Abstract
Type III and type VI secretion systems (T3SSs and T6SSs, respectively) are critical virulence determinants in several Gram-negative pathogens. In Burkholderia pseudomallei, the T3SS-3 and T6SS-1 clusters have been implicated in bacterial virulence in mammalian hosts. We recently discovered a regulatory cascade that coordinately controls the expression of T3SS-3 and T6SS-1. BsaN is a central regulator located within T3SS-3 for the expression of T3SS-3 effectors and regulators for T6SS-1 such as VirA-VirG (VirAG) and BprC. Whereas T6SS-1 gene expression was completely dependent on BprC when bacteria were grown in medium, the expression inside host cells was dependent on the two-component sensor-regulator VirAG, with the exception of the tssAB operon, which was dependent primarily on BprC. VirAG and BprC initiate different transcriptional start sites within T6SS-1, and VirAG is able to activate the hcp1 promoter directly. We also provided novel evidence that virAG, bprC, and tssAB are critical for T6SS-1 function in macrophages. Furthermore, virAG and bprC regulator mutants were avirulent in mice, demonstrating the absolute dependence of T6SS-1 expression on these regulators in vivo.
INTRODUCTION
Many bacterial pathogens employ secretion systems to facilitate their survival in hosts. One of the better characterized secretion systems is the type III secretion system (T3SS), important for virulence in many Gram-negative pathogens (11). The putative gene clusters of the less well characterized type VI secretion system (T6SS) have been identified in over one-fourth of sequenced Gram-negative bacterial genomes (9, 15). However, the function of T6SS appears to be more diverse than that of T3SS, with recent papers describing its importance in increasing bacterial fitness in the interactions with other bacterial species in the environment (14), besides its importance in virulence in some bacteria.
Burkholderia pseudomallei is a Gram-negative bacterial pathogen possessing six copies of T6SSs (28). It is the causative agent of melioidosis, an infectious disease endemic in Northern Australia and Southeast Asia. The T6SS-1 cluster was first identified in infected macrophages through in vivo expression technology (IVET) (28). In Burkholderia mallei, a species closely related to B. pseudomallei, the T6SS cluster homologous to B. pseudomallei T6SS-1 has been shown to be critical for virulence in a hamster infection model (27) and is important for intracellular growth, actin polymerization, and formation of multinucleated giant cells (MNGCs) in vitro (8). One of the genes in the B. pseudomallei T6SS-1 cluster was reported to be necessary for virulence in mice (23). A recent paper described a B. pseudomallei T6SS-1 hcp1 deletion mutant with attenuated virulence in hamsters and reduced intracellular growth and cytotoxicity, as well as a lack of MNGC formation in vitro (6).
We have recently identified a novel regulatory cascade controlling the coordinate expression of T3SS-3 and T6SS-1 of B. pseudomallei grown in medium where the T3SS-3 regulator BsaN controls not only the expression of T3SS-3 effectors but also T6SS-1 regulators (32). We found that the regulation of T6SS-1 genes is dependent on BprC, located within T3SS-3, but not VirAG (32). VirA-VirG belongs to the two-component sensor-regulator system. When overexpressed in trans in B. mallei or B. pseudomallei, VirAG upregulates T6SS expression in LB medium (6, 27). The apparent paradox is resolved in this study, as we show that T6SS-1 expression inside host cells is controlled primarily by VirA-VirG whereas BprC is the predominant regulator when bacteria are grown in medium. An exception is the tssAB operon, which is controlled primarily by BprC even when bacteria are inside host cells. We further elucidate the use of alternate transcriptional start sites by VirAG versus BprC and demonstrate the direct regulatory activity of VirA-VirG on the hcp1 promoter. We also examine and compare the phenotypes of the virAG and bprC or tssAB deletion mutants in macrophages and in mice and present novel evidence of the critical role of bprC or tssAB in T6SS-1 function in vitro and in vivo.
MATERIALS AND METHODS
Bacterial strains and culture conditions. Bacterial strains and plasmids used are listed in Table 1. Mutants created were derived from a clinical strain of B. pseudomallei named KHW (19). Antibiotics added to LB medium were as follows (in μg/ml): for Escherichia coli, ampicillin (Ap), 100; chloramphenicol (Cm), 25; kanamycin (Km), 25; tetracycline (Tc), 10; trimethroprim (Tm), 30; and zeocin (Zeo), 25; and for B. pseudomallei, Km, 250; gentamicin (Gm), 50; Tc, 40; Tm, 60; and Zeo, 1,000. Plasmids were transferred conjugally from E. coli SM10 to B. pseudomallei on membrane filters with E. coli donors (≈108) and B. pseudomallei recipients (≈108) from overnight cultures. Filters were incubated at 37°C on nonselective LB agar for 3 h before transferring filters onto selective medium.
Table 1.
List of strains and plasmids used in this study
| Plasmid or strain | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Plasmids | ||
| pBAKA | Conjugative suicide vector based on asdPa, containing pheS | 3 |
| pFRTT1 | pGEM contains FRT sites and tetRA sequence, Apr Tcr | 32 |
| pKO-tssAB | pBAKA containing tetRA cassette flanked by upstream and downstream sequences of tssAB, Tcr | This study |
| pFLP-AB5 | Contains an FLP recombinase gene, pheS+ as counterselection in 1% chlorophenylalanine, Tpr | 3 |
| pMLBAD | Broad-host-range vector containing inducible promoter Pbad, Tmr | 18 |
| pMLvirA | pMLBAD containing virA ORF from KHW, Tmr | This study |
| pMLvirG | pMLBAD containing virG ORF from KHW, Tmr | This study |
| pMLvirAG | pMLBAD containing virA and virG ORFs from KHW, Tmr | This study |
| pMLbprC | pMLBAD containing bprC ORF from KHW, Tmr | This study |
| pRW50 | Low-copy-number vector containing a promoterless lacZ gene, Tcr | 21 |
| pRWtssA | tssA-lacZ transcriptional fusion, pRW50 containing tssA and upstream sequence, Tcr | This study |
| pRWhcp1 | hcp1-lacZ transcriptional fusion, pRW50 containing hcp1 and upstream sequence, Tcr | This study |
| Bacterial strains | ||
| E. coli | ||
| DH5α | Cloning host | 26 |
| SM10 | Donor strain for conjugation | 29 |
| E1345 | EPMax10B-pir116-Δasd::Gm | 3 |
| E1354 | EPMax10B-pir116-Δasd-mob-Km-trp::Gm | 3 |
| B. pseudomallei | ||
| KHW | B. pseudomallei wild-type strain, Gmr | 19 |
| ΔbspR mutant | KHW ΔbpsR::zeo, codon 13-205 of BPSS1105 was replaced by zeo cassette, Zeor | 32 |
| ΔbprP mutant | KHW ΔbprP::FRT, codon 11-311 of BPSS1553 was deleted | 32 |
| ΔbsaN mutant | KHW ΔbsaN::FRT, codon 14-236 of BPSS1546 was deleted | 32 |
| ΔbprC mutant | KHW ΔbprC::zeo, codon 209-353 of BPSS1520 was replaced by zeo cassette, Zeor | 32 |
| ΔvirAG mutant | KHW ΔvirAG::FRT, codon 1-233 of BPSS1494 and codon 56-614 of BPSS1495 were deleted | 32 |
| ΔT3SS-3 mutant | KHW ΔT3SS-3::zeo, BPSS1520-to-BPSS1552 region was replaced with zeo cassette, Zeor | 33 |
| ΔT6SS-1reg mutant | KHW Δ(bimE+virAG+ T6SS-1)::FRT, BPSS1493-to-BPSS1511 region was deleted; previously named ΔT6SS5 | 33 |
| ΔtssAB mutant | KHW ΔtssAB::FRT, BPSS1496-BPSS1497 region was deleted | This study |
Abbreviations: FRT, FLP recombination target; ORF, open reading frame; Apr, ampicillin resistant; Gmr, gentamicin resistant; Kmr, kanamycin resistant; Tcr, tetracycline resistant; Tmr, trimethoprim resistant.
Cell culture and infection of macrophages.
RAW 264.7 mouse macrophage-like cells were obtained from the American Type Culture Collection and maintained in Dulbecco modified Eagle medium (Invitrogen) containing 10% heat-inactivated fetal bovine serum (DMEM) in a humidified 37°C, 5% CO2 tissue culture incubator. Before infection, cells were seeded in 12-well dishes at 5 × 105 cells/well and grown overnight until approximately 80% confluent. RAW 264.7 cells were infected with bacteria at an indicated multiplicity of infection (MOI), and cells were centrifuged at 500 × g for 10 min at room temperature. To block phagocytic uptake, cells were treated with cytochalasin D (2 μg/ml) for 2 h before infection.
Cell infection and measurement of gene expression by real-time PCR.
RAW 264.7 cells (0.5 × 105 cells/well) were seeded and grown overnight in DMEM in 12-well plates. Previously, we have found that the expression levels for both T3SS-3 and T6SS-1 were generally higher when the bacteria were grown in RPMI 1640 medium (RPMI) (Invitrogen) than in DMEM. Therefore, RPMI was used in all infection experiments. RAW 264.7 cells were changed to RPMI prior to infection and infected with bacteria at an MOI of 100:1. Bacterial and host cell RNAs were isolated from infected RAW 264.7 cells or RPMI using TRIzol (Invitrogen). DNA was removed from the RNA samples by treatment with Turbo DNase (Applied Biosystems). cDNA was synthesized using 1 μg of RNA and the High Capacity reverse transcription reagent kit (Applied Biosystems). Transcripts were quantified using SYBRgreener qPCR Supermix for iCycler (Invitrogen) in a Bio-Rad iQ5 machine. Real-time PCR primers are listed in Table 2. The relative RNA level of a particular gene in mutant strains was normalized to that of wild type using the threshold cycle (2−ΔΔCT) method (20) with 16S rRNA as a reference gene. The 16S rRNA was very abundant and fluctuated little when B. pseudomallei was grown in medium or during infection. The housekeeping gene dnaK was used as an internal control. We have previously shown that expression did not vary between different mutants (32). Results were reported as means with standard deviations of triplicate samples.
Table 2.
List of real-time PCR primers for this study
| Gene | Sequence (5′–3′) |
|---|---|
| 16S rRNA | GGCTAGTCTAACCGCAAGGA |
| TCCGATACGGCTACCTTGTT | |
| dnaK | CGCAGATCGAAGTGACCTT |
| ATCTTCTCGATCTCGGCTTC | |
| bspR | GCTCGGCTACTACATGGTGTC |
| AGCAGCAACTCGGTGTTCATC | |
| bprP | CGGCGACATCGTCACCAC |
| GGTGATAACTGCCTTCCGTGAC | |
| bsaN | AATAAATCGGCGCTGGTTATCGGC |
| AGCAATTTCGCCGCCTCGAATAAC | |
| bicA | ATAGATGCCGTCCATCAGGT |
| CGACGTGAACATAGACGACA | |
| bopE | TCCTTCGCTTCGCTGAAGATCG |
| ATTCGGCCGGCAAGTCTACG | |
| bprC | GCGGAACAGCCGATAGAG |
| CATCGAGCAGCATCTTCATC | |
| virG | CCCCATAGCGTCTCCACCTC |
| GATCCGAAGCATCCCGAACTG | |
| tssA | GTCGACAAGGACGACTTCAA |
| GAGCGTGAGCTGGAGGTT | |
| hcp1 | CACATCCTCGCCTTCAA |
| AGATGATGGAAGAGTTCGAGA | |
| vgrG | CTCACGTCCGGCAACAAGTTC |
| TTGCCGCCCATCGACACC | |
| bimA | TACACGAACCAGCGCATCG |
| GGCGACGCCGGAATATGC |
Deletion of tssAB by allelic exchange.
The deletion mutant was first generated by replacement of tssAB sequence with an antibiotic-resistant cassette. Approximately 1-kb fragments upstream and downstream of tssAB were amplified from genomic DNA and cloned into pBAKA vector in E. coli strain E1345 (3). The FRT-flanked tet cassette from pFRTT1 (32) was inserted between the gene fragments to generate pKO-tssAB. The plasmid was electroporated into E. coli E1354 and conjugated into strain KHW. Homologous recombination was then selected for retention of tet marker on LB plates containing tetracycline (40 μg/ml). Transconjugants were then grown in the counterselection medium (M9 medium + 1% chlorophenylalanine) for 3 days to counterselect the pheS gene in the pBAKA plasmid backbone (3). Successful double-crossover clones were screened by the retention of tet marker and the loss of tssAB sequence by PCR. Deletion of the chromosomally integrated tet marker was achieved by introduction of FLP recombinase on a curable plasmid, pFLP-AB5, as described previously (3). PCR primers for plasmid construction are listed in Table 3.
Table 3.
List of PCR primers used for constructing plasmids
| Primer name | Sequence (5′–3′)a | PCR amplification |
|---|---|---|
| BprC for | AAGCTTACGTGTTGCGCGACATTC | Forward primer for bprC ORFb |
| BprC rev | AAGCTTCGGAATCGACATTCATCGT | Reverse primer for bprC ORF |
| VirA for | GGTACCTATTCGCCGACCTCTGAGAC | Forward primer for virA, virAG ORFs |
| VirA rev | AAGCTTCGACAGGTAATTGCAAAGCA | Reverse primer for virA ORF |
| VirG for | GGTACCGGCGCAGTACAGGAGAGGAC | Forward primer for virG ORF |
| VirG rev | AAGCTTCCGACGAGCAAATCCTTG | Reverse primer for virA, virAG ORFs |
| PtssA for | TTGAATTCATGGAATTTCCGTGCATAGG | Forward primer for tssA promoter and ORF |
| PtssA rev | TTGAATTCAAGCTTCCATGGTCGTCTGTTCC | Reverse primer for tssA promoter and ORF |
| Phcp1 for | GAATTCAAGCTCGAAAAGCGCTAGG | Forward primer for hcp1 promoter and ORF |
| Phcp1 rev | AAGCTTATCAGCCATTCGTCCAGTTT | Reverse primer for hcp1 promoter and ORF |
| TssAB-up for | GAATTCGATGCACATCGACAGGATC | Forward primer for upstream of tssAB |
| TssAB-up rev | GCGATAGACGATGTTGACC | Reverse primer for upstream of tssAB |
| TssAB-dn for | ATCCAGAACAAGATCCCGA | Forward primer for downstream of tssAB |
| TssAB-dn rev | AAGCTTTTCCAACGAGTATCCCGAA | Reverse primer for downstream of tssAB |
Restriction sites are underlined.
ORF, open reading frame.
RLM-RACE.
The transcriptional start (+1) sites of the promoters were mapped by RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) (4). The RLM-RACE was performed using the GeneRacer kit (Invitrogen) according to the manufacturer's instructions. The B. pseudomallei RNA was isolated as described above.
Enzyme assays.
β-Galactosidase assays were performed and values were calculated as previously described (36). E. coli DH5α strains containing both promoter-lacZ and Pbad regulator plasmids were grown in 1 ml of RPMI (10 μg/ml tetracycline, 15 μg/ml trimethoprim, 0.1% l-arabinose) for 24 h at 37°C in 24-well tissue culture plates. Two hundred microliters of each sample was used per assay. Results were reported as means with standard deviations of triplicate samples.
Immunofluorescence staining and confocal microcopy.
RAW 264.7 cells (2.5 × 105 cells/well) were seeded and grown overnight on 13-mm coverslips in 24-well plates. Cells were infected with various B. pseudomallei strains at an MOI of 10:1. After 2 h of infection, kanamycin (250 μg/ml) was added to the culture medium. At specific time points, cells were washed and fixed with 1% paraformaldehyde in phosphate-buffered saline (PBS) for 5 min. Prior to staining, infected cells were washed three times with PBS and subsequent staining was performed at room temperature at an antibody dilution of 1:100 in PBS containing 0.05% saponin (Sigma), 10% fetal bovine serum (Gibco), 10 mM HEPES (Sigma), and 10 mM glycine (Sigma). Infected cells were stained with B. pseudomallei lipopolysaccharide (LPS)-specific rabbit polyclonal antibodies (provided by Ganjana Lertmemongkolchai, Khon Kaen University, Thailand) for 45 min. Alexa Fluo 488 phalloidin (Invitrogen) was simultaneously added to these cells to stain for actin fibers. Cells were then washed several times and incubated with Alexa Fluor 405 goat anti-rabbit IgG (Invitrogen) for 45 min. Following extensive washing in PBS, coverslips were rinsed with water and mounted on glass slides using Prolong Gold antifade reagent (Invitrogen). Laser scanning confocal microscopy was performed with an LSM 710 Zeiss imaging system using a 63× oil objective (Carl Zeiss MicroImaging Inc.). Images were acquired using the LSM 710 Zen 2010 software (Carl Zeiss MicroImaging Inc.). To quantify actin tail formation, five representative fields were taken for each mutant and the percentage of bacteria forming actin tails was calculated by dividing the total number of bacteria counted by the number of bacteria forming tails × 100.
Giemsa staining of infected cells.
RAW 264.7 cells (2.5 × 105 cells/well) were infected and treated as described above for the immunostaining experiment. At 10 h postinfection, cells were washed once with PBS and fixed with 100% methanol (Sigma) for 1 min. Cells were then rinsed once with water and air dried before staining with Giemsa stain for 20 min. After staining, cells were washed once with water and air dried before they were examined for the presence of MNGCs under a light microscope.
Intracellular survival and multiplication of B. pseudomallei in cells.
RAW 264.7 cells (2.5 × 105 cells/well) were seeded and grown overnight in a 24-well plate. Cells were infected with an MOI of 0.1:1. At 1 h postinfection, infected cells were washed once with PBS and incubated in fresh culture medium containing kanamycin (250 μg/ml) to kill off extracellular bacteria. Infected cells were washed three times with PBS and lysed at 2 and 24 h postinfection with 0.1% (vol/vol) Triton X-100, and serial dilutions of the lysates were plated onto tryptic soy agar (TSA) agar and incubated at 37°C for 24 h. Colony counts were then used to calculate bacterial loads.
Animal infection.
Female, 6- to 8-week-old BALB/c mice were purchased from Laboratory Animals Centre (National University of Singapore). Infection of mice was carried out in an animal biosafety level 3 (BSL3) facility with a protocol approved by the institutional IACUC committees. Mice were infected with 50 to 100 CFU intranasally (19). Animals were then monitored daily for survival.
RESULTS
T6SS-1 expression inside macrophages is dependent on bacterial internalization.
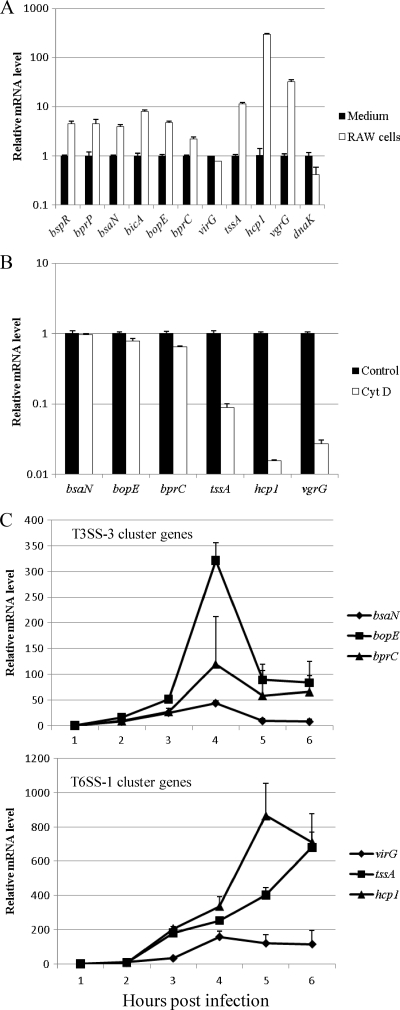
To investigate how T6SS-1 is regulated during infection, we used an in vitro infection model of macrophage-like cell line RAW 264.7. Cells were infected with B. pseudomallei wild-type strain KHW at an MOI of 100:1. Bacterial RNA was isolated from the infected monolayer at 5 h postinfection, and mRNA transcript levels were measured by real-time PCR. Compared to B. pseudomallei grown in culture medium (RPMI), transcript levels of T6SS-1 genes tssA (BPSS1496), hcp1 (BPSS1498), and vgrG (BPSS1503) were all significantly higher in bacteria associated with RAW 264.7 cells (Fig. 1A). We also examined the expression of T3SS-3 regulators and found that bspR, bprP, bsaN, and bprC were elevated in expression. In contrast, the expression levels of T6SS-1 response regulator virG and the housekeeping gene dnaK were similar in medium and in cells. The data suggest that expression of T6SS-1 is activated by host signals during infection. We have previously shown that cytochalasin D effectively blocks the phagocytic uptake of B. pseudomallei by RAW 264.7 cells (13). To determine whether the internalization of bacteria by phagocytosis was necessary for the activation of T6SS-1, RAW 264.7 cells were treated with cytochalasin D for 2 h prior to infection with B. pseudomallei. The relative transcript levels of T3SS-3 genes bsaN, bopE, and bprC were similar (<2-fold difference) between infected macrophages treated with cytochalasin D and those without, but the transcript levels of T6SS-1 genes tssA, hcp1, and vgrG were more than 10 times lower in infected macrophages treated with cytochalasin D than in those that were untreated (Fig. 1B). The data confirm that the activation of T3SS-3 requires only a bacterium-macrophage contact but the activation of T6SS-1 requires internalization of bacteria. After infection, expression of both T3SS-3 and T6SS-1 genes increased steadily. The expression of T3SS-3 cluster genes and virG peaked at 4 h postinfection, whereas the expression of tssA and hcp1 did not peak until 5 h after infection (Fig. 1C).
Fig. 1.
Expression of T3SS-3 and T6SS-1 genes in medium and during infection. (A) Transcript levels of T3SS-3 and T6SS-1 genes of B. pseudomallei during infection of RAW 264.7 cells (RAW cells) were measured by real-time PCR and normalized to bacteria grown in RPMI 1640 medium (Medium). The dnaK gene is used as a house-keeping gene control. (B) Transcript levels of T3SS-3 and T6SS-1 genes of B. pseudomallei during infection of cytochalasin D-treated RAW 264.7 cells (CytD) were measured by real-time PCR and normalized to bacteria from infection of untreated RAW 264.7 cells (Control). (C) Transcript levels of T3SS-3 cluster and T6SS-1 cluster genes of B. pseudomallei during infection of RAW 264.7 cells at each time point were measured by real-time PCR and normalized to those at the 1-h time point.
T6SS-1 expression inside macrophages is absolutely dependent on VirAG and less on BprC.
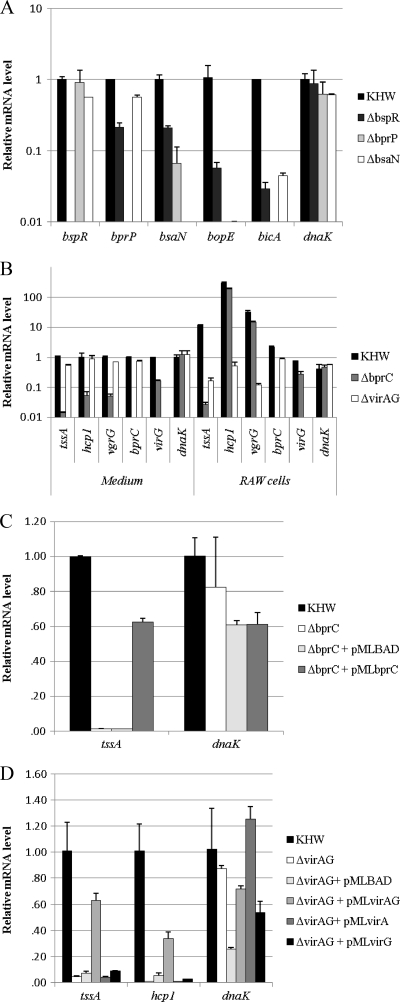
The regulatory cascade controlling the expression of T3SS-3 initiates from BspR and includes downstream transcriptional regulators BprP and BsaN (32). BsaN is required for the expression of T3SS-3 genes and virAG, a putative two-component system in the T6SS-1 cluster. In bacteria grown in culture medium, expression of T6SS-1 was controlled by BprC, an AraC regulator inside the T3SS-3 cluster, and was not affected by deletion of virAG (32). To investigate the role of these regulators in the activation of T3SS-3 and T6SS-1 during infection, we compared the gene expression levels of B. pseudomallei wild-type and ΔbspR, ΔbprP, ΔbsaN, ΔbprC, and ΔvirAG regulator mutant strains during infection of RAW 264.7 cells. Similar to that observed in the culture medium, expression of bsaN and T3SS-3 genes was dependent on BspR, BprP, and BsaN (Fig. 2A). However, the expression profiles of T6SS-1 genes in ΔbprC and ΔvirAG mutants were dramatically different inside RAW 264.7 cells from those in culture medium. When cultured in medium, the expression of T6SS-1 genes tssA, hcp1, and vgrG in the ΔbprC strain was more than 10 times lower than that in the wild-type strain whereas expression levels in the ΔvirAG mutant were comparable to those in the wild-type strain (Fig. 2B). But in RAW 264.7 cells, only tssA was not expressed whereas the other two downstream genes (hcp1 and vgrG) were still highly expressed in the ΔbprC mutant (Fig. 2B). The expression of tssA in the ΔbprC mutant could be complemented by providing bprC in trans on a plasmid (pMLbprC) (Fig. 2C). In the ΔvirAG mutant, the T6SS-1 genes tssA, hcp1, and vgrG were not expressed inside RAW 264.7 cells (Fig. 2B). The expression of the T6SS-1 genes in the virAG mutant could be restored by virAG on a plasmid (pMLvirAG) but not by an individual gene (pMLvirA or pMLvirG) (Fig. 2D). These results demonstrate that the intracellular activation of T6SS-1 is completely dependent upon the two-component system VirAG, which requires intracellular signals for its activity. BprC is needed only for activation of a subset of T6SS-1 genes (tssAB).
Fig. 2.
Expression of T3SS-3 and T6SS-1 genes in B. pseudomallei wild-type strain (KHW) and regulator mutants. (A) Transcript levels of regulators and T3SS-3 genes in ΔbspR, ΔbprP, and ΔbsaN strains during infection of RAW 264.7 cells were measured by real-time PCR and normalized to those of KHW. (B) Transcript levels of regulators and T6SS-1 genes in KHW, ΔbprC, and ΔvirAG strains in RPMI medium (Medium) and during infection of RAW 264.7 cells (RAW cells). (C) The expression of tssA in the ΔbprC mutant could be complemented by providing bprC in trans on a plasmid (pMLbprC). (D) The expression of the T6SS-1 genes in the virAG mutant could be restored by virAG on a plasmid (pMLvirAG) but not by an individual gene (pMLvirA or pMLvirG).
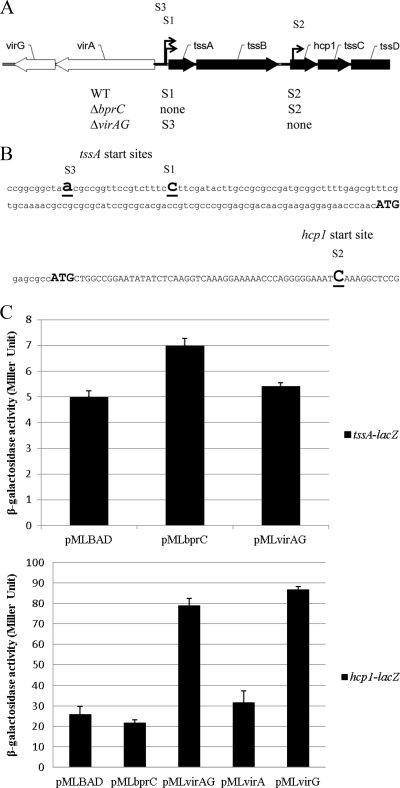
VirAG directly drives transcription of the hcp1 operon.
T6SS-1 cluster genes (BPSS1496 to BPSS1511) of B. pseudomallei are all coded on the same strand on chromosome 2 and organized in an operon-like structure (tss operon). Our data show that the first gene, tssA, and the downstream genes (hcp1 and vgrG) were differentially regulated. In order to identify promoters of this tss operon, the transcriptional start sites of the promoters (+1) were mapped by RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) in the wild-type (KHW) and ΔbprC and ΔvirAG strains. There were two apparent transcriptional start sites (S1 and S2) in the wild-type strain (Fig. 3A). One was upstream of tssA, and the other initiated inside the downstream gene hcp1. In the ΔbprC mutant, there was no transcript from the tssA promoter as expected, but the hcp1 promoter was still functional; conversely in the ΔvirAG mutant, the hcp1 promoter was not active, but tssA was still transcribed but initiated at another site (S3).
Fig. 3.
Transcriptional analysis of T6SS-1. (A) Transcriptional organization of T6SS-1 genes in KHW, ΔbprC, and ΔvirAG strains. S1, S2, and S3 indicate start sites. Arrows indicate transcription direction. WT, wild type. (B) Start sites of transcription. Bold and underlined letters indicate start site (+1). Bold ATGs indicate initial codon of proteins. (C) Effect of bprC and virAG on the expression of tssA-lacZ and hcp1-lacZ fusions in E. coli. Values given are means ± standard deviations.
To determine whether these regulators directly interact with the promoters, we used an E. coli system comprising two plasmids. The regulators were cloned downstream of the Pbad promoter of pMLBAD (18), which is inducible by arabinose. The promoters were cloned upstream of the promoterless lacZ gene in pRW50 (21). The ability of BprC or VirAG to directly activate tssA or hcp1 expression was examined by providing bprC or virAG in trans and measuring β-galactosidase activities arising from the expression of tssA-lacZ or hcp1-lacZ. Neither bprC nor virAG could activate tssA-lacZ (Fig. 3B). However, expression of hcp1-lacZ was dramatically induced by virAG or virG, but it was not affected by virA or bprC (Fig. 3C). This shows that VirG is acting directly on the hcp1 promoter.
T6SS-1 is important for the optimal growth of B. pseudomallei in RAW 264.7 cells.
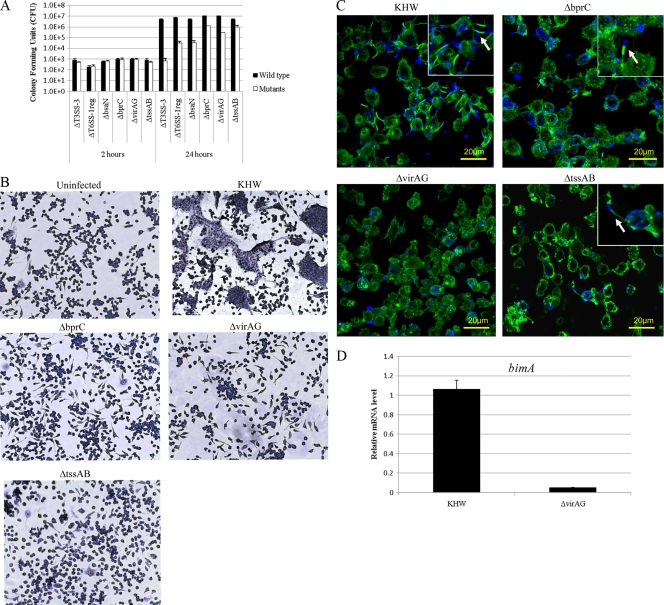
Previous studies showed that T3SS-3 contributes to early endosomal escape and replication within RAW 264.7 cells (7) and J774A.1 cells (22). Thus, we assessed the relative importance of T6SS-1 compared to T3SS-3 for survival and replication of B. pseudomallei inside RAW 264.7 cells. TssA and TssB, which are controlled primarily by BprC in infected macrophages, exhibited high homology with VipA/VipB. In Vibrio cholerae, VipA and VipB form a complex that has been shown to be essential for type VI protein secretion (5). To examine the importance of these genes in T6SS-1 function, we created a ΔtssAB mutant. We also included a T3SS-3 locus deletion mutant (ΔT3SS-3) and a mutant that we named ΔT6SS-1reg for comparison. ΔT6SS-1reg has bimE or tssA as named by Shalom et al (28) (BPSS1493), virAG (BPSS1494 to BPSS1495), and the T6SS-1 locus (BPSS1496 to BPSS1511) deleted. The initial internalizations of all the mutants were at levels comparable to those of the wild type at 2 h postinfection (Fig. 4A). At 24 h postinfection, both ΔbprC mutant- and ΔtssAB mutant-infected cells showed a 10-fold decrease in bacterial numbers compared to wild-type-infected cells (Fig. 4A). This shows that the BprC-controlled TssA and TssB are components of the T6SS-1 required for exerting its full function. The ΔvirAG mutant-infected cells showed a 50-fold decrease, whereas the ΔbsaN mutant-infected cells showed a greater-than-100-fold decrease in bacterial loads compared to wild-type-infected cells (Fig. 4A). The ΔT6SS-1reg mutant-infected cells showed an intermediate phenotype between the ΔbsaN and ΔvirAG mutants. The more severe phenotype of the ΔvirAG mutant than of bprC and ΔtssAB mutants indicates the importance of T6SS-1 and perhaps some of the bim genes for the optimal growth of B. pseudomallei inside host cells, since virAG controls more than just the T6SS-1 locus. ΔT6SS-1reg resembles the ΔvirAG mutant but could be a little more severe than the latter because the T6SS-1 locus is completely deleted. The most severe phenotypes exhibited by the ΔbsaN and ΔT3SS-3 mutants would be due to the disruption of T3SS-3 in these mutants. Taken together, our results show that besides T3SS-3, T6SS-1 is essential for intracellular growth. Possibly, the two secretion systems cooperate to confer optimal growth and survival of B. pseudomallei inside host cells.
Fig. 4.
Intracellular replication, MNGC formation, and actin tail formation of T6SS-1 mutants. (A) Intracellular replication of B. pseudomallei in RAW 264.7 cells. Cells were infected at an MOI of 0.1:1. Intracellular bacterial loads were quantified at 2 and 24 h postinfection by plate counting. The values are the means ± standard deviations of biological triplicates. (B) Light micrographs of Giemsa-stained RAW 264.7 cells infected with B. pseudomallei. The monolayers were visualized at 10 h postinfection and were infected at an MOI of 10:1. (C) Confocal micrographs of actin tail polymerization by various B. pseudomallei strains. Cells were infected with KHW and ΔbprC, ΔvirAG, and ΔtssAB mutants for 8 h at an MOI of 10:1. Bacteria were stained blue with rabbit anti-B. pseudomallei LPS and anti-rabbit IgG Alexa Fluor 405; actin was stained green with Alexa Fluor 488 phalloidin. The arrow indicates actin tail formation at one pole of the bacteria. (D) The expression levels of the bimA gene in the ΔvirAG mutant measured by real-time PCR.
ΔbprC, ΔvirAG, and ΔtssAB mutants are unable to induce MNGC formation in RAW 264.7 cells.
B. pseudomallei induces MNGC formation following infection of RAW 264.7 cells, and this has been shown to be a phenotype of T6SS-1, as the hcp1 deletion mutant could not induce MNGC formation (5). We tested our various T6SS-1 mutants for their ability to form MNGCs in RAW 264.7 cells. In contrast to the wild-type-infected cells which formed MNGCs at 10 h postinfection, ΔbprC, ΔvirAG, and ΔtssAB mutants were unable to induce MNGC formation (Fig. 4B). This defect is also not observed at 24 h postinfection. Our result is consistent with a recent finding in B. mallei that one of the T6SS genes, tssE, is required for MNGC formation (27). These data demonstrate that MNGC formation is also dependent on the function of tssAB.
The ΔvirAG mutant demonstrates a greater defect in actin-based motility than does the tssAB mutant in RAW 264.7 cells.
B. pseudomallei could stimulate actin polymerization within the cytoplasm of host cells, resulting in the formation of actin tail at one end of the bacterial pole (30). We assessed the abilities of the various T6SS-1 mutants to form actin tails in RAW 264.7 cells. Cells infected with wild-type, ΔvirAG, ΔbprC, and ΔtssAB bacteria were fixed, stained for actin tail, and examined by fluorescence microscopy at 8 h postinfection. Unlike the wild type, ΔvirAG did not exhibit actin tail polymerization (Fig. 4C). For the ΔbprC and ΔtssAB mutants, actin tail formation was present in only 1% of total bacteria, compared to 10% of total bacteria observed in the wild-type-infected cells (Fig. 4C). This reduction is not due to a difference in intracellular bacterial numbers, because at 8 h postinfection, the numbers of bacteria were comparable between the mutant- and wild-type-bacterium-infected cells (data not shown). Our findings indicate that tssAB is not absolutely required for bacterial actin-based motility but greatly facilitates the process. The difference between the ΔvirAG and ΔtssAB mutants in the ability to form actin tail could be due to the requirement of virAG for bimA expression (Fig. 4D).
T6SS-1 regulators are required for B. pseudomallei virulence in mice.
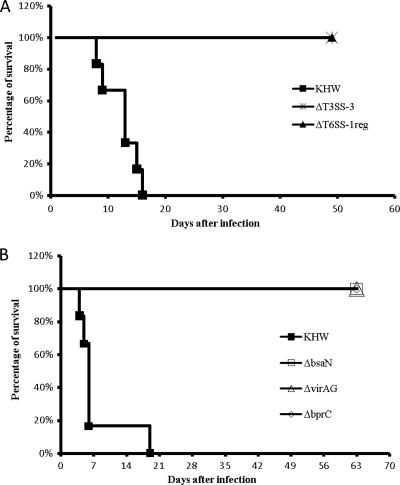
To assess the role of T6SS-1 regulators in B. pseudomallei virulence, we used an intranasal infection model in BALB/c mice (19) and compared the infection of these mutants with the ΔT3SS-3 and ΔT6SS-1reg mutants. Three groups each with six BALB/c mice were infected with ∼100 CFU of B. pseudomallei KHW, ΔT3SS-3, and ΔT6SS-1reg strains intranasally. Mice were monitored daily for 7 weeks. As expected, all mice infected with wild-type bacteria died within 3 weeks and all mice infected with ΔT3SS-3 survived when the experiment ended. The ΔT3SS-3 mutant is essentially a T3SS-3 and T6SS-1 double mutant. All mice infected with ΔT6SS-1reg also survived the duration of the experiment (Fig. 5A). In a separate experiment, mice infected with ∼50 CFU of ΔbsaN, ΔvirAG, ΔbprC mutant, or wild-type KHW were monitored daily for 9 weeks. All mutant-infected mice appeared to be healthy and survived up to 9 weeks, when they were sacrificed (Fig. 5B). Thus, the regulators bsaN, bprC, and virAG are essential for B. pseudomallei's virulence in mice.
Fig. 5.
Survival curves of mice infected with wild-type B. pseudomallei (KHW) or the ΔT3SS-3, ΔT6SS-1reg, ΔbsaN, ΔvirAG, or ΔbprC mutant. (A) Three groups of BALB/c mice with six mice per group were infected with ∼100 CFU of the T3SS-3 or T6SS-1reg mutant or the wild-type strain by the intranasal route. Mice were monitored daily for 7 weeks. (B) Four groups of BALB/c mice with six mice per group were infected with ∼50 CFU of the ΔbsaN, ΔvirAG, or ΔbprC mutant strain or KHW. Mice were monitored daily for 9 weeks.
DISCUSSION
T3SS and T6SS are critical virulence determinants in several Gram-negative pathogens. B. pseudomallei harbors three T3SSs and six T6SSs, and its T3SS-3 and T6SS-1 clusters have been implicated in bacterial virulence in mammalian hosts (6, 23, 31, 34). T3SS-3 and T6SS-1 are activated by host signals, as expression levels of both secretion systems are significantly elevated upon infection of murine RAW 264.7 macrophages. Cytochalasin D, an inhibitor of phagocytosis, blocks the expression of T6SS-1 but does not affect T3SS-3 expression. This is consistent with an earlier report documenting the induction of the T6SS-1 locus upon invasion of macrophages (28). Our results indicate that, whereas T3SS-3 is turned on after the bacteria encounter host cells, T6SS-1 is activated only after the bacteria enter the host cells. However, expression levels of both T3SS-3 and T6SS-1 genes increase rapidly upon infection, suggesting that these secretion systems are necessary for bacterial survival and replication immediately after infection. Nevertheless, the expression of T3SS-3 is a prerequisite for T6SS-1 expression because expression of T6SS-1 is dependent upon bsaN, which is located inside the T3SS-3 cluster.
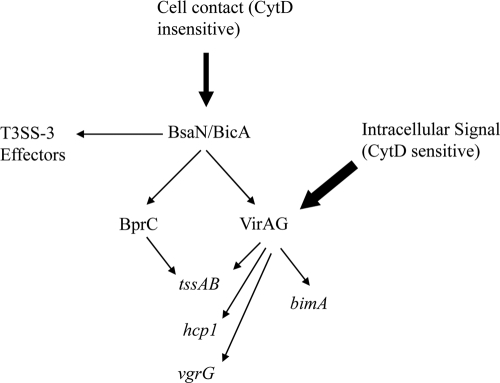
We have recently discovered a regulatory cascade that coordinately controls the expression of T3SS-3 and T6SS-1 (32). BsaN is a central regulator located within T3SS-3 for the expression of T3SS-3 effectors as well as another AraC regulator, BprC. BprC in turn regulates the expression of T6SS-1 genes. BsaN is also required for expression of the two-component system genes virAG within T6SS-1, but the deletion of virAG had no effect on T6SS-1 genes when bacteria were grown in medium. However, we have demonstrated in this study that the intracellular activation of T6SS-1 is completely dependent on VirAG, whereas BprC is required only for a subset of genes (tssAB) (Fig. 6). Previous studies have relied completely on the overexpression of virAG to demonstrate the transcriptional activation of the B. mallei T6SS cluster homologous to B. pseudomallei T6SS-1 (27) as well as the hcp1 gene in B. pseudomallei (6). Our data provide the physiological context for the virAG control of T6SS-1 expression during infection. It is very likely that VirAG exerts its effects on T6SS-1 genes by sensing host intracellular signals, somewhat akin to Salmonella's two-component system PhoPQ, which has been shown to activate T3SS SPI2 genes by sensing antimicrobial peptides (2) and acidic pH (24) inside host phagosomes. It is possible that signals are relayed through VirA, which in turn causes the activation and binding of VirG to the relevant promoters to initiate transcription. We found that the T6SS-1 locus has multiple transcriptional start sites, with one of them inside the predicted open reading frame of the hcp1 gene. There are no apparent sigma70- or sigma54-dependent promoters located upstream of these start sites. Thus, it seems that when bacteria are in culture medium, transcription originates only upstream of tssA and reads into the downstream gene cluster. Once bacteria are inside host cells, transcription originates both from upstream of tssA (S1) and from inside the hcp1 coding region (S2). Although there is a second start codon (ATG) after the S2 transcriptional start site, there are no apparent ribosome binding sites. There is evidence that the translation of Hcp1 initiates from the annotated start codon, resulting in a protein of around 18 kDa (6). Thus, it is likely that the promoter is used to enhance the expression of other genes downstream of hcp1. Sensing and activation of VirAG could lead to the use of this promoter to upregulate genes downstream of hcp1 to activate T6SS-1 function. As tssAB expression inside host cells is dependent on both virAG and bprC, one could speculate that virAG and bprC cooperate to allow tssAB transcription to initiate from a more efficient S1 site rather than a suboptimal S3 site.
Fig. 6.
Model of regulation of T6SS-1. Bold arrows indicate the input of environmental signals. Normal arrows point to the targets of regulators. CytD, cytochalasin D.
For an intracellular pathogen, being able to survive and replicate within the host cells is critical. T3SS-3 was previously shown to be involved with intracellular survival and replication of B. pseudomallei (31). We showed that T6SS-1 is also important for intracellular growth, as the ΔvirAG regulator mutant exhibited significant growth defects whereas the ΔbprC or ΔtssAB mutant demonstrated a less severe defect. The importance of T6SS-1 in intracellular growth is also shown in B. mallei (8). Likewise, in other pathogens such as Edwardsiella tarda and Francisella tularensis, T6SS has been shown to influence the intracellular growth within phagocytic cells (10, 25). However, our data differ from those of Shalom et al., who demonstrated that the T6SS-1 mutant of B. pseudomallei did not exhibit defective intracellular growth (28). Burtnick et al. (6) showed a delayed growth phenotype for the B. pseudomallei hcp1 mutant, as the numbers of intracellular wild-type and hcp1 mutant bacteria were different at 12 h but similar by 18 h after infection. The authors attributed the similarity at 18 h postinfection to the lysis of wild-type-infected cells and release of bacteria into the antibiotic-containing extracellular medium. In our study, we used a lower MOI of 0.1 in which cell death was minimal for the wild-type-infected cells at 24 h postinfection (<5% cytotoxicity) and still observed a significant growth defect. Thus, both T3SS-3 and T6SS-1 are essential for optimal intracellular growth inside macrophages. However, further studies will be required to determine the specific role and mechanism of each system in intracellular replication.
A hallmark of B. pseudomallei infection in phagocytic cells is the induction of cell fusion, which results in MNGC formation (16). This phenomenon had been observed in the tissues of melioidosis patients (35), suggesting its relevance in vivo. In addition, B. pseudomallei is able to polymerize host cell actin to facilitate its intracellular movement and intercellular spread (16). This is modulated by BimA, an autotransported protein that is localized at one end of the bacterial pole to induce actin polymerization (30). By doing so, both events could contribute to dissemination to surrounding host cells and lead to persistence within the host. Previously, the T3SS-3 ΔbsaZ mutant has been shown to be able to exhibit late-endosomal escape and induce MNGC and actin tail formation at late infection time points such as 18 h postinfection in RAW 264.7 cells (7). This indicates that T3SS is involved in early endosomal escape but not required to induce MNGC and actin tail formation. ΔvirAG, ΔbprC, and ΔtssAB were unable to induce MNGC formation even at 24 h postinfection. In addition, ΔvirAG was unable to induce actin polymerization, and this correlates with our results showing that bimA expression is under VirAG control. As the T6SS-1 mutant of B. mallei was shown to be able to undergo vacuolar escape as well as the wild-type bacteria did (8), the defects seen in MNGC and actin tail formation were not due to confinement in the vacuolar vesicle. On the other hand, the ΔbprC and ΔtssAB mutants could still induce actin tail formation but at a greatly reduced rate compared to that of the wild-type bacteria. Overall, we show that although both MNGC formation and actin-based motility phenotypes are attributed to T6SS-1, they are likely not dependent on each other, as tssAB is required for MNGC formation but not absolutely required for actin-based motility.
Since the ΔtssAB mutant exhibits phenotypes similar to those of the ΔvirAG mutant, which is deficient in T6SS-1 expression, it demonstrates that TssA and TssB are essential components of the T6SS-1. This is expected because many T6SSs of other pathogens encode TssA/TssB homologs which have been demonstrated to be essential for T6SS secretion activity. In V. cholerae, VipA and VipB form a complex of tubules and serve as substrates for ClpV, a member of the AAA+ protein family, whose members bind and hydrolyze ATP, to energize T6SS to secrete Hcp and VgrG2 (5). In addition, BcsKc and BcsLb of Burkholderia cenocepacia are also homologs of TssA and TssB and were postulated to play a role similar to that of VipA/B (1). However, the tssB homolog in B. mallei, which is the species most closely related to B. pseudomallei, is not required for hcp1 secretion (27). Our ΔtssAB mutant demonstrated a less severe phenotype than did the ΔvirAG mutant in terms of intracellular replication and actin tail formation. This likely means that virAG controls additional genes that are not directly related to T6SS secretory function but could contribute to these two phenotypes. For example, virAG controls bimA, which contributes to actin tail motility and may also affect intracellular replication. Nevertheless, tssAB is still essential for T6SS-1 function because bprC, crucial in regulating TssA and TssB, is absolutely required for bacterial virulence in mice. So, our animal experiments show that the regulation by bprC and virAG of bacterial virulence in vitro is also similarly reflected in vivo.
In summary, we have shown that T6SS-1 gene expression increases significantly and is dependent on BprC and VirAG inside host cells (Fig. 6). Most of the T6SS-1 genes are controlled by VirAG but not BprC. Only the tssAB operon requires both regulators and is essential for T6SS-1 function. Although T3SS-3 is important in facilitating early phagosomal escape into the cytosol, evasion from autophagy (12), and bacterial replication in the cytosol, T6SS-1 likely becomes critical during the time that the bacterium is transiting from the phagosome to the cytosol. VirAG could be sensing phagosomal signals to activate T6SS-1 so that once the bacterium gets into the cytosol, T6SS-1 helps in ensuring its optimal survival and replication, as well as spreading to other cells through MNGC formation and actin tail motility. Cell fusion and cell-to-cell spreading could provide an optimal intracellular niche for the bacteria to thrive and protect them from being killed by the antimicrobial substances in the extracellular milieu. The cooperation of T3SS-3 and T6SS-1 shows some resemblance to the SPI1 and SPI2 T3SS in Salmonella, where the former is responsible for invasion and the latter for survival inside the cells and for bacterial spread (17), although the regulation and mechanism of action are different in the two species. The importance and relevance of T6SS-1 regulation by VirAG and BprC in infected macrophages are also reproduced in mouse infection models, demonstrating the absolute reliance of T6SS-1 function on the regulation by VirAG and BprC in mammalian hosts.
ACKNOWLEDGMENTS
We thank T. T. Hoang (University of Hawaii at Manoa) for plasmids pBAKA and pFLP-AB5, M. A. Valvano (University of Western Ontario) for pMLBAD plasmid, S. J. Busby (University of Birmingham) for pRW50 plasmid, and G. Lertmemongkolchai (Khon Kaen University, Thailand) for B. pseudomallei anti-LPS antibody.
This work is supported by grant T208A3105 from the Ministry of Education and grant NMRC/1221/2009 from the National Medical Research Council.
Footnotes
Published ahead of print on 13 June 2011.
REFERENCES
- 1. Aubert D., MacDonald D. K., Valvano M. A. 2010. BcsKC is an essential protein for the type VI secretion system activity in Burkholderia cenocepacia that forms an outer membrane complex with BcsLB. J. Biol. Chem. 285:35988–35998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bader M. W., et al. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472 [DOI] [PubMed] [Google Scholar]
- 3. Barrett A. R., et al. 2008. Genetic tools for allelic replacement in Burkholderia species. Appl. Environ. Microbiol. 74:4498–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bensing B. A., Meyer B. J., Dunny G. M. 1996. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc. Natl. Acad. Sci. U. S. A. 93:7794–7799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonemann G., Pietrosiuk A., Diemand A., Zentgraf H., Mogk A. 2009. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28:315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burtnick M. N., et al. 2011. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79:1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burtnick M. N., et al. 2008. Burkholderia pseudomallei type III secretion system mutants exhibit delayed vacuolar escape phenotypes in RAW 264.7 murine macrophages. Infect. Immun. 76:2991–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burtnick M. N., DeShazer D., Nair V., Gherardini F. C., Brett P. J. 2010. Burkholderia mallei cluster 1 type VI secretion mutants exhibit growth and actin polymerization defects in RAW 264.7 murine macrophages. Infect. Immun. 78:88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cascales E. 2008. The type VI secretion toolkit. EMBO Rep. 9:735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chong A., et al. 2008. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect. Immun. 76:5488–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coburn B., Sekirov I., Finlay B. B. 2007. Type III secretion systems and disease. Clin. Microbiol. Rev. 20:535–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cullinane M., et al. 2008. Stimulation of autophagy suppresses the intracellular survival of Burkholderia pseudomallei in mammalian cell lines. Autophagy 4:744–753 [DOI] [PubMed] [Google Scholar]
- 13. Hii C. S., et al. 2008. Interleukin-8 induction by Burkholderia pseudomallei can occur without Toll-like receptor signaling but requires a functional type III secretion system. J. Infect. Dis. 197:1537–1547 [DOI] [PubMed] [Google Scholar]
- 14. Hood R. D., et al. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jani A. J., Cotter P. A. 2010. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe 8:2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kespichayawattana W., Rattanachetkul S., Wanun T., Utaisincharoen P., Sirisinha S. 2000. Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infect. Immun. 68:5377–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knodler L. A., Steele-Mortimer O. 2003. Taking possession: biogenesis of the Salmonella-containing vacuole. Traffic 4:587–599 [DOI] [PubMed] [Google Scholar]
- 18. Lefebre M. D., Valvano M. A. 2002. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 68:5956–5964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu B., Koo G. C., Yap E. H., Chua K. L., Gan Y. H. 2002. Model of differential susceptibility to mucosal Burkholderia pseudomallei infection. Infect. Immun. 70:504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 21. Lodge J., Fear J., Busby S., Gunasekaran P., Kamini N. R. 1992. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol. Lett. 74:271–276 [DOI] [PubMed] [Google Scholar]
- 22. Muangsombut V., et al. 2008. Inactivation of Burkholderia pseudomallei bsaQ results in decreased invasion efficiency and delayed escape of bacteria from endocytic vesicles. Arch. Microbiol. 190:623–631 [DOI] [PubMed] [Google Scholar]
- 23. Pilatz S., et al. 2006. Identification of Burkholderia pseudomallei genes required for the intracellular life cycle and in vivo virulence. Infect. Immun. 74:3576–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Prost L. R., et al. 2007. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol. Cell 26:165–174 [DOI] [PubMed] [Google Scholar]
- 25. Rao P. S., Yamada Y., Tan Y. P., Leung K. Y. 2004. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol. Microbiol. 53:573–586 [DOI] [PubMed] [Google Scholar]
- 26. Sambrook J., Maniatis T., Fritsch E. F. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 27. Schell M. A., et al. 2007. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 64:1466–1485 [DOI] [PubMed] [Google Scholar]
- 28. Shalom G., Shaw J. G., Thomas M. S. 2007. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153:2689–2699 [DOI] [PubMed] [Google Scholar]
- 29. Simon R., Priefer U., Pühler A. 1983. A broad range mobilization system for in vitro genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology (NY) 1:784–791 [Google Scholar]
- 30. Stevens J. M., et al. 2005. Actin-binding proteins from Burkholderia mallei and Burkholderia thailandensis can functionally compensate for the actin-based motility defect of a Burkholderia pseudomallei bimA mutant. J. Bacteriol. 187:7857–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stevens M. P., et al. 2003. A Burkholderia pseudomallei type III secreted protein, BopE, facilitates bacterial invasion of epithelial cells and exhibits guanine nucleotide exchange factor activity. J. Bacteriol. 185:4992–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun G. W., et al. 2010. Identification of a regulatory cascade controlling type III secretion system 3 gene expression in Burkholderia pseudomallei. Mol. Microbiol. 76:677–689 [DOI] [PubMed] [Google Scholar]
- 33. Tan K. S., et al. 2010. Suppression of host innate immune response by Burkholderia pseudomallei through the virulence factor TssM. J. Immunol. 184:5160–5171 [DOI] [PubMed] [Google Scholar]
- 34. Warawa J., Woods D. E. 2005. Type III secretion system cluster 3 is required for maximal virulence of Burkholderia pseudomallei in a hamster infection model. FEMS Microbiol. Lett. 242:101–108 [DOI] [PubMed] [Google Scholar]
- 35. Wong K. T., Puthucheary S. D., Vadivelu J. 1995. The histopathology of human melioidosis. Histopathology 26:51–55 [DOI] [PubMed] [Google Scholar]
- 36. Zhang X., Bremer H. 1995. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J. Biol. Chem. 270:11181–11189 [DOI] [PubMed] [Google Scholar]