Abstract
Expanded trinucleotide repeats are responsible for a number of neurodegenerative diseases, such as Huntington disease and myotonic dystrophy type 1. The mechanisms that underlie repeat instability in the germ line and in the somatic tissues of human patients are undefined. Using a selection assay based on contraction of CAG repeat tracts in human cells, we screened the Prestwick chemical library in a moderately high-throughput assay and identified 18 novel inducers of repeat contraction. A subset of these compounds targeted pathways involved in the management of DNA supercoiling associated with transcription. Further analyses using both small molecule inhibitors and small interfering RNA (siRNA)-mediated knockdowns demonstrated the involvement of topoisomerase 1 (TOP1), tyrosyl-DNA phosphodiesterase 1 (TDP1), and single-strand break repair (SSBR) in modulating transcription-dependent CAG repeat contractions. The TOP1-TDP1-SSBR pathway normally functions to suppress repeat instability, since interfering with it stimulated repeat contractions. We further showed that the increase in repeat contractions when the TOP1-TDP1-SSBR pathway is compromised arises via transcription-coupled nucleotide excision repair, a previously identified contributor to transcription-induced repeat instability. These studies broaden the scope of pathways involved in transcription-induced CAG repeat instability and begin to define their interrelationships.
INTRODUCTION
Expansion of CAG·CTG repeats in specific human genes is associated with several neurodegenerative diseases that cause neuron dysfunction or death. These diseases include Huntington disease (HD), myotonic dystrophy type 1 (DM1), and several spinocerebellar ataxias (SCAs) (11, 23, 39). Long CAG repeat tracts in disease genes tend to be unstable in the germ line and in many somatic tissues, giving rise to contractions (fewer repeat units) and expansions (more repeat units), but usually with a distinct bias toward expansions. In the germ line, expansions lead to earlier disease onset and increased severity in affected individuals (40), while expansions in specific types of neuron may exacerbate the disease phenotype (13, 40, 49). Treatments designed to prevent repeat expansion or to promote repeat contraction would be welcome, but no such treatments exist, and their development will depend on a better understanding of the mechanisms responsible for repeat instability (34).
In the past 20 years, numerous studies using bacteria, yeast, flies, human cells, and mice as model systems have identified potentially important roles for various DNA-based processes— including DNA replication, recombination, DNA repair, and transcription—in driving repeat instability (29, 35, 40). Each of these processes exposes single strands of repeats, which can form secondary structures such as hairpins and slipped-strand duplexes (10, 41), which are thought to be the key intermediates that trigger repeat instability. As if that were not enough diversity, additional identified contributors to instability include epigenetic modifications, chromatin structure, and local sequence effects (4, 7). Thus, the molecular details of repeat instability, especially the number of relevant pathways and the interconnections among them, are not yet clear.
This embarrassment of mechanistic riches precludes any simple answers to the question of what drives repeat instability in human germ line and somatic tissues. However, it also raises the critical question of whether the list of repeat-altering processes is complete. In an attempt to identify additional modulators of repeat instability, we took an unbiased approach: screening for reagents in the Prestwick chemical library that stimulate repeat contraction. Although a few genotoxic chemicals and DNA metabolic inhibitors have previously been tested in human cells (13–16, 18, 43, 57), a chemical screen is not feasible with common methods for detecting changes in repeat length such as small-pool PCR. To carry out a chemical screen, we modified the selective assay for CAG repeat contraction in human cells that we have previously used to probe various aspects of repeat instability (6, 16, 27, 28, 30–32, 36, 37).
We chose to screen the Prestwick chemical library (880 compounds in the version we tested) because it represents a diverse range of chemical structures and covers most therapeutic areas (45). We felt that a broad screen might provide additional insight into potential disease-causing processes. Moreover, since we assay for enhancement of repeat contraction by mostly U.S. Food and Drug Administration-approved drugs, we could potentially identify useful drugs that might specifically stimulate repeat contraction. As described here, our chemical screen identified topoisomerase 1 (TOP1), tryrosyl-DNA phosphodiesterase (TDP1), and single-strand break repair (SSBR) as important regulators of repeat instability. These components operate in a transcription-dependent pathway—the TOP1-TDP1-SSBR pathway—that normally acts to suppress repeat instability. When this pathway is compromised, the frequencies of CAG contraction rise due to the involvement of the transcription-coupled nucleotide excision repair (TC-NER) pathway of repair, which normally acts to enhance repeat instability.
MATERIALS AND METHODS
Cell culture.
FLAH25 cells were derived from the human fibrosarcoma HT1080 cell line. Construction of this cell line was described previously (28). FLAH25 cells carry a spontaneous nonreverting deletion of the single endogenous HPRT gene; an integrated plasmid that constitutively expresses rtTA protein, which is the reverse tetracycline repressor protein fused to the HSV VP16 activation domain (17); and a single copy of the HPRT minigene with a CAG95 tract embedded in its single intron. The expression of the HPRT minigene in FLAH25 cells is driven by the promoter, pCMV-mini, which is suppressed by rtTA, but can be induced 22-fold by doxycycline (28). FLAH25 cells were grown at 37°C with 5% CO2 in Dulbecco modified Eagle medium/F-12 medium supplemented with 10% fetal bovine serum and 1% minimal essential medium nonessential amino acids.
HPRT+ selection.
To select for HPRT+ cells, 500,000 FLAH25 cells were plated on 10-cm dishes in HAT medium (0.1 mM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymine [Sigma]) plus doxycycline (2.0 μg/ml) for 2 weeks, with the addition of fresh doxycycline (2.0 μg/ml) after the first week of selection. Colonies were stained with Coomassie bright blue, and their numbers were counted. Contraction frequencies, which were calculated as the number of HPRT+ colonies divided by the number of viable cells, are the average of at least four experiments. The number of viable cells was calculated by multiplying the number of cells plated by the plating efficiency, which was determined by plating 200 cells on each 10-cm dish in the presence doxycycline, but without selection, for 2 weeks.
Real-time RT-PCR.
Total RNA was prepared from about two million cells using RNeasy minikits (Qiagen). For the induction of HPRT minigene expression, doxycycline was added 1 day before RNA was extracted. For real-time reverse transcription-PCR (RT-PCR), 50 ng of total RNA per reaction was assayed using the SYBR green RT-PCR kit (Qiagen). Real-time RT-PCR primers are shown in Table 1 . For all genes, results were normalized to the concentration of β-actin mRNA in the same sample, which was also determined by real-time RT-PCR (42). The doxycycline-induced increase in HPRT gene expression in cells that were also treated with a specific small interfering RNA (siRNA) was measured relative to the HPRT mRNA level in vimentin siRNA-treated cells (the control cells) that had not been exposed to doxycycline. The percentage of knockdown of target gene mRNA was determined by comparison with the target gene mRNA level in vimentin siRNA-treated control cells, which was defined as 100%. The conditions for real-time RT-PCR were 50°C for 30 min and 95°C for 15 min, followed by 45 cycles of 94°C for 15 s, 52 or 55°C (depending on the primers) for 30 s, and 72°C for 30 s. The relative levels of mRNA were calculated by comparing number of cycles (generally between 14 and 26 cycles) at which the PCR products became detectable above the basal threshold.
Table 1.
Sequences of real-time RT-PCR primers and siRNAs used in this study
| Gene | Real-time RT-PCR primer |
Primer no.a and siRNA | |
|---|---|---|---|
| Forward | Reverse | ||
| β-Actin | AGAGAGGCATCCTCACCCTG | CATGAGGTAGTCAGTCAGGT | |
| HPRT | CGGCTACAAGGACGACTCTAG | TTGATGTAATCCAGCAGGTCAGC | |
| PARP1 | 1, GUGGCGAAGAAGAAAUCUA | ||
| TDP1 | GATGATGAGCTGCAACCAG | CTGAAGAAACAAGCGTCCC | 1, CCACCUUUCCUGUGCCAUAUGAUUU |
| TAAAACAGTATCCACCAGAGTTCA | CCATCAGCAATTCGTGGG | 2, GGAUAUGGAACAUUCCUUAUGUCAA | |
| TOP1 | GAAGCGGATTTCCGATTGAAT | AGGTTCATCTTTAATTTGTGGTGG | 1, GAGACGAAUCAUGCCCGAGGAUAUA |
| GACAAACATAAAGACAGAGACAAGG | AGGTTCATCTTTAATTTGTGGTGG | 2, GAUGAAAGUCCGGCAGAGAGCUGUA | |
| Vimentin | 1, GAAUGGUACAAAUCCAAGU | ||
| XPA | GCGGCGGCTTTAGAGCAAC | GCGGCGGCTTTAGAGCAAC | 1, GCUACUGGAGGCAUGGCUAb |
| XRCC1 | CTCTACCTCATCCTTCTGGC | GCCATCATTCCCAATGTCC | 1, GGAAGAUCCUUCAGGGUGUGGUAGU |
| CATGTCCCCTTCCGAGAG | GATCCGGCTGAAGAAGAGAG | 2, CAGUUUGUGAUCACAGCACAGGAAU | |
Chemical and siRNA treatments.
For chemical treatments, 100,000 FLAH25 cells were plated onto 96-well plates and allowed to adhere overnight to the plates. Individual chemicals from the Prestwick chemical library (Prestwick Chemicals) were added to each well (1 μl of chemical/well to a final concentration of ∼2.5 μM). Since the chemicals were dissolved in dimethyl sulfoxide (DMSO), we used an equal concentration of DMSO as the control treatment. For the screen, the cells were treated for three cycles in 96-well plates, with each cycle consisting of 1 day of treatment with chemical, followed by 1 day of recovery in fresh medium without chemical. Doxycycline was present at all times. After recovery at the end of the third cycle, the cells were plated onto six-well plates, and HPRT+ colonies were selected. After 14 days, plates were stained, and the HPRT+ colonies were counted. Chemicals that gave more than five colonies in the initial screen were counted as positive. A subset of those chemicals (ones that were easily obtained in larger quantities) was then tested at 10 μM in our standard assay for repeat contraction (Table 2, round 2). For chemicals that we investigated further, as described in the figures, we determined the concentration dependence of cell killing and selected a concentration that killed ca. 50 to 70% of the cells after 3 days of continuous treatment. For round 2 in Table 2 and for the experiments reported in the figures, we plated one million cells in each 10-cm dish on day −1. Beginning on day 0, the cells were treated for 3 days in the presence of chemical and doxycycline and then allowed to recover for 1 day in the presence of fresh medium lacking the chemical but containing doxycycline. Because all of the chemicals were dissolved in DMSO, the same concentration of DMSO was added to the FLAH25 cells as a negative control. On day 3, the medium was replaced with fresh medium lacking chemicals, and after one additional day the cells were replated for HRPT+ colony selection.
Table 2.
Positive compounds in the screen of the Prestwick chemical library
| Compound | Function | Contraction frequency (10−6) |
|
|---|---|---|---|
| Round 1 | Round 2 | ||
| DMSO | Organic solvent (control) | 3.0 | 8.9 |
| Acacetin | Anticancer drug, TOP1 inhibitor | 80 | 40 |
| Acyclovir | Inhibits viral DNA polymerase | 20 | 136 |
| Amikacin | Aminoglycoside antibiotic | 13 | 87 |
| Amphotericin B | Polyene antifungal drug | 53 | |
| Amprolium | Antiprotozoal agent | 33 | 76 |
| Antipyrine | Analgesic and antipyretic agent | 27 | |
| Betulinic acid | Anticancer agent | 67 | 75 |
| Butirosin | Aminoglycoside antibiotic | 20 | 52 |
| Chinchonine | Antimalaria agent | 27 | |
| Hydroflumethiazide | Thiazide diuretic | 60 | 78 |
| Mefloquine | Antimalarial agent | 33 | |
| Metformin | Antimalarial agent | 27 | |
| Oleandomycin | Macrolide antibiotic | 20 | |
| Syrosingopine | Antipsychotic and antihypertensive | 53 | |
| Thiamine | Vitamin B1 | 67 | 143 |
| Trihexyphidyl | Muscarinic antagonist | 13 | |
| Vidarabine | Antiviral drug | 47 | 98 |
| Vitexin | Flavonoid with antioxidant activity | 27 | 38 |
The siRNAs (Dharmacon or Invitrogen) used in the present study are listed in Table 1. For siRNA treatments, about 250,000 FLAH25 cells were plated in each 10-cm dish on day −3. On day −2, cells were transfected with siRNAs at a final concentration of 200 nM, using Oligofectamine (Invitrogen). Treatments with 200 nM vimentin-siRNA served as controls. For siRNA knockdowns, the specific siRNA was transfected at 100 nM, together with vimentin-siRNA at 100 nM to bring the final siRNA concentration to 200 nM. For double knockdowns, each of the two siRNA was transfected at 100 nM. On day 0, the cells were again transfected with siRNA, and cultures were then grown in the presence or absence of doxycycline. We evaluated knockdown of target gene expression and HPRT transcription by analyzing total RNA isolated on day 1.
Statistics.
Statistical analyses of significance were conducted using a Student t test to compare the means and standard deviations derived from six to eight experiments for each of the treatments in Fig. 2 to 5.
Fig. 2.
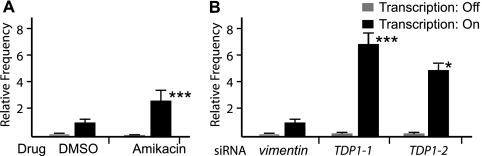
Treatments that interfere with TOP1. (A) Chemical treatments. Acacetin (10 μM) and camptothecin (10 μM) were compared to the DMSO control in the presence (■) or absence (▩) of doxycycline. Relative contraction frequencies were normalized to 1 for DMSO treatment in the presence of doxycycline (8.0 × 10−6 ± 0.8 × 10−6). (B) siRNA treatments. Two TOP1 siRNAs were compared to control siRNA against vimentin in the presence (■) or absence (▩) of doxycycline. Relative contraction frequencies were normalized to 1 for vimentin siRNA in the presence of doxycycline (7.4 × 10−6 ± 1.5 × 10−6). In all cases, error bars indicate standard deviations, and the statistical significance is indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Fig. 5.
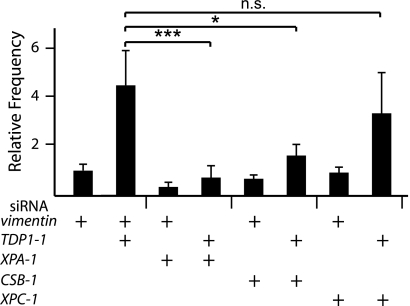
Combination treatments with siRNAs. FLAH25 cells were treated with combinations of siRNAs against vimentin (the control), TDP1, and XPA. Relative contraction frequencies were normalized to 1 for vimentin siRNA (9.5 × 10−6 ± 2.9 × 10−6). In all cases, error bars show the standard deviations, and the statistical significance is indicated (n.s., not significant; *, P < 0.05; ***, P < 0.001).
RESULTS
Chemical screen identifies modulators of CAG repeat instability.
Previously, we developed a CAG-contraction assay in FLAH25 human cells that consists of a selection marker, the HPRT (hypoxanthine-guanine phosphoribosyl transferase) minigene, containing a (CAG)95 repeat tract driven by an inducible promoter (Fig. 1A) (28). Using this assay, we can detect large CAG contraction events that reduce the size of the repeat tract from 95 to 38 units or fewer. When transcription through the CAG repeat is induced 20-fold, large contractions occur at a frequency of about 1 in 105 cells. With this background, we can use 96-well plates to screen reagents for those that stimulate repeat contraction, as illustrated in Fig. 1B. We chose to screen the Prestwick chemical library because it includes a broad diversity of structures with a wide range of pharmacological effects, it consists largely of compounds with verified biological mechanisms or pharmacological targets, and 85% of these chemicals have been marketed in the United States or Europe (45). We expected that the diversity represented in the library would not only maximize our chance of identifying new repeat-destabilizing processes but also that the extensive database of information would make it easier to associate chemicals that stimulate contraction with cellular pathways that modulate repeat stability.
Fig. 1.
Screening the Prestwick chemical library. (A) Selection assay for CAG repeat contractions. Large CAG repeat tracts are spliced aberrantly, preventing expression of functional HRPT and making the cells HAT sensitive (HATS). When the repeat tract is 38 units or fewer, sufficient correctly spliced message is produced to permit expression of enough HPRT to become HAT resistant (HATR). (B) Design of chemical screen. FLAH25 cells were plated at near confluence in 96-well plates, treated for 3 days with chemicals in the presence of doxycycline, and then replated for HAT selection in six-well plates.
To screen the chemical library, we plated HPRT− FLAH25 cells at near confluence in 96-well plates and allowed them to adhere overnight. Individual compounds were added to the wells (∼2.5 μM) in the presence of doxycycline to induce transcription. After treatment, the cells were replated into six-well plates and grown in HAT medium for 14 days to select for HPRT+ colonies. Compounds that gave at least 5-fold more HPRT+ colonies than the DMSO control were counted as positive. Using this method, we screened 880 compounds and identified 18 that increased CAG repeat contraction frequencies (Table 2, round 1). For the subset of these compounds that was easy to obtain, we confirmed these initial findings in our standard large-plate format (Table 2, round 2). These compounds included a variety of structures (see Fig. S1 in the supplemental material), with a wide range of pharmacological actions. In contrast to our expectations, the information on most of these compounds did not immediately suggest a pathway by which they might alter repeat stability.
Topoisomerase 1 inhibitors stimulate CAG repeat contraction.
Among the group of 18 chemicals that increased repeat instability, we identified two flavones, acacetin and vitexin. Several flavones, including acacetin, have been reported to inhibit TOP1 (1), but with other known or potential activities (19). For this reason, we also tested camptothecin, a highly specific and well-defined inhibitor of TOP1, which relaxes supercoiled DNA by nicking and religating a single strand of the duplex (25, 54). Acacetin and camptothecin interfere with TOP1 in different ways, but both inhibit the religation step in the reaction (3). Because TOP1 plays a critical role in regulating the supercoiling associated with RNA transcription (54), we initially tested to see whether stimulation of repeat instability was dependent or independent of transcription. As shown in Fig. 2A, neither drug significantly enhanced repeat contraction in the absence of doxycycline, the inducer of transcription through the CAG repeat. We confirmed that HPRT+ colonies were CAG contractions by isolating and analyzing repeat lengths in individual colonies (data not shown). These results indicate that proper regulation of DNA supercoiling during transcription is critical for preventing repeat instability.
To confirm its role in repeat instability, we used siRNA to knock down TOP1 activity. These siRNAs reduced TOP1 mRNA levels by 71 and 69% (Table 3), and they significantly increased CAG contractions (Fig. 2B). The lesser effect of siRNAs relative to the chemical inhibitors may reflect the residual levels of TOP1 after siRNA knockdown, or it may be a consequence of the mechanistic difference: the reduction of activity versus trapping of a reaction intermediate. In any case, these results confirm that TOP1 modulates transcription-induced CAG repeat contraction.
Table 3.
siRNA knockdowns and their effects on cells
| siRNA | Cell no., 106 (%)a | Proliferation rate (%)b | Plating efficiency (%)c | HPRT mRNA (fold)d | % Target mRNAe |
|---|---|---|---|---|---|
| Vimentin | 4.6 (100) | 0.82 (100) | 100 | 23 | |
| TOP1-1 | 2.3 (50) | 0.66 (81) | 98 | 31 | 29 |
| TOP1-2 | 2.4 (52) | 0.67 (82) | 87 | 25 | 31 |
| TDP1-1 | 1.7 (36) | 0.58 (71) | 111 | 30 | 34 |
| TDP1-2 | 1.6 (35) | 0.57 (70) | 106 | 32 | 37 |
| XRCC1-1 | 4.7 (103) | 0.83 (101) | 108 | 33 | 24 |
| XRCC1-2 | 4.9 (106) | 0.84 (102) | 109 | 21 | 28 |
| PARP1-1 | 3.5 (75) | 0.76 (93) | 61 | ND |
Cell number refers to the total number of cells present per plate after 6 days of siRNA treatment. Each plate began with an initial cell population of 150,000 cells. Treatment with vimentin siRNA was used as the reference and was defined as 100%.
The proliferation rate is the number of cell doublings per day required to increase the cell population at the beginning of the siRNA treatment (150,000 cells per plate) to the number present at the end of the 6-day treatment. For vimentin siRNA treatment, for example, the initial population of cells increased to 4.6 × 106 cells after 6 days, a 31-fold increase, which corresponds to 4.9 population doublings per 6 days, or 0.82 doublings per day. The proliferation rate during treatment with vimentin siRNA was used as the reference and was defined as 100%.
The plating efficiency was measured at the time cells were replated for selection for HPRT− cells. It is the percentage of colonies that formed when 200 cells were plated in nonselective medium at the time of plating. The absolute plating efficiency for vimentin siRNA-treated cells was 76%. This plating efficiency was used as the reference and was defined as 100%.
Induced HPRT mRNA levels in cells treated with specific siRNAs in the presence of doxycycline are expressed relative to the amount of HPRT mRNA in cells treated with vimentin siRNA in the absence of induction by doxycycline. In all cases the mRNA was first normalized to the β-actin mRNA in the same sample. HPRT mRNA levels were not determined (ND) in the samples treated with PARP1 siRNA.
The target mRNA is the amount of target gene mRNA present in cells treated with the specific siRNA relative to that present in cells treated with vimentin siRNA. For example, the amount of TOP1 mRNA present after treatment with TOP1-2 siRNA was 31% of the amount of TOP1 mRNA present in the vimentin siRNA-treated control. In all cases, the mRNA was first normalized to the β-actin mRNA in the same sample.
Tyrosyl-DNA phosphodiesterase inhibitors promote repeat contraction.
We identified two aminoglycoside antibiotics, amikacin and butirosin, in our initial screen (Table 2). Several aminoglycoside antibiotics, including amikacin, have recently been shown to inhibit the human enzyme, tyrosyl-DNA phosphodiesterase (TDP1) (26), which is responsible for removing TOP1 that has become unproductively attached to DNA (5). As shown in Fig. 3A, amikacin treatment of FLAH25 cells gave a modest stimulation of contraction frequency in the presence of the transcription inducer doxycycline, but none in its absence. We confirmed the involvement of TDP1 using siRNA knockdown. Treatment of FLAH25 cells with two siRNAs, which reduced TDP1 mRNA levels by 63 and 66% (Table 3), stimulated CAG contraction severalfold above cells treated with a control siRNA (Fig. 3B). These results indicate that TDP1, presumably in its primary role as scavenger of trapped TOP1 molecules, normally acts to repress transcription-induced CAG repeat instability.
Fig. 3.
Treatments that interfere with TDP1. (A) Chemical treatments. Amikacin (100 μM) was compared to the DMSO control in the presence (■) or absence (▩) of doxycycline. Relative contraction frequencies were normalized to 1 for DMSO treatment in the presence of doxycycline (8.0 × 10−6 ± 0.8 × 10−6). (B) siRNA treatments. Two TDP1 siRNAs were compared to control siRNA against vimentin in the presence (■) or absence (▩) of doxycycline. Relative contraction frequencies were normalized to 1 for vimentin siRNA in the presence of doxycycline (7.4 × 10−6 ± 1.5 × 10−6). In all cases, error bars show the standard deviations, and the statistical significance is indicated (*, P < 0.05; ***, P < 0.001).
Interference with SSBR enhances CAG repeat instability.
TDP1 is a component of the large multiprotein complex that carries out single-strand break repair (SSBR) (2), suggesting that the SSBR pathway might also modulate CAG repeat contraction. In addition, our chemical screen identified betulinic acid, an inhibitor of another component of this complex, DNA polymerase β, which fills in gaps in the final stage of SSBR (Fig. 4A). To examine the involvement of SSBR, we used siRNAs to knock down X-ray repair cross-complementing protein 1 (XRCC1), the major scaffolding protein involved in the complex, and poly-ADP ribose polymerase (PARP1), which binds to single-strand nicks and recruits XRCC1 and DNA polymerase β (2). Treatment with these siRNAs decreased XRCC1 mRNA levels 72 and 76% (Table 3) and reduced PARP1 protein levels by more than 70% (data not shown). Knockdowns of these mRNAs significantly increased the frequency of CAG repeat contractions (Fig. 4B). These results implicate the SSBR complex, including XRCC1, PARP1, and DNA polymerase β, in addition to TOP1 and TDP1, in the suppression of transcription-induced CAG repeat contraction.
Fig. 4.
Treatments that interfere with SSBR. (A) Chemical treatments. Betulinic acid (10 μM) was compared to the DMSO control in the presence (■) or absence (▩) of doxycycline. Relative contraction frequencies were normalized to 1 for DMSO treatment in the presence of doxycycline (8.0 × 10−6 ± 0.8 × 10−6). (B) siRNA treatments. Two XRCC1 siRNAs and one PARP1 siRNA were compared to control siRNA against vimentin in the presence (■) or absence (▩) of doxycycline. Relative contraction frequencies were normalized to 1 for vimentin siRNA in the presence of doxycycline (7.4 × 10−6 ± 1.5 × 10−6). In all cases, error bars show the standard deviations, and the statistical significance is indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Inhibition of NER blocks the increase in repeat contractions induced by interfering with the TOP1-TDP1-SSBR pathway.
Collectively, our results show that chemicals and siRNAs that interfere with TOP1 or with the cell's ability to correct stalled TOP1 intermediates via TDP1 and SSBR increase the frequency of CAG repeat contractions. These results indicate that TOP1, TDP1, and SSBR normally act to suppress CAG repeat contractions. What then is responsible for the rise in the frequency of repeat contractions when the TOP1-TDP1-SSBR pathway is blocked? Is it simply the absence of this pathway, or do the intermediates “spill over” into an alternative pathway of DNA repair that removes the lesions, but does so in a way that leads to contractions?
We have previously demonstrated that transcription-coupled nucleotide excision repair (TC-NER) normally functions to increase transcription-induced CAG repeat contractions (28, 32). If TC-NER were the cause of the increase in CAG contractions that arise by inhibiting the TOP1-TDP1-SSBR pathway, then we should be able to block the increase by simultaneously knocking down both pathways. To test this prediction, we knocked down TDP1 in combination with components of NER. To distinguish between the contributions of global genome NER (GG-NER) and TC-NER, we tested XPC, which is specifically required for GG-NER, and CSB, which is specific for TC-NER. In addition, we tested XPA, which is a central and critical component in both NER subpathways. As shown in Fig. 5, the stimulation observed with TDP1 knockdown was eliminated by the simultaneous knockdown of TDP1 and CSB or XPA but not significantly affected by the knockdown of TDP1 and XPC. These results indicate that the increased CAG repeat instability observed when the TOP1-TDP1-SSBR pathway was blocked is due to the TC-NER pathway, which normally functions to stimulate transcription-induced CAG repeat contractions (28, 32).
DISCUSSION
We devised a moderately high-throughput chemical screen to provide an unbiased search for new pathways that modulate CAG repeat instability. We chose to screen the Prestwick chemical library because it contains a broad diversity of chemical structures with a wide range of pharmacological effects (45). As with most screens, ours was blind to certain chemicals that alter repeat instability. First, the screen was designed to detect only those chemicals that increased the frequency of contractions. Chemicals that decrease contraction frequency would not have been detected. Second, chemicals that stimulated repeat contraction but were too toxic or were ineffectual at the single concentration tested would also have been missed. Camptothecin, which is in the library but was missed in the initial screen, may be an example of a compound that was tested below its effective concentration, but we have not investigated that possibility further. In addition, our screen may be biased toward modulators that affect transcription, since transcription through the repeat was induced throughout the screen. Even with these limitations, the screen identified 18 chemicals that substantially increased CAG repeat contraction. Five of these—acacetin, vitexin, amikacin, butirosin, and betulinic acid—provided initial clues that led to the identification of the TOP1-TDP1-SSBR repair pathway as a modulator of CAG repeat contraction. These clues were confirmed by camptothecin treatment and siRNA knockdown. The other 13 chemicals identified in the screen may conceal links to repeat instability that are not obvious at present, but may be revealed with additional experiments.
Supercoiling has been shown to play a role in the instability of CGG, GAA, and CAG repeats in bacteria (38), but not previously in mammalian cells. In bacteria the link has been ascribed to the effects of negative supercoiling on enhancing the formation of repeat-induced non-B DNA structures (38), which are thought to constitute the key common event leading to changes in repeat-tract length (24, 40, 56). The negative supercoiling that develops behind a transcribing RNA polymerase (54) would provide a natural connection between transcription and repeat instability and could explain our results with TOP1 inhibitors, especially with siRNA knockdowns of TOP1, which might be expected to increase supercoiling stress (9).
Supercoiling by itself, however, does not readily accommodate the results with inhibitors of TDP1 and SSBR. The TDP1-SSBR connection suggests that formation of irreversible TOP1 cleavage complexes may be the source of the CAG repeat instability observed in our studies. If an RNA polymerase runs into a TOP1-DNA cleavage intermediate on the template strand, the intermediate will be converted to an irreversible cleavage complex that is unable to religate the DNA and release TOP1 (44). In the normal course of events, these complexes are resolved by the action of TDP1 and the SSBR pathway (44). Thus, we favor the interpretation that TOP1, TDP1, and SSBR normally act to suppress CAG repeat instability by restricting the consequences of irreversible TOP1-DNA complex formation. In the presence of TOP1 inhibitors, the frequency of irreversible complexes rises, with a concomitant rise in repeat instability. When TDP1 or the SSBR pathway is inhibited, the frequency of irreversible complexes also rises, resulting in an increase in repeat instability. How such irreversible TOP1-DNA intermediates lead to CAG repeat instability is not clear. In any case, the normal action of the TOP1-TDP1-SSBR pathway suppresses repeat instability, and when this pathway is compromised, expanded CAG repeats become more unstable.
In addition to defining a new pathway—the TOP1-TDP1-SSBR pathway—for CAG repeat instability, we have addressed the fundamental question of what causes the increase in repeat instability when this pathway is compromised by chemical or siRNA treatments. By knocking down the TC-NER pathway at the same time we interfered with the TOP1-TDP1-SSBR pathway, we showed that we could block the increase in repeat contractions. We previously demonstrated that the TC-NER pathway acts to increase transcription-induced CAG contractions; that is, when TC-NER components were knocked down, contraction frequencies were substantially reduced (28, 32). Thus, it appears that TOP1-DNA irreversible cleavage complexes are removed by TC-NER, when the preferred repair pathway is unavailable, as summarized in Fig. 6.
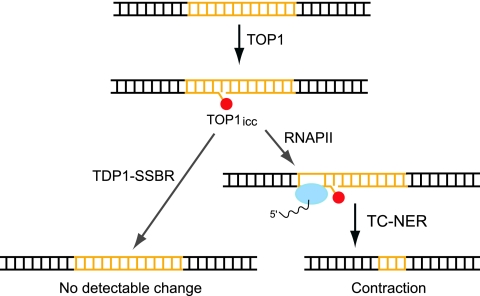
Fig. 6.
Proposed relationship between the TOP1-TDP1-SSBR and the TC-NER pathways. The TOP1-DNA irreversible cleavage complexes (TOP1icc) that arise normally in the course of TOP1 action are usually taken care of by TDP1 and SSBR, which prevent the formation of the large CAG contractions our system is able to detect. When the TDP1-SSBR pathway is compromised, however, RNA polymerase II stalls at the damage, eliciting involvement of TC-NER, which promotes CAG repeat contractions, as described previously (28, 32).
It will be important to determine the relevance of the TOP1-TDP1-SSBR pathway to the CAG repeat instability that occurs in human patients. We are currently testing the effects of the TOP1-TDP1-SSBR pathway in a SCA1 mouse model (55), which allows both germ line and somatic instability to be assessed in an organism that displays patterns of repeat instability similar to those in human patients. There is generally good agreement between results in our assay in human cells and those in mouse models (29–31). Among the 20 genes shown to affect CAG repeat contraction in human cells (6, 27, 28, 31, 32, 37), six have been tested in a mouse model and also shown to modulate CAG repeat instability, including MSH2 (22, 47, 48), MSH3 (52), PMS2 (12), DNMT1 (6), CSB (20), and XPA (L. Hubert et al., unpublished data). Similarly, among those genes with little effect on CAG contraction in human cells (28), three have been tested in mouse models and shown to have little effect on CAG repeats, including MSH6 (52), XPC (8), and FEN1 (50, 51). Interestingly, only in the case of the OGG1 glycosylase discussed below do the results differ, with deficiencies having no effect on CAG contraction in human cells (32), but large effects in mice (21). Overall, the otherwise good agreement between these two assays suggests that screening candidates in human cells will be a productive way to select genes to test in mice.
The TOP1-TDP1-SSBR pathway is similar to the repair process that operates during base excision repair (BER), except that TOP1-TDP1 are replaced by a variety of glycosylases and apurinic-apyrimidinic endonuclease 1 (APE1), which control the initial steps in BER: removing the damaged base and breaking the single strand (46). As in the TOP1-TDP1-SSBR pathway, the final stage of BER—repair of the broken strand—involves XRCC1, DNA polymerase β, and DNA ligase 3, among others (46). The most extensively studied glycosylase is OGG1. It is responsible for excising 8-oxo-guanine, a common base damage caused by reactive oxygen species (53). In cell extracts and in mice, it has been shown that OGG1 normally acts to promote CAG repeat expansions (21, 33). Thus, in the absence of OGG1, CAG repeats become more stable. Strikingly, mouse models of Huntington disease that are genetically deficient for OGG1 have much more stable CAG repeats in brain than do OGG1-positive mice (21). It is curious that the TOP1-TDP1-SSBR pathway acts to suppress CAG instability, while the OGG1-APE1-BER pathway acts to stimulate repeat instability.
There are several possible reasons for these differences. First, it could be a trivial consequence of the selective detection of large contractions in our study versus the analysis of frequent events in the OGG1 studies (21, 33). Thus, it is possible that defects in OGG1 promote large contractions, even as they reduce expansions, but that contractions occur at frequencies that are too low to detect by small-pool PCR. Experiments in our selective system, however, have shown that siRNA knockdowns of OGG1 and APE1 do not change the frequency of transcription-induced CAG contractions (32). A second possibility is that the distinct effects on CAG repeat instability are a consequence of the differences in mechanism. The action of TOP1-TDP1 generates clean ends with 3′ hydroxyls and 5′ phosphates, which are conducive to religation (44); it is only when the process is interrupted that the repeat tracts are affected. In contrast, OGG1-APE1 leaves ends with 3′ hydroxyls and 5′ sugar phosphates (33). Although DNA polymerase β can remove this obstruction, it can also displace the strand, allowing a CAG hairpin to form, presumably as a precursor to expansion (33). In the absence of OGG1, the troublesome ends would not be generated and the repeats would, therefore, be more stable. Resolving these differences may prove enlightening for our understanding of CAG repeat instability.
In summary, by using a selectable CAG contraction system in human cells to screen the Prestwick chemical library, we have made two novel findings. First, we have identified the TOP1-TDP1-SSBR pathway as a modulator of transcription-induced CAG repeat instability in human cells. Although supercoiling has been shown to destabilize triplet repeats in bacteria, this report is the first that identifies the repair of transcription-induced DNA damage—presumably, the TOP1 irreversible cleavage complexes—as a critical determinant of repeat stability. Second, we have shown that TC-NER drives CAG repeat instability not only when transcription through the repeat is induced but also when the TOP1-TDP1-SSBR pathway is compromised. These studies substantially enrich our understanding of transcription-induced CAG repeat instability. Also, as a result of this work we have discovered a set of bioactive small molecules that modulate CAG repeat instability. These compounds may serve as tools for dissecting the molecular mechanism of CAG repeat instability and as aids in the development of preventive and therapeutic approaches for repeat-associated diseases.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zaowen Chen and Jason Shohet for help with real-time RT-PCR. We thank Michele Washington for characterization of the siRNA against PARP1 and the Western blot to determine PARP1 knockdown in cells. We thank Motonari Uesugi for advice and help with the Prestwick chemical library. We thank members of the Wilson lab for helpful discussions.
This study was supported by NIH grant 1F31HG004918 to L.H., Natural Sciences and Engineering Research Council of Canada postgraduate scholarship D to V.D., and NIH grant GM38219 to J.H.W.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 31 May 2011.
REFERENCES
- 1. Boege F., et al. 1996. Selected novel flavones inhibit the DNA binding or the DNA religation step of eukaryotic topoisomerase I. J. Biol. Chem. 271:2262–2270 [DOI] [PubMed] [Google Scholar]
- 2. Caldecott K. W. 2008. Single-strand break repair and genetic disease. Nat. Rev. Genet. 9:619–631 [DOI] [PubMed] [Google Scholar]
- 3. Capranico G., et al. 2007. The effects of camptothecin on RNA polymerase II transcription: roles of DNA topoisomerase I. Biochimie 89:482–489 [DOI] [PubMed] [Google Scholar]
- 4. Cleary J. D., Pearson C. E. 2003. The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenet. Genome Res. 100:25–55 [DOI] [PubMed] [Google Scholar]
- 5. Dexheimer T. S., Antony S., Marchand C., Pommier Y. 2008. Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anticancer Agents Med. Chem. 8:381–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dion V., Lin Y., Hubert L., Jr., Waterland R. A., Wilson J. H. 2008. Dnmt1 deficiency promotes CAG repeat expansion in the mouse germline. Hum. Mol. Genet. 17:1306–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dion V., Wilson J. H. 2009. Instability and chromatin structure of expanded trinucleotide repeats. Trends Genet. 25:288–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dragileva E., et al. 2009. Intergenerational and striatal CAG repeat instability in Huntington's disease knock-in mice involve different DNA repair genes. Neurobiol. Dis. 33:37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. French S. L., et al. 2011. Distinguishing the roles of topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol. Cell. Biol. 31:482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gacy A. M., Goellner G., Juranic N., Macura S., McMurray C. T. 1995. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 81:533–540 [DOI] [PubMed] [Google Scholar]
- 11. Gatchel J. R., Zoghbi H. Y. 2005. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 6:743–755 [DOI] [PubMed] [Google Scholar]
- 12. Gomes-Pereira M., Fortune M. T., Ingram L., McAbney J. P., Monckton D. G. 2004. Pms2 is a genetic enhancer of trinucleotide CAG·CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum. Mol. Genet. 13:1815–1825 [DOI] [PubMed] [Google Scholar]
- 13. Gomes-Pereira M., Monckton D. G. 2006. Chemical modifiers of unstable expanded simple sequence repeats: what goes up, could come down. Mutat. Res. 598:15–34 [DOI] [PubMed] [Google Scholar]
- 14. Gomes-Pereira M., Monckton D. G. 2004. Chemically induced increases and decreases in the rate of expansion of a CAG*CTG triplet repeat. Nucleic Acids Res. 32:2865–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gorbunova V., et al. 2003. Selectable system for monitoring the instability of CTG/CAG triplet repeats in mammalian cells. Mol. Cell. Biol. 23:4485–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gorbunova V., Seluanov A., Mittelman D., Wilson J. H. 2004. Genome-wide demethylation destabilizes CTG·CAG trinucleotide repeats in mammalian cells. Hum. Mol. Genet. 13:2979–2989 [DOI] [PubMed] [Google Scholar]
- 17. Gossen M., Bujard H. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U. S. A. 89:5547–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hashem V. I., et al. 2004. Chemotherapeutic deletion of CTG repeats in lymphoblast cells from DM1 patients. Nucleic Acids Res. 32:6334–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsu Y. L., Kuo P. L., Lin C. C. 2004. Acacetin inhibits the proliferation of Hep G2 by blocking cell cycle progression and inducing apoptosis. Biochem. Pharmacol. 67:823–829 [DOI] [PubMed] [Google Scholar]
- 20. Kovtun I. V., Johnson K. O., McMurray C. T. 2011. Cockayne syndrome B protein antagonizes OGG1 in modulating CAG repeat length in vivo. Aging 3:509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kovtun I. V., et al. 2007. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 447:447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kovtun I. V., McMurray C. T. 2001. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 27:407–411 [DOI] [PubMed] [Google Scholar]
- 23. La Spada A. R., Taylor J. P. 2010. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 11:247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lenzmeier B. A., Freudenreich C. H. 2003. Trinucleotide repeat instability: a hairpin curve at the crossroads of replication, recombination, and repair. Cytogenet. Genome Res. 100:7–24 [DOI] [PubMed] [Google Scholar]
- 25. Leppard J. B., Champoux J. J. 2005. Human DNA topoisomerase I: relaxation, roles, and damage control. Chromosoma 114:75–85 [DOI] [PubMed] [Google Scholar]
- 26. Liao Z., Thibaut L., Jobson A., Pommier Y. 2006. Inhibition of human tyrosyl-DNA phosphodiesterase by aminoglycoside antibiotics and ribosome inhibitors. Mol. Pharmacol. 70:366–372 [DOI] [PubMed] [Google Scholar]
- 27. Lin Y., Dent S. Y., Wilson J. H., Wells R. D., Napierala M. 2010. R loops stimulate genetic instability of CTG·CAG repeats. Proc. Natl. Acad. Sci. U. S. A. 107:692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin Y., Dion V., Wilson J. H. 2006. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 13:179–180 [DOI] [PubMed] [Google Scholar]
- 29. Lin Y., Hubert L., Jr., Wilson J. H. 2009. Transcription destabilizes triplet repeats. Mol. Carcinog. 48:350–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin Y., Leng M., Wan M., Wilson J. H. 2010. Convergent transcription through a long CAG tract destabilizes repeats and induces apoptosis. Mol. Cell. Biol. 30:4435–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin Y., Wilson J. H. 2009. Diverse effects of individual mismatch repair components on transcription-induced CAG repeat instability in human cells. DNA Repair 8:878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin Y., Wilson J. H. 2007. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 27:6209–6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Y., et al. 2009. Coordination between polymerase beta and FEN1 can modulate CAG repeat expansion. J. Biol. Chem. 284:28352–28366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lopez-Castel A., Cleary J. D., Pearson C. E. 2010. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell. Biol. 11:165–170 [DOI] [PubMed] [Google Scholar]
- 35. Mirkin S. M. 2007. Expandable DNA repeats and human disease. Nature 447:932–940 [DOI] [PubMed] [Google Scholar]
- 36. Mittelman D., et al. 2009. Zinc-finger directed double-strand breaks within CAG repeat tracts promote repeat instability in human cells. Proc. Natl. Acad. Sci. U. S. A. 106:9607–9612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mittelman D., Sykoudis K., Hersh M., Lin Y., Wilson J. H. 2010. Hsp90 modulates CAG repeat instability in human cells. Cell Stress Chaperones 15:753–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Napierala M., Bacolla A., Wells R. D. 2005. Increased negative superhelical density in vivo enhances the genetic instability of triplet repeat sequences. J. Biol. Chem. 280:37366–37376 [DOI] [PubMed] [Google Scholar]
- 39. Orr H. T., Zoghbi H. Y. 2007. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30:575–621 [DOI] [PubMed] [Google Scholar]
- 40. Pearson C. E., Edamura K. N., Cleary J. D. 2005. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 6:729–742 [DOI] [PubMed] [Google Scholar]
- 41. Pearson C. E., Wang Y. H., Griffith J. D., Sinden R. R. 1998. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n. (CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 26:816–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pineiro E., et al. 2003. Mutagenic stress modulates the dynamics of CTG repeat instability associated with myotonic dystrophy type 1. Nucleic Acids Res. 31:6733–6740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pommier Y. 2006. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6:789–802 [DOI] [PubMed] [Google Scholar]
- 45. Poroikov V. V., Filimonov D. A. 2002. How to acquire new biological activities in old compounds by computer prediction. J. Comput. Aided Mol. Des. 16:819–824 [DOI] [PubMed] [Google Scholar]
- 46. Robertson A. B., Klungland A., Rognes T., Leiros I. 2009. DNA repair in mammalian cells: base excision repair: the long and short of it. Cell. Mol. Life Sci. 66:981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Savouret C., et al. 2003. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 22:2264–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Savouret C., et al. 2004. MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol. Cell. Biol. 24:629–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shelbourne P. F., et al. 2007. Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum. Mol. Genet. 16:1133–1142 [DOI] [PubMed] [Google Scholar]
- 50. Spiro C., McMurray C. T. 2003. Nuclease-deficient FEN-1 blocks Rad51/BRCA1-mediated repair and causes trinucleotide repeat instability. Mol. Cell. Biol. 23:6063–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van den Broek W. J., Nelen M. R., W. van der Heijden G., Wansink D. G., Wieringa B. 2006. Fen1 does not control somatic hypermutability of the (CTG)n*(CAG)n repeat in a knock-in mouse model for DM1. FEBS Lett. 580:5208–5214 [DOI] [PubMed] [Google Scholar]
- 52. van den Broek W. J., et al. 2002. Somatic expansion behavior of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 11:191–198 [DOI] [PubMed] [Google Scholar]
- 53. van Loon B., Markkanen E., Hubscher U. 2010. Oxygen as a friend and enemy: how to combat the mutational potential of 8-oxo-guanine. DNA Repair 9:604–616 [DOI] [PubMed] [Google Scholar]
- 54. Wang J. C. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 3:430–440 [DOI] [PubMed] [Google Scholar]
- 55. Watase K., et al. 2002. A long CAG repeat in the mouse Sca1 locus replicates SCA1 features and reveals the impact of protein solubility on selective neurodegeneration. Neuron 34:905–919 [DOI] [PubMed] [Google Scholar]
- 56. Wells R. D. 2007. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 32:271–278 [DOI] [PubMed] [Google Scholar]
- 57. Yang Z., Lau R., Marcadier J. L., Chitayat D., Pearson C. E. 2003. Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type 1 disease locus in human cells. Am. J. Hum. Genet. 73:1092–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.