Abstract
CRL4Cdt2 is a cullin-based E3 ubiquitin ligase that promotes the ubiquitin-dependent proteolysis of various substrates implicated in the control of cell cycle and various DNA metabolic processes such as DNA replication and repair. Substrates for CRL4Cdt2 E3 ubiquitin ligase include the replication licensing factor Cdt1 and the cyclin-dependent kinase (Cdk) inhibitor p21. Inhibition of this E3 ligase leads to serious abnormalities of the cell cycle and cell death. The ubiquitin-conjugating enzyme (UBC) involved in this important pathway, however, remains unknown. By a proteomic analysis of Cdt2-associated proteins and an RNA interference-based screening approach, we show that CRL4Cdt2 utilizes two different UBCs to target different substrates. UBCH8, a member of the UBE2E family of UBCs, ubiquitylates and promotes the degradation of p21, both during the normal cell cycle and in UV-irradiated cells. Importantly, depletion of UBCH8 by small interfering RNA (siRNA) increases p21 protein level, delays entry into S phase of the cell cycle, and suppresses the DNA damage response after UV irradiation. On the other hand, members of the UBE2G family of UBCs (UBE2G1 and UBE2G2) cooperate with CRL4Cdt2 to polyubiquitylate and degrade Cdt1 postradiation, an activity that is critical for preventing origin licensing in DNA-damaged cells. Finally, we show that UBCH8, but not UBE2G1 or UBE2G2, is required for CRL4Cdt2-mediated ubiquitylation and degradation of the histone H4 lysine 20 monomethyltransferase Set8, a previously identified CRL4Cdt2 substrate, as well as for CRL4Cdt2-dependent monoubiquitylation of PCNA in unstressed cells. These findings identify the UBCs required for the activity of CRL4Cdt2 on multiple substrates and demonstrate that different UBCs are involved in the selective ubiquitylation of different substrates by the same E3 complex.
INTRODUCTION
Protein ubiquitylation is an essential posttranslational modification that plays a significant role in all aspects of cell physiology. The covalent attachment of the small 76-amino-acid ubiquitin moiety to target proteins and the subsequent assembly of polyubiquitin chains frequently trigger the targeted proteolysis of the substrate proteins via the 26S proteasome (11). Protein polyubiquitylation involves three distinct and consecutive enzymatic steps: (i) ATP-dependent activation of ubiquitin by an E1 ubiquitin-activating enzyme, (ii) the transfer of activated ubiquitin to an E2 ubiquitin-conjugating enzyme via a thioester intermediate, and (iii) the covalent attachment of ubiquitin to a specific lysine residue of the target protein substrate by an E3 ubiquitin ligase (13, 32). A long-standing question is whether a given E3 cooperates with different E2s to target different substrates.
Cullin ring-based E3 ubiquitin ligases (CRLs) constitute one of the largest families of E3 ubiquitin ligases and are important for cell cycle regulation, differentiation, transcription, apoptosis, and tumorigenesis (16). Among these, the CRL4Cdt2 E3 ubiquitin ligase is emerging as a master regulator of cell cycle progression and genomic stability (1). CRL4Cdt2 is comprised of a scaffold protein subunit (Cul4A/B), an adapter protein (DDB1 [DNA damage-specific protein 1]), a ring finger protein (Rbx1/Roc1), and the substrate recognition DCAF (DDB1 and Cul4-associated factor) subunit Cdt2. We and others have recently demonstrated that CRL4Cdt2 promotes the ubiquitylation and subsequent degradation of the replication factor Cdt1 (6, 14, 15, 17–19, 21, 28, 33, 34, 36), the Cdk inhibitor p21 (4, 22), and Set8 (3, 10, 29, 40) in an unperturbed cell cycle and in response to UV irradiation. Additional substrates for CRL4Cdt2 include PCNA (41), Xic1 (20), the Schizosaccharomyces pombe ribonucleotide reductase inhibitor Spd1 (25), the Caenorhabditis elegans polymerase eta (polη) (21), the Drosophila melanogaster transcription factor E2F1 (38), and possibly the tumor suppressor p53 (7).
Despite the significance of the targeted proteolysis of these proteins via CRL4Cdt2, the ubiquitin-conjugating enzyme (UBC) that cooperates with CRL4Cdt2 to promote substrate polyubiquitylation remains unidentified. UBCH6 (also known as UBE2E1), UBCH8 (UBE2E2), and UBCH9 (UBE2E3), are members of the UBE2E group of UBCs and have recently been shown to stably associate with the DDD complex comprised of DDB1, DET1 (de-etiolated 1), and DDA1 (Det1, DDB1-associated 1) (31). Here we report that UBCH8 and members of the UBE2G family, UBE2G1 and UBE2G2, cooperate with CRL4Cdt2 in promoting the polyubiquitylation and subsequent degradation of p21 and Cdt1, respectively. This unexpected finding suggests that E2s play an important role not only in promoting substrate ubiquitylation but also in dictating the specificity for substrate ubiquitylation, a function that has been attributed exclusively to the E3 ubiquitin ligase.
MATERIALS AND METHODS
Cell culture and transfections.
HCT116p53−/− cells (9) (a generous gift from Fred Bunz) were cultured in McCoy's 5A medium containing 10% fetal bovine serum (FBS) in a 37°C incubator with 5% CO2. Transfections were performed using Lipofectamine 2000 (Invitrogen) reagent for plasmid and RNAiMAX (Invitrogen) for small interfering RNA (siRNA) transfection according to the manufacturer's instructions. UV irradiation (20 J/m2) was carried out 72 h after siRNA transfections using UV Stratalinker (Stratagene). After irradiation, the cells were incubated in fresh medium for the indicated times, washed with phosphate-buffered saline (PBS), and lysed directly with SDS sample buffer.
Plasmids and antibodies.
UBCH8 (GI:2274932), UBE2G1 (GI:13489085), and UBE2G2 (GI:321117276) cDNAs were PCR amplified from a cDNA library using the primers indicated below, and cloned between the BamHI and NotI restriction digestion sites in the pEFMN vector to express myc-tagged protein in mammalian cells or the pGEX vector to express N-terminal glutathione S-transferase (GST)-tagged protein in bacteria. To generate stable cell lines expressing myc-tagged UBCH8 (Myc-UBCH8), pEFMN-UBCH8 was cotransfected with pPUR (Clontech), which confers puromycin resistance. One day after transfection, the cells were replated in selection medium containing 2 μg/ml puromycin (Sigma). Anti-Cdt1 and anti-Cdt2 antibodies were described previously (36). Anti-p21 (C-19; Santa Cruz Biotechnology), anti-UBCH8 (Proteintech Group, Inc.), anti-UBE2G1 (Sigma), anti-β-actin (Sigma), and anti-tubulin (Santa Cruz Biotechnology) antibodies were purchased from the indicated companies.
The primers for cloning were as follows: UBCH8 forward primer, 5′CCGGATCCATGTCCACTGAGGCACAAAGAG3′; UBCH8 reverse primer, 5′AACCTTGCGGCCGCCTATGTGGCGTACCGCTTG3′; UBE2G1 forward primer, 5′CCGGATCCATGACGGAGCTGCAGTCGG3′; UBE2G1 reverse primer, 5′AACCTTGCGGCCGCTCACTCAAAAGCAGTCTCTTGGC3′; UBE2G2 forward primer, 5′CCGGATCCATGGCGGGGACCGCG3′; and UBE2G2 reverse primer, 5′AACCTTGCGGCCGCTCACAGTCCCAGAGACTTCTGGACG3′.
Identification of Cdt2-interacting proteins and mass spectrometry analysis.
293T cells were stably transfected, either with control vector or with an N-terminal 3×FLAG- and eight-histidine-tagged full-length Cdt2, using retroviral transduction (4). Stable Cdt2-expressing 293T cells were UV irradiated with 25 J/m2 and incubated for 3 h in the presence of MG132 (20 μg/ml) prior to harvest. The cells were washed with cold PBS, lysed in FLAG buffer (100 mM Tris [pH 8.0], 100 mM NaCl, 5 mM EDTA, 5% glycerol, 0.1% NP-40, 1 mM dithiothreitol [DTT], phenylmethylsulfonyl fluoride [PMSF], pepstatin, leupeptin, NaF, and sodium orthovanadate), washed 5 times in the same buffer, and eluted with 3×FLAG peptide (250 μg/ml). Eluates were incubated with nickel beads overnight in nickel buffer (100 mM Tris [pH 8.0], 100 mM NaCl, 5% glycerol, 0.1% NP-40, 5 mM β-mercaptoethanol, 5 mM imidazole, PMSF, pepstatin, leupeptin, NaF, sodium orthovanadate, and protease inhibitors) and eluted with 500 mM imidazole in nickel buffer. The eluted proteins were precipitated by the addition of trichloroacetic acid (TCA) and analyzed by mass spectrometry as previously described (47).
siRNA screening for UBCs involved in the UV-induced degradation of Cdt1.
HCT116p53−/− human colon cancer cells were transfected with siRNA oligonucleotides targeting the various ubiquitin-conjugating enzymes (UBCs) (Table 1 ) and irradiated with UV (20 J/m2) 3 days posttransfection. The cells were harvested 30 min after irradiation, the point at which we see maximal decrease of Cdt1, and cellular proteins were analyzed by SDS-PAGE and Western blotting with anti-Cdt1 or anti-β-actin.
Table 1.
UBC genes and the corresponding siRNA list
| Name | NCBI reference sequence | siRNA | Target sequence |
|---|---|---|---|
| UBE2G1 | NM_003342 | UBE2G1-A | TGTTGATGCTGCGAAAGAA |
| UBE2G1 | NM_003342 | UBE2G1-B | GGGAAGATAAGTATGGTTA |
| UBE2G2 | NM_003343 | UBE2G2-A | ATGATGACTTAATGTCGAA |
| UBE2G2 | NM_003343 | UBE2G2-B | TGACGAAAGTGGAGCTAAC |
| UBCH6 | NM_003341 | UBCH6 | GTATTGCCACTCAGTATAT |
| UBCH8 | NM_152653 | UBCH8-A | GCAAGAACCAGAAAGAGAA |
| UBCH8 | NM_152653 | UBCH8-B | ACTTGAAAGATTTGGGATT |
| UBCH9 | NM_006357 | UBCH9 | ATCTATCACTGCAACATCA |
| UBE2A | NM_003336 | UBE2A | TGATGTGTCTTCCATTCTA |
| UBE2B | NM_003337 | UBE2B | ATAGACAACTGGTCTGTTA |
| UBE2C | NM_007019 | UBE2C | GGTATAAGCTCTCGCTAGA |
| UBE2D1 | NM_003338 | UBE2D1 | TACTGTATGTGTTGTCTAA |
| UBE2D2 | NM_003339 | UBCH5BC | CAGTAATGGCAGCATTTGT |
| UBE2D2 | NM_003339 | UBE2D2 | CCAACCAGATTAAACTCTA |
| UBE2D3 | NM_003340 | UBE2D3 | TGATGTAAAGTTCGAAAGA |
| UBE2D4 | NM_015983 | UBE2D4 | GCATCTGCCTTGATATCCT |
| UBE2F | NM_080678 | UBE2F | AGGAGGACTTCCGGAATAA |
| UBE2H | NM_003344 | UBE2H | ACTGTGTGTCTAGATGTAA |
| UBE2I | NM_003345 | UBE2I | AATTCGAACCACCATTATT |
| UBE2J1 | NM_016021 | UBE2J1 | GAACTGGCTAGGCAAATAA |
| UBE2J2 | NM_194315 | UBE2J2 | ATATTCTCGAGTGGTTCAA |
| UBE2L3 | NM_003347 | UBE2L3 | GCTGATGTTACCACAGTAA |
| UBE2L6 | NM_004223 | UBE2L6 | TGATCAAATTCACAACCAA |
| UBE2 M | NM_003969 | UBE2 M | AGCCAGTCCTTACGATAAA |
| UBE2N | NM_003348 | UBE2N | CTATCTAGCTTGTGTGTCA |
| UBE2O | NM_022066 | UBE2O | ACATCGACTGTGCCGTCAA |
| UBE2Q1 | NM_017582 | UBE2Q1 | TCATCTCCGACCTGTGTAA |
| UBE2Q2 | NM_173469 | UBE2Q2 | TACAGATCACAGAGTTATA |
| Cdc34 | NM_004359 | Cdc34-A | CGCAGAACGTCAGGACCAT |
| Cdc34 | NM_004359 | Cdc34-B | GGAAGTGGAAAGAGAGCAA |
| UBE2R2 | NM_017811 | UBE2R2 | TGTGAGGACTATCCTATTA |
| UBE2S | NM_014501 | UBE2S | ATGGCGAGATCTGCGTCAA |
| UBE2T | NM_014176 | HSPC150 | AGAGAGAGCTGCACATGTT |
| UBE2U | NM_152489 | UBE2U | ACAGGCCATTACAAATGAA |
| UBE2V1 | NM_021988 | UBE2V1 | AAGCAAACTGAGTGATGAA |
| UBE2V2 | NM_003350 | UBE2V2 | AGTTGTACTTCAAGAGCTA |
| UBE2W | NM_001001481 | UBE2W | CGACCACCGGATAATTCTT |
| UBE2Z | NM_023079 | UBE2Z | ATGTTCGTTGTACCTGATA |
| UEV3 | NM_018314 | UEV3 | CGATGGACCTTGAAATCTT |
| TSG101 | NM_006292 | TSG101 | AGTAGCCGAGGTTGATAAA |
| HIP2 | NM_005339 | HIP2 | CTCTCCGCACGGTATTATT |
| FTS | NM_022476 | FTS | GAATTTACCTTGGTTGTGA |
| BIRC6 | NM_016252 | BIRC6 | TCATTGCCTTACTCACATA |
Expression and purification of recombinant UBE2 proteins.
pGEX-UBCH8 was transformed into Escherichia coli (TOP10) to express glutathione S-transferase (GST)-tagged protein. Bacteria were grown in LB medium at 30°C until the optical density at 600 nm (OD600) was approximately 0.5. Expression of protein was induced by the addition of 0.05 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to the culture medium, and growth continued overnight at 18°C. Bacteria were collected by centrifugation at 3,000 rpm for 10 min, sonicated in buffer containing 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA, and 1 mM DTT, and incubated on ice for 10 min before the addition of 1% Triton X-100. Soluble lysates were cleared of cell debris by centrifugation at 13,000 rpm for 20 min, and the cleared lysates were rotated with glutathione Sepharose 4B (GE Healthcare) for 1 h at 4°C. The beads were washed five times with PBS containing 1% Tween 20 (Sigma), and the GST-tagged proteins were eluted with elution buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 10% glycerol, and 10 mM reduced glutathione.
In vitro ubiquitylation reaction.
The in vitro ubiquitylation of p21 was performed essentially as described previously (4), but with the addition of 100 ng of recombinant UBCH5C (Boston Biochem) or GST-UBCH8.
Quantitative real-time PCR (RT-PCR).
Total RNA was extracted with Trizol reagent (Invitrogen), and cDNA was synthesized using SuperScript III first-strand synthesis system (Invitrogen) according to the manufacturer's instructions. Real-time PCRs were performed by using an ABI Prism 7000 sequence detection system. The mRNA levels of the relevant genes were normalized to the mRNA of β-actin. The primers used for the amplification reactions were as follows: actin forward primer, 5′CCAGATCATGTTTGAGACCTTCAAC3′; actin reverse primer, 5′CCAGAGGCGTACAGGGATAGC3′; p21 forward primer, 5′CTGGAGACTCTCTGCAGGGTCGAAA3′; p21 reverse primer, 5′GATTAGGGCTTCCTCTTGGAGAA3′; Cdt1 forward primer, 5′AGACTGCGGTCGGT3′; Cdt1 reverse primer, 5′CCGGGGTGGATTTCTTTATC3′; UBCH6 forward primer, 5′AGGATCCGTGTATGAGGGTG3′; UBCH6 reverse primer, 5′TGGTTAGTGCTGGACTCCAAT3′; UBCH9 forward primer, 5′CCCAGCAGAAGAAAAACACC3′; UBCH9 reverse primer, 5′TGCAATTAGGAGGAGGATCAA3′; UBE2G2 forward primer, 5′TTTGAATGGGAGGCATTGAT3′; and UBE2G2 reverse primer, 5′ATCTTTGGGGGACTTAACGG3′.
RESULTS
UBCH8-conjugating enzyme associates with the CRL4Cdt2 E3 ubiquitin ligase to promote p21 ubiquitylation and proteolysis.
To identify the ubiquitin-conjugating enzyme(s) [UBC(s)] that support CRL4Cdt2 ubiquitin ligase activity, we searched for UBCs that may specifically associate with the CRL4Cdt2 E3 ubiquitin ligase complex using tandem affinity purification of Cdt2-interacting proteins, coupled with mass spectrometric analysis. To accomplish this, we generated 293T cells stably expressing FLAG- and histidine-tagged full-length human Cdt2 using retroviral transduction. FLAG-tagged Cdt2 (FLAG-Cdt2) was expressed at levels equal to or less than endogenous Cdt2 (data not shown). Purification of Cdt2 and Cdt2-associated proteins was performed as outlined in Materials and Methods. In parallel, 293T cells mock transduced with empty retroviral vector was used to control for background interactions. As expected, the major peptides identified using this approach were those of Cdt2, DDB1, Cul4A, and Cul4B (Table 2), confirming that Cdt2 is a stable component of the CRL4 ubiquitin ligase complex. Peptides corresponding to the ubiquitin-conjugating enzymes UBCH6, UBCH8, and UBCH9 were also identified. Multiple attempts to detect these UBCs by immunoprecipitation of Cdt2 or Cul4A/B and Western blotting, however, failed to detect the interaction (data not shown), suggesting that the association of these UBCs with CRL4Cdt2 is substoichiometric and can be detected only by sensitive large-scale methods like mass spectrometry.
Table 2.
Partial list of Cdt2-associated proteins, identified by mass spectrometric analysis
| Protein | No. of peptides |
|---|---|
| Damage-specific DNA-binding protein 1 (DDB1) | 31 |
| Denticleless protein homolog (CDT2) | 20 |
| Cullin-4B (CUL4B) | 5 |
| COP9 signalosome complex subunit (COPS4) | 4 |
| Cullin-4A (CUL4A) | 3 |
| RING-box protein 1 (RBX1) | 3 |
| Ubiquitin-conjugating enzyme UBCH6 | 3 |
| Ubiquitin-conjugating enzyme UBCH8 | 3 |
| Ubiquitin-conjugating enzyme UBCH9 | 3 |
We next tested whether the UBCH6, UBCH8, and UBCH9 ubiquitin-conjugating enzymes are required for the activity of the CRL4Cdt2 E3 ubiquitin ligase on the Cdk inhibitor p21, a previously identified substrate of CRL4Cdt2 (4, 27), in the p53-deficient human colon cancer HCT116 cells (HCT116p53−/−). The use of this cell line eliminates any complications from the p53-dependent induction of p21 and the possibility that p53 could itself be a substrate of CRL4Cdt2 (7). Individual UBCH6, -8, and -9 proteins were depleted by small interfering RNA (siRNA) to test their effect on p21 steady-state protein levels. Two siRNAs (si-UBCH8-A and si-UBCH8-B) targeting UBCH8 effectively decreased the UBCH8 protein level (Fig. 1 A, top panel). Consistent with previous results, UV irradiation downregulated p21 in control cells transfected with a negative-control siRNA targeting the luciferase gene (si-GL2) (Fig. 1A), an activity that is mediated via the CRL4Cdt2 E3 ubiquitin ligase (4, 27). Importantly, the depletion of UBCH8 by si-UBCH8-A or -B prevented UV-induced degradation of p21 (Fig. 1A, top panel). On the other hand, depletion of UBCH6 and UBCH9 by siRNAs did not prevent p21 degradation postradiation (Fig. 1A, bottom panel), although both siRNAs effectively decreased their target mRNAs (Fig. 1B). siRNA directed against UBCH8 mildly decreased the mRNA levels of UBCH6 or UBCH9, two related UBCs, likely because of conserved sequences among these family members (Fig. 1B and data not shown). However, because the specific depletion of UBCH6 or UBCH9 did not affect p21 degradation postradiation, we conclude that the reduction of UBCH6 or UBCH9 after UBCH8 depletion does not contribute to the stabilization of p21.
Fig. 1.
UBCH8, but not UBCH6 or UBCH9, is required for p21 degradation. (A) p21 in HCT116p53−/− cells is not degraded upon UV irradiation in the absence of UBCH8. Depletion of UBCH8 by two siRNA oligonucleotides (siUBCH8-A and siUBCH8-B) prevents UV-induced degradation of p21. Control cells were transfected with siRNA against luciferase (GL2). The Western blot shows the steady-state levels of p21 protein in UV-irradiated cells (+) and nonirradiated cells (−) as indicated. LC is a nonspecific band that cross-reacts with the anti-p21 antibody and is used as a loading control. (B) UBCH6 or UBCH9 mRNAs are decreased by their cognate siRNAs. The mRNA level of UBCH6 or UBCH9 was determined by quantitative real-time PCR, normalized to β-actin mRNA in the same sample and expressed as a percentage of the level in the si-GL2 sample. (C) UBCH8, but not UBE2G1 or UBE2G2, regulates p21 protein stability after UV irradiation. HCT116p53−/− cells were transfected with siRNA as indicated and treated with cycloheximide (CHX) for 10 min before UV irradiation. The Western blot shows the level of p21 protein at the indicated time points. Smaller quantities of protein lysates were loaded from the siUBCH8-A- or the si-UBE2G-transfected cells to ensure that the p21 signal at 0 min was comparable to that in the si-GL2-transfected cells (because si-UBCH8 increases the basal levels of p21; see text for details). The top blot shows a long exposure (exp.), and the middle blot shows a short exposure. Anti-β-actin was used as a loading control. (D) Quantitation of the p21 protein and the loading control signals was performed using GeneTools software. The plot shows the ratio of p21 to β-actin after normalizing the ratio at 0 min to 100.
To ascertain that p21 protein was indeed stabilized in irradiated cells by si-UBCH8, we measured the half-life of the protein after adding cycloheximide to inhibit new protein synthesis. In control transfected cells, the half-life of p21 was 30 min in cells irradiated with UV, and this was prolonged to about 120 min in cells depleted of UBCH8 (Fig. 1C and D). The p21 half-life in UV-irradiated cells transfected with siRNA against another pair of UBCs, UBE2G1 and UBE2G2 (see below) was relatively unchanged (Fig. 1C and D). Since we and others have already demonstrated that the ubiquitylation of p21 in UV-irradiated cells is dependent on CRL4Cdt2 (4, 27), these results indicate that UBCH8 is the E2 that cooperates with CRL4Cdt2 to promote p21 degradation in UV-irradiated cells. As an aside, the decrease in basal level of p21 after depletion of UBCH9 was reproducible but accompanied by a decrease in p21 mRNA (Fig. 1A, bottom panel, and data not shown). We suspect that UBCH9 directly or indirectly plays a role in the regulation of the p21 promoter and did not follow this further.
The CRL4Cdt2 ubiquitin ligase promotes the degradation of p21 also in unperturbed cells during the S phase of the cell cycle (4, 27). Indeed, in several experiments, the depletion of UBCH8 increased the basal levels of p21 protein relative to loading control in unirradiated cells (Fig. 1C, 2A, and 2G), without affecting the steady-state levels of p21 mRNA (Fig. 2 C). Furthermore, the half-life of p21 was prolonged in UBCH8-depleted unirradiated cells to >90 min (Fig. 2A and B). Thus, UBCH8 cooperates with CRL4Cdt2 to degrade p21 also in unirradiated cells. The significant stabilization of p21 in UBCH8-depleted cells after UV irradiation (Fig. 1C and D) and the modest stabilization of p21 in unirradiated cells (Fig. 2A and B) is consistent with the fact that p21 degradation postradiation is mediated exclusively via CRL4Cdt2 (4), while its stability in unirradiated asynchronous cells is regulated by multiple E3s, CRL4Cdt2 in S phase (4), SCFSkp2 (SCF stands for Skp1, Cul1, and F-box proteins) during G1/S phase (8, 35, 43, 46), and APC/CCdc20 in G2/M phase (5).
Fig. 2.
UBCH8 destabilizes p21 during the normal cell cycle. (A) The half-life of p21 protein is increased by knockdown of UBCH8. siRNA-transfected cells were treated with cycloheximide (CHX) for the indicated time. The level of p21 was analyzed by Western blotting. Smaller quantities of protein lysates were loaded from the si-UBCH8-A-transfected cells to ensure that the signal at 0 min was comparable to that in the si-GL2-transfected cells. (B) Quantitation of the p21 protein and the loading control signals was performed using GeneTools software. The ratio of p21 to β-actin was plotted after normalizing the ratio at 0 h to 100. (C) p21 mRNA is not affected by knockdown of UBCH8. The p21 mRNA levels in siRNA-transfected cells were determined by quantitative real-time PCR, normalized to β-actin, and the ratio was expressed relative to the control si-GL2 sample. (D) UBCH8 overexpression destabilizes p21 protein. The p21 level in Myc-UBCH8-overexpressing HCT116p53−/− cells was determined by Western blotting. Three independent clones of cells were analyzed. β-Actin is used as loading control. (E) Quantitation of the p21 protein and the loading control signals was performed using GeneTools software. The ratio of p21 to β-actin was plotted. (F) The basal level of p21 mRNA is not significantly decreased by overexpression of Myc-UBCH8 as determined by quantitative real-time PCR in the same clones of cells as shown in panel B. The mRNA level of p21 was normalized to β-actin and expressed relative to the ratio in vector-transfected control cells. (G) Vector-control or Myc-UBCH8-expressing HCT116p53−/− cells were transfected with siRNA against the open reading frame (ORF) (si-UBCH8-A) or the 3′UTR (si-UBCH8-B) of UBCH8. Immunoblots are shown with the indicated antibodies. The asterisk indicates a degradation product of overexpressed UBCH8. For the UBCH8 blot, a lower exposure is shown for lanes 4 to 6 to prevent saturation of signal. β-Actin is shown to control for protein loading.
Because the downregulation of UBCH8 stabilized p21 protein in unperturbed cells, we wondered whether the level of UBCH8 could be rate-limiting for determining p21 levels in unperturbed cells. To test this hypothesis, we generated clones of HCT116p53−/− cells that stably overexpressed myc-tagged UBCH8 (Myc-UBCH8). Three independent clones stably expressing UBCH8 exhibited lower levels of p21 protein (Fig. 2D and E). The 80% decrease of p21 protein was associated with a 20 to 30% reduction of p21 mRNA (Fig. 2F), suggesting that the decrease of protein is by a posttranscriptional mechanism. To test whether the reduction of p21 was a direct consequence of Myc-UBCH8 overexpression, the cells were transfected with si-UBCH8-A, targeting endogenous and ectopically expressed UBCH8, or with si-UBCH8-B, targeting the 3′ untranslated region (3′UTR) of UBCH8 (Fig. 2G). si-UBCH8-A reduced endogenous UBCH8 protein and elevated p21 protein in control cells and similarly reduced both endogenous and ectopic UBCH8 and elevated p21 protein in UBCH8-overexpressing cells (Fig. 2G, lanes 2 and 5). On the other hand, targeting the 3′UTR of UBCH8 by si-UBCH8-B elevated the levels of p21 only in control cells but not in cells overexpressing UBCH8 lacking the 3′UTR (Fig. 2G, lanes 3 and 6). Thus, (i) UBCH8 appears rate-limiting for p21 degradation, and (ii) the stabilization of p21 by si-UBCH8 was specifically due to a decrease of UBCH8 and not due to any off-target activity of the siRNA duplexes. Collectively, these results demonstrate that UBCH8 is necessary and sufficient to promote p21 proteolysis in vivo, both in UV-irradiated and unperturbed cells.
UBCH8 functions with CRL4Cdt2 to promote p21 ubiquitylation in vitro.
To test whether UBCH8 is directly responsible for p21 ubiquitylation, we assayed whether UBCH8 could promote ubiquitylation in vitro. Recombinant GST-UBCH8 was expressed and purified from bacteria (Fig. 3 A). Overexpressed CRL4Cdt2 complex in 293T cells (Myc-DDB1, Myc-Cul4A, FLAG-Cdt2, and FLAG-Rbx1) was immunoprecipitated with anti-Myc antibody to purify a functional CRL4Cdt2 E3 ubiquitin ligase (Fig. 3A). The substrate, p21, along with the associated accessory protein, PCNA, was immunopurified with anti-p21 antibody from 293T cells. For a positive control, we used UBCH5C, because it is an E2 that is used promiscuously in many ubiquitylation reactions in vitro. The ubiquitylation of p21 is expected to produce slower moving bands that react with anti-p21 antibody. In the presence of ATP, ubiquitin, and ubiquitin-activating enzyme E1, UBCH8, similar to UBCH5C, cooperated with CRL4Cdt2 to promote p21 ubiquitylation (Fig. 3B; see Fig. S3 in the supplemental material). UBCH5C appears to specifically stimulate the monoubiquitylation of p21. This result demonstrates that UBCH8 functions in vitro as an ubiquitin-conjugating enzyme for p21 ubiquitylation via CRL4Cdt2.
Fig. 3.
UBCH8 functions with CRL4Cdt2 to promote p21 ubiquitylation in vitro. (A) (Left) Purified GST-tagged recombinant UBCH8 protein was resolved by SDS-PAGE and stained with Coomassie blue. (Middle and right) The CRL4Cdt2 complex immunopurified from 293T cells transfected with expression vectors of Myc-Ddb1, Myc-Cul4A, FLAG-Cdt2, and FLAG-Rbx1. The Western blot shows Ddb1 and Cul4A (anti-Myc [middle panel]) or RBX1, Cdt2 (anti-FLAG [right panel]) in the anti-Myc immunoprecipitates. IgG is the immunoglobulin light chain. The positions of molecular weight (MW) markers (in thousands) are shown to the left of the leftmost blot. IP, immunoprecipitation; IB, immunoblotting. (B) Recombinant UBCH8 polyubiquitylates p21 in vitro. The indicated recombinant E2s were incubated in ubiquitin ligation reaction mixture containing immunoprecipitated CRL4Cdt2, p21 substrate, E1 enzyme, and ubiquitin. UBCH5C is a positive control for ubiquitin conjugation. The ubiquitylation products of p21 are evident by the formation of the higher-molecular-weight bands detected by anti-p21 blotting.
UBCH8 does not promote CRL4Cdt2-mediated and UV-induced Cdt1 protein degradation.
In addition to p21, the CRL4Cdt2 E3 ubiquitin ligase polyubiquitylates the replication licensing factor Cdt1 both in S-phase cells and after genotoxic stress (4, 6, 14, 15, 17, 28, 33, 36). We thus tested whether UBCH8 is also required for promoting Cdt1 degradation. As expected, UV irradiation of HCT116p53−/− cells rapidly degraded the Cdt1 protein (Fig. 4 A). Unlike the case for p21, however, the depletion of UBCH8 did not prevent Cdt1 degradation (Fig. 4A). Because CRL4Cdt2 associated with UBCH6 and UBCH9 in addition to UBCH8 in our mass spectrometry analysis (Table 2), we tested whether these proteins are involved in Cdt1 degradation postradiation. We knocked down UBCH6 and UBCH9 either individually (Fig. 4A) or in combination with UBCH8 (Fig. 4B and C) by siRNA before irradiating cells. The degradation of Cdt1 following UV irradiation was not impaired even in cells depleted of all three UBCs combined (Fig. 4B and C). This result is consistent with the previously published report showing that although UBCH6, -8, and -9 associate with the DDD complex, they are not required for the targeted degradation of Cdt1 postradiation (31). Thus, UBCs other than UBCH6, -8, and -9 must be involved in the targeted ubiquitylation and degradation of Cdt1 via the CRL4Cdt2 E3 ubiquitin ligase.
Fig. 4.
UBCH6, UBCH8, and UBCH9 are not required for UV-induced Cdt1 degradation. (A and B) The degradation of Cdt1 after UV irradiation in HCT116p53−/− cells depleted of UBCH6, UBCH8, or UBCH9 (UBCH6/8/9)is not impaired. Western blot analysis of Cdt1 steady-state level following UV irradiation in cells depleted of the indicated ubiquitin-conjugating enzymes either individually (A) or in combination (B) by siRNA. Cells transfected with siRNA against luciferase (GL2) are used as a control. The results demonstrate that UBCH6, UBCH8, and UBCH9 are not required for efficient degradation of Cdt1 postradiation. (C) Quantitation of the Cdt1 protein and β-actin shown in panel B was performed using GeneTools software. The ratio of Cdt1 to β-actin was plotted after normalizing the ratio at 0 min to 100.
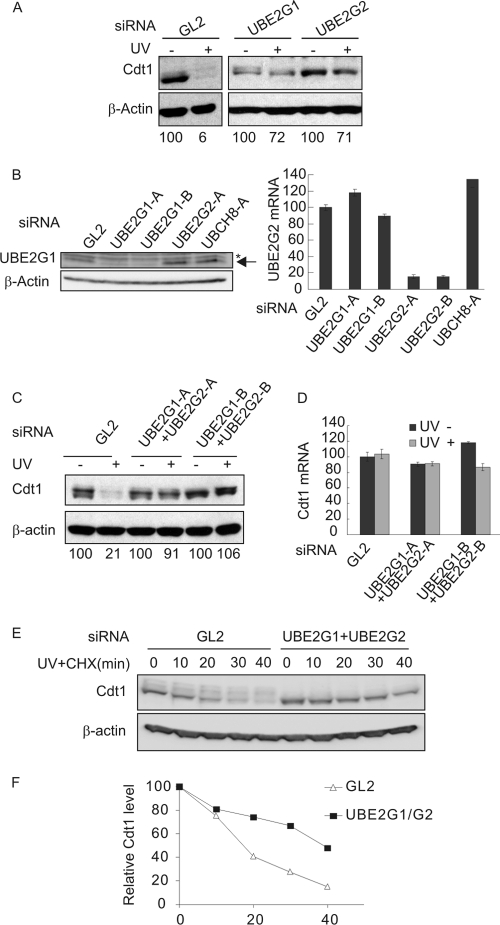
UBE2G1/2-conjugating enzymes are required for UV-induced Cdt1 protein degradation.
To identify the UBC that functions with the CRL4Cdt2 in promoting Cdt1 ubiquitylation in UV-irradiated cells, we conducted an RNA interference (siRNA) screen of proteins containing the highly conserved catalytic core (UBC) domain (Fig. 5 A; see Fig. S1 in the supplemental material). Out of the 36 UBC targets, knockdown of UBE2G1 or UBE2G2 partially prevented Cdt1 degradation in response to UV (Fig. 5A). Furthermore and as expected, knockdown of Nedd-8-conjugating enzyme UBE2M also stabilized Cdt1, confirming the requirement of neddylation of cullin 4 for Cdt1 degradation (Fig. S1) (12, 30).
Fig. 5.
UBE2G1 and UBE2G2 are required for UV-induced Cdt1 degradation. (A) HCT116p53−/− cells depleted of UBE2G1 or UBE2G2 are deficient in their ability to degrade Cdt1 postradiation. The results of Western blot analysis of the Cdt1 steady-state level following UV irradiation in cells depleted of the indicated ubiquitin-conjugating enzymes by siRNA are shown. The degree of Cdt1 degradation postradiation was determined by quantitating the signal intensity of Cdt1 (normalized to β-actin) and expressed as a percentage of that in unirradiated cells of the corresponding siRNA transfection. (B) Efficient knockdown of UBE2G1 or UBE2G2 by the cognate siRNAs. HCT116p53−/− cells were transfected with siRNA targeting the indicated proteins for 72 h before harvesting. (Left) The protein level of UBE2G1 was determined by Western blotting. The lowest band is the specific band. (Right) The mRNA level of UBE2G2 was determined by quantitative real-time PCR, normalized to that of β-actin and expressed relative to that of the control si-GL2 sample. (C) Coknockdown of UBE2G1 and UBE2G2 by two separate siRNAs (-A or -B for each gene) prevents the degradation of Cdt1 after UV irradiation. The levels were measured as described above for panel A. The extent of Cdt1 degradation postradiation was determined as described above for panel A. (D) Cdt1 mRNA is not increased by knockdown of UBE2G1 and UBE2G2. The levels were measured as described above for panel B. (E) The half-life of Cdt1 protein after UV irradiation is increased by knockdown of UBE2G1 and UBE2G2. (F) Quantitation of the Cdt1 protein and β-actin was performed using GeneTools software. The ratio of Cdt1 to β-actin was plotted after normalizing the ratio at 0 min to 100.
Western blotting of UBE2G1 and quantitative real-time PCR of UBE2G2 mRNAs showed that two separate siRNAs (siUBE2G2-A and siUBE2G2-B, respectively) specifically knocked down the levels of the corresponding protein and mRNA, respectively (Fig. 5B). Because individual knockdown of these highly homologous E2s each had a significant stabilizing effect of Cdt1, we hypothesize that UBE2G1 and UBE2G2 may be dependent on each other for their activity. Codepletion of UBE2G1 and UBE2G2 from HCT116 cells also prevented UV-induced Cdt1 degradation, and this was confirmed using a second set of siRNA duplexes targeting these two UBCs (Fig. 5C). The mRNA of Cdt1 in UBE2G1- and UBE2G2-depleted cells was unchanged by this treatment (Fig. 5D). Importantly, the half-life of Cdt1 protein in UV-irradiated cells showed that it is increased from 15 min to 40 min following the knockdown of UBE2G1 and UBE2G2 (Fig. 5E and F).
UBE2G1 and UBE2G2 are not required for CRL4Cdt2-mediated degradation of p21 protein.
Since we have already implicated UBCH8 in the degradation of p21 by CRL4Cdt2, we next asked whether UBE2G1 or UBE2G2 could also ubiquitylate and degrade p21. Although the individual or combined depletion of UBE2G1 and UBE2G2 significantly increased the basal level of p21 protein (Fig. 6A and D, 0 h), this increase was likely due to the 2- to 5-fold increase of p21 mRNA (Fig. 6B and C, left panel). The half-life of p21 protein was not prolonged in cells depleted of UBE2G1 or UBE2G2 or both (Fig. 6D and E and data not shown). Knockdown of UBE2G1 and UBE2G2 did not prevent the degradation of p21 following UV irradiation (Fig. 1C, right panel). Interestingly, p21 mRNA was similarly induced in cells depleted of Cdt2 (Fig. 6C, right panel), suggesting that UBE2G1/G2 and CRL4Cdt2 may repress p21 mRNA expression through an unidentified mechanism. From these results, we conclude that although UBE2G1 and UBE2G2 promote the degradation of Cdt1, they do not promote the degradation of p21 in cycling or UV-irradiated cells.
Fig. 6.
UBE2G1 and UBE2G2 regulate p21 transcriptionally but do not affect its protein stability. (A) Basal level of p21 protein increased after knockdown of UBE2G1 or UBE2G2. The level was measured as described in the legend of Fig. 1A. (B) p21 mRNA induced by knockdown of UBE2G1 or UBE2G2. The p21 mRNA level in siRNA-transfected cells was determined by quantitative real-time PCR, normalized to β-actin, and the ratio was expressed relative to the control si-GL2 sample. (C) The basal level of p21 mRNA is induced by siRNA-mediated depletion of UBE2G1 and UBE2G2 (left) or Cdt2 (right). The levels were determined as described above for panel B. (D) The half-life of p21 protein after UV irradiation is not significantly altered by knockdown of UBE2G1 and UBE2G2. Smaller quantities of protein lysates were loaded from the si-UBE2G1/G2-transfected cells to ensure that the p21 signal at 0 min was comparable to that in the si-GL2-transfected cells. Quantitation of the p21 protein and the loading control signals was performed using Scion image software. (E) The ratio of p21 to the signal from the loading control (LC) was plotted after normalizing the ratio at 0 h to 100.
Cdc34 does not play a role in the degradation of Cdt1 or p21 upon UV irradiation.
Cdc34 is the UBC that cooperates with various SCF (Skp1, Cul1, and F-box protein) ubiquitin ligase complexes, including SCFSkp2, SCFβ-TrCP2, and SCFCdc4. Cdc34 has a unique and conserved acidic tail that promotes its association with SCF (23, 24, 26, 39, 44). In addition, Cdc34 has been reported to elongate the ubiquitin chain on IκBα after the addition of the first ubiquitin by another E2 (45). Thus, we wished to test whether Cdc34 cooperates with UBCH8 and UBE2G1/G2 for the elongation of ubiquitin chains via CRL4Cdt2. Since SCFSkp2 regulates the ubiquitylation of p21 and Cdt1 in a cell cycle-dependent manner, we specifically focused on the degradation of the two proteins by UV irradiation, because that degradation was uniquely promoted via the CRL4Cdt2 ubiquitin ligase complex.
Two homologs of Saccharomyces cerevisiae Cdc34 (Cdc34 and Ube2R2) were codepleted by siRNA transfections, and the degradation of Cdt1 or p21 after UV irradiation was examined by immunoblotting (see Fig. S2 in the supplemental material). Two siRNAs targeting Cdc34 (si-Cdc34-A or si-Cdc34-B) were combined with si-Ube2R2 to decrease both proteins (see Fig. S2A in the supplemental material). Consistent with reports that Cdc34 cooperates with SCFSkp2 to destabilize p21 protein during the normal cell cycle, codepletion of Cdc34 and Ube2R2 increased the basal p21 protein level (Fig. S2A). However, degradation of p21 or Cdt1 after UV irradiation was not prevented by the depletion of Cdc34 and UBE2R2 (Fig. S2B and S2C). These results demonstrate that Cdc34, unlike UBCH8 or UBE2G1/G2 does not have a role in the CRL4Cdt2-mediated polyubiquitylation of p21 or Cdt1 after UV irradiation.
UBCH8 is required for cell cycle regulation and the DNA damage response to UV irradiation.
CRL4Cdt2 has already been shown to promote p21 degradation in S phase (4, 27) so that depletion of UBCH8 would be expected to interfere with S-phase progression by causing the accumulation of this Cdk inhibitor. Indeed, the depletion of UBCH8 from HCT116p53−/− cells by siRNA resulted in the accumulation of cells in the G1 phase of the cell cycle and a corresponding decrease of cells in S phase (Fig. 7A, top panel). These changes in the cell cycle were dependent on p21 induction, as they were reversed by codepleting UBCH8-depleted cells of p21 (Fig. 7A, top panel). Similar results were obtained in the p53-positive U2OS osteocarcinoma-derived cells (Fig. 7A, bottom panel).
Fig. 7.

UBCH8 is required for normal cell cycle progression and the response to DNA damage. (A) Depletion of UBCH8 inhibits the transition from G1 to S through the stabilization of p21. HCT116p53−/− cells (top) or U2OS cells (bottom) were treated with the indicated siRNA for 72 h, and the cell cycle profile was determined by propidium iodide staining and fluorescence-activated cell sorting (FACS) analysis. (B) si-UBCH8-treated cells reduces phospho-H2AX (P-H2AX) and prevents its induction following UV irradiation, and this is alleviated by coknockdown of p21. Cells transfected with the indicated siRNAs for 72 h were collected 30 min after the UV treatment (20 J/m2) by direct lysis in SDS sample buffer for immunoblotting with the indicated antibodies.
UV-induced DNA damage is sensed by replication forks in S phase. In p53-deficient cells, p21 is not induced by DNA damage, but we wondered whether the increased basal levels of p21 in cells depleted of UBCH8 would suppress the DNA damage response by decreasing the S-phase population. Both the basal and UV-induced phospho-H2AX levels (a marker of DNA damage) were indeed decreased when UBCH8 is knocked down and basal levels of p21 increased (Fig. 7B, GL2 and UBCH8-A lanes). Following the coknockdown of p21, however, the DNA damage response was restored. Thus, UBCH8 is required for the UV-induced DNA damage response, primarily because it keeps p21 levels low and allows more cells to enter S phase.
UBCH8 is required for UV-induced degradation of Set8 and the monoubiquitylation of PCNA in unstressed cells.
CRL4Cdt2 promotes the ubiquitylation of two additional substrates, Set8, the histone H4 lysine 20 (H4K20) monomethyltransferase (3, 10, 29, 40) and PCNA (41). Whereas CRL4Cdt2 promotes the polyubiquitylation of Set8 both during the S phase of the cell cycle and after UV irradiation (3, 10, 29, 40), it promotes the monoubiquitylation of PCNA in unstressed cells to promote translesion DNA synthesis (41). Thus, we tested whether UBCH8 or UBE2G1/G2 are required for the ubiquitylation of these substrates. Although the depletion of UBCH8 or UBE2G1/G2 by siRNA increased the basal level of Set8 (data not shown), only the depletion of UBCH8 inhibited the degradation of Set8 upon UV irradiation (Fig. 8A). In addition, the monoubiquitylation of PCNA in unstressed cells was specifically decreased by knocking down of UBCH8, but not by the combined knockdown of UBE2G1 and UBE2G2 (Fig. 8B and C). Together, these results demonstrate that UBCH8 is required for the ubiquitylation of Set8 and PCNA in addition to p21.
Fig. 8.

UBCH8 promotes UV-induced Set8 degradation and increases the monoubiquitylation of PCNA in unstressed cells. (A) HCT116p53−/− cells transfected with siRNA against UBCH8 (top) or UBE2G1 and UBE2G2 together (bottom), irradiated with UV (20 J/m2), and collected at the indicated times by direct lysis in SDS sample buffer for Western blotting with Set8 or β-actin. Smaller quantities of protein lysates were loaded from the siUBCH8-A- or the si-UBE2G-transfected cells to ensure that the Set8 signal at 0 min was comparable to that in the si-GL2-transfected cells. (B) Immunoblot of PCNA shows ubiquitylated PCNA (Ub-PCNA) and unmodified PCNA. U2OS cells were treated with siUSP1 to detect basal PCNA monoubiquitylation along with the indicated siRNA for 72 h. (C) The histogram displays the ratio of monoubiquitylated PCNA to total PCNA normalized to the ratio in control si-GL2-transfected cells.
DISCUSSION
Different UBCs cooperate with CRL4Cdt2 to promote the selective polyubiquitylation of p21 or Cdt1.
The CRL4Cdt2 E3 ubiquitin ligase complex is emerging as a critical regulator of several key cell cycle proteins whose degradation is essential for normal S-phase cell cycle progression (1). These include the Cdk inhibitor p21 and the replication initiation factor Cdt1. The identity of the ubiquitin-conjugating enzyme responsible for the covalent attachment of the polyubiquitin chains to these substrates via CRL4Cdt2 was unknown. In this study, we report the first ubiquitin-conjugating enzymes that cooperate with CRL4Cdt2 and show that UBCH8 selectively promotes the CRL4Cdt2-dependent ubiquitylation and degradation of p21, both in unperturbed cells and in response to UV irradiation. UBCH8 also promotes the CRL4Cdt2-mediated monoubiquitylation of PCNA in unstressed cells and the UV-induced ubiquitylation and degradation of Set8 (Fig. 8), two recently identified substrates for CRL4Cdt2. UBCH8, however, did not affect the stability or the degradation of the replication licensing factor Cdt1. Instead, we found that the ubiquitin-conjugating enzymes UBE2G1 and UBE2G2 promote Cdt1 degradation in UV-irradiated cells. Although the depletion of UBE2G1/G2 also increased the steady-state level of p21 mRNA and protein via a p53-independent mechanism, they did not affect the stability of p21 protein (Fig. 6D and E) and are therefore not involved in CRL4Cdt2-mediated p21 degradation. Our findings provide an example of an ubiquitin ligase complex, CRL4Cdt2, cooperating in vivo with different ubiquitin-conjugating enzymes to promote the ubiquitylation of different substrates (Fig. 9). An interesting question that arises is whether an UBC helps specify substrate recognition by an E3.
Fig. 9.

Differential utilization of UBCs by CRL4Cdt2 to promote Cdt1 and p21 ubiquitylation and degradation in cycling and DNA-damaged cells. UBCH8 and UBE2G1/UBE2G2 cooperate with CRL4Cdt2 to target ubiquitylation (Ub) of p21, Cdt1, Set8, and PCNA. p21 mRNA expression is independently suppressed by CRL4Cdt2, in cooperation with UBE2G1/UBE2G2.
UBCH8 is present in two functionally distinct complexes in the cells.
UBCH6, UBCH8, and UBCH9 have been reported to associate with the DDD (DET1, DDA1, and DDB1) complex (31). UBCH9 associated with this complex, however, is not active, and the DDD complex negatively regulates the polyubiquitin chain assembly by Cul4A in vitro (31). Our results demonstrate that UBCH8 is required for the activity of CRL4Cdt2, another complex that contains DDB1, on various substrates, including p21, Set8, and PCNA. These findings suggest that UBCH8 associates with at least two different DDB1-containing complexes, the DDD complex, where it is not able to promote ubiquitin elongation, and with CRL4Cdt2, where it is required for substrate-ubiquitin conjugation. Although UBCH8 (and other UBE2E family members) associates stably with the DDD complex (31), its association with the CRL4Cdt2 complex is labile as indicated by our failure to detect UBCH8 in CRL4Cdt2 complexes by standard immunoprecipitation (IP)/Western blot experiments. We propose that the DDD complex may function to sequester the UBCH8 to inhibit its association with an active ubiquitin ligase complex. We suspect that the transient association of UBCH8 with active CRL4Cdt2 E3 ubiquitin ligase and its sequestration by inactive DDD complex may be important to allow for the interaction of other ubiquitin-conjugating enzymes, such as UBE2G1/G2, with the same E3 ubiquitin ligase complex to promote the ubiquitylation of a different substrate like Cdt1. If true, such a mechanism would imply that upstream signals, such as those evoked by UV irradiation or during S phase of the cell cycle could regulate the proper assembly of the specific UBC with the E3 ubiquitin ligase complexes to target ubiquitylation of a specific substrate. The fact that different E2s interact with a given E3 adds another layer of complexity in the targeted ubiquitylation and degradation of substrates. The absence of UBE2G1/G2 from our proteomic analysis of Cdt2-associated proteins is in accordance with this hypothesis given that the CRL4Cdt2 promotes Cdt1 ubiquitylation within minutes of UV irradiation (not hours like p21), a condition that was not employed in our analysis of Cdt2-associated proteins.
Role of UBCH8 in regulating p21.
While many transcriptional factors have been implicated in the control of p21 (2), the regulation of this protein by polyubiquitylation has been controversial (4, 5, 8, 22, 37, 42), with some groups showing that p21 can be directly targeted to the proteasome without any polyubiquitylation and others showing evidence of polyubiquitylation. Our report on the role of CRL4Cdt2 in degrading p21 provided strong evidence that p21 is indeed regulated by polyubiquitylation (4, 22). In this paper, we add to the evidence of polyubiquitylation being a major regulator of p21 level by implicating an E2, UBCH8, in this pathway. While we cannot rule out the possibility that the level of p21 is also regulated by direct recruitment of the protein to proteasomes without any polyubiquitylation, the fact that varying the level of an E2 (UBCH8) or an E3 (CRL4Cdt2) (4) alters the half-life of p21 and that these enzymes polyubiquitylate p21 in vitro strongly suggest that polyubiquitylation is an important mechanism to regulate the degradation of p21 in cells. CRL4Cdt2 in cooperation with UBCH8 may impact genomic stability by regulating the basal levels of p21 to ensure proper entry into S phase for sensing DNA damage. CRL4Cdt2-UBCH8 may also regulate the response to DNA damage via the monoubiquitylation of PCNA, an activity that is required not only for translesion DNA synthesis (TLS) but also for other DNA repair pathways.
Roles of other E2s in regulating p21.
Our results also suggest that the p21 protein level is regulated in unirradiated cells, not only by UBCH8 but also by at least two other E2s (Fig. 9). Cdc34, in conjunction with CRL1Skp2, regulates p21 levels. Such dual recognition of a substrate in unirradiated cells by CRL1 and CRL4 has also been reported for Cdt1 (33). Unexpectedly, another E2, UBE2G, represses p21 mRNA also through CRL4Cdt2. Thus, at least three E2s regulate p21 levels in unirradiated cells.
Despite this complexity, overexpression of UBCH8 clearly decreases p21 levels, and knockdown of UBCH8 stabilizes p21 sufficiently to affect the cell cycle and the phospho-H2AX response. There is a great emphasis in the field of polyubiquitylation on the regulation of the substrate by the E3, with the E2s being somewhat overlooked. However, the results reported here suggest that there may be occasions where variation in the E2 level regulate the stability of a substrate sufficiently to effect changes in cellular function.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants R01 CA60499 and CA89406 to A.D. and the Jonsson Comprehensive Cancer to J.A.W. at UCLA. T.A. was supported by the Cancer Training Grant T32CA009109 and by NCI grant (KCA140774A).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 31 May 2011.
REFERENCES
- 1. Abbas T., Dutta A. 2011. CRL4Cdt2: master coordinator of cell cycle progression and genome stability. Cell Cycle 10:241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abbas T., Dutta A. 2009. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer 9:400–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abbas T., et al. 2010. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell 40:9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abbas T., et al. 2008. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 22:2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amador V., Ge S., Santamaria P. G., Guardavaccaro D., Pagano M. 2007. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol. Cell 27:462–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arias E. E., Walter J. C. 2006. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 8:84–90 [DOI] [PubMed] [Google Scholar]
- 7. Banks D., et al. 2006. L2DTL/CDT2 and PCNA interact with p53 and regulate p53 polyubiquitination and protein stability through MDM2 and CUL4A/DDB1 complexes. Cell Cycle 5:1719–1729 [DOI] [PubMed] [Google Scholar]
- 8. Bornstein G., et al. 2003. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 278:25752–25757 [DOI] [PubMed] [Google Scholar]
- 9. Bunz F., et al. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497–1501 [DOI] [PubMed] [Google Scholar]
- 10. Centore R. C., et al. 2010. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell 40:22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Glickman M. H., Ciechanover A. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373–428 [DOI] [PubMed] [Google Scholar]
- 12. Gong L., Yeh E. T. 1999. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J. Biol. Chem. 274:12036–12042 [DOI] [PubMed] [Google Scholar]
- 13. Hershko A., Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479 [DOI] [PubMed] [Google Scholar]
- 14. Higa L. A., et al. 2006. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 5:1675–1680 [DOI] [PubMed] [Google Scholar]
- 15. Higa L. A., Mihaylov I. S., Banks D. P., Zheng J., Zhang H. 2003. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5:1008–1015 [DOI] [PubMed] [Google Scholar]
- 16. Hotton S. K., Callis J. 2008. Regulation of cullin RING ligases. Annu. Rev. Plant Biol. 59:467–489 [DOI] [PubMed] [Google Scholar]
- 17. Hu J., McCall C. M., Ohta T., Xiong Y. 2004. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6:1003–1009 [DOI] [PubMed] [Google Scholar]
- 18. Hu J., Xiong Y. 2006. An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J. Biol. Chem. 281:3753–3756 [DOI] [PubMed] [Google Scholar]
- 19. Jin J., Arias E. E., Chen J., Harper J. W., Walter J. C. 2006. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23:709–721 [DOI] [PubMed] [Google Scholar]
- 20. Kim D. H., et al. 2010. The CRL4Cdt2 ubiquitin ligase mediates the proteolysis of cyclin-dependent kinase inhibitor Xic1 through a direct association with PCNA. Mol. Cell. Biol. 30:4120–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim Y., Kipreos E. T. 2007. Cdt1 degradation to prevent DNA re-replication: conserved and non-conserved pathways. Cell Div. 2:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim Y., Starostina N. G., Kipreos E. T. 2008. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 22:2507–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kolman C. J., Toth J., Gonda D. K. 1992. Identification of a portable determinant of cell cycle function within the carboxyl-terminal domain of the yeast CDC34 (UBC3) ubiquitin conjugating (E2) enzyme. EMBO J. 11:3081–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lisztwan J., et al. 1998. Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45(SKP2): evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J. 17:368–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu C., et al. 2005. Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4-Ddb1-CSN ubiquitin ligase. EMBO J. 24:3940–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mathias N., Steussy C. N., Goebl M. G. 1998. An essential domain within Cdc34p is required for binding to a complex containing Cdc4p and Cdc53p in Saccharomyces cerevisiae. J. Biol. Chem. 273:4040–4045 [DOI] [PubMed] [Google Scholar]
- 27. Nishitani H., et al. 2008. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J. Biol. Chem. 283:29045–29052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishitani H., et al. 2006. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 25:1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oda H., et al. 2010. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol. Cell 40:364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Osaka F., et al. 1998. A new NEDD8-ligating system for cullin-4A. Genes Dev. 12:2263–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pick E., et al. 2007. Mammalian DET1 regulates Cul4A activity and forms stable complexes with E2 ubiquitin-conjugating enzymes. Mol. Cell. Biol. 27:4708–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pickart C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503–533 [DOI] [PubMed] [Google Scholar]
- 33. Ralph E., Boye E., Kearsey S. E. 2006. DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep. 7:1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sansam C. L., et al. 2006. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 20:3117–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sarmento L. M., et al. 2005. Notch1 modulates timing of G1-S progression by inducing SKP2 transcription and p27 Kip1 degradation. J. Exp. Med. 202:157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Senga T., et al. 2006. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J. Biol. Chem. 281:6246–6252 [DOI] [PubMed] [Google Scholar]
- 37. Sheaff R. J., et al. 2000. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell 5:403–410 [DOI] [PubMed] [Google Scholar]
- 38. Shibutani S. T., et al. 2008. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev. Cell 15:890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Silver E. T., Gwozd T. J., Ptak C., Goebl M., Ellison M. J. 1992. A chimeric ubiquitin conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): implications for the structure, function and evolution of the E2s. EMBO J. 11:3091–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tardat M., et al. 2010. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 12:1086–1093 [DOI] [PubMed] [Google Scholar]
- 41. Terai K., Abbas T., Jazaeri A. A., Dutta A. 2010. CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol. Cell 37:143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Touitou R., et al. 2001. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 20:2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang W., Nacusi L., Sheaff R. J., Liu X. 2005. Ubiquitination of p21Cip1/WAF1 by SCFSkp2: substrate requirement and ubiquitination site selection. Biochemistry 44:14553–14564 [DOI] [PubMed] [Google Scholar]
- 44. Wu K., Chen A., Tan P., Pan Z. Q. 2002. The Nedd8-conjugated ROC1-CUL1 core ubiquitin ligase utilizes Nedd8 charged surface residues for efficient polyubiquitin chain assembly catalyzed by Cdc34. J. Biol. Chem. 277:516–527 [DOI] [PubMed] [Google Scholar]
- 45. Wu K., Kovacev J., Pan Z. Q. 2010. Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol. Cell 37:784–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu Z. K., Gervais J. L., Zhang H. 1998. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc. Natl. Acad. Sci. U. S. A. 95:11324–11329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zamudio J. R., et al. 2009. Trypanosoma brucei spliced leader RNA maturation by the cap 1 2′-O-ribose methyltransferase and SLA1 H/ACA snoRNA pseudouridine synthase complex. Mol. Cell. Biol. 29:1202–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.