Abstract
The cohesin complex holds sister chromatids together and is essential for chromosome segregation. Recently, cohesins have been implicated in transcriptional regulation and insulation through genome-wide colocalization with the insulator protein CTCF, including involvement at the imprinted H19/Igf2 locus. CTCF binds to multiple imprinted loci and is required for proper imprinted expression at the H19/Igf2 locus. Here we report that cohesins colocalize with CTCF at two additional imprinted loci, the Dlk1-Dio3 and the Kcnq1/Kcnq1ot1 loci. Similar to the H19/Igf2 locus, CTCF and cohesins preferentially bind to the Gtl2 differentially methylated region (DMR) on the unmethylated maternal allele. To determine the functional importance of the binding of CTCF and cohesins at the three imprinted loci, CTCF and cohesins were depleted in mouse embryonic fibroblast cells. The monoallelic expression of imprinted genes at these three loci was maintained. However, mRNA levels for these genes were typically increased; for H19 and Igf2 the increased level of expression was independent of the CTCF-binding sites in the imprinting control region. Results of these experiments demonstrate an unappreciated role for CTCF and cohesins in the repression of imprinted genes in somatic cells.
INTRODUCTION
A number of mammalian genes termed imprinted genes are expressed exclusively from one parental allele, and many of these genes are important for embryonic development. The aberrant expression of these genes is associated with human cancers and genetic diseases, such as Beckwith-Wiedemann syndrome (BWS), Silver-Russell syndrome, Prader-Willi syndrome, and Angelman syndrome (4). Imprinted gene expression requires the establishment of imprinting marks in the germ line and early embryo and, subsequently, the maintenance of these marks during cell division in somatic tissues. To date, approximately 100 imprinted genes have been identified, and many are located in 1-Mb clusters that harbor differentially methylated regions (DMRs) (for a review, see reference 2). Within a cluster, imprinted gene expression is coregulated by a cis-acting regulatory element, termed an imprinting control region (ICR), that is also a DMR. There are two major mechanisms that regulate imprinted gene clusters. One mechanism requires the transcription of a long noncoding RNA (ncRNA), which silences genes in cis, allowing the alternate allele to be expressed. This study focuses on the second mechanism, which employs allele-specific insulation mediated by the insulator protein CTCF.
CTCF is a highly conserved 11-zinc-finger protein, which was initially identified as a CCCTC binding factor in the chicken c-myc locus (36). CTCF is implicated in transcriptional activation, repression, and insulation (for a review, see reference 15). In addition, CTCF facilitates inter- and intrachromosomal interactions (68). CTCF binds to the H19/Igf2 ICR in a DNA methylation-sensitive manner (3, 21, 27, 55). In this case, CTCF mediates the insulator activity of the unmethylated maternal ICR by blocking the Igf2 promoter from engaging enhancers downstream of H19 that are shared by H19 and Igf2. The deletion or mutation of the four CTCF-binding sites within the ICR causes a paternalization of the maternal allele, Igf2 biallelic expression, and H19 repression (12). In addition to the H19/Igf2 locus, CTCF binds to DMRs at a number of imprinted loci, including Gtl2, Kcnq1/Kcnq1ot1, Grb10, and Rasgrf1 (16, 22, 29, 70). CTCF binding appears to be methylation sensitive at the KvDMR1 region in the Kcnq1/Kcnq1ot1 locus as well as the Grb10 and Rasgrf1 loci (16, 22, 70). Interestingly, the Kcnq1/Kcnq1ot1 locus and, possibly, the Gtl2 locus use the ncRNA mechanism to regulate imprinted expression (see below), whereas imprinting regulation at the Grb10 and Rasgrf1 loci remains unclear. Furthermore, the depletion of CTCF in mouse oocytes led to a reduction in Gtl2 and Grb10 RNA levels (65). It is intriguing that CTCF is present and may possibly function at imprinted loci that are regulated by different imprinting control mechanisms (insulator versus ncRNA). Nevertheless, it is still incompletely understood how CTCF manifests its insulator function at the H19/Igf2 locus and whether it has a similar function at other imprinted loci.
Recently, we and others reported the genome-wide colocalization of CTCF and cohesin complex subunits, including that at the H19 ICR (43, 48, 54, 67). Those studies suggested that cohesins bind to the DNA through a consensus sequence similar to that of CTCF. Cohesins are required for sister chromatid cohesion during cell division. The mitotic cohesin complex consists of four subunits, SMC1/SMC1A, SMC3, RAD21/SCC1, and SCC3 (SA1/SA2), which were previously proposed to form a ringlike structure and encircle the sister chromatids during mitosis (19). Recent studies of Saccharomyces cerevisiae and higher organisms suggested that cohesins are also involved in transcriptional regulation, chromatin structure, and development (for a review, see reference 9), likely through their ability to interact with chromosomes. Drosophila melanogaster SMC1, RAD21, and SCC3 were suggested previously to prevent enhancer-promoter interactions at the cut locus (10, 47), indicating that cohesins function as insulator proteins. Taken together, the genome-wide colocalization of CTCF and cohesins as well as the noncanonical roles of cohesins in transcription regulation provide a clue for the possible mechanism underlying the function of CTCF in chromosome biology.
We demonstrated that the binding of cohesin complex subunits to the H19 ICR requires the CTCF-binding sites (54), suggesting that CTCF and cohesins may function in concert to regulate the imprinted expression of H19 and Igf2. Additionally, RNA interference (RNAi) experiments with HeLa cells and a human breast epithelial cell line (HB2 cells) revealed similar expression changes of the H19/Igf2 locus genes when CTCF knockdown (KD) and cohesin KD cells were compared (42, 67). Those studies suggested that cohesins are involved in the imprinted regulation of the H19/Igf2 locus. However, HeLa cells and HB2 cells express low levels of H19 and Igf2; thus, the maintenance of imprinted expression may not be tightly regulated. Moreover, the changes observed by these studies could be due to indirect effects rather than a perturbed function of cohesins in imprinting.
To investigate further the role of CTCF and cohesins in genomic imprinting and insulator function, we used a primary embryonic cell type, mouse embryonic fibroblasts (MEFs), and expanded our studies to two DMRs at additional CTCF-bound imprinted loci, the Gtl2 DMR and KvDMR1. We show that cohesin complex subunits also bind these DMRs in MEFs, and similar to the H19/Igf2 locus, CTCF and cohesins preferentially bind to the unmethylated allele of the Gtl2 DMR. However, CTCF and cohesins bind biallelically to KvDMR1. To investigate the roles of CTCF and cohesins in these imprinted loci, we performed RNAi experiments targeting CTCF and two cohesin complex subunits in MEFs and found that the Igf2 expression level is elevated but remains imprinted, suggesting an indirect effect of cohesin depletion at the H19/Igf2 locus. We propose that cohesins are involved in the regulation of proper expression levels of imprinted genes rather than having a direct role in imprinting.
MATERIALS AND METHODS
Isolation and culture of F1 hybrid MEFs.
MEFs were isolated from individual mouse 14.5-day-postcoitus (dpc) embryos generated from crosses between C57BL/6 (B6) (for both the wild-type and the H19ΔR/+ mutant) females and males with Mus musculus castaneus (C) chromosome (Chr) 7, distal 12 and X, in a B6 background [B6(CAST7P12X)], as previously described (64). The H19+/ΔR MEFs were generated from crosses between female with Mus musculus castaneus (C) chromosome 7 in a B6 background [B6(CAST7)] and B6 males as previously described (40).
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) assays were carried out by using a chromatin immunoprecipitation assay kit (catalog numbers 17-295 and 17-610; Upstate) according to the manufacturer's instructions and as previously described (64). Approximately 1 × 106 MEFs were used in each immunoprecipitation, which resulted in 40 to 100 μg of chromatin-immunoprecipitated (ChIPed) DNA. All PCR primers and conditions are listed in Tables 1 and 2.
Table 1.
Primers and enzymes used for allele-specific analysis by PCRsa
| Region or gene | Primer or probe (sequence) | Temp (°C) | SNP B/C | Enzyme | Product size(s) (bp) |
|---|---|---|---|---|---|
| IG-DMR | IG-DMR207 (TACGGAGATGTGCTGTGGAC) | 62 | A/G | NcoI | B, 104, 95, 37 |
| IG-DMR442 (CTCGCTAGTTCACGGAGGTC) | C, 199, 37 | ||||
| Gtl2 DMR | Gtl2 1915F (TGAAACCTGTTGGGCG) | 61 | T/C | HaeII | B, 109, 70, 48, 17 |
| Gtl2 2158R (CAATGGGAGGGGTACAGATG) | C, 97, 70, 48, 17, 12 | ||||
| KvDMR CTCF BS1 | KvDMR 2701F (CCCACCGAAGTAATCCAAAA) | 62 | C/T | Hpy188I | B, uncut, 398 |
| KvDMR 3098R (TCAGCTAGGAAGGGATGAGG) | C, 292, 106 | ||||
| KvDMR CTCF BS2 | KvDMR 2258F (CTGAGAAGCCAAGTGGATCG) | 62 | A/G | MfeI/Tsp509I | B, 214, 82 |
| KvDMR 2553R (CCACCAGCCTCAGCATATTT) | C, uncut, 296 | ||||
| H19 | HI3 (CCTCAAGATGAAAGAAATGGT) | 55 | G/A | NA, real-time PCR | NA, real-time PCR |
| HE5 (AACACTTTATGATGGAACTGC) | |||||
| Probe Mut (CCACCTGTCGTCCATCTCC-FL) | |||||
| Probe Anc (RED640-TCTGAGGGCAACTGGGTGTGG-P) | |||||
| HE2 (TGATGGAGAGGACAGAAGGG) | 55 | G/A | Cac8I | B, 173, 62 | |
| HE4 (TTGATTCAGAACGAGACGGAC) | C, uncut, 235 | ||||
| Igf2 | Igf18 (ATCTGTGACCTCTTGAGCAGG) | 58 | Tsp509I | B, 180, 20 | |
| Igf20 (GGGTTGTTTAGAGCCAATCAA) | C, 165, 20, 15 | ||||
| Gtl2 | Gtl3 (CCAAAGCCATCATCTGGAATC) | 55 | T/? | SfcI | B, 250, 87 |
| Gtl4 (CAGCCCTGTGAGGTAGGAAC) | C, uncut, 337 | ||||
| Dlk1 | Dlk2up (CTGGCTTTCTTCCCGCTGGAC) | 54 | T/C | DraIII | B, 212, 104 |
| Dlk317dn (GACACAGCCAGGGGCAGTTA) | C, uncut, 316 |
SNP, single-nucleotide polymorphism; B, B6 maternal allele; C, paternal allele; NA, not applicable.
Table 2.
Primers and PCR conditions used for real-time PCRs
| Region or gene | Primer | Temp (°C) | Primer concn (μM) | MgCl2 concn (mM) |
|---|---|---|---|---|
| IG-DMR | IG-DMR207 (TACGGAGATGTGCTGTGGAC) | 62 | 0.3 | 1.25 |
| IG-DMR442 (CTCGCTAGTTCACGGAGGTC) | ||||
| Gtl2 DMR | Gtl2 1998F (TGGTTGGGCTATTGGAGTCT) | 61 | 0.5 | 3 |
| Gtl2 2158R (CAATGGGAGGGGTACAGATG) | ||||
| KvDMR Ctrl-1 | KvDMR 3025F (TACAGGATTGTACGACCTAG) | 63 | 0.3 | 3.5 |
| KvDMR 3220R (AGAGCCAGGGGCATACTCAT) | ||||
| KvDMR CTCF BS-1 | KvDMR 2644F (ACCATGCAGAGAAAAGCACA) | 56 | 0.4 | 5 |
| KvDMR 2844R (CTAGCCGTTGTCGCTAGGAG) | ||||
| KvDMR CTCF BS-2 | KvDMR 2258F (CTGAGAAGCCAAGTGGATCG) | 62 | 0.2 | 3 |
| KvDMR 2553R (CCACCAGCCTCAGCATATTT) | ||||
| KvDMR Ctrl-2 | KvDMR 1762F (CACTCACCTTGGGACTCGAC) | 58 | 0.6 | 4 |
| KvDMR 2007R (AGAAGCAGAGGTGATTCGTG) | ||||
| Ctcf | CTCF5F1 (GCCAGCAGGGACACATACAAG) | 56 | 0.5 | 3.5 |
| CTCF5R1 (GCTTTCGCAAGTGGACACC) | ||||
| Smc1 | Smc1 3767 (CAAGTACCCAGATGCCAACC) | 55 | 0.4 | 2 |
| Smc1 3983 (CGATCCATGATAGGGGGTAA) | ||||
| Rad21 | Rad21 2815 (CAAGGCTGCACACTCCTGTA) | 55 | 0.3 | 2.5 |
| Rad21 3045 (CCCCATAAAAGTGCCAACAC) | ||||
| H19 | HE2 (TGATGGAGAGGACAGAAGGG) | 55 | 0.4 | 4.5 |
| HE4 (TTGATTCAGAACGAGACGGAC) | ||||
| Igf2 | Igf2f (CGCTTCAGTTTGTCTGTTCG) | 58 | 0.25 | 2.4 |
| Igf2r (GCAGCACTCTTCCACGATG) | ||||
| Gtl2/Meg3 | Meg3f (TTGCTGTTGTGCTCAGGTTC) | 60 | 0.4 | 2 |
| Meg3r (ATCCTGGGGTCCTCAGTCTT) | ||||
| Dlk1 | Dlk1f (CGGGAAATTCTGCGAAATAG) | 60 | 0.4 | 1.25 |
| Dlk1r (TGTGCAGGAGCATTCGTACT) | ||||
| Arpp0 | Arpp0#72L (TCCCACTTACTGAAAAGGTCAAG) | 55 | 0.4 | 4.5 |
| Arpp0#72R (TCCGACTCTTCCTTTGCTTC) |
siRNA treatments.
MEFs were cultured in 24-well plates and treated every 48 h with two to three sequential small interfering RNA (siRNA) treatments. Cells were trypsinized and plated at 30 to 40% confluence. A total of 40 to 80 pM siRNAs was mixed with 1 μl Lipofectamine 2000 (Invitrogen) and 100 μl Opti low-serum medium (Invitrogen) and added to the medium (high-glucose Dulbecco's modified Eagle's medium [DMEM] plus 10% fetal bovine serum [FBS]) upon cell plating. The following siRNAs were used: control siRNA (catalog number 12935-300; Invitrogen), CTCF siRNA (Stealth Ctcf-MSS203343; Invitrogen), Smc1 siRNA (Smc1a-MSS216158; Invitrogen), and Rad21 (RAD21 On-Target Plus siRNA J-058531-12; Dharmacon). For each experiment, at least three biological replicates were generated and analyzed.
Western blots.
Cells were lysed with TNE buffer (100 mM Tris [pH 7.4], 1% NP-40, 10 mM EDTA) with a 1:100 dilution of proteinase inhibitor cocktail (Sigma) and 1 mM dithiothreitol (DTT). Lysates were mixed with 5× loading buffer, denatured by heating at 95°C for 10 min, and fractionated on an 8% SDS-PAGE gel. The protein was transferred onto a nitrocellulose membrane (Bio-Rad), blocked in 3% nonfat milk (Bio-Rad) in Tris-buffered saline (TBS)-Tween (TBST) (0.05% Tween 20 in 1× TBS), and probed with primary and secondary antibodies (horseradish peroxidase [HRP]-conjugated anti-mouse and anti-rabbit antibody; GE). The blot was visualized by using chemiluminescence (ECL Plus; GE). Quantification was performed by using ImageJ software (NIH).
RNA extraction and reverse transcription.
Total RNA from MEFs was extracted by using an RNeasy Microkit (Qiagen) according to the manufacturer's instructions. The reverse transcription (RT) reaction, primed with random primers, was performed by using Superscript II or III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. For each sample, the minus-RT control was done without Superscript II or III and confirmed to be negative by PCR of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene.
Real-time PCRs and statistical analysis.
Real-time PCR analysis was carried out with the LightCycler real-time PCR system (Roche), as previously described (64), with a few modifications. Reaction mixtures were set up in duplicates or triplicates by using Ready-To-Go PCR beads (Amersham) with a 5-min initial incubation with TaqStart antibody (Clontech), followed by the addition of 0.2 to 0.6 μM primers (Table 2), 1× EvaGreen (Biotium), and 1.25 to 5 mM MgCl2. Data analysis was performed by using Light Cycler 4.0 software using the Relative Quantification program (Monocolor) to determine the ratio of each ChIP sample relative to input or cDNA levels relative to the acidic ribosomal phosphoprotein P0 (Arpp0), β-actin, or GAPDH gene (64, 66). For statistical analysis, the two-tail paired Student t test was used, and differences with a P value of <0.05 were considered significant.
Allele-specific analysis of ChIPed DNA by PCRs and restriction enzyme digestions.
All PCRs were carried out with Ready-To-Go PCR beads (Amersham) using 0.3 μM each primer (Table 1) and 0.1 μCi of [32P]dCTP. Products were digested (Table 1) and resolved on 7% or 12% polyacrylamide gels. Gels were exposed to phosphorscreens and scanned on a Typhoon Trio Phosphorimager (GE). The relative band intensities were quantified by using ImageJ software (NIH).
Allele-specific RNA analysis.
H19 RNA assays were conducted on cDNA using the LightCycler real-time PCR system (Roche Molecular Biochemicals) as described previously (46). Igf2 and Gtl2 RNA assays were conducted on cDNA by allele-specific restriction digests as previously described (66). Dlk1 RNA assays were conducted, with some modifications, as previously described (20). The digested PCR products were resolved by 7% or 12% polyacrylamide gel electrophoresis. The gels were stained with ethidium bromide. The contribution of each parental allele to the total RNA was determined by using Quantity One software (Bio-Rad). All primers used are listed in Table 1.
Antibodies.
The following antibodies were used in ChIP and Western blotting (WB): anti-histone H3 (ab1791 [ChIP, 5 μg]; Abcam), anti-acetylated histone H3 (AcH3) (catalog number 06-599 [ChIP, 5 μg]; Millipore), anti-dimethyl-histone H3 lysine 4 (H3K4me2) (catalog number 07-030 [ChIP, 5 μl]; Millipore), anti-CTCF (catalog number 07-729 [ChIP, 5 μl; WB, 1:1,000]; Millipore), anti-SMC1 (catalog number A300-055A [ChIP, 5 μl; WB, 1:1,000]; Bethyl), anti-RAD21 (catalog number A300-080A [ChIP, 5 μl; WB, 1:500]; Bethyl), and anti-α-tubulin (catalog number T6199 [WB, 1:2,000]; Sigma).
RESULTS
Colocalization of CTCF and cohesins at the Gtl2 DMR and the KvDMR1 region.
To determine whether CTCF and cohesins interact and function similarly at multiple imprinted loci, we investigated binding at two independent imprinted loci. Gtl2 encodes a long ncRNA with multiple spliced forms of unknown function (45, 51). Gtl2 resides in the Dlk1-Dio3 imprinting cluster, which was initially identified in a uniparental disomy (UPD) of distal mouse chromosome (Chr) 12 deficiencies (56) and UPDs of the orthologous human Chr 14q32 (23). The locus contains three paternally expressed protein-coding genes (Dlk1, Rtl1, and Dio3) and a number of maternally expressed ncRNAs (Fig. 1A). It was proposed that these ncRNAs can make up a large polycistronic transcription unit that represses the protein-coding genes on the maternal chromosome. In human and mouse, the intergenic DMR (IG-DMR), which is methylated in the male germ line, functions as the ICR of the cluster (25, 34, 35). There is a second postfertilization-derived paternally methylated Gtl2 DMR, spanning the promoter, exon 1, and part of intron 1 of Gtl2 (56). The maternal deletion of the Gtl2 DMR in human results in the regional silencing of imprinted genes (24). The Gtl2 DMR contains a CTCF-binding site (29, 45), raising the possibility that cohesins bind to the region.
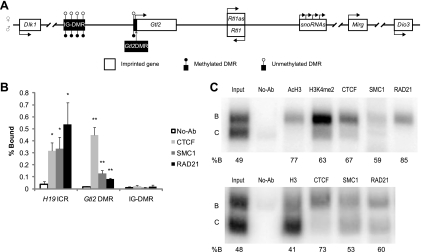
Fig. 1.
CTCF and cohesins colocalize at the Gtl2 DMR in MEFs. (A) Schematic showing the Dlk1-Dio3 imprinted locus (not to scale). The transcriptional and methylation statuses of the maternal and paternal chromosomes are represented at the top and bottom of the DNA strand, respectively. (B) CTCF and cohesins colocalize at the Gtl2 DMR but not the IG-DMR. The H19 ICR is included for comparison. Shown are real-time PCR results from at least three independent ChIP experiments. *, P < 0.05; **, P < 0.01. P values show differences between no-antibody (No-Ab) controls and antibodies against CTCF, SMC1, or RAD21. Error bars indicate standard errors of data from at least three biological replicates. (C) Allele-specific analysis of ChIPed DNA by PCR showing preferential binding of CTCF and cohesins on the maternal Gtl2 DMR. Two independent experiments are shown. Histone H3, AcH3, and H3K4me2 are included as positive controls. B, maternal allele; C, paternal allele. The percentage of protein binding on the B allele over total binding (B plus C) is indicated at the bottom. At least three experiments were performed, and similar results were obtained.
To investigate the binding of CTCF and cohesins at the Gtl2 DMR, we used quantitative real-time PCR following chromatin immunoprecipitation (ChIP) in F1 hybrid MEFs (see Materials and Methods). At the Gtl2 DMR, antibodies against CTCF, SMC1, and RAD21 precipitated significantly more chromatin (P < 0.01) than the no-antibody negative control. However, no enrichment of CTCF and cohesins was seen at the IG-DMR (Fig. 1B), consistent with the absence of CTCF-binding sites (45). CTCF bound to the Gtl2 DMR at a level similar to that of the H19 ICR, whereas not as much SMC1 and RAD21 bound to the Gtl2 DMR as what bound to the H19 ICR (Fig. 1B), indicating that the affinities of cohesins differ between the Gtl2 DMR and the H19 ICR. Therefore, cohesins may interact with CTCF differently and exhibit disparate functions at these two loci.
To determine if the binding of CTCF and cohesins is exclusive to one parental allele, we performed allele-specific PCR analysis of DNA isolated from ChIP. Because the CTCF-binding site resides near the Gtl2 promoter, and active histone modifications were reported previously to be associated with the maternal allele at this region (5), we included two antibodies against acetylated histone H3 (AcH3) and dimethyl-histone H3 lysine 4 (H3K4me2) as controls. AcH3 and H3K4me2 preferentially bind the transcribed, maternal allele (Fig. 1C, top), as do CTCF and the two cohesin complex subunits (Fig. 1C). An antibody against histone H3 was also included; H3 was present at approximately equal levels on the parental alleles (Fig. 1C, bottom). The binding of CTCF on the unmethylated maternal allele is consistent with results of experiments showing a methylation-sensitive binding pattern (15). In conclusion, CTCF and cohesins colocalize in an allele-specific manner at the Gtl2 DMR, similar to the H19 ICR.
The Kcnq1/Kcnq1ot1 imprinted locus contains maternally expressed genes and one paternally expressed long ncRNA, Kcnq1ot1, which is an antisense transcript of Kcnq1 (Fig. 2A). The loss of imprinted expression at this locus is the most frequent alteration in individuals with BWS (reviewed in reference 44). This locus uses primarily the ncRNA mechanism to regulate imprinted expression. The maternally methylated KvDMR1 is the ICR of the locus and includes the Kcnq1ot1 promoter (53). On the paternal allele, where KvDMR1 is unmethylated, Kcnq1ot1 is expressed, which results in the silencing of the adjacent genes. The paternal deletion of KvDMR1 causes a loss of expression of Kcnq1ot1 and results in the activation of genes in cis in embryonic and extraembryonic tissues (17, 32). The transcription of the intact Kcnq1ot1 mRNA is required for the imprinted expression of the entire locus (39, 52, 58), with one exception: Shin and colleagues showed previously that Cdkn1c imprinting was maintained on a Kcnq1ot1 truncation allele in several fetal and neonatal tissues (52). Fitzpatrick et al. additionally identified two CTCF-binding sites in KvDMR1 (16). These data, together with evidence that KvDMR1 has insulator and silencer activities in vitro (16, 38, 57), indicate that CTCF may form an insulator at KvDMR1 as a secondary mechanism in the Kcnq1/Kcnq1ot1 imprinted locus.
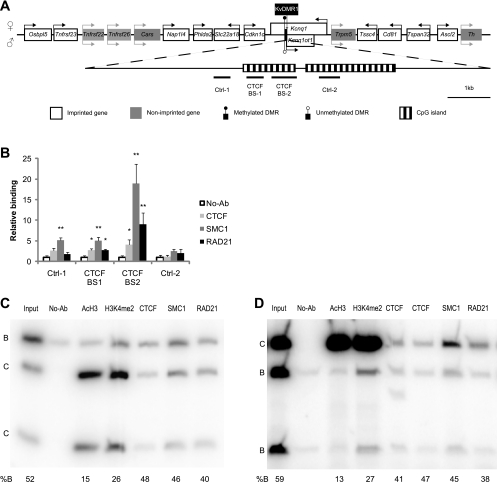
Fig. 2.
CTCF and cohesins colocalize at KvDMR1 in MEFs. (A) Schematic showing the Kcnq1/Kcnq1ot1 locus. The transcriptional and methylation statuses of the maternal and paternal chromosomes are represented at the top and bottom of the DNA strand, respectively. The KvDMR1 region is enlarged at the bottom. The regions analyzed in B, C, and D are indicated below the diagram. (B) ChIPs followed by real-time PCR. *, P < 0.05; **, P < 0.01. P values show differences between no-antibody (No-Ab) controls and antibodies against proteins of interest. The data are normalized to the no-antibody controls of each PCR. Error bars indicate standard errors of data from at least three biological replicates. (C and D) Allele-specific analysis of ChIPed DNA by PCR showing binding of CTCF and cohesins in KvDMR1. (C) CTCF BS-1; (D) CTCF BS-2. AcH3 and H3K4me2 are included as controls. B, maternal allele; C, paternal allele. The percentage of protein binding on the B allele over total binding (B plus C) is indicated at the bottom. At least three experiments were performed, and similar results were obtained.
We examined CTCF and cohesin occupancy at KvDMR1, including the two previously reported CTCF-binding sites (CTCF-binding site 1 [CTCF BS-1] and CTCF BS-2) (16) and two negative controls (control 1 [Ctrl-1] and Ctrl-2). KvDMR1 has two CpG islands, as shown in Fig. 2A. The two CTCF-binding sites reside in the first CpG island; Ctrl-1 is located about 150 bp upstream of the first CpG island, whereas Ctrl-2 is in the second CpG island. Following ChIP, real-time PCR analysis showed that CTCF, SMC1, and RAD21 associated with both CTCF BS-1 and -2 but not Ctrl-2 (Fig. 2B). SMC1 was also detected at Ctrl-1 (Fig. 2B), most likely because Ctrl-1 is only about 200 bp away from CTCF BS-1. Allele-specific analyses revealed that the allelic bias of CTCF binding at KvDMR1 in MEFs was very weak, in contrast to AcH3 and H3K4me2, which were strongly biased toward the paternal allele. Compared to the input DNA, CTCF antibody precipitated 5 to 20% more of the paternal allele than the maternal allele. Similarly, the two cohesin complex subunits showed a small bias (∼5%) toward the paternal allele (Fig. 2C and D). Our results are in contrast with those of Fitzpatrick and colleagues, who showed that CTCF binding at KvDMR1 was exclusively on the paternal allele in C57BL/6 × SD7 F1 hybrid MEFs (16). These contrasting results could be reflective of mouse strain differences.
Functional analysis of CTCF and cohesins in MEFs.
The results described above demonstrated that CTCF and cohesins colocalize at three imprinted DMRs. To assess the functional significance of the binding of CTCF and cohesins in regulating the expression of imprinted genes, we performed RNAi experiments. F1 hybrid MEFs were treated with siRNAs targeting Ctcf, Smc1, and Rad21. An siRNA that does not target any known transcripts in mouse was included as a control. To achieve a better knockdown (KD) efficiency, we tested sequential siRNA treatments by treating the cells every 48 h. Maximum depletion (∼75% to 90%) of targeted proteins occurred by 72 h and was maintained for an additional 72 h, with two to three treatments of siRNAs (data not shown). Accordingly, we conducted most of the experiments with two or three sequential siRNA treatments and harvested cells at 96 h to 144 h. Because no obvious growth or proliferation defects were observed in the siRNA-treated cells, any changes in these cells were likely due to the altered function of CTCF and cohesin complex subunits in transcriptional regulation, rather than cell division.
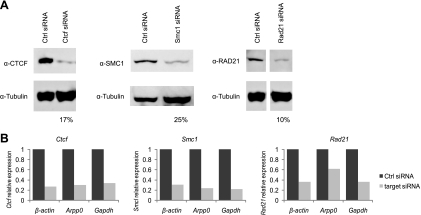
The RNAi approach using two sequential treatments of one siRNA against each transcript successfully reduced the amount of the protein encoded by the targeted mRNA (Fig. 3A). In general, the siRNAs targeting Ctcf and Rad21 had higher KD efficiencies than did the siRNA targeting Smc1 (Fig. 3A). Furthermore, because there are thousands of CTCF- and cohesin-binding sites in the genome (43, 48, 67), it was possible that the depletion of CTCF and cohesins would affect housekeeping genes. Therefore, we examined the mRNA levels of three housekeeping genes (the β-actin, acidic ribosomal phosphoprotein P0 [Arpp0], and GAPDH genes) in the siRNA-treated cells (Fig. 3B) and found that the target mRNA was reduced to 30 to 40% compared to the control siRNA-treated cells. Because no effect on the relative transcript abundance of housekeeping genes was observed, only the Arpp0 gene was used as the control in subsequent experiments.
Fig. 3.
Successful depletion of CTCF, SMC1, and RAD21 from MEFs. (A) Western blots showing 75% to 90% depletion of CTCF and cohesin complex subunits by siRNA treatments. MEFs were treated twice (48 h apart), and cell lysates were collected at 96 h. α-Tubulin was used as a control. (B) Ctcf, Smc1, and Rad21 mRNA levels were normalized to three housekeeping genes, the β-actin, Arpp0, and GAPDH genes.
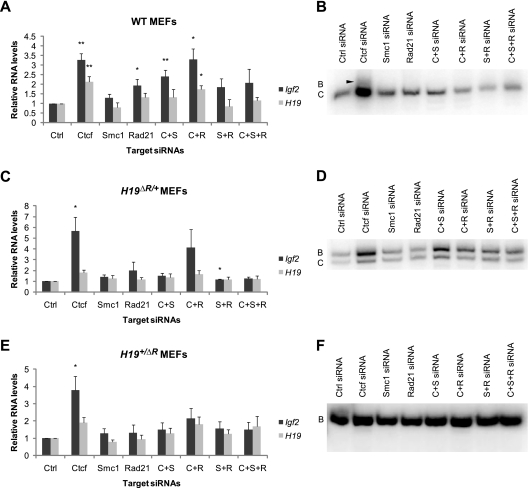
We first characterized the effect of the depletion of CTCF and cohesins at the H19/Igf2 locus. We hypothesized that CTCF and cohesins work in concert at the H19 ICR as insulator proteins and that the depletion of CTCF and/or cohesin complex subunits would reduce the insulator activity, resulting in increased Igf2 transcription levels due to expression from the normally repressed maternal allele. The deletion of the H19 ICR or CTCF-binding sites from the maternal chromosome caused a biallelic expression of Igf2 and a reduction of H19 RNA levels, indicating competition between H19 and Igf2 for the shared enhancers on the maternal chromosome (12, 59, 61). Therefore, we expected to observe reduced H19 expression levels in CTCF- and/or cohesin-depleted cells. To test this hypothesis, we treated MEFs with individual siRNAs against CTCF and cohesins as well as with different combinations of siRNAs (Fig. 4A and 5A). As expected, Ctcf siRNA-treated cells had elevated Igf2 expression levels (P < 0.01), approximately 3-fold relative to those of control siRNA-treated cells. Rad21 siRNA treatments also led to increased Igf2 expression levels (P < 0.05), whereas Smc1 siRNA had no significant effect (P > 0.05), probably due to an insufficient KD. Surprisingly, H19 levels were mostly unaffected, and in Ctcf siRNA treatments, the levels were elevated (Fig. 5A). Next, we examined the allelic expression of Igf2 and H19. Unexpectedly, most cells maintained imprinted Igf2 and H19 expressions (Fig. 5B and data not shown). When biallelic Igf2 expression was observed, the maternal expression level was very low (Fig. 5B, arrowhead). Because Igf2 levels were increased ∼3-fold compared to those of controls, we concluded that the enrichment was not from the normally silent maternal allele but rather from the normally active paternal allele. One possible explanation is that the residual amount of CTCF is sufficient to maintain imprinting.
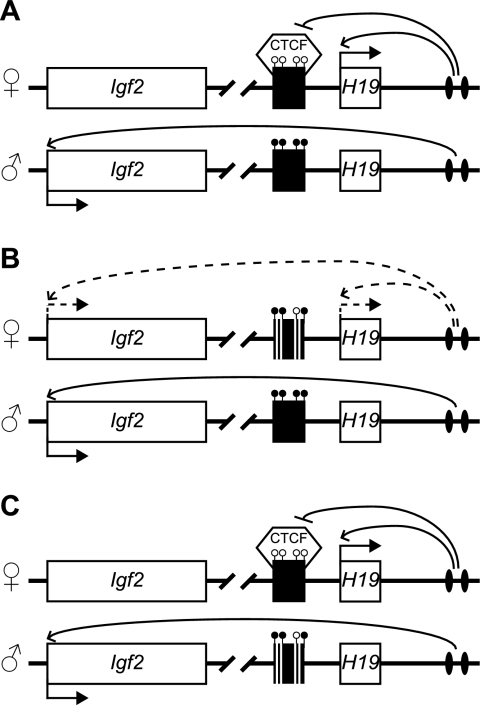
Fig. 4.
MEFs used for functional analysis of CTCF and cohesins. Schematics show the H19/Igf2 locus (not to scale). The black rectangle represents the H19 ICR. Open lollipops represent unmethylated DNA; closed lollipops represent methylated DNA. (A) On the wild-type locus, the ICR exhibits paternal-specific methylation and contains binding sites for CTCF. On the maternal allele, CTCF binds to the ICR and blocks the Igf2 promoter from accessing the 3′ shared enhancers. On the paternal allele, the ICR is methylated, and H19 transcription is repressed. Because CTCF binding is methylation sensitive, the ICR cannot act as an insulator on the paternal allele, allowing Igf2 expression. (B and C) In the H19ΔR MEFs, four CTCF-binding sites in the ICR are deleted (white bars). (B) Upon maternal transmission of the mutant allele, the insulator activity is lost because CTCF cannot bind to the ICR, Igf2 exhibits biallelic expression, and the H19 expression level is reduced (12). (C) Upon paternal transmission of the mutant allele, H19 remains monoallelically expressed, and the Igf2 expression level is similar to that of the wild-type (12).
Fig. 5.
Depletion of CTCF and cohesins associated with elevated Igf2 expression levels. (A) Relative RNA levels of H19 and Igf2 in siRNA-treated wild-type (WT) MEFs. *, P < 0.05; **, P < 0.01. P values show differences between CTCF or cohesin siRNA treatments and the control siRNA treatment. H19 and Igf2 RNA levels are normalized to Arpp0. The relative value of the control siRNA treatment is set to 1. Error bars indicate standard errors. C, Ctcf siRNA; S, Smc1 siRNA; R, Rad21 siRNA. (B) Allele-specific analysis of Igf2 RNA in wild-type MEFs. Samples were collected at 144 h after three sequential treatments with siRNAs. Only the second lane had a low level of maternal Igf2 expression detected (arrowhead). B, maternal allele; C, paternal allele. (C) H19 and Igf2 relative RNA levels in siRNA-treated H19ΔR/+ MEFs. *, P < 0.05. The P value shows the difference between CTCF siRNA treatments and the control siRNA treatment. H19 and Igf2 RNA levels are normalized to Arpp0. The relative value of the control siRNA treatment was set to 1. Error bars indicate standard errors. (D) Allele-specific analysis of Igf2 RNA in H19ΔR/+ MEF cells. Samples were collected at 96 h after two sequential treatments of siRNAs. All samples have similar amounts of B (maternal) and C (paternal) alleles. (E) H19 and Igf2 relative RNA levels in siRNA-treated H19+/ΔR MEFs. *, P < 0.05. The P value shows the difference between CTCF or cohesin siRNA treatments and the control siRNA treatment. H19 and Igf2 RNA levels are normalized to Arpp0. The relative value of the control siRNA treatment was set to 1. Error bars indicate standard errors. (F) Allele-specific analysis of Igf2 RNA in H19+/ΔR MEFs. Samples were collected at 120 h after two sequential treatments of siRNAs. All samples have monoallelic expression of the B (paternal) allele.
To test whether the above-described phenotype depends on the CTCF-binding sites in the H19 ICR, we conducted siRNA treatments of MEFs that harbor the H19ΔR mutant allele (12). When the mutant allele was transmitted maternally, the insulator function at the maternal H19 ICR was disrupted due to the deletion of the four CTCF-binding sites at the endogenous locus (Fig. 4B), leading to biallelic Igf2 expression (12) (Fig. 5D). In contrast, when the mutant allele was transmitted paternally, the normal imprinted expression of H19 and Igf2 was maintained (12) (Fig. 4C and 5F). If the phenotype in the wild-type cells (Fig. 5A and B) depended on the maternal CTCF-binding sites in the H19 ICR, we would expect to see no changes in H19 and Igf2 expression in the H19ΔR/+ cells. Contrary to our expectation, the depletion of CTCF resulted in increased Igf2 expression levels in KD H19ΔR/+ MEFs (Fig. 5C). When both SMC1 and RAD21 were depleted in these cells, the Igf2 expression level was increased (Fig. 5C). Furthermore, an allele-specific analysis showed that Igf2 expression remained biallelic (Fig. 5D) (i.e., there was no difference compared to control siRNA-treated cells), indicating that the activation was on both alleles in the H19ΔR/+ MEFs. In H19+/ΔR MEFs, the depletion of CTCF also caused elevated Igf2 expression levels but not a loss of imprinting (Fig. 5E and F). We conclude that CTCF has repressive functions on Igf2 expression independent of the CTCF-binding sites in the H19 ICR. The role of cohesins in the repression of Igf2 was not as pronounced and likely indirect.
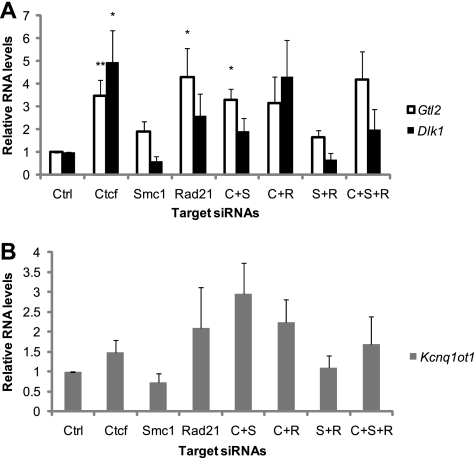
We also examined five imprinted genes in the Dlk1-Dio3 and the Kcnq1/Kcnq1ot1 loci in the CTCF KD and cohesin KD MEFs. No loss of imprinting was observed for Dlk1, Gtl2, Kcnq1, Kcnq1ot1, and Cdkn1c (data not shown). Although the expression levels of Dlk1 and Gtl2 were not affected in most siRNA treatments, when a difference was observed, it was always an increase in expression levels (Fig. 6A). No significant difference in the Kcnq1ot1 expression level was observed (Fig. 6B).
Fig. 6.
Depletion of CTCF and cohesins leads to changes in Gtl2 and Dlk1 expression but not in Kcnq1ot1 RNA levels. (A) Relative RNA levels of Gtl2 and Dlk1 in siRNA-treated MEF cells. *, P < 0.05; **, P < 0.01. P values reflect the differences between CTCF or cohesin siRNA treatments and the control siRNA treatment. (B) Relative RNA levels of Kcnq1ot in siRNA-treated MEF cells. No significant change was seen between CTCF or cohesin siRNA treatments and the control siRNA treatment. For both panels, the levels of imprinted gene RNA were normalized to Arpp0. The relative value of the control siRNA treatment was set to 1. Error bars indicate standard errors.
DISCUSSION
The first evidence for noncanonical functions of cohesins came from human diseases associated with mutations in cohesins and cohesin-related genes. Mutations in the NIPBL gene (the human ortholog of Scc2, which loads cohesins onto chromatin [7]) and, subsequently, in the SMC1A and SMC3 genes of Cornelia de Lange syndrome (CdLS) patients were described previously (8, 30, 62). Surprisingly, some CdLS individuals do not have severe defects in sister chromatid cohesion (28), suggesting that cohesins are involved in gene regulation and development independent of their mitotic role. Following this discovery, a number of studies revealed that cohesins were required in neuronal morphogenesis and embryonic development in model organisms (for a review, see reference 1). Although cohesins can function as transcriptional activators at some genes, they may repress the expression of other genes (1). The possible mechanism(s) by which cohesins carry out distinct functions at different genes remained unknown until the genome-wide colocalization of cohesins and CTCF was described (43, 48, 67). Consistent with the idea that CTCF and cohesins function cooperatively in transcriptional regulation, we found a colocalization of CTCF and cohesins at three imprinted loci and observed similar changes in imprinted gene RNA levels between CTCF KD and cohesin KD cells (Fig. 5 and 6). However, the CTCF KD had a much stronger effect than did the cohesin KD (Fig. 5 and 6). Therefore, we cannot rule out the possibility that cohesins do not have direct roles at imprinted loci and function independently of CTCF. In addition to CTCF, other partners of cohesins in gene regulation have been described, including the Polycomb protein complex PRC2 (49), the Mediator complex (26), and estrogen receptor (ER) alpha (50). The Mediator- and ER-associated sites are largely depleted of CTCF-associated cohesin-binding sites (26, 50), suggesting that cohesins function through distinct cofactors. Nevertheless, the colocalization of the binding of CTCF and cohesins at imprinted loci and the similar effects seen upon KD suggest that CTCF and cohesins cooperate in regulating the expression of imprinted genes.
CTCF's role at the H19 ICR as an insulator protein regulating genomic imprinting is well established (for a review, see reference 2). When CTCF-binding sites in the H19 ICR are deleted from the maternal chromosome, the insulator activity is lost, resulting in the biallelic expression of Igf2 and reduced H19 expression levels (12). However, we did not observe a loss of insulation at the H19 ICR in MEFs when CTCF was depleted by RNAi (Fig. 4C and E), nor did we see a reduction in H19 RNA levels. This finding could be attributed to our KD experiments not reducing CTCF below a threshold level required to maintain insulation at the H19 ICR in MEFs. Alternatively, CTCF may be required only for the establishment, rather than the maintenance, of Igf2 imprinting. Our previous work, however, argues against this idea. The conditional deletion of the entire H19 ICR from the mouse neonatal liver led to a loss of imprinting, suggesting that the ICR is required to maintain Igf2 repression on the maternal allele (60). Nevertheless, the presence of CTCF prior to the RNAi treatment could be sufficient for establishing a chromatin state that is able to maintain imprinted gene expression. The fact that we observed increased expression levels of Igf2 and H19 RNAs upon the depletion of CTCF (Fig. 5A) suggests that CTCF has dual functions in the regulation of the H19/Igf2 locus, namely, insulation and transcriptional repression.
Similar experiments involving the depletion of CTCF and cohesin complex subunits by siRNA or short hairpin RNA (shRNA) treatments have been reported in the literature. Wendt et al. showed a reduction of H19 RNA levels and increased Igf2 RNA levels upon the depletion of both CTCF and RAD21 in HeLa cells (67). However, neither a statistical analysis of the mRNA changes nor an allele-specific expression analysis was performed in that study. Furthermore, Nativio et al. reported no changes in H19 imprinted expression or total H19 RNA in human HB2 cells. The authors described elevated and biallelic expression of Igf2 after RAD21 KD, but the level was so low prior to KD that it was impossible to determine whether basal transcription was monoallelic or biallelic (42). The disparity between data from these studies and data from our study could originate from differences in cell types and species (mouse versus human). A recent study using MEFs by Yao et al. showed reduced H19 and increased Igf2 expression levels upon CTCF depletion by shRNA (69). However, the reduction of the H19 expression level was not statistically significant, and those authors did not show the allelic expression profile of Igf2, leaving the underlying mechanism for the phenotypes unclear. Thus, the role of cohesins in insulator function at the H19 ICR remains uncertain.
Despite the disparate observations in our study and the ones described above (42, 67, 69), one phenotype is strikingly similar, namely, elevated Igf2 RNA levels after the depletion of CTCF and cohesins, which is independent of the CTCF-binding sites in the H19 ICR (Fig. 5). ChIP assays with various human and mouse tissues revealed multiple CTCF-binding sites at the Igf2 locus (6, 11, 31, 33, 71); CTCF appears preferentially bound to the maternal allele of Igf2 DMR1 and the P2 and P3 promoters (31, 33). Although those observations suggest that CTCF is involved in the maternal-specific repression of Igf2, our data argue against this idea. Additional CTCF-binding sites at the mouse Igf2 locus have been reported. One report stated that CTCF binds to Igf2 DMR0, which is largely placenta specific (41), on both alleles (33), whereas we detected no binding in MEFs (data not shown). In concordance with the latter observation, we saw no consistent enhanced expression from the Igf2 P0 promoter in KD MEFs (data not shown). Other sites, which are located 5.2 kb and 1.7kb upstream of the Igf2 DMR0, have not been tested allelically (6, 71), but SMC1 also binds to these sites in MEFs (Gene Expression Omnibus [GEO] accession number GSM560355) (26). Finally, CTCF and SMC3 bind upstream of human Igf2 DMR0 on both parental alleles (42). We hypothesized that the CTCF-binding sites in the H19 ICR have a higher affinity for CTCF and/or cohesins than the binding sites at Igf2 and that the depletion of CTCF and RAD21 causes a loss of binding at Igf2 but not the H19 ICR. Therefore, only the repressive function but not the insulator function is disturbed upon siRNA treatment. To test this idea, we performed ChIP experiments on MEFs treated with control and Ctcf siRNAs. In Ctcf siRNA-treated MEFs, CTCF, SMC1, and RAD21 remained bound at the H19 ICR and at both sites upstream of Igf2 DMR0 (data not shown). Therefore, it is unlikely that CTCF functions through these binding sites to repress Igf2 expression.
Although the role of CTCF at the H19/Igf2 locus is well established, its functions at the Gtl2 DMR and KvDMR1 remain to be elucidated. The colocalization of CTCF and cohesins at these DMRs suggests that the proteins function together. Although we did not observe a loss of imprinting in any genes tested following the depletion of CTCF and cohesin in MEFs, mRNA levels of imprinted genes were either increased or unaltered (Fig. 4C and E and 5), suggesting a role in the repression of some genes at these loci. An elucidation of the function at these loci is complicated by the fact that CTCF and cohesins bind to thousands of genomic sites. The mechanism(s) by which CTCF and cohesins impact gene expression at imprinted loci other than the H19/Igf2 locus is unclear. Interestingly, the depletion of both CTCF and cohesins similarly affects the genes in the H19/Igf2 and Dlk1/Gtl2 imprinted clusters (Fig. 5A and 6A).
The above-mentioned results suggest the following possible mechanisms. First, CTCF and cohesins function in concert to regulate a transcriptional activator or repressor. The removal of CTCF and cohesins would increase the activity of an activator (or diminish the activity of a repressor), which would lead to the overexpression of imprinted genes in the same cluster. Second, given that CTCF mediates inter- and intrachromosomal interactions (for a review, see reference 68) and is enriched at nuclear lamina-associated domains (18), CTCF and cohesins would participate in the higher chromosome organization that is required to localize gene loci to certain domains in the nucleus. The loss of CTCF and cohesins may therefore impair this chromatin organization, leading to elevated expression levels of imprinted genes. Third, CTCF and cohesins could function through the imprinted gene network (IGN). The IGN was first described in a microarray analysis that suggested a coregulation of imprinted genes (63). Interestingly, H19, Igf2, and Dlk1 expression levels are co-upregulated or downregulated in Zac1-overexpressed or -depleted tissues, respectively (63). Additional evidence for the IGN came from a study showing the coordinated downregulation of 11 imprinted genes (including H19, Igf2, Dlk1, and Gtl2) independent of DNA methylation during postnatal growth deceleration in various mouse tissues (37). Furthermore, Cdkn1c, an imprinted gene in the Kcnq1/Kcnq1ot cluster, was also identified as a member of the IGN in those two studies (37, 63). It is possible that CTCF and cohesins play a role in the IGN because of their binding patterns in various related genes. However, the mechanism underlying IGN gene cross talk and coregulation remains to be elucidated.
The complexity of CTCF and cohesins is further highlighted due to their cell-type-specific binding patterns. ChIPs targeting CTCF and cohesins in various cell types revealed different binding profiles (14, 26, 49, 50). A previous analysis by Essien et al. suggested that CTCF-binding sites at the imprinted loci described here had a relatively low affinity for CTCF (14). These low-occupancy CTCF-binding sites tend to be cell type specific and developmentally regulated (14). Similarly, cohesins also have different binding profiles between mouse embryonic stem cells and MEFs (26) as well as among different human tissues (50) and Drosophila cell lines (49). Those observations suggest that CTCF and cohesins have distinct functions in different cell types at a subset of targets. To illustrate further the roles of CTCF and cohesins in genomic imprinting, more cell types need to be investigated. By comparing the binding profiles of CTCF and cohesins at imprinted loci in different cell types, and subsequently correlating these with the imprinted gene expression patterns, one would be able to determine which binding sites are controlling imprinting and which are controlling transcriptional levels.
In conclusion, our results have provided a systematic view of the colocalization of CTCF and cohesins at imprinted gene loci. Distinct binding profiles of CTCF and cohesins at different imprinted DMRs indicate locus-specific functions. Furthermore, by performing our allele-specific analysis, we have uncovered new roles for CTCF in the transcription regulation of the active allele of imprinted genes. Our findings also suggest a role for cohesins in regulating the expression of imprinted genes.
ACKNOWLEDGMENTS
We thank D. N. Filippova for the siRNA protocol. We thank the members of Bartolomei laboratory for their helpful discussion of this work and critical reading of the manuscript.
This research was supported by the National Institutes of Health (grants HD042026 to M.S.B. and R.M.S., GM51270 to M.S.B., and HD022681 to R.M.S.) and the Medical Research Council, United Kingdom (A.C.F.-S.).
Footnotes
Published ahead of print on 31 May 2011.
REFERENCES
- 1. Barbero J. L. 2009. Cohesins: chromatin architects in chromosome segregation, control of gene expression and much more. Cell. Mol. Life Sci. 66:2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartolomei M. S. 2009. Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev. 23:2124–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bell A. C., Felsenfeld G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482–485 [DOI] [PubMed] [Google Scholar]
- 4. Butler M. G. 2009. Genomic imprinting disorders in humans: a mini-review. J. Assist. Reprod. Genet. 26:477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carr M. S., Yevtodiyenko A., Schmidt C. L., Schmidt J. V. 2007. Allele-specific histone modifications regulate expression of the Dlk1-Gtl2 imprinted domain. Genomics 89:280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen X., et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133:1106–1117 [DOI] [PubMed] [Google Scholar]
- 7. Ciosk R., et al. 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5:243–254 [DOI] [PubMed] [Google Scholar]
- 8. Deardorff M., et al. 2007. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am. J. Hum. Genet. 80:485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dorsett D. 2007. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma 116:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dorsett D., et al. 2005. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development 132:4743–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Du M., et al. 2003. Insulator and silencer sequences in the imprinted region of human chromosome 11p15.5. Hum. Mol. Genet. 12:1927–1939 [DOI] [PubMed] [Google Scholar]
- 12. Engel N., Thorvaldsen J. L., Bartolomei M. S. 2006. CTCF-binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum. Mol. Genet. 15:2945–2954 [DOI] [PubMed] [Google Scholar]
- 13. Reference deleted.
- 14. Essien K., et al. 2009. CTCF-binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol. 10:R131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Filippova G. N. 2008. Genetics and epigenetics of the multifunctional protein CTCF. Curr. Top. Dev. Biol. 80:337–360 [DOI] [PubMed] [Google Scholar]
- 16. Fitzpatrick G. V., et al. 2007. Allele-specific binding of CTCF to the multipartite imprinting control region KvDMR1. Mol. Cell. Biol. 27:2636–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fitzpatrick G. V., Soloway P. D., Higgins M. J. 2002. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 32:426–431 [DOI] [PubMed] [Google Scholar]
- 18. Guelen L., et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453:948–951 [DOI] [PubMed] [Google Scholar]
- 19. Haering C. H., Farcas A., Arumugam P., Metson J., Nasmyth K. 2008. The cohesin ring concatenates sister DNA molecules. Nature 454:297–301 [DOI] [PubMed] [Google Scholar]
- 20. Hagan J. P., O'Neill B. L., Stewart C. L., Kozlov S. V., Croce C. M. 2009. At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1. PLoS One 4:e4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hark A. T., et al. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405:486–489 [DOI] [PubMed] [Google Scholar]
- 22. Hikichi T., Kohda T., Kaneko-Ishino T., Ishino F. 2003. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic Acids Res. 31:1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kagami M., et al. 2005. Segmental and full paternal isodisomy for chromosome 14 in three patients: narrowing the critical region and implication for the clinical features. Am. J. Med. Genet. 138A:127–132 [DOI] [PubMed] [Google Scholar]
- 24. Kagami M., et al. 2010. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet. 6:e1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kagami M., et al. 2008. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat. Genet. 40:237–242 [DOI] [PubMed] [Google Scholar]
- 26. Kagey M. H., et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467:430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanduri C., et al. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10:853–856 [DOI] [PubMed] [Google Scholar]
- 28. Kaur M., et al. 2005. Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am. J. Med. Genet. 138A:27–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kernohan K. D., et al. 2010. ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev. Cell 18:191–202 [DOI] [PubMed] [Google Scholar]
- 30. Krantz I. D., et al. 2004. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. Genet. 36:631–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kurukuti S., et al. 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. U. S. A. 103:10684–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lewis A., et al. 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 36:1291–1295 [DOI] [PubMed] [Google Scholar]
- 33. Li T., et al. 2008. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol. Cell. Biol. 28:6473–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin S., et al. 2007. Differential regulation of imprinting in the murine embryo and placenta by the Dlk1-Dio3 imprinting control region. Development 134:417–426 [DOI] [PubMed] [Google Scholar]
- 35. Lin S., et al. 2003. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 35:97–102 [DOI] [PubMed] [Google Scholar]
- 36. Lobanenkov V. V., et al. 1990. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5:1743–1753 [PubMed] [Google Scholar]
- 37. Lui J. C., Finkielstain G. P., Barnes K. M., Baron J. 2008. An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295:R189–R196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mancini-DiNardo D., Steele S. J. S., Ingram R. S., Tilghman S. M. 2003. A differentially methylated region within the gene Kcnq1 functions as an imprinted promoter and silencer. Hum. Mol. Genet. 12:283–294 [DOI] [PubMed] [Google Scholar]
- 39. Mancini-Dinardo D., Steele S. J. S., Levorse J. M., Ingram R. S., Tilghman S. M. 2006. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 20:1268–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mann M. R. W., et al. 2003. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol. Reprod. 69:902–914 [DOI] [PubMed] [Google Scholar]
- 41. Moore T., et al. 1997. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc. Natl. Acad. Sci. U. S. A. 94:12509–12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nativio R., et al. 2009. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 5:e1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Parelho V., et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132:422–433 [DOI] [PubMed] [Google Scholar]
- 44. Pateras I. S., Apostolopoulou K., Niforou K., Kotsinas A., Gorgoulis V. G. 2009. p57KIP2: “Kip”ing the cell under control. Mol. Cancer Res. 7:1902–1919 [DOI] [PubMed] [Google Scholar]
- 45. Paulsen M., et al. 2001. Comparative sequence analysis of the imprinted Dlk1-Gtl2 locus in three mammalian species reveals highly conserved genomic elements and refines comparison with the Igf2-H19 region. Genome Res. 11:2085–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reese K. J., Lin S., Verona R. I., Schultz R. M., Bartolomei M. S. 2007. Maintenance of paternal methylation and repression of the imprinted H19 gene requires MBD3. PLoS Genet. 3:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rollins R. A., Korom M., Aulner N., Martens A., Dorsett D. 2004. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell. Biol. 24:3100–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rubio E. D., et al. 2008. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. U. S. A. 105:8309–8914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schaaf C. A., et al. 2009. Regulation of the Drosophila Enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS One 4:e6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schmidt D., et al. 2010. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 20:578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schuster-Gossler K., Bilinski P., Sado T., Ferguson-Smith A., Gossler A. 1998. The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev. Dyn. 212:214–228 [DOI] [PubMed] [Google Scholar]
- 52. Shin J., Fitzpatrick G. V., Higgins M. J. 2008. Two distinct mechanisms of silencing by the KvDMR1 imprinting control region. EMBO J. 27:168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smilinich N. J., et al. 1999. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc. Natl. Acad. Sci. U. S. A. 96:8064–8069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stedman W., et al. 2008. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 27:654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Szabó P. E., Tang S. E., Rentsendorj A., Pfeifer G. P., Mann J. R. 2000. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 10:607–610 [DOI] [PubMed] [Google Scholar]
- 56. Takada S., et al. 2000. Delta-like and Gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr. Biol. 10:1135–1138 [DOI] [PubMed] [Google Scholar]
- 57. Thakur N., Kanduri M., Holmgren C., Mukhopadhyay R., Kanduri C. 2003. Bidirectional silencing and DNA methylation-sensitive methylation-spreading properties of the Kcnq1 imprinting control region map to the same regions. J. Biol. Chem. 278:9514–9519 [DOI] [PubMed] [Google Scholar]
- 58. Thakur N., et al. 2004. An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol. Cell. Biol. 24:7855–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Thorvaldsen J. L., Duran K. L., Bartolomei M. S. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 12:3693–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Thorvaldsen J. L., Fedoriw A. M., Nguyen S., Bartolomei M. S. 2006. Developmental profile of H19 differentially methylated domain (DMD) deletion alleles reveals multiple roles of the DMD in regulating allelic expression and DNA methylation at the imprinted H19/Igf2 locus. Mol. Cell. Biol. 26:1245–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Thorvaldsen J. L., Mann M. R. W., Nwoko O., Duran K. L., Bartolomei M. S. 2002. Analysis of sequence upstream of the endogenous H19 gene reveals elements both essential and dispensable for imprinting. Mol. Cell. Biol. 22:2450–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tonkin E. T., Wang T., Lisgo S., Bamshad M. J., Strachan T. 2004. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 36:636–641 [DOI] [PubMed] [Google Scholar]
- 63. Varrault A., et al. 2006. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev. Cell 11:711–722 [DOI] [PubMed] [Google Scholar]
- 64. Verona R. I., Thorvaldsen J. L., Reese K. J., Bartolomei M. S. 2008. The transcriptional status but not the imprinting control region determines allele-specific histone modifications at the imprinted H19 locus. Mol. Cell. Biol. 28:71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wan L., et al. 2008. Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Development 135:2729–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Weaver J. R., et al. 2010. Domain-specific response of imprinted genes to reduced DNMT1. Mol. Cell. Biol. 30:3916–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wendt K. S., et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451:796–801 [DOI] [PubMed] [Google Scholar]
- 68. Williams A., Flavell R. A. 2008. The role of CTCF in regulating nuclear organization. J. Exp. Med. 205:747–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yao H., et al. 2010. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 24:2543–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yoon B., et al. 2005. Rasgrf1 imprinting is regulated by a CTCF-dependent methylation-sensitive enhancer blocker. Mol. Cell. Biol. 25:11184–11190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yoon Y. S., et al. 2007. Analysis of the H19ICR insulator. Mol. Cell. Biol. 27:3499–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]