Abstract
In pancreatic β cells, elevated glucose concentrations stimulate mitochondrial oxidative metabolism to raise intracellular ATP/ADP levels, prompting insulin secretion. Unusually low levels of expression of genes encoding the plasma membrane monocarboxylate transporter, MCT1 (SLC16A1), as well as lactate dehydrogenase A (LDHA) ensure that glucose-derived pyruvate is efficiently metabolized by mitochondria, while exogenous lactate or pyruvate is unable to stimulate metabolism and hence insulin secretion inappropriately. We show here that whereas DNA methylation at the Mct1 promoter is unlikely to be involved in cell-type-specific transcriptional repression, three microRNAs (miRNAs), miR-29a, miR-29b, and miR-124, selectively target both human and mouse MCT1 3′ untranslated regions. Mutation of the cognate miR-29 or miR-124 binding sites abolishes the effects of the corresponding miRNAs, demonstrating a direct action of these miRNAs on the MCT1 message. However, despite reports of its expression in the mouse β-cell line MIN6, miR-124 was not detectably expressed in mature mouse islets. In contrast, the three isoforms of miR-29 are highly expressed and enriched in mouse islets. We show that inhibition of miR-29a in primary mouse islets increases Mct1 mRNA levels, demonstrating that miR-29 isoforms contribute to the β-cell-specific silencing of the MCT1 transporter and may thus affect insulin release.
INTRODUCTION
Glucose metabolism in pancreatic β cells is specialized to efficiently couple glucose oxidation to an increase in ATP/ADP ratio, critical for stimulating insulin secretion (37). Alternative metabolic pathways that could interfere with glucose sensing are suppressed by specifically “disallowing” expression of certain genes in β cells. These disallowed genes include those encoding lactate dehydrogenase A (LDHA), which converts pyruvate to lactate (25, 39, 40), and MCT1 (SLC16A1) (14, 15, 17, 40, 45, 46), a plasma membrane monocarboxylate transporter. Both of these genes are widely expressed in other tissues but display very low expression levels in β cells (32, 40). This modification seems likely to serve a 2-fold role: first, to avoid inappropriate stimulation of oxidative metabolism, and hence insulin release, in response to circulating pyruvate or lactate; and second, to prevent the loss of glucose-derived pyruvate from β cells.
The effects of inappropriate overexpression of MCT1 are observed in the rare genetic disorder physical exercise-induced hypoglycemia (32). In this condition, autosomal dominant mutations in the MCT1 (SLC16A1) promoter lead to increased transcription of the MCT1 gene sufficient to overcome the β-cell-specific block on expression (31). During strenuous physical exercise, pyruvate and lactate produced by anaerobic metabolism in skeletal muscle are released into the bloodstream. The presence of MCT1 then appears to allow the circulating pyruvate/lactate to enter β cells, where it acts as a substrate for mitochondrial oxidation leading to an increased cytosolic ATP/ADP ratio. This triggers insulin release despite the absence of elevated blood glucose levels, resulting in hypoglycemia.
Given the critical importance of disallowing MCT1 expression in β cells, we were interested in the mechanism by which this widely expressed gene is so specifically silenced. Although mouse β cells express very low levels Mct1 mRNA, luciferase assays have demonstrated low but significant activity of exogenous Mct1 promoter sequences when transfected into these cells (31). This suggests that additional epigenetic or posttranscriptional mechanisms are responsible for further suppressing Mct1 expression in the β cell.
DNA methylation is an epigenetic modification of DNA which can regulate gene expression. In eukaryotes, DNA methylation occurs on cytidine residues of CG dinucleotides (CpG) (29). High levels of DNA methylation at gene promoters are associated with gene silencing. DNA methylation may contribute to silencing genes both in a tissue-specific manner and also for aberrantly silencing tumor suppressor genes in cancer (3).
MicroRNAs (miRNAs) are short 19- to 21-nucleotide (nt) RNAs expressed as hairpin precursors which, following processing by Dicer, can bind to sites mainly within the 3′ untranslated region (UTR) of target genes. This interaction can either block translation or can destabilize the mRNA leading to destruction of the message (6, 30, 42). A number of miRNAs have previously been implicated in β cell function. miR-375 is specifically expressed in islets and is reportedly the most abundant miRNA in β cells (35). This miRNA plays roles both in regulating insulin secretion (35) and in islet development (19, 36). miR-7 is also abundantly and specifically expressed in β cells (5, 9). miR-124 is reportedly expressed both in β cell lines (35) and in mouse islets (1) and is thought to regulate both the development (1) and the secretory function (26) of islets. miR-9 has also been shown to regulate insulin secretion by controlling the expression of granuphilin (34).
Here, we have investigated whether DNA methylation or microRNAs contribute to β-cell-specific silencing of Mct1. We show that whereas DNA methylation is unlikely to be responsible for regulating Mct1 expression in β cells, miR-29a, miR-29b, and miR-124 all target Mct1. Of these, the miR-29 isoforms are highly expressed in islets and contribute to silencing Mct1 in β cells. The maintained expression of miR-29 isoforms in the β cell thus seems likely to be required for the normal and selective stimulation of insulin secretion by glucose.
MATERIALS AND METHODS
Cell culture.
The cell lines mhAT3F (22), MIN6 (28), and HEK293 (13) and isolated mouse islets were cultured as described previously (1).
DNA methylation analysis.
MIN6 cells previously grown in medium containing 25 mM glucose were maintained at 3 mM glucose for 16 h prior to culture at 3 or 30 mM glucose in the presence or absence of 100 μM 5-azacytidine (ACT) for 6 h. Cells were harvested in Trizol, and RNA was purified according to the manufacturer's instructions. RNA was quantified using a Nanodrop-1000 spectrophotometer and then reverse transcribed using a high-capacity cDNA reverse transcription (RT) kit (Applied Biosystems) with random primers. Real-time PCR was performed using Power SYBR green PCR Mastermix (Applied Biosystems) on a 7500 fast real-time PCR machine (Applied Biosystems). Mct1 mRNA was quantified relative to cyclophilin A by using the threshold cycle (ΔΔCT) method. Results are the means of four experiments, analyzed for significant differences by Student's t test with Bonferroni correction for multiple testing.
Genomic DNA (gDNA) was prepared from MIN6 and mhAT3F cells using proteinase K digestion, phenol-chloroform extraction, and isopropanol precipitation (38). Purified gDNA was subjected to bisulfite conversion using the EpiTect bisulfite kit (Qiagen) and then amplified by PCR using primers designed with MethPrimer (http://www.urogene.org/methprimer/) (24). (All primer sequences are given in a table in the supplemental material.) Products were ligated into pCR2.1 by TA cloning, and the inserts were sequenced. Sequence reads were analyzed using BiQ Analyzer (4) (http://biq-analyzer.bioinf.mpi-inf.mpg.de) to exclude poorly converted sequences and multiple copies of a single clone. Five independent clones were included for each analysis. For bisulfite sequencing of islets, DNA was prepared using a protocol based on the method of Millar et al. (27). Ten islets isolated from a CD1 mouse were placed in 10 μl lysis buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA, 1 mM SDS, 280 μg/ml proteinase K), heated to 37°C for 90 min and 98°C for 15 min, and then cooled on ice. The whole 20-μl incubation was used as the input for the bisulfite conversion reaction, resulting in approximately 750 cell equivalents per PCR.
Bioinformatic analysis of microRNAs.
Data containing the raw counts for mature miRNA detected in various mouse and human tissues and cell lines by large-scale cloning and sequencing were obtained from the supplemental data of Landgraf et al. (21). To exclude cancerous samples, libraries were only included with malignancy code 1. Libraries containing fewer than 100 counts were also excluded. The miRNA expression profile of the mouse MIN6 β cell line was compared to that of other mouse samples. Islet α and β cells have relatively similar miRNA profiles: for example, both highly express miR-375. As we aimed to assess the degree to which miRNAs expressed in β cells are also expressed in nonislet cells, the α-TC1 library was also excluded from the analysis. The following libraries were therefore included: MIN6 pancreatic β cells, brain, glomeruli, heart, kidney, kidney-collecting duct, liver, lung, midbrain, ovary, placenta, skin, spermatogonia-C18, and testis. The miRNA expression profile of human islets was also compared to those of all tissue samples fulfilling the criteria described above, which comprised cerebellum, frontal cortex, midbrain, hippocampus, liver, heart, spleen, pituitary, thyroid, ovary, testis, uterus, placenta, epididymis, and prostata.
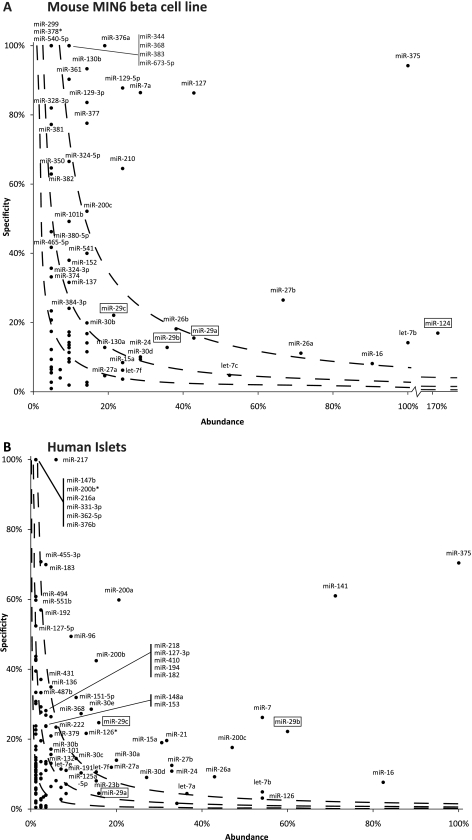
Counts for each miRNA were converted to counts per million (CPM) miRNAs detected in each library. The abundance of miRNAs in β cells/islets was calculated as a percentage of the abundance of miR-375 as this has been widely reported to be the most abundant miRNA in β cells and islets (5, 35). Specificity of expression to β cells was calculated by dividing the CPM for an miRNA in the β cell/islet library by the total CPM for that miRNA in all libraries included from that species. A combined score for abundance and specificity was calculated by multiplying the percentage scores for each. Lines dividing the quartiles of the ranked distribution of this combined score are marked on the graphs in Fig. 2.
Fig. 2.
miRNA expression in MIN6 mouse β cells (A) and human islets (B) relative to other tissues. The abundance of miRNAs is presented as the percentage of expression relative to miR-375. Specificity represents that percentage of miRNA found in β cells/islets relative to the total level found across all tissue types of the appropriate species. The miRNAs were ranked according to the combined abundance and specificity scores, and the quartiles of this rank distribution are marked by dashed lines on the graph. miR-29 isoforms and miR-124 are highlighted with boxes.
DNA constructs.
miRNA expression vectors were created by first amplifying the coding sequence of selected miRNAs along with 230- to 300-bp flanking sequences on either side by PCR using mouse genomic DNA as template. A list of primers used is given in the supplemental material. The resulting PCR products were inserted between the BamHI and XbaI sites of pcDNA3.1(+).
Firefly luciferase-expressing vectors were constructed as follows. A BglII-to-NheI fragment containing the herpes simplex virus thymidine kinase (TK) promoter was inserted between BglII and HindIII sites of pGL3 Basic to create pGL3-TK, in which firefly luciferase cDNA, followed by a simian virus 40 (SV40)-derived polyadenylation signal, is expressed from the TK promoter. Regions covering the entire human and mouse MCT1 3′ UTR were amplified by PCR and inserted between XbaI and BamHI sites of pGL3-TK to replace the SV40 polyadenylation signal and create pGL3-TK-HsM3 and pGL3-TK-MmM3, respectively. Putative microRNA binding sites were mutated by amplifying the whole vector excluding the 8-bp seed region from a pair of primers containing an additional restriction enzyme site, using high-fidelity Phusion polymerase (Finnzyme). The product was digested using that restriction enzyme and ligated to recircularize the plasmid such that the seed region was replaced with the restriction enzyme site.
Luciferase assay of microRNA binding.
The effect of overexpressed miRNAs on the Mct1 3′ UTRs was assessed in HEK293 cells. Cells grown in 24-well plates were transfected with the following plasmids using Lipofectamine 2000 (Invitrogen): 50 ng firefly luciferase reporter plasmid, 50 ng pRL-TK control plasmid, and 300 ng miRNA expression plasmid. In tests of multiple miRNAs, the following mixture of DNA was used: 25 ng pRL-TK, 25 ng firefly luciferase reporter plasmid, 250 ng each miRNA expression plasmid, and sufficient empty miRNA expression vector (pcDNA3.1) to make the total up to 800 ng. Cells were harvested 48 h later and assayed with the dual-luciferase assay system (Promega). Two levels of controls were used to exclude from the results variation due to transfection efficiency and nonspecific effects of miRNAs. First, the firefly luciferase signal was normalized to the Renilla luciferase signal for each reading. Second, each miRNA was assessed in parallel with firefly plasmids containing a 3′ UTR (pGL3-TK-HsM3 or pGL3-TK-MmM3) or just the SV40 polyadenylation signal (pGL3-TK) and the signal from the former normalized to the latter. All results were then presented as a percentage of the signal obtained with an empty miRNA expression vector. All results are the mean of at least three independent experiments. Significant downregulation of normalized luciferase expression was identified using one-tailed Student's t test with Bonferroni correction for multiple tests.
The effect of endogenous β cell miRNAs on the Mct1 3′ UTR were assessed by luciferase assay in MIN6 cells. Cells grown in 24-well plates were transfected with 50 ng pRL-TK control plasmid along with 50 ng of firefly luciferase plasmid containing either the wild-type mouse Mct1 3′ UTR (pGL3-TK-MmM3) or a plasmid in which the miR-29 or miR-124 binding sites had been mutated by site-directed mutagenesis.
Effects of miRNA overexpression on endogenous Mct1.
Mouse hepatoma-derived mhAT3F cells, grown in 12-well plates, were transiently transfected with 1.6 μg of various miRNA expression vectors or an empty vector control, using Lipofectamine 2000. Cells were harvested 72 h later in TRIzol (Invitrogen), and total RNA was prepared according to the manufacturer's instructions. Mct1 mRNA levels were quantified relative to cyclophilin A using an Applied Biosystems high-capacity reverse transcription kit followed by quantitative PCR (qPCR) using Power SYBR green master mix (Applied Biosystems) running on a 7500 fast real-time PCR system (Applied Biosystems). Data are means of three independent experiments, and significant downregulation of the Mct1 level was identified using one-tailed Student's t test with Bonferroni's correction for multiple tests.
mhAT3F cells grown in 6-well plates were stably transfected with 4 μg linearized miRNA expression vector or empty expression vector, using Lipofectamine 2000. Twenty-four hours after transfection, cells were passaged into 75-cm2 flasks and then transferred to selective medium containing 250 μg/ml G418 24 h later. Stably transfected cells were kept as a polyclonal mixture resulting from presumably many individual stable transfection events and were maintained in 125 μg/ml G418.
Protein samples were separated by SDS-PAGE, transferred to nitrocellulose membrane, and then probed with a rabbit anti-rat Mct1 antibody followed by a horseradish peroxidase (HRP)-conjugated secondary antibody. The signal was detected using Supersignal West Dura substrate (Pierce) on a Chemidoc XRS imager (Bio-Rad). Blots were reprobed with mouse anti-β-tubulin antibody followed by an HRP-conjugated secondary antibody.
Detection of microRNA expression by stem-loop reverse transcription-PCR.
Mature microRNAs were specifically detected using a modified version of the stem-loop RT-PCR protocol described by Chen et al (7). miRNA expression during luciferase assays in HEK293 cells was confirmed by performing RT-PCR on heat-treated cell lysates. Cells grown in 24-well plates were transfected as for the luciferase assay and harvested 48 h later in 100 μl phosphate-buffered saline (PBS) supplemented with 10 mM EDTA and then heated to 95°C for 5 min, diluted with an equal volume of water. For mhAT3F cells, total RNA was purified using TRIzol as described above and then diluted to 2 ng/μl. RNA samples were reverse transcribed using stem-loop primers designed to specifically transcribe mature miRNA molecules at a concentration of 50 mM in 15-μl reaction mixtures also containing 1× RT buffer (P/N; 4319981 [Applied Biosystems]), 1 mM deoxynucleoside triphosphate (dNTP) mix, 2.67 U/μl Multiscribe (Applied Biosystems), and 0.63 U/μl RNase inhibitor (Applied Biosystems), which were incubated at 16°C for 30 min, 42°C for 30 min, and then 85°C for 5 min. The cDNA was amplified by PCR with a pair of primers, one of which was complementary to part of the reverse transcription stem-loop primer and the other of which was complementary to the miRNA in 50-μl reaction mixtures containing 1 × Phire reaction buffer (Finnzyme), 0.2 mM dNTP mix, 0.5 μM (each) forward and reverse primers, 3.5 μl reverse transcription reaction mixture, and 1 μl Phire hot-start polymerase (Finnzyme). The products were visualized by agarose gel electrophoresis. Controls without the reverse transcriptase enzyme confirmed that the primers were specific to miRNAs and could not amplify the expression vectors (data not shown). As the recombinant Moloney murine leukemia virus (rMo-MuLV) reverse transcriptase recognizes either RNA or DNA as a template, DNA oligonucleotides corresponding to the mature miRNA were used a positive control for each miRNA assay at a concentration of 1 pM in the reverse transcription reaction. As a loading control, the U6 small nuclear RNA was also amplified using stem-loop RT-PCR from each sample.
Measurement of miRNAs using quantitative stem-loop RT-PCR.
Levels of endogenous miRNAs were measured in total RNA samples with relative quantification by quantitative RT-PCR (qRT-PCR) using standard curves. Total RNA was prepared from three cultures of MIN6 cells and from islets isolated from three male C57BL/6 mice aged 26 weeks. Five-point standard curves were prepared from DNA oligonucleotides corresponding to mature miRNAs in the range of 200 pM to 20 fM (miR-375), 100 pM to 10 fM (miR-29a, miR-29b, miR-29c, and miR-7a), or 1 pM to 100 aM (miR-124) (final concentration in reverse transcription reaction). DNA oligonucleotides were used in place of RNA oligonucleotides for the standard curve as they are both less expensive and more stable. In test experiments with miRNA-29a and miR-124 sequences, the two template types produced very similar results (see the figure in the supplemental material). RNA samples were diluted to 10 ng/μl, and separate reverse transcription reactions, as described above, were performed for each sample and standard curve point. Reverse transcription reaction mixtures were diluted 4-fold, and 5 μl was used in 20-μl qPCRs using Power SYBR green master mix (Applied Biosystems) with forward and reverse primers at 0.2 μM. Reactions were run on a 7500 fast real-time PCR system (Applied Biosystems), and individual miRNAs were first quantified by using the standard curves and then presented as a percentage of the miR-375 expression of that sample to permit comparison with the miRNA quantification by large-scale cloning. To test the specificity of the miR-29 isoform assays, 1 pM DNA oligonucleotide template for each miR-29 isoform was tested in each miR-29 isoform assay. miR-29a and miR-29b assays failed to detect either of the other two isoforms; however, the miR-29c assay detected the miR-29a oligonucleotide at 22% ± 1.19% (mean ± standard deviation [SD]) of the level at which it detected the miR-29a oligonucleotide.
miRNA knockdown with LNA inhibitors.
Pancreatic islets were isolated from C57BL/6 mice, cultured overnight, and then transfected with locked nucleic acid (LNA) miRNA inhibitors (Exiqon) targeting either miR-29a or miR-29b. Islets grown in 3 ml complete medium in 60-mm petri dishes were transfected with 25 nM LNA using 12 μl TransIT-TKO (Mirus) according to the manufacturer's instructions. Islets were harvested 48 h later, and total RNA was purified using TRIzol. Data are the means of three independent experiments performed on islets isolated from three separate mice.
RESULTS
DNA methylation at the promoter CpG island of Mct1 is not responsible for β-cell-specific silencing.
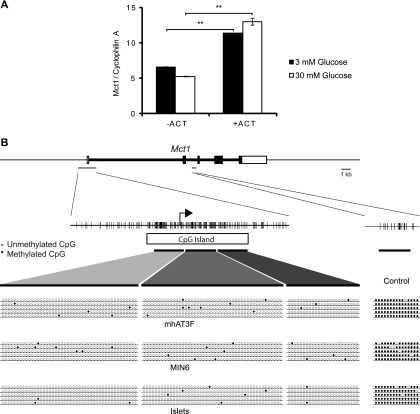
Initially, the DNA methylation inhibitor 5-azacytidine (ACT) was used to investigate the potential involvement of DNA methylation in Mct1 expression in the mouse β cell line MIN6 (28). Cells were “starved” for 16 h in culture medium containing 3 mM glucose and then treated with 1 mM ACT for 6 h in medium containing 3 or 30 mM glucose. Mct1 mRNA was quantified by quantitative reverse transcription-PCR (qRT-PCR) using SYBR green. In both high- and low-glucose media, ACT treatment increased Mct1 mRNA levels (P < 0.01) (Fig. 1 A). These data indicate that, either through direct or indirect mechanisms, DNA methylation may be acting to suppress Mct1 expression in β cells. To identify whether this was a direct effect of DNA methylation of the Mct1 promoter, bisulfite sequencing (8) was performed and the methylation status at this locus was assessed.
Fig. 1.
The Mct1 promoter is not methylated in pancreatic β or hepatoma cells. (A) Mct1 mRNA quantified relative to cyclophilin A by qRT-PCR from MIN6 cells treated with DNA methylation inhibitor 5-azacytidine (ACT) in 3 or 30 mM glucose. (B) Bisulfite sequencing of the CpG island associated with the Mct1 promoter in the liver cell line mhAT3F, β cell line MIN6, and in isolated pancreatic islets. The arrow marks the transcriptional start site, and individual CpGs are marked with vertical dashes. The thick dark lines represent the regions amplified by PCR for sequencing, with the results from five independent clones for each cell type shown below. Open circles represent unmethylated CpG and closed circles methylated ones. As a positive control for the bisulfite sequencing, an internal region of the Mct1 gene was also analyzed.
Unmethylated cytidines were converted to uracil by bisulfite conversion of genomic DNA from both MIN6 cells and a mouse liver cell line, mhAT3F, which expresses a high level of Mct1. Three contiguous regions of the Mct1 promoter covering almost the entire CpG island were amplified by PCR, and the methylation status of individual CpGs was determined by sequencing (Fig. 1B).
The promoter region displayed a very low level of methylation in both cell lines, with no differences apparent between the two (Fig. 1B). To determine whether the methylation status of the β cell line MIN6 accurately represented the in vivo situation in primary β cells, the same analysis was also performed on isolated mouse islet tissue. The Mct1 promoter was examined in freshly isolated pancreatic islets and was again found to display a very low level of DNA methylation. To control for the possibility that demethylation might occur during the procedure, another region of the Mct1 gene, expected to be highly methylated, was also analyzed. A region was chosen which was distant from a CpG island such that its CpGs were likely to be methylated. In both cell lines and in isolated islets, this region was indeed found to be highly methylated, confirming the validity of the analysis of the Mct1 promoter (Fig. 1B). We therefore conclude that the CpG island associated with the Mct1 promoter is not methylated in β cells and therefore that DNA methylation at this locus is not responsible for the β-cell-specific silencing of Mct1.
miRNA expression analysis in mouse β cells and human islets.
We next investigated whether miRNAs may contribute to silencing Mct1 expression in β cells. To this end, we needed to identify both miRNAs that specifically target Mct1 and also determine which miRNAs are expressed in β cells or islets. Both the abundance and specificity of expression are expected to affect the ability of a particular miRNA to perform β-cell-specific functions. While previous analyses have described the most abundant miRNAs in β cells (35) or those most differentially expressed between two tissues (5), a comprehensive analysis comparing the β cell miRNA complement with those of a range of other tissues has not yet been published. We therefore aimed to generate in silico a measure of both abundance and specificity of miRNA expression in β cells.
This analysis was performed using publically available data from large-scale cloning and sequencing of miRNAs in various tissues from both humans and mice (21). Data from human islets or mouse MIN6 β cells were compared to miRNA expression in other nonmalignant, differentiated, adult tissues of the same species. In Fig. 2, the abundance of individual miRNAs is presented as a percentage of miR-375 expression, as this has previously been identified as the most abundant miRNA in islets and β cells (5, 35). The specificity of miRNA expression was determined as the percentage of the total normalized counts for a particular miRNA detected in all tissues that were found in β cells/islets (Fig. 2). Finally, to combine these two measures, we ranked miRNAs according to the product of the abundance and specificity scores, as a predictor of how likely a miRNA is to perform β cell- or islet-specific functions. The quartiles (Q) of this distribution are also presented (Fig. 2, dashed lines). Thus, miRNAs in Q4 have the highest combination of abundance and specificity and those in Q1 the lowest. This analysis agrees with previous work which identified miR-375 and miR-7 as islet-specific miRNAs (5, 35), but also scores all other miRNAs detected in mouse MIN6 β cells or human islets on their likelihood of contributing to β-cell/islet-specific phenotypes.
MicroRNAs are predicted to target Mct1.
To identify miRNAs capable of targeting Mct1, we bioinformatically identified potential miRNA binding sites in the Mct1 3′ UTR and then experimentally tested whether the putative sites were indeed functional. Since miRNAs bind their targets with incomplete homology, and the requirements for active binding have not been well characterized, the currently available tools have a relatively poor predictive value. Algorithms for predicting miRNA binding sites in a single mRNA often produce a large number of false positives. These were reduced considerably using tools which searched for miRNA binding sites conserved between orthologous genes in several species. The Human MicroRNA Targets tool (http://cbio.mskcc.org/cgi-bin/mirnaviewer/mirnaviewer.pl) (18), which uses the miRanda algorithm, identified five miRNAs with binding sites conserved between the human, mouse, and rat Mct1 3′ UTRs. The PicTar algorithm (http://pictar.mdc-berlin.de) (20), searching for binding sites conserved across vertebrate orthologues, identified six miRNAs showing considerable overlap with the previous algorithm (Table 1).
Table 1.
miRNAs predicted to target Mct1a
| miRNA | Result with prediction tool: |
Quartile in: |
||
|---|---|---|---|---|
| miRanda | PicTar | Mouse MIN6 β cell line | Human islets | |
| miR-9* | + | + | ||
| miR-29a | − | + | Q3 | Q3 |
| miR-29b | + | + | Q3 | Q4 |
| miR-29c | − | + | Q3 | Q4 |
| miR-124 | + | + | Q4 | |
| miR-183 | + | − | Q4 | |
| miR-320 | − | + | Q1 | |
| miR-377 | + | − | Q4 | |
Listed are miRNAs predicted have conserved binding sites in the Mct1 3′ UTR by the miRanda and PicTar algorithms. The quartile within the distribution of the combined abundance and specificity score of the individual miRNAs in both MIN6 cells and human islets are also given.
A different version of the miRanda algorithm (http://www.microrna.org) (2), which includes sites not highly conserved between species, predicted 160 miRNAs targeting human MCT1 and 83 targeting mouse Mct1. While this number of miRNAs was unfeasible to validate experimentally, we noted that both miR-375 and miR-7 were predicted to target human MCT1. Since these have previously both been identified as specifically and highly expressed in pancreatic islets (5, 35), they were included in the list of miRNAs which we went on to examine experimentally.
All three miR-29 isoforms and miR-124 target Mct1.
To identify miRNAs capable of directly targeting the Mct1 3′ UTR, we assessed our candidate miRNAs by luciferase assay. This assay quantified the ability of overexpressed miRNAs to reduce expression of firefly (Photinus pyralis) luciferase from an mRNA bearing the Mct1 3′ UTR.
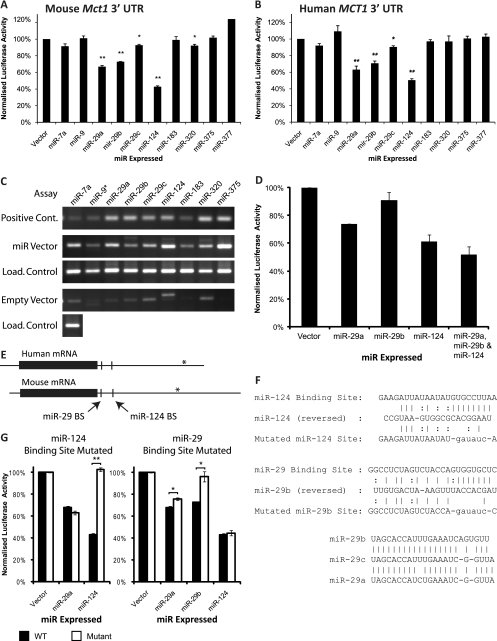
miRNA expression plasmids were constructed by amplifying the corresponding miRNA gene with short flanking sequences from mouse genomic DNA and then cloning this downstream of a polymerase II promoter in a mammalian expression vector (pcDNA3.1). Expression of the correct mature miRNA from these vectors was confirmed by RT-PCR using stem-loop primers (Fig. 3 C). miR-124 and miR-375 were also expressed by cloning annealed oligonucleotides into the miRNA expression vector pcDNA 6.2 EmGFP-miR. In both cases, very similar results were produced from the different types of vector, so results from different vectors expressing the same miRNA were averaged.
Fig. 3.
Reporter-based assays of the impact of miRNAs targeting the Mct1 3′ UTR in HEK293 cells. Luciferase assays were performed to assess the effect of miRNA overexpression on mouse (A) or human (B) Mct1 3′ UTR. The level of firefly luciferase activity, normalized against Renilla luciferase, is presented as a percentage of signal obtained with an empty miRNA expression vector. (C) Mature miRNAs expressed in samples from a parallel transfection were detected by reverse transcription using stem-loop primers followed by PCR. Assays for each miRNA were performed on samples transfected with the appropriate miRNA expression vector and on an empty vector control. U6 snRNA was assayed in all samples as a loading (load.) control. As a positive control for each miRNA assay, 1 pM synthetic DNA oligonucleotide corresponding to the mature miRNA sequence was assayed in parallel. (D) Luciferase assay of the combined effects of miR-29a, miR-29b, and miR-124 expression on mouse Mct1 3′ UTR. (E) Mct1 mRNA from humans and mice with open reading frames marked with solid boxes. The locations of putative miR-29 and miR-124 binding sites (BS) are also indicated. In both species, cDNAs with shorter 3′ UTRs are reported in databases, and the sites of alternative transcription termination sites are marked with asterisks. (F) Base pair binding between miRNAs and putative miRNA binding sites in the mouse Mct1 3′ UTR. The binding sites were mutated by replacing the 8-bp seed region with a restriction enzyme site. An alignment of the three miR-29 isoforms is also shown. (G) Luciferase assay of mouse Mct1 3′ UTR with the putative miRNA binding sites mutated.
The whole 3′ UTR of mouse Mct1 was cloned downstream of the firefly luciferase coding sequence, whose expression was driven by the herpes simplex virus thymidine kinase promoter (TK). A second vector used the same TK promoter to express Renilla reniformis luciferase, followed by an SV40 polyadenylation signal. The two vectors were cotransfected into HEK293 cells along with various miRNA expression vectors, and the ratio of the two luciferases was detected by dual-luciferase assay. To exclude any confounding effects of miRNAs binding to the luciferase open reading frames (ORFs), data were normalized to a parallel experiment that examined the effect of miRNA expression on firefly luciferase followed by an SV40 polyadenylation signal rather than the Mct1 3′ UTR (Fig. 3A). We also performed the same analysis using the human MCT1 3′ UTR on the basis that evolutionary conserved interactions are more likely to be biologically meaningful (Fig. 3B).
A one-tailed Student's t test identified five miRNAs which significantly reduced expression of firefly luciferase by targeting the mouse Mct1 3′ UTR (Fig. 3A). Four of these also significantly targeted the human MCT1 3′ UTR (Fig. 3B). miR-124 exerted the largest effect, decreasing luciferase expression by at least 50% in assays of both mouse and human sequences (P < 0.00001). miR-29a reduced expression by ≥33% (P < 0.01), and miR-29b reduced expression by ≥27% (P < 0.01) in both cases. A smaller effect was produced by miR-29c which reduced expression by ≥7% (P < 0.05). Transfection with miR-7 produced a small but nonsignificant decrease in luciferase activity for both human and mouse Mct1 sequences. Thus, 4 out of the 10 miRNAs tested significantly target both human and mouse Mct1 3′ UTRs: miR-124 and the three isoforms of miR-29. Of these, miR-29a, miR-29b, and miR-124 were capable of reducing expression by over one-quarter.
To test the combined effect of multiple miRNAs on the mouse Mct1 3′ UTR, miR-29a, miR-29b, and miR-124 were tested individually and together by luciferase assay (Fig. 3D). This confirmed that the combined effect of the three miRNAs was greater than the effect of each miRNA individually.
miR-29a, miR-29b, and miR-124 target Mct1 directly: mutational analysis of binding.
The controls used in the above luciferase assays excluded from the analysis any effects of miRNA expression on the promoter or coding sequence of either luciferase variant. The assay therefore identified direct interactions between the miRNA and Mct1 3′ UTR. To provide further confirmation of the specificity of the interaction and to confirm the precise binding site, we mutated the predicted binding sites and repeated the luciferase assay.
The predicted binding sites for both miR-29b and miR-124 are found in the first 150 bp of the 3′ UTRs (Fig. 3E). PicTar predicted that miR-29a and miR-29c may also potentially target the miR-29b site. The 6 to 8 nucleotides at the 3′ end of miRNA binding sites, termed the “seed region” (10), are highly complementary to the miRNA sequence and critical for binding. In both of the putative miR-29 and miR-124 binding sites, the 8 bp of the seed region were replaced with a restriction enzyme site with minimal complementarity to the miRNA sequence (Fig. 3F). Mutation of the putative miR-124 site abolished the effect of miR-124 while leaving the action miR-29a unaffected (Fig. 3G). This confirmed that the former region is an miR-124 binding site.
Mutation of the putative miR-29 binding site led to the unexpected finding that while this virtually abolished the action of miR-29b, miR-29a was still able to target the mutated 3′ UTR albeit with an efficacy reduced by ∼25% (P < 0.05) (Fig. 3G). This finding confirmed that this region forms an miR-29b binding site, and while miR-29a does bind this site, it also acts through binding at other sites within the Mct1 3′ UTR.
miR-29a, miR-29b, and miR-124 target endogenous Mct1 mRNA.
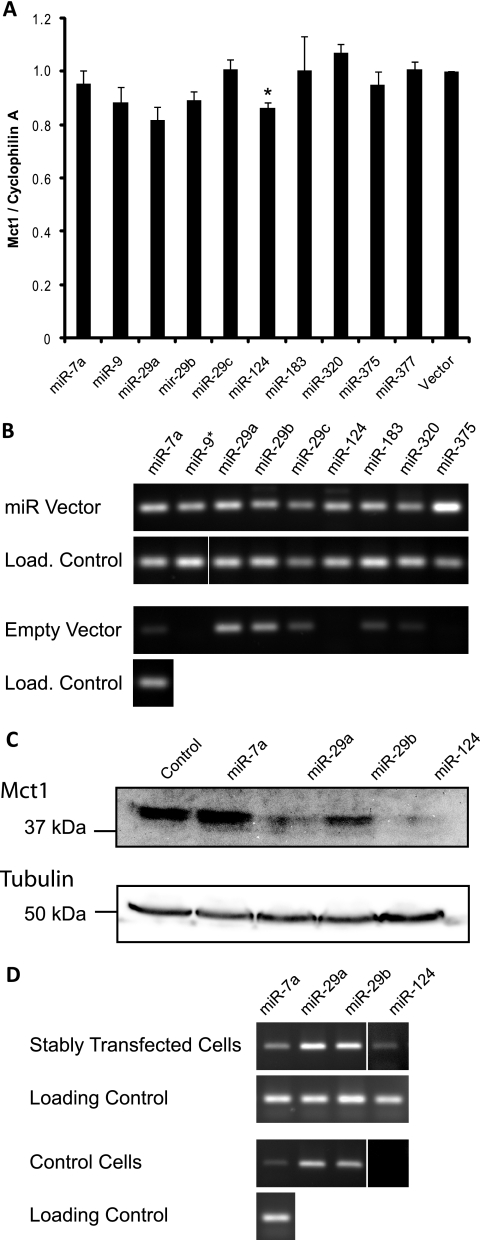
As an independent assay of miRNA function, we also assessed the ability of each of the above miRNAs to target endogenous Mct1 mRNA. miRNAs predicted to target Mct1 were overexpressed in a mouse liver cell line, mhAT3F, which expresses a high level of Mct1 consistent with high levels of Mct1 protein expression in this tissue (15, 31). The effect on endogenous Mct1 mRNA abundance was quantified by qRT-PCR (Fig. 4A). Overexpression of miR-124 produced a statistically significant decrease in the endogenous Mct1 mRNA level. This confirms that miR-124 is able to target naturally occurring Mct1 mRNA and that this results in mRNA degradation. Although the results for miR-29a and -29b fell below the level of statistical significance, the pattern of effects observed in the luciferase assay (Fig. 3A and B) were broadly replicated. miR-29a and miR-29b thus both tended to cause decreases in Mct1 mRNA levels, with miR-29a exerting a larger effect than miR-29b. This suggests that miR-29a and miR-29b may target Mct1 at least partially through mRNA degradation.
Fig. 4.
Effect of miRNA overexpression on endogenous Mct1 in the mhAT3F hepatocyte cell line. (A) The endogenous Mct1 mRNA level in mhAT3F cells was quantified by qRT-PCR following transient transfection with miRNA expression vectors. (B) Overexpression of mature miRNAs was confirmed by RT-PCR with stem-loop primers. Assays for each miRNA were performed on samples transfected with the appropriate miRNA expression vector and also on an empty vector control. U6 snRNA was also assayed in each sample as a loading (Load.) control. (C) mhAT3F cells were stably transfected with miRNA expression vectors or an empty expression vector as a control. Endogenous Mct1 protein detected by Western blotting, and the blot was reprobed for β-tubulin as a loading control. (D) Mature miRNAs were detected by RT-PCR with stem-loop primers in cells stably transfected with miRNA expression vectors relative to the empty vector control. U6 snRNA was assayed in all samples as a loading control. Note that the contrast for lane 4 (miR-124) was enhanced equally in both stably transfected and control cells with respect to lanes 1 to 3 to allow visualization of the band.
To overcome problems caused by limited transfection efficiency, mhAT3F cells were stably transfected with expression vectors for miRNAs shown to target Mct1 (miR-29a, miR-29b, and miR-124), miRNAs shown not to target Mct1 (miR-7a), or an empty vector control. Expression of mature miRNAs in the stably transfected cell lines was confirmed by RT-PCR using stem-loop primers (Fig. 4D). As in the transient transfection experiments some endogenous expression of miR-29a and miR-29b was detected in the control cell line consistent with low expression of miR-29 detected in human liver tissue. Stable transfection with each of the miRNA expression vectors led to a moderate increase in expression of that miRNA. The level of Mct1 protein in the stable cell lines was detected by Western blotting of whole-cell lysates (Fig. 4C). The anti-Mct1 antibody detected a single band of ∼43 kDa that was greatly reduced in samples expressing miR-29b and virtually abolished in samples expressing miR-29a or miR-124. It is interesting to note that these dramatic changes in Mct1 protein level are achieved by relatively modest expression of miRNAs, partly because continued drug selection presumably ensured consistent miRNA expression across the whole-cell population.
miR-29 downregulates Mct1 in primary mouse islets.
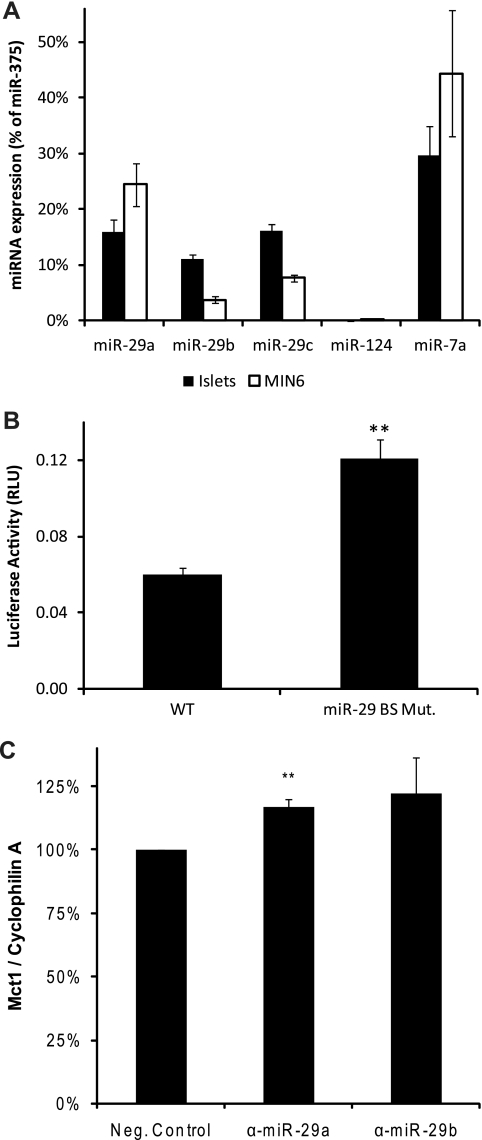
Having confirmed that miR-29a, miR-29b, and miR-124 can specifically and significantly target Mct1, we examined which of these may act in primary β cells within islets. All three members of the miR-29 family fall within the top two quartiles in both MIN6 cells and human islets. Indeed, miR-29b is the fourth most abundant miRNA in human islets. This indicates that miR-29 isoforms are both abundant and enriched in islets and are therefore highly likely to contribute to the islet phenotype. Although miR-124 was abundantly present in MIN6 cells, it was not detected in human islets. To confirm these data from large-scale sequencing, we measured expression levels of miR-29 isoforms and miR-124 in both MIN6 cells and mouse pancreatic islets by qRT-PCR with stem-loop primers, and their expression was calculated relative to that of miR-375 (Fig. 5A). Expression of all three miR-29 isoforms was confirmed in both mouse islets and MIN6 cells. In islets, miR-29a and miR-29c were detected at >15% of the miR-375 level, and miR-29b was detected at >10%. The levels of the three isoforms were more variable in MIN6 cells, ranging from miR-29a at 24% to miR-29b at 4%. While miR-124 was abundantly detected in the MIN6 cells used for large-scale cloning (21), the level detected in MIN6 cells grown for these experiments by qRT-PCR was substantially lower at just 0.24% ± 0.032% (mean ± standard error [SE]) of the miR-375 level. Indeed, in the study that provided the data for the bioinformatic analysis (21), Landgraf et al. reported failing to detect mature miR-124 in mouse or human islets and considered the presence of miR-124 in MIN6 cells to be an artifact of cell immortalization. In agreement with this finding, levels of miR-124 in islets, as given by our qRT-PCR analysis, were below the level of detection. Thus, the lowest point of the standard curve for miR-124 which detected a higher signal than the no-template control was 1 fM, equivalent to 0.19% of the mean level of miR-375 detected in the islet samples. We therefore conclude that, in the mouse islets studied here, the miR-124 level is <0.19% of the miR-375 level, and at this level we consider miR-124 unlikely to play a significant role in β cells, by far the most abundant islet cell type.
Fig. 5.
(A) Mature miRNAs expressed in mouse islets and the MIN6 β cell line were measured by qRT-PCR using stem-loop primers. Quantification is presented as a percentage of the miR-375 level to facilitate comparison with Fig. 2. (B) Luciferase assay of endogenous miRNA function in MIN6 β cells. MIN6 cells were transfected with firefly luciferase plasmids containing the mouse Mct1 3′ UTR along with control Renilla luciferase plasmid. Expression from the plasmid containing the wild-type (WT) 3′ UTR was compared with that of mutants in which the miR-29 binding site was disrupted. The increase in expression caused by these mutations indicates the extent to which endogenous levels of miR-29 reduce Mct1 expression. RLU, relative light units. (C) Isolated mouse islets were transfected with LNA miRNA inhibitors, and Mct1 mRNA levels were measured relative to cyclophilin A. α-miR-29 and α-miR-29b, anti-miR-29a and -29b LNAs, respectively.
To assess the effect of endogenous β cell miRNAs on the Mct1 3′ UTR, we compared levels of luciferase expression in MIN6 β cells from constructs containing the wild-type Mct1 3′ UTR with those containing mutated miR-29b binding sites (Fig. 5B). Destruction of the miR-29 binding site doubled expression of luciferase (P < 0.01), showing that the levels of miR-29 isoforms in β cells are sufficient to downregulate Mct1.
Finally, locked nucleic acid (LNA) miRNA inhibitors designed against miR-29a and miR-29b were used to investigate the effect of miR-29 isoforms on Mct1 expression in isolated mouse islets. Since it was not practical to obtain sufficient islet tissue to detect changes in the level of this nonabundant protein by Western (immuno-) blotting, the effect of its miRNA-mediated knockdown on Mct1 mRNA concentrations was analyzed by qRT-PCR. Transfection with the miR-29a inhibitor produced a 17% increase in the Mct1 mRNA level (P < 0.01) (Fig. 5C). While the miR-29b inhibitor produced a slightly greater average increase in Mct1 expression, the variability between experiments meant that this effect did not reach statistical significance.
Treatment of whole isolated islets is predicted to transfect efficiently only the outer cell layer (11), and measurement of mRNA, rather than protein levels, will not detect any translational inhibition by miRNAs. Despite these technical limitations, we were still able to detect that the blockade of miR-29a action led to a significant increase in Mct1 expression. Thus, Mct1 expression is regulated by miR-29a in primary mouse islets.
DISCUSSION
The principal aim of this study was to elucidate the molecular mechanism(s) through which MCT1/SLC16A1 expression is selectively and potently inhibited in the pancreatic β cell (17, 40, 46). While recent studies describing mutations in the MCT1 promoter that lead to exercise-induced hyperinsulinism in humans (31) have implicated transcriptional inhibition as an important mechanism, it is likely that such mutations affect MCT1 expression globally rather than interfering with the β-cell-specific repression of transcription. We therefore sought here to examine both epigenetic changes, which may be involved acting in cis, and posttranscriptional mechanisms, including the targeting of Mct1 mRNA by miRNAs.
Regulation of Mct1 in β cells by DNA methylation.
Although promoter-associated CpG islands are generally held to remain constitutively hypomethylated, recent genome-wide studies have revealed that up to 8% of promoter-associated CpG islands are methylated in at least one tissue, such that methylation may be involved in developmental silencing at least of a subset of genes (16, 44). However, whereas global inhibition of DNA methylation increased expression of Mct1, we found that the CpG island associated with the Mct1 start site is hypomethylated in β cells and islets. Despite this apparent contradiction, DNA methylation may conceivably be acting at many levels rather than directly at the Mct1 promoter to decrease its expression. For example, DNA methylation at other loci may regulate the expression of transcription factors which in turn regulate the Mct1 promoter. So while DNA methylation may play a role in β-cell-specific Mct1 silencing, this is unlikely to be via altered Mct1 transcription.
Regulation of Mct1 expression in β cells by miRNAs.
We used various functional assays to assess which miRNAs were capable of targeting Mct1. The luciferase assay of the effect of miRNA overexpression on the Mct1 3′ UTR provides a quantitative means of detecting direct interactions between miRNA and 3′ UTR. The striking similarity between the results from luciferase assays using the mouse and human Mct1 3′ UTRs suggests that miRNA regulation of Mct1 is well conserved between these species and likely to perform an important biological function. Several miRNAs that showed significant abundance and selectivity in β cells (i.e., in Q4), including miR-377 (predicted to interact with MCT1 3′ UTR by two of the bioinformatic tools used), as well as miR-7a and miR-375 (interaction predicted by a single tool), were not found to interact directly with Mct1 mRNA, highlighting the importance of validating all miRNA-binding predictions with biochemical assays. However, we show that miR-29a, miR-29b, and miR-124 can all target endogenous Mct1, resulting in decreased expression at the protein level. miR-124 was also found to significantly decrease the mRNA level in mhAT3F cells, and consistent but nonsignificant decreases were also caused by miR-29a and miR-29b, suggesting that these miRNAs act at least partly through mRNA degradation.
Through mutation of the mouse 3′ UTR, we confirmed the identities of both the miR-29b and miR-124 binding sites. Of note, the binding site for miR-124 we describe here was also described recently by Li et al. (23) as contributing to Mct1 upregulation in medulloblastoma cells. Using these mutated 3′ UTRs, we showed that at endogenous levels within clonal β cells, these miRNAs produce a large decrease in the expression of Mct1 through binding to the miR-29 binding site we have mapped in the Mct1 3′ UTR.
Unexpectedly, we found that miR-29a was still able substantially to affect message stability or translation via the Mct1 3′ UTR after mutation of the identified miR-29 site, a maneuver that substantially blocked miR-29b action and was the predicted target for miR-29a. We hypothesize that whereas miR-29b binds to the cognate site, miR-29a also binds to another site (or possibly sites) in the 3′ UTR. Since miR-29c shares features with both miR-29a and miR-29b (Fig. 3F), we suspect that the latter produces its small effect by binding to either the mir-29a or miR-29b binding sites. While each of these miRNAs is expressed in other tissues, the simultaneous and abundant expression of all three in a single tissue is unique to pancreatic islets. Thus, the coordinated action of the three miR-29 isoforms is likely to contribute to the highly efficient β-cell-specific silencing of Mct1.
The present investigation as to whether miRNAs contribute to β-cell-specific silencing of Mct1 required an understanding of miRNA expression in the β cell. The likelihood that a particular miRNA will contribute to a β-cell-specific phenotype is determined by both the magnitude and specificity of its expression in β cells. While there have been several reports of the relative abundance of miRNAs in β cells, there has been only limited analysis of specificity. However, the large-scale sequencing of miRNAs purified from a range of tissues (21) has provided a large amount of publically available data allowing us to perform a more comprehensive analysis of β cell miRNA expression. Our analyses covered 14 mouse tissues, including the mouse β cell line MIN6 and 16 human tissues, including pancreatic islets.
The miRNAs with high combined specificity and abundance show many similarities between the two samples. In addition to miR-375 and miR-7a/miR-7, members of the miR-8 family (including miR-200a, miR-200b, miR-200c, and miR-141) and miR-29 family (miR-29a, miR-29b, and miR-29c) have high combined specificity and abundance in both samples. In total, there are 15 miRNAs found in the top two quartiles of the combined abundance and specificity distribution in both MIN6 cells and human islets: let-7b, miR-7/miR-7a, miR-16, miR-24, miR-26a, miR-27b, miR-29a, miR-29b, miR-29c, miR-30b, miR-30d, miR-127-3p, miR-200c, miR-368, and miR-375.
Whereas the β cell line used here represents a single defined cell type, it is nonetheless a transformed cell line whose miRNA expression profile may have altered with time. In contrast, human and mouse islets contain a mixture of the five cell types. While β cells make up the majority of islet cells, and thus β cell miRNAs will be well represented, the expression profile obtained will be influenced by the other cell types present. Given the different natures of the samples, it is not possible to draw detailed conclusions about species differences in miRNA expression, yet similarities between the samples particularly in the region of high abundance and specificity suggest that miRNA expression is likely to be well conserved between the species. This view is supported by the demonstration that the miR-29 and miR-124 binding sites are well conserved between human and mouse Mct1.
Although miR-29 isoforms are highly expressed in human islets, they are also expressed in other tissues, so the question of whether they are likely to contribute to β-cell-specific silencing must be addressed. First, the level of expression in islets is very high, where miR-29b is the fourth most abundant miRNA detected. Indeed the combined expression of all three miR-29 isoforms is close (92%) to that of miR-375. For the human tissues for which data were obtained, the expression level of both miR-29b alone and the combined expression of all miR-29 isoforms is second highest in islets, being more abundant only in pituitary cells. While the levels of miR-29 isoforms measured by qRT-PCR in this study were lower than those from the large-scale sequencing study, all three isoforms still showed significant levels of expression, with a combined expression substantially greater than that of miR-7a, previously identified as a major islet miRNA (5).
While miR-124 was confirmed to significantly and specifically target Mct1, we failed to detect any expression of this miRNA in primary mouse islets and only very low levels in MIN6 cells. These data contrast strongly with the report of high levels of miR-124 in MIN6 cells by Landgraf et al (21). It seems likely that clonal differences between the MIN6 cells used in the two studies are most likely to account for the difference in miR-124 levels measured. miR-124 levels in mouse islets were below the level of detection of our assay, producing a similar signal to the no-template control. This contrasts with previous reports, which did detect miR-124 expression in mouse (1) and human (12) islets. Baroukh et al (1). used RT-PCR with conventional primers (as opposed to stem-loop primers) which would detect miRNA primary transcripts and pre-miRNAs but not mature miRNA. The signal obtained in this earlier study may therefore represent immature miR-124, which may not be fully processed into mature miR-124. However, as miR-124 expression was not quantified in mouse islets, it is also possible that the earlier results correspond to a low level of mature miR-124. Fred et al. (12) quantify miR-124 in human islets by qRT-PCR using TaqMan probes. However, as these authors only quantified relative changes in miR-124 expression upon various treatments, this study gives little indication of the amount of miR-124 relative to other miRNAs. Importantly, the high average CTs obtained for miR-124 indicated “a low abundance for miR-124a” (12), consistent with our own findings. It should be emphasized that, in the present study, we performed relative quantification of mature miR-124 and miR-375 in mouse islets by qRT-PCR using stem-loop primers with DNA oligonucleotide standard curves, allowing us to quantify miRNAs relative to each other. This revealed that the abundance of miR-124 is <0.19% of the miR-375 level. This measurement does, however, represent a single time point, and miRNA levels may be affected by a variety of stimuli (e.g., glucose). At this level, we conclude that miR-124 is unlikely to affect Mct1 expression significantly in β cells, but cannot exclude the possibility that under conditions where miR-124 levels were increased, it may also regulate Mct1 expression.
We also show here that miR-29a is able to significantly regulate Mct1 expression in a hepatocyte cell line, in MIN6 cells, and, importantly, primary mouse islets. The latter observation demonstrates the likely functional relevance of this interaction in mature β cells. We have previously predicted (40, 46) that low levels of MCT1 are a prerequisite in the β cell to ensure that exogenous pyruvate does not inappropriately stimulate insulin secretion and also to prevent the possibility of glucose carbons being lost as pyruvate. Changes in miR-29 that impact MCT1 level would therefore be expected to affect either or both of these parameters. In the present study, we were unable to detect a significant impact on basal or glucose-stimulated insulin secretion of miR-29a or miR-29b suppression either in clonal β cells (which show abnormal stimulation of secretion) (40) or in primary islets despite, in the latter case, observing significant changes in Mct1 mRNA (Fig. 5C). We suspect, however, that the extent of the changes in Mct1 level we were able to induce using the tools currently available to us to modulate miR-29a or miR-29b are below the limits likely to impact secretion from the intact islet, given that only a small percentage of cells are likely to be affected. Furthermore, we cannot exclude the possibility that these miRNAs target other proteins whose expression may influence, and in part compensate for, the increase in MCT1 level. Importantly, in unpublished studies in which we have induced MCT1 expression selectively in the β cells of adult transgenic mice, we have observed only modest effects on glucose-stimulated insulin secretion and a 2- to 3-fold stimulation of insulin secretion by exogenous pyruvate (T. J. Pullen and G. A. Rutter, unpublished data). It seems likely, therefore, that a full exploration of the impact of miR-29a and miR-29b levels will await the generation of analogous cell-type-specific knockout or transgenic mice.
While the present studies were under revision, Thorrez et al (41). also provided a bioinformatic prediction that several miRNAs may regulate Mct1, although no direct experimental evidence of interactions was given. This recent report also supported previous findings from van Arensbergen et al. (43) demonstrating that the Mct1 promoter from islets or purified β cells is enriched in repressive histone modifications. It therefore appears that miRNA action is one of multiple independent mechanisms that contribute to the deep repression of Mct1 expression in β cells.
Conclusion.
We demonstrate that Mct1 expression is silenced in pancreatic β cells at least in part by miRNAs selectively present in this cell type. The presence of these miRNAs seems likely to complement other, nonredundant transcriptional mechanisms which must also play an important role given the impact of mutations in the MCT1 promoter that result in exercise-induced hyperinsulinism. Given the importance of Mct1 in controlling insulin secretion, and its dysregulation in some forms of type 2 diabetes (33), regulation of the expression of the latter gene by exogenous miRNAs may provide a new therapeutic strategy for some forms of type 2 diabetes.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants to G.A.R. from the Wellcome Trust (Programme 081958/Z/07/Z), MRC (G0401641), National Institutes of Health (ROI DKO71962-01), and EU FP6 grant Save beta (G.A.R. and G.K.).
We thank Maz Wilson (University of Bristol) for the kind gift of the anti-MCT1 antibody and Andrew P. Halestrap (University of Bristol) for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 6 June 2011.
REFERENCES
- 1. Baroukh N., et al. 2007. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J. Biol. Chem. 282:19575–19588 [DOI] [PubMed] [Google Scholar]
- 2. Betel D., Wilson M., Gabow A., Marks D. S., Sander C. 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res. 36:D149–D153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6–21 [DOI] [PubMed] [Google Scholar]
- 4. Bock C., et al. 2005. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 21:4067–4068 [DOI] [PubMed] [Google Scholar]
- 5. Bravo-Egana V., et al. 2008. Quantitative differential expression analysis reveals miR-7 as major islet microRNA. Biochem. Biophys. Res. Commun. 366:922–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carthew R. W., Sontheimer E. J. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen C., et al. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clark S. J., Statham A., Stirzaker C., Molloy P. L., Frommer M. 2006. DNA methylation: bisulphite modification and analysis. Nat. Protoc. 1:2353–2364 [DOI] [PubMed] [Google Scholar]
- 9. Correa-Medina M., et al. 2009. MicroRNA miR-7 is preferentially expressed in endocrine cells of the developing and adult human pancreas. Gene Expr. Patterns 9:193–199 [DOI] [PubMed] [Google Scholar]
- 10. Didiano D., Hobert O. 2006. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 13:849–851 [DOI] [PubMed] [Google Scholar]
- 11. Diraison F., et al. 2004. Over-expression of sterol-regulatory-element-binding protein-1c (SREBP1c) in rat pancreatic islets induces lipogenesis and decreases glucose-stimulated insulin release: modulation by 5-aminoimid-azole-4-carboxamide ribonucleoside (AICAR). Biochem. J. 378:769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fred R. G., Bang-Berthelsen C. H., Mandrup-Poulsen T., Grunnet L. G., Welsh N. 2010. High glucose suppresses human islet insulin biosynthesis by inducing miR-133a leading to decreased polypyrimidine tract binding protein-expression. PLoS One 5:e10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Graham F. L., Smiley J., Russell W. C., Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59–72 [DOI] [PubMed] [Google Scholar]
- 14. Halestrap A. P., Meredith D. 2004. The SLC16 gene family-from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflugers Arch. 447:619–628 [DOI] [PubMed] [Google Scholar]
- 15. Halestrap A. P., Price N. T. 1999. The proton-linked monocarboxylate transporter (MCT) family: structure, function and regulation. Biochem. J. 343:281–299 [PMC free article] [PubMed] [Google Scholar]
- 16. Illingworth R., et al. 2008. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 6:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ishihara H., Wang H., Drewes L. R., Wollheim C. B. 1999. Overexpression of monocarboxylate transporter and lactate dehydrogenase alters insulin secretory responses to pyruvate and lactate in beta cells. J. Clin. Invest. 104:1621–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. John B., et al. 2004. Human microRNA targets. PLoS Biol. 2:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kloosterman W. P., Lagendijk A. K., Ketting R. F., Moulton J. D., Plasterk R. H. 2007. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 5:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krek A., et al. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37:495–500 [DOI] [PubMed] [Google Scholar]
- 21. Landgraf P., et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levrat F., et al. 1993. Influence of the content in transcription factors on the phenotype of mouse hepatocyte-like cell lines (mhAT). Exp. Cell Res. 209:307–316 [DOI] [PubMed] [Google Scholar]
- 23. Li K. K., et al. 2009. miR-124 is frequently down-regulated in medulloblastoma and is a negative regulator of SLC16A1. Hum. Pathol. 40:1234–1243 [DOI] [PubMed] [Google Scholar]
- 24. Li L. C., Dahiya R. 2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18:1427–1431 [DOI] [PubMed] [Google Scholar]
- 25. Liang Y., et al. 1996. Glucose metabolism and insulin release in mouse beta HC9 cells, as model for wild-type pancreatic beta-cells. Am. J. Physiol. 270:E846–E857 [DOI] [PubMed] [Google Scholar]
- 26. Lovis P., Gattesco S., Regazzi R. 2008. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by micro-RNAs. Biol. Chem. 389:305–312 [DOI] [PubMed] [Google Scholar]
- 27. Millar D. S., Warnecke P. M., Melki J. R., Clark S. J. 2002. Methylation sequencing from limiting DNA: embryonic, fixed, and microdissected cells. Methods 27:108–113 [DOI] [PubMed] [Google Scholar]
- 28. Miyazaki J., et al. 1990. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology 127:126–132 [DOI] [PubMed] [Google Scholar]
- 29. Mueller W. C., von Deimling A. 2009. Gene regulation by methylation. Recent Results Cancer Res. 171:217–239 [DOI] [PubMed] [Google Scholar]
- 30. Murchison E. P., Hannon G. J. 2004. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 16:223–229 [DOI] [PubMed] [Google Scholar]
- 31. Otonkoski T., et al. 2007. Physical exercise-induced hypoglycemia caused by failed silencing of monocarboxylate transporter 1 in pancreatic beta cells. Am. J. Hum. Genet. 81:467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Otonkoski T., et al. 2003. Physical exercise-induced hyperinsulinemic hypoglycemia is an autosomal-dominant trait characterized by abnormal pyruvate-induced insulin release. Diabetes 52:199–204 [DOI] [PubMed] [Google Scholar]
- 33. Parton L. E., et al. 2006. Limited role for SREBP-1c in defective glucose-induced insulin secretion from Zucker diabetic fatty rat islets: a functional and gene profiling analysis. Am. J. Physiol. Endocrinol. Metab. 291:E982–E994 [DOI] [PubMed] [Google Scholar]
- 34. Plaisance V., et al. 2006. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J. Biol. Chem. 281:26932–26942 [DOI] [PubMed] [Google Scholar]
- 35. Poy M. N., et al. 2004. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432:226–230 [DOI] [PubMed] [Google Scholar]
- 36. Poy M. N., et al. 2009. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc. Natl. Acad. Sci. U. S. A. 106:5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rutter G. A. 2004. Visualising insulin secretion. The Minkowski Lecture 2004. Diabetologia 47:1861–1872 [DOI] [PubMed] [Google Scholar]
- 38. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39. Schuit F., et al. 1997. Metabolic fate of glucose in purified islet cells. Glucose-regulated anaplerosis in beta cells. J. Biol. Chem. 272:18572–18579 [DOI] [PubMed] [Google Scholar]
- 40. Sekine N., et al. 1994. Low lactate dehydrogenase and high mitochondrial glycerol phosphate dehydrogenase in pancreatic beta-cells. Potential role in nutrient sensing. J. Biol. Chem. 269:4895–4902 [PubMed] [Google Scholar]
- 41. Thorrez L., et al. 2011. Tissue-specific disallowance of housekeeping genes: the other face of cell differentiation. Genome Res. 21:95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Valencia-Sanchez M. A., Liu J., Hannon G. J., Parker R. 2006. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20:515–524 [DOI] [PubMed] [Google Scholar]
- 43. van Arensbergen J., et al. 2010. Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing beta-cells to adopt a neural gene activity program. Genome Res. 20:722–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weber M., et al. 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 37:853–862 [DOI] [PubMed] [Google Scholar]
- 45. Zhao C., Rutter G. A. 1998. Overexpression of lactate dehydrogenase A attenuates glucose-induced insulin secretion in stable MIN-6 beta-cell lines. FEBS Lett. 430:213–216 [DOI] [PubMed] [Google Scholar]
- 46. Zhao C., Wilson M. C., Schuit F., Halestrap A. P., Rutter G. A. 2001. Expression and distribution of lactate/monocarboxylate transporter isoforms in pancreatic islets and the exocrine pancreas. Diabetes 50:361–366 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.