P66 binds integrins (5, 10-13), this was not the focus of interest in this work, so more-detailed analyses of pathways of potential interest in terms of pathogenesis were analyzed.
Table 2.
Clusters of genes with changes in expression common to lifted and adherent Ea.hy926 cells infected with p66+ versus Δp66 B. burgdorferi
| KEGG pathwaya | Lifted cellsb |
Adherent cellsb |
||
|---|---|---|---|---|
| No. of genes | P value | No. of genes | P value | |
| Focal adhesionc | 46 | 1.42E−09 | 10 | 4.89E−03 |
| Regulation of actin cytoskeleton | 39 | 5.77E−06 | 9 | 1.85E−02 |
| MAPK signaling pathwayc | 47 | 1.68E−05 | 15 | 3.1E−04 |
| Colorectal cancerc | 20 | 3.60E−05 | 7 | 1.01E−03 |
| Gliomac | 16 | 9.27E−05 | 4 | 3.38E−02 |
| Fc epsilon receptor I signaling pathway | 18 | 1.27E−04 | 7 | 6.85E−04 |
| VEGF signaling pathwayc | 16 | 5.27E−04 | 5 | 1.16E−02 |
| Natural killer cell-mediated cytotoxicityc | 23 | 6.28E−04 | 6 | 3.27E−02 |
| Jak-STAT signaling pathwayc | 26 | 7.65E−04 | 9 | 2.96E−03 |
| Chronic myeloid leukemiac | 16 | 8.66E−04 | 6 | 3.38E−03 |
| GnRH signaling pathwayc | 19 | 1.03E−03 | 5 | 4.12E−02 |
| Dorsoventral axis formation | 8 | 1.97E−03 | 3 | 1.41E−02 |
| Phosphatidylinositol signaling systemc | 14 | 4.33E−03 | 6 | 3.38E−03 |
| Pyrimidine metabolism | 16 | 8.16E−03 | 5 | 3.26E−02 |
| Sphingolipid metabolism | 8 | 1.64E−02 | 3 | 3.59E−02 |
| Alanine and aspartate metabolism | 7 | 2.22E−02 | 3 | 2.27E−02 |
| Inositol phosphate metabolismc | 9 | 2.51E−02 | 4 | 1.56E−02 |
| Cell cyclec | 17 | 2.95E−02 | 6 | 2.26E−02 |
| T cell receptor signaling pathwayc | 14 | 4.03E−02 | 5 | 3.53E−02 |
A complete list of KEGG pathways can be found at http://www.genome.jp/kegg/pathway.html.
P values considered to be statistically significant for lifted and adherent cells were <0.05 and <0.01, respectively, due to differences in the arrays used.
These pathways were also significantly different (P < 0.01) in lifted human microvascular endothelial cells (HMECs) infected with p66+ B. burgdorferi versus Δp66 B. burgdorferi.
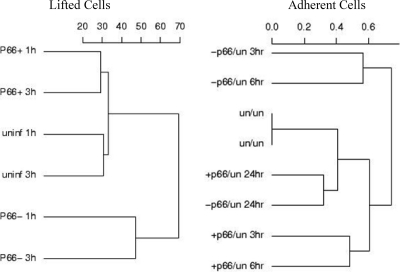
The differences in gene expression varied with infection time, but in many cases, they were consistent between the shorter infection time periods of 1 to 3 h. At the 24-h time point (at which time only adherent cells were analyzed), the differences between all the samples were minimal, indicating that the initial responses to the p66+ bacteria versus Δp66 bacteria are lost over time, perhaps due to downregulation of the cellular responses to the bacteria. An overview of gene expression changes is diagrammed in Fig. 1, which summarizes the differences between infection conditions (infected with p66+ bacteria versus Δp66 bacteria versus not infected) and time points. In the lifted cells, the notable result is that the differences between the cells infected with p66+ bacteria and the uninfected controls are less than those between the cells infected with Δp66 bacteria and the uninfected cells. In the adherent cells, the cells infected with Δp66 bacteria are again the most different from the uninfected controls at the earlier time points, but the differences in gene expression between the cells in any infection condition are minimal after 24 h of infection.
Fig. 1.
Analysis of gene expression in Ea.hy926 cells in response to B. burgdorferi cells that express P66 and that do not express P66. The overall expression levels of genes represented on the microarrays were compared for the three conditions (uninfected [uninf and un], infected with p66+ B. burgdorferi [P66+ and +p66], and infected with Δp66 B. burgdorferi [P66− and −p66]). Cluster analysis was performed using the Cluster and Treeview programs and based on the subset of genes that were statistically significantly different based on the different infection conditions. Scales were arbitrarily set by the program.
The pathways that were common to infection of both adherent and lifted cells are listed in Table 2. Two of these pathways, the focal adhesion pathway and regulation of actin cytoskeleton pathway, contain many common genes, and so are considered together. The relevance of some pathways (e.g., colorectal cancer) to B. burgdorferi infection is not clear, but these pathways include a number of signaling intermediates common to the other pathways. Additional KEGG pathways with no obvious connection to infection also were illuminated, but similarly, they contain genes (e.g., those encoding signaling proteins) that are common to many pathways. Pathways that were also significantly different between the p66+ versus Δp66 bacterial infection conditions in a different endothelial cell line, HMEC, are indicated but not discussed here.
Focal adhesion and actin cytoskeleton pathways are affected by P66.
The regulation of actin cytoskeleton and focal adhesion pathways were two KEGG pathways that were significantly enriched in both adherent and lifted cells for genes differentially expressed in response to p66+ or Δp66 B. burgdorferi. While both pathways were significantly affected by the presence versus absence of P66, the specific genes that were significantly affected in the adherent versus lifted cells were generally different, demonstrating the importance of the context of the cells during infection. While an apparent total of 85 genes were differentially regulated in the focal adhesion and regulation of actin cytoskeleton pathways in the lifted cells, the actual number of genes affected is smaller, due to the significant overlap of the two pathways. Focal adhesion kinase (FAK) (or protein tyrosine kinase 2 [PTK2]), ROCK1, Rac1, myosin, and β-actin (ACTB) were all upregulated at least 2-fold in the cells infected with Δp66 bacteria at 3 h postinfection compared to uninfected cells (Table 3 ). Compared to uninfected cells, the cells infected with p66+ bacteria had nonsignificant changes of expression in these genes at the same time point. The ratios of gene expression changes in the cells infected with p66+ bacteria compared to cells infected with Δp66 bacteria for multiple genes whose products are involved in the regulation of actin and in focal adhesion were, however, statistically significantly different. For select genes, these results were validated by quantitative reverse transcriptase PCR (qRT-PCR) (Table 3). This would suggest that rearrangement of the cytoskeleton should be occurring in cells infected with B. burgdorferi and is affected by the interaction of P66 with its receptor, again consistent with integrin ligation.
Table 3.
Genes belonging to selected significantly differentially expressed KEGG pathways in Ea.hy926 cells infected with p66+ versus Δp66 B. burgdorferi bacteriaa
| Pathway | Gene name(s)b | Protein encoded | Fold change comparing the followingc: |
Predicted effect(s) or note(s)d | ||
|---|---|---|---|---|---|---|
| p66+ and uninfected | Δp66 and uninfected | p66+ and Δp66 | ||||
| Regulation of actin cytoskeleton/focal adhesion | ARHGEF1A | Rho guanine nucleotide exchange factor (GEF) 1 | −1.6 | NS | −1.5 | Decreased actin polymerization +P66 |
| GSNA | Gelsolin | −1.8 | NS | −2.0 | Decreased actin polymerization +P66 | |
| ACTN4A | Actinin, alpha 4 | NS | −1.6 | 1.5 | Decreased actin polymerization −P66 | |
| PI3K-R2, PI3K-R3, and PI3K-CGA,L | Phosphoinositide-3-kinases | NS | 2.2 | −2.0 | Increased inflammation −P66, decreased actin polymerization, increased vessel permeability +P66 | |
| TAOK2A | TAO kinase 2, PSK2 | −1.7 | NS | −1.9 | Decreased MAPK p38 signaling, decreased inflammatory cytokine induction +P66 | |
| RAC1L | Rho family, small GTP-binding protein Rac1 | NS | 2.76 | −2.01 | Increased actin polymerization, −P66, decreased JNK activation, decreased cortical F actin, increased vessel permeability +P66 | |
| ROCK1L* | Rho-associated, coiled-coil containing protein kinase 1 | −1.63 | NS | −2.20 | Increased actin polymerization −P66 | |
| ACTBL* | β-Actin | NS | 2.63 | −2.41 | Increased actin polymerization −P66 | |
| PPP1R12AL | Protein phosphatase 1, regulatory (inhibitor) subunit 12A | −1.52 | 2.03 | −1.88 | Increased actin polymerization −P66 | |
| MYL6L | Myosin, light chain 6, alkali, smooth muscle and nonmuscle | NS | 2.06 | −1.87 | Increased actin polymerization −P66 | |
| PTK2L | Protein tyrosine kinase 2, FAK | NS | 1.97 | −2.4 | Increased actin polymerization −P66 | |
| PRKCB1L | Protein kinase C, beta 1 | NS | NS | −1.82 | Decreased MAPK signaling, focal adhesion, immune response +P66 | |
| VAV2L | Rho family GEF | NS | 2.26 | −1.77 | Increased Rho activation −P66 | |
| Jak-STAT, MAPK, VEGF, and other signaling pathways | STAT1L, STAT4L, STAT6A | Signal transducers and activators of transcription | −1.7 (1 and 6), 5.81 (4) | NS (1 and 6), −2.18 (4) | −2.0 (1 and 6), 12.66 (4) | Decreased immune response +P66 for STAT1 and STAT6, opposite for STAT4 |
| IL11RAA | Interleukin 11 receptor, alpha | NS | 1.9 | −2.2 | Increased immune response −P66 | |
| PI3K-R2, PI3K-R3, and PI3K-CGA,L | Phosphoinositide-3-kinases | NS | 2.2 | −2.0 | Increased inflammation −P66, decreased actin polymerization +P66, increased vessel permeability +P66 | |
| TAOK2A | TAO kinase 2, PSK2 | −1.7 | NS | −1.9 | Decreased MAPK p38 signaling, decreased inflammatory cytokine induction +P66 | |
| PLA2-G2A, PLA2-G2E, and PLA2-G5A,L | Phospholipase A2, groups II and V | 1.7 | −1.6 | 2.7 | Increased inflammation +P66, decreased inflammation −P66 | |
| RAC1L | Rho family, small GTP-binding protein Rac1 | NS | 2.76 | −2.01 | Increased actin polymerization, −P66, decreased JNK activation, decreased cortical F actin, increased vessel permeability +P66 | |
| MAPK8, JNKL* | Mitogen-activated protein kinase 8; Jun N-terminal kinase | −1.56 | NS | −2.05 | Decreased JNK signaling, inflammation +P66 | |
| MAPK14, p38L | Mitogen-activated protein kinase 10 | NS | 1.70 | −2.3 | Increased inflammation −P66 | |
| RapGEF2L | Rap guanine nucleotide exchange factor (GEF) 2 | 1.55 | 2.69 | −1.74 | Decreased MAPK signaling +P66 | |
| PRKCB1L | Protein kinase C, beta 1 | NS | NS | −1.82 | Decreased MAPK signaling, focal adhesion, immune response +P66 | |
| CACNG1, CACNG4, and CACNG5L | Calcium channel γ subunits | NS | 1.80 | −1.84 | Similar changes for all three; increased Ca2+ signaling −P66 | |
| CACNB3L | Calcium channel β subunit | NS | 3.16 | −3.72 | Increased Ca2+ signaling −P66 | |
| HSP72L | Heat shock protein 72 | NS | 1.99 | −1.92 | Increased proinflammatory cytokine production −P66, increased protection against stress −P66 | |
| Defense response, response to wounding, stress | NOS2AA | Nitric oxide synthase 2A (inducible) | −1.9 | 2.6 | −5.0 | Decreased immune response +P66, increased immune response −P66 |
| response | TNIP1A | TNFAIP3-interacting protein 1 | 1.8 | NS | 1.8 | Decreased immune response +P66 |
| F2RL3A | Coagulation factor II receptor-like 3 | −1.5 | 1.6 | −2.5 | Decreased immune response +P66, increased immune response −P66 | |
| CEACAM1 A | Carcinoembryonic antigen-related cell adhesion molecule 1 | NS | −1.8 | 1.9 | Decreased cell adhesion, integrin signaling −P66 | |
| PLA2-G2A, PLA2-G2E, and PLA2-G5A,L | Phospholipase A2, groups II and V | 1.7 | −1.6 | 2.7 | Increased inflammation +P66, decreased vessel permeability +P66, decreased inflammation −P66 | |
| PRKCB1L | Protein kinase C, beta 1 | NS | NS | −1.82 | Decreased MAPK signaling, focal adhesion, immune response +P66, increased vessel permeability +P66 | |
| HSP72L | Heat shock protein 72 | NS | 1.99 | −1.92 | Increased proinflammatory cytokine production −P66, increased protection against stress −P66 | |
| PI3K-R2, PI3K-R3, and PI3KCGA,L | Phosphoinositide-3-kinases | NS | 2.2 | −2.0 | Increased inflammation −P66, decreased actin polymerization +P66, increased vessel permeability +P66 | |
| MAPK14, p38L | Mitogen-activated protein kinase 10 | NS | 1.70 | −2.3 | Increased inflammation −P66 | |
| IL11RAA | Interleukin 11 receptor, alpha | NS | 1.9 | −2.5 | Increased immune response −P66 | |
| MAPK8, JNKL* | Mitogen-activated protein kinase 8; Jun N-terminal kinase | −1.56 | NS | −2.05 | Decreased JNK signaling, inflammation +P66 | |
Ea.hy926 cells were infected with B. burgdorferi at an MOI of 10 or left uninfected. Bacterial cell-host cell contact was not facilitated by centrifugation. For genes represented more than once on each array, all the data were averaged. Genes listed in one pathway are included again in different pathways when appropriate.
Adherent cells are indicated by a superscript A letter, and lifted cells are indicated by a superscript L letter. Cells that are both adherent and lifted cells are indicated by both superscript A and L letters. Some genes were validated using quantitative reverse transcriptase PCR (qRT-PCR); these genes are indicated with an asterisk, and the qRT-PCR values are shown in parentheses.
The fold change for value for the following three comparisons is shown: p66+ B. burgdorferi-infected cells and uninfected cells (p66+ and uninfected), Δp66 B. burgdorferi-infected cells and uninfected cells (Δp66 and uninfected), and p66+ B. burgdorferi-infected cells and Δp66 B. burgdorferi-infected cells (p66+ and Δp66). The data shown are the average fold changes for three independent experiments at one or more time points. NS, not significant. Parenthetical 1, 4, and 6 refer to STAT1 to STAT6, respectively.
The predicted effect or note in the presence of P66 (+P66) or in the absence of P66 (−P66) is shown.
Eight genes involved in actin cytoskeleton dynamics were differentially expressed in adherent cells infected with p66+ bacteria versus Δp66 bacteria. Five of these showed a similar pattern in which cells infected with the p66+ bacteria showed downregulation. For example, mRNA levels for RhoGEF1 and gelsolin were decreased ≥1.6 fold in response to p66+ bacteria, which would suggest a decrease in overall actin polymerization. In cells infected with the Δp66 mutant bacteria, actinin alpha 4 was decreased ≥1.6 fold, which would potentially lead to decreased focal adhesion complex formation. The ratios of the change in expression in the comparison of cells infected with p66+ bacteria and Δp66 bacteria were consistent with significant differences in the actin dynamics of cells infected with p66+ bacteria versus Δp66 bacteria.
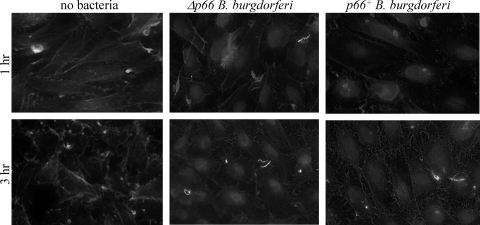
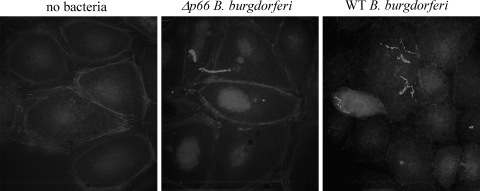
To determine whether B. burgdorferi might be affecting the actin cytoskeleton at the functional as well as the mRNA level, adherent Ea.hy926 cells were infected with p66+ or Δp66 B. burgdorferi under conditions similar to those used to generate mRNA for the microarray analyses. The cells were fixed, permeabilized, and then stained with phalloidin, which stains filamentous actin. Anti-Lyme spirochete antiserum was used to stain the bacteria. As shown in Fig. 2, both the p66+ bacteria and the Δp66 bacteria alter the actin in the endothelial cell layers. The p66+ bacteria lead to greater loss of stress fibers and cortical F-actin (phalloidin) staining and to the development of a lace-like network in the cells. Less-extensive rearrangements were seen after infection with the Δp66 bacteria. These changes were, like the gene expression changes, subtle and transient and did not result in gross loss of endothelial layer integrity.
Fig. 2.
P66 affects endothelial cell morphology. Ea.hy926 cells were plated in tissue culture-treated glass chamber slides and then infected at an MOI of 10 with p66+ or Δp66 B. burgdorferi HB19. The infections were allowed to proceed at 37°C under 5% CO2 for 1 h or 3 h, after which the cells were washed to remove unbound bacteria and fixed in paraformaldehyde. After permeabilization in 0.1% Triton X-100, the samples were stained with antibody against the B. burgdorferi plus fluorescein isothiocyanate (FITC)-phalloidin. In the endothelial cells, cortical actin is perturbed and stress fibers are less apparent after infection with the p66+ bacteria. All panels are shown at the same magnification.
To determine whether any of the Rho family GTPases might be involved in these changes, their activation states in infected cells were determined. No significant differences were seen for Rho, Rac, or CDC42 in any of the conditions tested: uninfected or infected with p66+ bacteria or with Δp66 bacteria at any time point through 24 h infection (data not shown). It is possible that other GTPases are more affected by the presence of P66 than are the three recognized by the reagents used. Nevertheless, it is apparent that the presence of P66 does affect how the endothelial cells respond to the bacteria at the mRNA level and beyond.
Signaling pathways that are altered by P66.
The adherent and lifted cells also showed significant changes in the mitogen-activated protein (MAP) kinase, Jak-STAT, phosphatidylinositol, and vascular endothelial growth factor (VEGF) signaling pathways, all of which are involved in cellular responses to changes in the environment, as well as responses to infection. Several genes in these pathways are also included in the focal adhesion and regulation of actin cytoskeleton pathways and so have already been mentioned above but are listed in each applicable section of Table 3.
In the lifted cells, the transcript levels of two MAP kinases, a Rap guanine nucleotide exchange factor (Rap GEF) and Rac1, were decreased in the cells infected with the p66+ bacteria compared to the cells infected with Δp66 bacteria (Table 3), suggesting that P66 is decreasing signaling in the MAP kinase pathways leading to inflammation. In support of this, a protein kinase C subunit, phosphatidylinositol (PI) 3-kinase forms, and calcium channel subunits, as well as three forms of STAT (signal transducer and activator of transcription), were also decreased in the comparison of cells infected with p66+ bacteria versus Δp66 bacteria, again suggesting that the cells are not activating as robust an immune response if P66 is expressed by the bacteria. It should be noted, however, that the proinflammatory STAT4 mRNA level was significantly higher in the comparison of cells infected with p66+ bacteria versus Δp66 bacteria. This form, STAT4, is expressed at low levels and is activated by alpha interferon (IFN-α) in endothelial cells (9). The only IFN-α form that was differentially expressed, IFN-α8, was downregulated approximately 1.8-fold in the comparison of cells infected with p66+ bacteria versus Δp66 bacteria (not shown), so these preliminary results will require further investigation at additional levels but overall may suggest a downregulation of normal cell signaling.
Several genes in the MAP kinase (MAPK) and other signaling pathways were also differentially expressed in the adherent cells (Table 3). TAO kinase 2 (TAOK2) was downregulated in the cells infected with p66+ bacteria and is involved in actin cytoskeleton signaling as well as Jun N-terminal protein kinase (JNK) activation. This alteration would be predicted to decrease MAPK p38 signaling and the induction of inflammatory cytokines in the presence of P66. In both adherent and lifted cells, heat shock protein 72 (HSP72) was increased in response to the Δp66 bacteria but not to the p66+ bacteria. This result suggests an increase in proinflammatory cytokine production and protection against stress in the cells infected with the Δp66 bacteria. PI 3-kinases were also affected by the expression of P66 by B. burgdorferi (Table 3). Three forms showed consistent increases in expression in response to the Δp66 bacteria. Particular forms of phospholipase A2, however, were found to be upregulated in response to the p66+ bacteria and downregulated in response to the Δp66 bacteria in both adherent and lifted cells in comparison to the uninfected controls. Phospholipase A2 enzymes (PLA2s) are both secreted and cytosolic and participate in generation of inflammatory responses through production of arachidonic acid and so may contribute to the inflammation seen with particular manifestations of Lyme disease.
Effects of P66 on the VEGF signaling pathway.
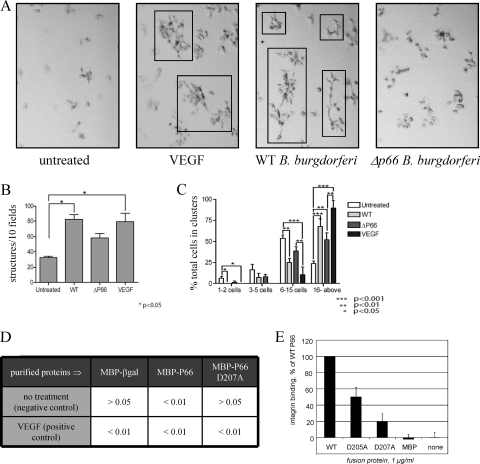
Modulation of the VEGF signaling pathway, in which several of the aforementioned gene products participate, was intriguing in light of B. burgdorferi being an arthropod-borne pathogen. VEGF was originally known as vascular permeability factor, so to determine whether the p66+ versus the Δp66 B. burgdorferi display differences in the ability to cross endothelial cell layers, Ea.hy926 macrovascular endothelial cells were grown to confluence on 3-μm-pore-size inserts. The bacteria were added to the cells in the upper chamber, and migration across the cell layer was quantified by counting bacteria in both the upper and lower chambers over time for up to 72 h. In this system, no statistically significant differences were seen (L. C. Ristow and J. Coburn, data not shown). In a second measure of VEGF signaling effects on endothelial cells in culture, the same cell line was plated on Matrigel, an extracellular matrix that allows the cells to respond to growth factor stimuli in a more physiologically relevant setting than is possible using culture in plastic dishes. In addition, the cells may form multicellular structures thought to model angiogenesis when cultured on Matrigel and stimulated with VEGF (31). In this setting, B. burgdorferi that express P66 stimulated growth of the cells in a manner that mimicked VEGF, while the Δp66 B. burgdorferi had a smaller effect (Fig. 3).
Fig. 3.
B. burgdorferi promotes growth and structure formation by endothelial cells in Matrigel. Cells were plated at low density on either conventional plastic tissue culture plates or on the same culture plates precoated with Matrigel. After 2 days of incubation, the cells were treated with recombinant vascular endothelial growth factor (VEGF), B. burgdorferi, or the medium control and followed visually over several days of additional incubation. (A) Representative micrographs from day 5 postinfection or posttreatment. Multicellular structures distinct from clumps are boxed. Wild-type (WT) B. burgdorferi and Δp66 B. burgdorferi were examined. (B) Quantification of multicellular structures on day 5. At earlier time points, there were no statistically significant differences between the values for the different conditions. On day 5, the number of structures in the cells infected with WT B. burgdorferi or in the cells treated with VEGF was significantly higher (P < 0.05) than that of the untreated cells. (C) Quantification of overall endothelial cell growth to form multicellular clusters and structures. The growth and formation of structures do not depend solely on the presence of P66 expression, as the Δp66 bacteria stimulate growth and structure formation by the endothelial cells, but to a lesser degree than do the wild-type bacteria. While in the bacterial infections, P66 was not the only determinant of endothelial cell growth, when purified proteins were tested in similar experiments, the integrin binding activity of P66 was critical (panels D and E). (D) P values in comparisons of purified maltose-binding protein (MBP)-P66 fusion proteins and control proteins in comparisons to VEGF and no additions (positive and negative controls, respectively). βgal, β-galactosidase. (E) Integrin αIIbβ3 binding activity of MBP-P66 carrying site-directed changes to two aspartic acid residues in the previously identified integrin-binding domain. E. coli never resulted in structure formation or endothelial cell growth but did kill the cells within 3 days regardless of the presence or absence of Matrigel (not shown).
To determine whether the stimulation of endothelial cell growth was due to P66 or other bacterial factors, purified MBP-P66 and the control MBP were tested for the ability to promote growth of the cells. While in the bacterial infections P66 was not the only determinant of endothelial cell responses, purified MBP-P66 alone did promote growth of the cells, while MBP alone and MBP-P66 D207A (which is defective in β3 integrin binding) were less effective (Fig. 3). We recently found that OspC of B. burgdorferi also stimulated endothelial cell growth (4), so while this property is not unique to P66, the effect of P66 appears to be correlated with β3 integrin binding activity. For B. burgdorferi, the more relevant test of vascular permeability in response to intradermal infection of mice with wild-type and mutant strains will be pursued in future studies, but the cell culture model demonstrates that this is a direction of potential interest.
Immune response signaling affected by P66.
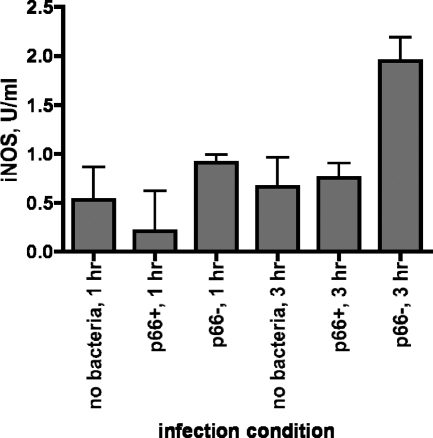
Several of the genes that were differentially regulated in both adherent and lifted endothelial cells are involved in the immune response as well as in the signaling pathways as described above. In addition to the genes described earlier, HSP72, which is involved in the response to a variety of stresses and promotes production of proinflammatory cytokines via the JNK pathway, was decreased in response to the p66+ bacteria versus Δp66 bacteria. This may alter the type/balance of immune response to infection. Likewise, the downregulation of the transcription factor STAT6, p38 MAPK, JNK, coagulation factor II receptor-like 3, interleukin 11 (IL-11) receptor, tumor necrosis factor alpha (TNF-α)-interacting protein, PI 3-kinase, and especially nitric oxide synthase 2A (NOS2A) would all decrease the response to the p66+ bacteria compared to the response to the Δp66 bacteria. This was verified at the protein level for NOS2A, which is also known as NOS2 and iNOS (Fig. 4). This form of nitric oxide synthase is induced by mammalian cells in response to a variety of stresses, including bacterial infection. The cells infected with the Δp66 bacteria showed increased iNOS protein levels in comparison to the uninfected controls. In contrast, the cells infected with the p66+ bacteria showed no increase in iNOS protein, suggesting that P66 might repress induction of iNOS in response to other bacterial components.
Fig. 4.
Inducible nitric oxide synthase (iNOS) induction by p66+ versus Δp66 B. burgdorferi. Ea.hy926 endothelial cells were incubated with either p66+ or Δp66 B. burgdorferi at an MOI of 10 or with no bacteria. After 1 or 3 h, the cell layers were washed and lysed, and iNOS protein levels in cell lysates were measured using a commercially available ELISA kit. The values are the means plus standard deviations (error bars) of three replicate samples from a single representative experiment. At the 3-h time point, comparing the value for Δp66 B. burgdorferi-infected cells to the value for p66+ B. burgdorferi-infected cells yielded a P value of <0.02, and comparing the value for Δp66 B. burgdorferi-infected cells to the value for the uninfected control at the 3-h time point yielded a P value of 0.03. At 1 h, there were no significant differences, and there was no significant difference between the uninfected and the p66+B. burgdorferi-infected cell samples at 3 h.
Similarities and differences of HEK 293 epithelial cells and endothelial cells.
In addition to the endothelial cells, comparisons between responses to the p66+ and Δp66 B. burgdorferi were made between HEK 293 epithelial cells, which express no β3-chain integrins, and a derivative expressing αvβ3, after infection with p66+ B. burgdorferi versus Δp66 B. burgdorferi. Although 293 cells do not express β3 integrins, P66 does also display some binding to integrin α3β1 and has been demonstrated to affect signaling through α3β1 in human chondrocytes. The two epithelial cell lines showed many fewer statistically significant differences in gene expression in response to the different bacterial strains (Table 4). These data demonstrated that the responses to B. burgdorferi infection in these cells are quite different from the responses in the endothelial cells. An additional difference between the epithelial and endothelial cells is that there was no overlap of genes that were differentially expressed in both the lifted and adherent epithelial cells.
Table 4.
Clusters of genes with changes in expression common to lifted and adherent 293 and 293+αvβ3 cells infected with p66+ versus Δp66 B. burgdorferia
| Cell | KEGG pathwayb | Lifted cellsc |
Adherent cellsc |
||
|---|---|---|---|---|---|
| No. of genes | P value | No. of genes | P value | ||
| 293 | NAd | NA | NA | NA | NA |
| 293+αvβ3 | Cell communication | 3 | 0.0384 | 5 | 3.68E−02 |
| Pathogenic Escherichia coli infection (EHEC) | 2 | 0.0362 | 3 | 4.39E−02 | |
| Pathogenic Escherichia coli infection (EPEC) | 2 | 0.0362 | 3 | 4.39E−02 | |
Human kidney epithelial line HEK 293 (293 cells) and a derivative transfected to express the genes for the integrin αv and β3 subunits (293+αvβ3 cells) were examined.
KEGG pathways listed at http://www.genome.jp/kegg/pathway.html. EHEC, enterohemorrhagic Escherichia coli; enteropathogenic EPEC, Escherichia coli.
P values considered to be statistically significant for lifted and adherent cells were <0.05 and < 0.01, respectively, due to differences in the arrays used.
NA, not applicable. There were no pathways significantly differentially affected by infection with p66+ versus Δp66 B. burgdorferi in both adherent and lifted cells.
There are differences between the 293 and the 293+αvβ3 epithelial cell lines responses to infection with p66+ B. burgdorferi versus Δp66 B. burgdorferi, demonstrating that the integrin and the different physiology resulting from the expression of integrin αvβ3 affect how these cells respond to the bacteria. In fact, the 293+αvβ3 cells are morphologically quite distinct from the parental 293 cells, as they spread out more on tissue culture plastic ware and are more resistant to lifting with EDTA (unpublished observations). Nevertheless, the 293+αvβ3 cells did show changes in expression of genes that are predicted to affect the actin cytoskeleton (Table 5). Staining of these cells with phalloidin after infection revealed changes in the actin cytoskeleton (Fig. 5), demonstrating that although changes in expression of genes that affect cytoskeletal dynamics were not dramatic at the mRNA level, the integrin ligand P66 affects the mammalian cell at functional levels.
Table 5.
Genes significantly differentially expressed in 293 and 293+αvβ3 cells infected with p66+ versus Δp66 bacteriaa
| Cell line | Gene nameb | Protein encoded | Fold change comparing the followingc: |
Predicted effect(s) or note(s)d | ||
|---|---|---|---|---|---|---|
| p66+ and uninfected | Δp66 and uninfected | p66+ and Δp66 | ||||
| 293 | COLL11A1L | Collagen XI alpha 1 | NS | 3.05 | −2.83 | Increased cell-extracellular matrix (ECM) adhesion −P66 |
| COLL5A1L* | Collagen V alpha 1 | NS | 2.61 | −2.97 | Increased cell-ECM adhesion −P66 | |
| COLL9A1L | Collagen IX alpha 1 | NS | −3.28 | 3.58 | Decreased cell-ECM adhesion −P66 | |
| MAN1A2L | Mannosidase, alpha, class 1A, member 2 | NS | 2.86 | −2.72 | Fewer N-glycans on the cell surface −P66 | |
| MGAT1L | Mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyl transferase | NS | −2.65 | 2.67 | Fewer N-glycans on the cell surface −P66 | |
| ALG14L | Asparagine-linked glycosylation 14 homolog | NS | −2.50 | 2.84 | Fewer N-glycans on the cell surface −P66 | |
| LAMA4L | Laminin, alpha 4 | NS | −1.52 | 1.61 | Decreased cell-ECM adhesion −P66 | |
| PCDHB4A | Protocadherin beta 4 | 1.5 | NS | 1.9 | Increased cell-cell adhesion +P66 | |
| PPP1CBA | Protein phosphatase 1 | NS | 1.8 | −2.5 | Decreased actin polymerization −66 | |
| ACTBL* | β-Actin | NS | NS | −1.64 | Decreased actin polymerization +P66 | |
| SOS1L | Son of sevenless homolog 1 | NS | −1.94 | 2.31 | Decreased proliferation of cells −P66 | |
| THBS2L | Thrombospondin 2 | NS | 2.92 | −2.97 | Decreased angiogenesis −P66 | |
| TLN1A | Talin | 2.0 | NS | 1.8 | Increased actin polymerization +P66 | |
| TNFSF11A | Tumor necrosis family super family, member 11 | −1.8 | NS | −2.2 | Decreased immune response +P66 | |
| HEK 293+αvβ3 | HAPLN3A | Hyaluronan and proteoglycan link protein 3 | NS | −1.8 | −2.3 | Decreased cell adhesion −P66 |
| FGFR2A | Fibroblast growth factor receptor 2 | NS | 1.9 | −2.5 | Decreased actin polymerization +P66 | |
| VAV2A | Rho family GEF | NS | 2.2 | −2.0 | Increased Rho activation, actin polymerization −P66 | |
| JAK2A | Janus kinase 2 | NS | 2.0 | −2.3 | Increased immune response −P66 | |
| ULBP1A | UL16-binding protein 1 | NS | −1.6 | 2.1 | Increased immune response −P66 | |
| MEKKK5A | Mitogen-activated protein kinase kinase kinase 5 | NS | 1.5 | −2.0 | Increased immune response −P66 | |
| STAT5BA | Signal transducers and activators of transcription 5B | NS | 2.0 | −2.5 | Increased immune response −P66 | |
| ERK5A | Mitogen-activated protein kinase 7 | NS | −1.7 | 2.2 | Increased apoptosis −P66 | |
| EMID2L | Collagen α1-26 | −1.57 | NS | −1.61 | Decreased cell-ECM adhesion +P66 | |
| CTNNB1L | Catenin beta 1 | 2.5 | NS | 2.07 | Increased cell-cell adhesion +P66 | |
| SCYE1A | Small inducible cytokine subfamily E, 1 | NS | 1.6 | −2.0 | Increased immune response, apoptosis −P66 | |
Human kidney epithelial line HEK 293 (293 cells) and a derivative transfected to express the genes for the integrin αv and β3 subunits (293+αvβ3 cells) were infected with B. burgdorferi at an MOI of 10 or left uninfected. Bacterial cell-host cell contact was not facilitated by centrifugation. For genes represented more than once on each array, all the data were averaged.
Adherent cells are indicated by a superscript A letter, and lifted cells are indicated by a superscript L letter. Some genes were validated using quantitative reverse transcriptase PCR (qRT-PCR); these genes are indicated with an asterisk.
The fold change for values for the following three comparisons is shown: p66+ B. burgdorferi-infected cells and uninfected cells (p66+ and uninfected), Δp66 B. burgdorferi-infected cells and uninfected cells (Δp66 and uninfected), and p66+ B. burgdorferi-infected cells and Δp66 B. burgdorferi-infected cells (p66+ and Δp66). The data shown are the average fold changes for three independent experiments at any time point. NS, not significant.
The predicted effect or note in the presence of P66 (+P66) or in the absence of P66 (−P66) is shown.
Fig. 5.
P66 disrupts cortical actin in epithelial cells. HEK 293 cells transfected to express integrin αvβ3 were infected at an MOI of 10 with p66+ or Δp66 B. burgdorferi. The infections were allowed to proceed at 37°C under 5% CO2 for 1 h, after which the cells were washed to remove unbound bacteria and fixed in paraformaldehyde. After permeabilization in 0.1% Triton X-100, the samples were stained with anti-OspA mouse serum plus Alexa Fluor 350-phalloidin. Note that in the cells infected with the bacteria that express P66, the cortical rim of actin has largely dissipated, and the cells appear to have contracted. All panels are shown at the same magnification.
Unlike the EA.hy926 cells and the 293+αvβ3 cells, much of the 293 epithelial cell response appeared to be centered on changes that may affect the production of cell surface and extracellular matrix components. For example, in the lifted 293 cells, expression of collagen chain 11A1 is decreased, while collagen chains 5A1 and 9A1 are increased in the cells infected with the Δp66 bacteria. The level of transcript encoding laminin 4 is initially increased but then declines in the cells infected with the Δp66 bacteria, while the opposite pattern was observed for thrombospondin 1. In the adherent cells, β-laminin transcript increases in cells infected with the p66+ bacteria and decreases in cells infected with the Δp66 bacteria, and protocadherin 4 increases in cells infected with the p66+ bacteria. The 293 epithelial cells therefore undergo changes in gene expression consistent with rearrangement of their outer surfaces and extracellular matrices. This may result in altered exposure of molecules that participate in defense against the pathogen or altered availability of potential receptors used by the bacteria.
DISCUSSION
The B. burgdorferi outer membrane protein P66 was identified as an integrin ligand, but its role in B. burgdorferi infection is not yet known. Our hypothesis was that integrin binding might allow the bacteria to trigger changes in host cell biology that would promote bacterial survival and dissemination within the host. In contrast to invasin, a β1 integrin ligand of Yersinia pseudotuberculosis, P66 would not be expected to promote uptake of the bacteria by mammalian cells, as there is no current evidence that B. burgdorferi occupies an intracellular niche during mammalian infection. The significance of P66 is also predicted to be in the mammal rather than in the tick, as P66 is not produced by B. burgdorferi in unfed ticks but is expressed during mammalian infection and as the bacteria are acquired by, or transmitted by, the tick (14). P66 expression then correlates with factors specific to mammalian infection. Indeed, preliminary evidence suggests that P66 is required for mammalian infection (unpublished observations). A second consideration is that P66 was also identified as a channel-forming porin (33, 35), an activity that may also influence bacterial survival in the host. We therefore sought to determine whether the mammalian host cells would respond differently to B. burgdorferi that express P66 than to B. burgdorferi that that do not express P66. Taken together, our results suggest that the presence of P66 may decrease the human endothelial cell response to B. burgdorferi and affect the nature of the responses of endothelial and epithelial cells to these bacteria. In this work, several groups of genes that appear to be differentially expressed by human cells that have been infected by B. burgdorferi that produce the P66 protein and that do not produce the P66 protein were identified. These groups include actin cytoskeleton and focal adhesion, cell adhesion proteins, immune response/stress response genes, and signaling pathways.
Alterations in the actin cytoskeleton may have at least two functions during B. burgdorferi infection. One possibility is that manipulation of actin may allow increased migration of B. burgdorferi through various cell layers and tissues. A second possibility is that the effects on actin dynamics may alter the ability of immune cells to migrate to sites of infection. The presence of P66 was also shown to increase the expression of multiple cell surface adhesion proteins, which may again have two functions. In one scenario, this could serve to increase the availability of receptors for the bacteria. An alternative, but not mutually exclusive, possibility is that the changes may affect the ability of infiltrating immune cells to traffic to the site of infection.
We also observed a decrease in expression of genes involved in the immune response in cells infected with p66+ B. burgdorferi, and conversely, increased expression of genes relating to the immune response in cells infected with the Δp66 bacteria. This may be one factor that contributes to the ability of B. burgdorferi to establish persistent infection in immunocompetent animals in the absence of antibiotic therapy. We did, however, observe increases in expression of genes encoding certain proinflammatory molecules, such as phospholipases and metalloproteinases (data not shown), which may contribute to the inflammation in particular tissues seen in Lyme disease. The MAPK and Jak-STAT pathways, which contribute to effecting the immune response and inflammation downstream of integrins, also showed differences in the expression of the genes whose products participate in these signaling cascades. Together with changes seen in signaling and response pathways, particularly VEGF signaling, it is possible that the significance of P66 in B. burgdorferi infection is primarily due to subtle influences on host cell and tissue biology.
It is important to note that gene expression changes are not the only mechanism by which important changes can occur in how mammalian cells respond to bacteria. Many of the changes are immediate and short-lived alterations in the activities of signaling pathways that mediate the host response to the bacteria. However, modest changes in expression of signaling intermediates may have significant effects at the cellular level. Changes in expression of several genes within the same pathway also suggest that the host cell response can affect the outcome of the interaction over longer periods of time, particularly in the cases of pathogens such as B. burgdorferi that do not rapidly kill their hosts but need to maintain a persistent infection.
This work also emphasizes that, although changes in gene expression may be important, analysis of changes at the protein and cellular levels are also important to consider, due to the multiple layers of regulation of the activities of many proteins. This is particularly true for interactions of bacteria with host cells mediated by signaling molecules such as integrins. The invasin protein of Yersinia is a well-established example of how binding to integrins can contribute to bacterial virulence. In addition, for many years, the importance of bacterial toxins in manipulation of host cell biology has been appreciated, but B. burgdorferi has not been demonstrated to produce toxins. Manipulation of host cell biology through binding to integrins may therefore be a tool used by B. burgdorferi to its own advantage in facilitating establishment of persistent, disseminated infection in immunocompetent hosts.
ACKNOWLEDGMENTS
We thank Melisa Medrano, James McCarthy, Mark Fahey, Charlotte Dittman Majerczyk, Clarita Lefthand, and Yue Shao for their contributions to this work, Denis Guillemette and the staff at GEO, particularly Katherine Phillippy, for help with deposition of microarray data, and C.-J. Edgell and E. Ades for cell lines.
This work was supported by NIH grants AI051407 and AI059505 to J.C.
Footnotes
Published ahead of print on 16 May 2011.
REFERENCES
- 1. Ades E. W., et al. 1992. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J. Invest. Dermatol. 99:683–690 [DOI] [PubMed] [Google Scholar]
- 2. Aliprantis A. O., et al. 1999. Cell activation and apoptosis by bacterial lipoproteins through Toll-like receptor-2. Science 285:736–739 [DOI] [PubMed] [Google Scholar]
- 3. Alitalo A., et al. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169:3847–3853 [DOI] [PubMed] [Google Scholar]
- 4. Antonara S., Ristow L., McCarthy J., Coburn J. 2010. Effect of Borrelia burgdorferi OspC at the site of inoculation in mouse skin. Infect. Immun. 78:4723–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Behera A. K., et al. 2008. Borrelia burgdorferi BBB07 interaction with integrin alpha(3)beta(1) stimulates production of pro-inflammatory mediators in primary human chondrocytes. Cell. Microbiol. 10:320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brooks C. S., et al. 2005. Complement regulator-acquiring surface protein 1 imparts resistance to human serum in Borrelia burgdorferi. J. Immunol. 175:3299–3308 [DOI] [PubMed] [Google Scholar]
- 7. Casjens S., et al. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490–516 [DOI] [PubMed] [Google Scholar]
- 8. Centers for Disease Control Prevention 1995. Recommendations for test performance and interpretation from the Second National Conference on Serologic Diagnosis of Lyme Disease. Morb. Mortal. Wkly. Rep. 44:590–591 [PubMed] [Google Scholar]
- 9. Cho S. S., et al. 1996. Activation of STAT4 by IL-12 and IFN-alpha: evidence for the involvement of ligand-induced tyrosine and serine phosphorylation. J. Immunol. 157:4781–4789 [PubMed] [Google Scholar]
- 10. Coburn J., Chege W., Magoun L., Bodary S. C., Leong J. M. 1999. Characterization of a candidate Borrelia burgdorferi β3-chain integrin ligand identified using a phage display library. Mol. Microbiol. 34:926–940 [DOI] [PubMed] [Google Scholar]
- 11. Coburn J., Cugini C. 2003. Targeted mutation of the outer membrane protein P66 disrupts attachment of the Lyme disease spirochete, Borrelia burgdorferi, to integrin αvβ3. Proc. Natl. Acad. Sci. U. S. A. 100:7301–7306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coburn J., Leong J., Erban J. 1993. Integrin αIIbβ3 mediates binding of the Lyme disease agent, Borrelia burgdorferi, to human platelets. Proc. Natl. Acad. Sci. U. S. A. 90:7058–7063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coburn J., Magoun L., Bodary S. C., Leong J. M. 1998. Integrins αvβ3 and α5β1 mediate attachment of Lyme disease spirochetes to human cells. Infect. Immun. 66:1946–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cugini C., Medrano M., Schwan T. G., Coburn J. 2003. Regulation of expression of the Borrelia burgdorferi β3-chain integrin ligand, P66, in ticks and in culture. Infect. Immun. 71:1001–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Defoe G., Coburn J. 2001. Delineation of Borrelia burgdorferi p66 sequences required for integrin αIIbβ3 recognition. Infect. Immun. 69:3455–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dressler F., Whalen J. A., Reinhardt B. N., Steere A. C. 1993. Western blotting in the serodiagnosis of Lyme disease. J. Infect. Dis. 167:392–400 [DOI] [PubMed] [Google Scholar]
- 17. Edgell C. J., et al. 1990. Endothelium specific Weibel-Palade bodies in a continuous human cell line, EA.hy926. In Vitro Cell. Dev. Biol. 26:1167–1172 [DOI] [PubMed] [Google Scholar]
- 18. Edgell C. J., McDonald C. C., Graham J. B. 1983. Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc. Natl. Acad. Sci. U. S. A. 80:3734–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fraser C. M., et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586 [DOI] [PubMed] [Google Scholar]
- 20. Guo B. P., Brown E. L., Dorward D. W., Rosenberg L. C., Hook M. 1998. Decorin-binding adhesins from Borrelia burgdorferi. Mol. Microbiol. 30:711–723 [DOI] [PubMed] [Google Scholar]
- 21. Guo B. P., Norris S. J., Rosenberg L. C., Hook M. 1995. Adherence of Borrelia burgdorferi to the proteoglycan decorin. Infect. Immun. 63:3467–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hartmann K., et al. 2006. Functional characterization of BbCRASP-2, a distinct outer membrane protein of Borrelia burgdorferi that binds host complement regulators factor H and FHL-1. Mol. Microbiol. 61:1220–1236 [DOI] [PubMed] [Google Scholar]
- 23. Hellwage J., et al. 2001. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427–8435 [DOI] [PubMed] [Google Scholar]
- 24. Hirschfeld M., et al. 1999. Inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by Toll-like receptor 2. J. Immunol. 163:2382–2386 [PubMed] [Google Scholar]
- 25. Hovis K. M., et al. 2006. Selective binding of Borrelia burgdorferi OspE paralogs to factor H and serum proteins from diverse animals: possible expansion of the role of OspE in Lyme disease pathogenesis. Infect. Immun. 74:1967–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hynes R. O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell 110:673–687 [DOI] [PubMed] [Google Scholar]
- 27. Kraiczy P., Skerka C., Kirschfink M., Brade V., Zipfel P. F. 2001. Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL-1/reconectin and Factor H. Eur. J. Immunol. 31:1674–1684 [DOI] [PubMed] [Google Scholar]
- 28. Leong J. M., et al. 1998. Different classes of proteoglycans contribute to the attachment of Borrelia burgdorferi to cultured endothelial and brain cells. Infect. Immun. 66:994–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McDowell J. V., et al. 2003. Comprehensive analysis of the factor H binding capabilities of Borrelia species associated with Lyme disease: delineation of two distinct classes of factor H binding proteins. Infect. Immun. 71:3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Medigue C., Rechenmann F., Danchin A., Viari A. 1999. Imagene: an integrated computer environment for sequence annotation and analysis. Bioinformatics 15:2–15 [DOI] [PubMed] [Google Scholar]
- 31. Obeso J., Weber J., Auerbach R. 1990. A hemangioendothelioma-derived cell line: its use as a model for the study of endothelial cell biology. Lab. Invest. 63:259–269 [PubMed] [Google Scholar]
- 32. Parveen N., et al. 2006. Bgp, a secreted glycosaminoglycan-binding protein of Borrelia burgdorferi strain N40, displays nucleosidase activity and is not essential for infection of immunodeficient mice. Infect. Immun. 74:3016–3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pinne M., et al. 2007. Elimination of channel-forming activity by insertional inactivation of the p66 gene in Borrelia burgdorferi. FEMS Microbiol. Lett. 266:241–249 [DOI] [PubMed] [Google Scholar]
- 34. Probert W. S., Johnson B. J. 1998. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol. Microbiol. 30:1003–1015 [DOI] [PubMed] [Google Scholar]
- 35. Skare J. T., et al. 1997. The Oms66 (p66) protein is a Borrelia burgdorferi porin. Infect. Immun. 65:3654–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Verma A., Brissette C. A., Bowman A., Stevenson B. 2009. Borrelia burgdorferi BmpA is a laminin-binding protein. Infect. Immun. 77:4940–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weis J. J. 2002. Host-pathogen interactions and the pathogenesis of murine Lyme disease. Curr. Opin. Rheumatol. 14:399–403 [DOI] [PubMed] [Google Scholar]
- 38. Wooten R. M., et al. 2002. Toll-like receptor 2 is required for innate, but not acquired, host defense to Borrelia burgdorferi. J. Immunol. 168:348–355 [DOI] [PubMed] [Google Scholar]
- 39. Zhang B., Kirov S., Snoddy J. 2005. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33:W741–W748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang J. R., Hardham J. M., Barbour A. G., Norris S. J. 1997. Antigenic variation in Lyme disease Borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275–285 [DOI] [PubMed] [Google Scholar]
- 41. Zhang J. R., Norris S. J. 1998. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect. Immun. 66:3698–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang J. R., Norris S. J. 1998. Kinetics and in vivo induction of genetic variation of vlsE in Borrelia burgdorferi. Infect. Immun. 66:3689–3697 (Erratum, 67:468, 1999.) [DOI] [PMC free article] [PubMed] [Google Scholar]