Abstract
The accumulation of damaged or misfolded proteins, if unresolved, can lead to a detrimental consequence within cells termed proteotoxicity. Since cancerous cells often display elevated protein synthesis and by-product disposal, inhibition of the protein degradation pathways is an emerging approach for cancer therapy. However, the molecular mechanism underlying proteotoxicity remains largely unclear. We show here that inhibition of proteasomal degradation results in an increased oligomerization and activation of caspase-8 on the cytosolic side of intracellular membranes. This enhanced caspase-8 oligomerization and activation are promoted through its interaction with the ubiquitin-binding protein SQSTM1/p62 and the microtubule-associated protein light chain 3 (LC3), which are enriched at intracellular membranes in response to proteotoxic stress. Silencing LC3 by shRNA, or the LC3 mutants defective in membrane localization or p62 interaction fail to induce caspase-8 activation and apoptosis. Our results unveiled a previously unknown mechanism through which disruption of protein homeostasis induces caspase-8 oligomerization, activation, and apoptosis.
INTRODUCTION
Cellular proteins are degraded by two mechanistically connected processes: the ubiquitin-proteasomal pathway and the autophagolysosomal pathway (11, 16, 32, 35). Suppression of these protein degradation pathways leads to the accumulation of damaged or unwanted proteins, which, if unresolved, is detrimental to the cell, leading to a consequence known as proteotoxicity (24). Proteotoxicity plays an important role in numerous physiological and pathological conditions such as degenerative disorders and diabetes. Cancer cells, owing to their aberrant transcription/translation activity and protein disposal, may become more vulnerable to proteotoxicity. Indeed, inhibition of the proteasomal and autophagolysosomal degradation pathways is clinically used or under investigation for treating cancer (1, 4, 12, 18, 38, 54, 55).
The molecular machinery underlying proteotoxicity remains largely unclear. Several mechanisms have been proposed for the cytotoxic activity of proteasome inhibition, including stabilization of p53 (23) and the BH3-only proteins (37), cleavage of Mcl-1 (42), downregulation of XIAP and survivin (51), inhibition of NF-κB activity (3), and downregulation of the PI3K/Akt survival pathway (13). For the autophagolysosomal degradation pathway, it has been shown that lysosomal inhibitors such as chloroquine can lead to cell death via p53 (36). In addition, since both the proteasome and autophagolysosome pathways function as a safeguard system to degrade misfolded or unwanted proteins, a failure within either degradation pathway leads to endoplasmic reticulum (ER) stress (16), which can induce apoptosis through upregulation of the BH3-only proteins Puma and Bim (44, 46). However, the effectiveness of proteasome and lysosome inhibitors on a broad spectrum of malignancies, including cancers which are often defective in apoptosis, suggests a more fundamental biochemical mechanism for proteotoxicity (54, 55). In vitro cell culture studies have shown that ER stress and proteasome inhibitors can effectively induce cell death in p53−/− and bax−/− bak−/− cells (37, 52), indicating that the main apoptotic mediators p53, Bax, and Bak are dispensable for proteotoxicity. In the present study, we first treated a number of breast cancer cell lines with the proteasome inhibitor MG132 and observed that caspase-8 activation plays an important role in MG132-induced apoptosis. We went on to study the molecular mechanism and significance of caspase-8 activation in response to proteasome inhibition.
MATERIALS AND METHODS
Cell lines, culture, and transfection.
HEK293T, bax−/− bak−/− baby mouse kidney (BMK), MDA-MB-231, and MCF-7 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (FBS), 100 U of penicillin/ml, and 100 μg of streptomycin/ml (Invitrogen). MDA-MB-468 cells were cultured in RPMI 1640 supplemented with 10% FBS, 100 U of penicillin/ml, and 100 μg of streptomycin/ml.
Reagents and antibodies.
The following reagents were used: MG132 (Sigma catalog no. C2211), z-VAD (Calbiochem catalog no. 219007), Hoechst 33342 (Molecular Probes catalog no. H1399, used at 0.5 μg/ml), DAPI (4′,6′-diamidino-2-phenylindole; Sigma catalog no. D9542, used at 1 μg/ml), chloroquine (CQ; Sigma catalog no. C6628, used at 10 μM), tumor necrosis factor α (TNF-α; Invitrogen catalog no. PMC3014), cycloheximide (Calbiochem catalog no. 239763), Ni-NTA agarose (Qiagen catalog no. 133203974), protein A (Roche catalog no. 11719408001), protein G (Roche catalog no. 11719416001), trypsin (Sigma catalog no. T1426, used at 100 μg/ml), trypsin inhibitor (Sigma catalog no. T2327, used at 60 μg/ml), and a gold enhancement kit (Nanoprobes catalog no. 2113).
The following antibodies were used: PARP (Cell Signaling catalog no. 9542; 1:1,000 for Western blotting [WB]), caspase-8 (R&D Systems catalog no. AF1650; 1:1,000 for WB; Alexis Biochemicals catalog no. Alx-804-447; 1 μg per reaction for immunoprecipitation [IP], 1:200 for immunofluorescence [IF], and 1:100 for electron microscopy [EM]), caspase-3 (Cell Signaling catalog no. 9661; 1:1,000 for WB), tubulin (Sigma catalog no. T4026; 1:10,000 for WB), Flag (Sigma catalog no. F7425 [1:1,000 for WB] and catalog no. F3165 [1:1,000 for WB and 1:500 for IF]), hemagglutinin (HA; Rockland catalog no. 21345; 1:5,000 for WB, and 0.5 μg per reaction for IP), caspase-2 (Millipore catalog no. MAB3507; 1:500 for WB), caspase-9 (for mouse; Cell Signaling catalog no. 9504; 1:1,000 for WB), SQSTM1/p62 (Novus Biologicals catalog no. H00008878-MOL; 1:5,000 for WB), LC3 (MBL catalog no. PM036; 1:1,000 for WB), green fluorescent protein (GFP; Santa Cruz Biotechnology catalog no. sc-9996; 1:1,000 for WB, 1 μg per reaction for IP), rabbit GFP antibody (a gift from Zhenyu Yue, Mount Sinai School of Medicine; 1:2,000 for WB, 0.3 μg per reaction for IP), anti-Flag M2 affinity gel (Sigma catalog no. A2220), LAMP-1 (a gift from J. Thomas August, Department of Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine; clone 1D4B; 1:200 for WB), and complex IV subunit Vα monoclonal antibody (Invitrogen catalog no. 459120; 1:500).
Plasmids.
Human SQSTM1/p62 and human caspase-8 expression constructs were generated by using cDNA amplified by reverse transcriptase PCR and inserted into p3xFlag-CMV-10 (Sigma). The HA-tagged caspase-8 was generated by replacing the Flag tag in p3xFLAG-CMV-10 with an HA tag. The retroviral LPC-Flag-p62 and LPC-HA-caspase-8 constructs were generated by cloning Flag-p62 and HA-caspase-8 from the pCMV-10 constructs into the LPC vector. The LPC-GFP-LC3 construct was described previously (52). LC3 F52A and G120A mutants were generated by standard site-directed mutagenesis using the following oligonucleotides: for F52A, 5′-GCTGCTGTACCTGATCACGTGAATATGAGCG-3′ and 5′-CTTGGTCTTGTCCAGGACGGG-3′; for G120A, 5′-CGACAGCACTGGCTGTTACATACATG-3′ and 5′-CGAACGTCTCCTGGGAGGCATAG-3′. The primers used for caspase-8 mutant construct C360S were 5′-CAAAGTGTTTTTTATTCAGGCTaGTCAGGGGGATAACTACCAG-3′ and 5′-CTGGTAGTTATCCCCCTGACtAGCCTGAATAAAAAACACTTTG-3′ (the lowercase letter indicates the mutated site). Bimolecular fluorescence complementation (BiFC) constructs were made by fusing VN (Venus N1-173) and VC (Venus C155-239) to the C termini of caspase-8 mutants. A linker GGSGSGSS was inserted between the Venus tag and caspase-8. Venus and linker sequences were kindly provided by Michael A. Frohman. shRNAs were obtained from Sigma: for LC3, NM_025735, CATGAGCGAGTTGGTCAAGAT; for p62, NM_011018, CCGCATCTACATTAAAGAGAA; for mouse caspase-8, NM_009812, CCTCCATCTATGACCTGACA; and for human caspase-8, NM_001228, CCTGGCCGATGGTACTATTTA. The primers used for p62 mutant constructs were as follows: for D69A, 5′-CTTCCAGGCGCACTACCGCGcTGAGGACGGGGACTTGGTTGCC-3′ and 5′-GGCAACCAAGTCCCCGTCCTCAgCGCGGTAGTGCGCCTGGAAG-3′; for I431A, 5′-CATCGGAGCGGCTCTGGACACCgcCCAGTATTCAAAGCATCC-3′ and 5′-GGATGCTTTGAATACTGGgcGGTGTCCAGAGCCGCTCCGATG-3′; and for Δ321-342, 5′-GATCGCCTTGGAGTCCGAGGGGTCAAAAGAAGTGGACCCGTC-3′ and 5′-GACGGGTCCACTTCTTTTGACCCCTCGGACTCCAAGGCGATC-3′ (lowercase letters indicated mutated sites).
Bimolecular fluorescence complementation (BiFC) assay.
The pairs of VN and VC caspase-8 mutants were transfected into cells with Lipofectamine 2000. At 24 h posttransfection, the cells were plated into wells for treatment. The cells were harvested 18 h after treatment, and green the fluorescence of the cells was determined by flow cytometry.
Lentiviral and retroviral infection.
For lentiviral infection, 5 × 105 HEK293T cells were plated into a six-well plate. After 24 h, the cells were transfected with the packaging construct pCMV Δ8.91 (2 μg), a vesicular stomatitis virus glycoprotein (VSV-G) expression construct (1 μg), and the lentiviral expression construct (3 μg) by using Lipofectamine 2000. At 24 h posttransfection, the virus-containing cell culture supernatant was collected, filtered through 0.45-μm-pore-size nylon filter and subsequently used to infect target cells. The infection procedure was repeated every 12 h for a total of three rounds of infection. One day after the last infection, the cells were selected with 4 μg/ml of puromycin (InvivoGen). Retroviral infection was performed as described previously (17). Briefly, retrovirus was produced in HEK293T cells after transfection with LPC-retroviral constructs (2.5 μg) and helper viral construct (1.5 μg). The same infection procedure was used as for lentiviral infection, except that the viral supernatant was supplemented with 10 μg of Polybrene (Sigma)/ml.
His-Ub assay for mammalian cells.
A total of 3 × 106 cells were plated into a 10-cm culture dish. After overnight recovery, the cells were transfected with 2 μg of pMT107 plasmid encoding polyhistidine-tagged Ub (kindly provided by Erich R. Mackow, Stony Brook University). One day after transfection, the cells were collected, and the same number of cells was plated into multiple wells for treatment. After treatment, the cells were collected, washed twice with phosphate-buffered saline (PBS), and resuspended in 1 ml of buffer A (6 M guanidine-HCl, 0.1 M Na2HPO4-NaH2PO4, 10 mM imidazole; pH 8.0). After a 10-s sonication, each sample was added with 50 μl of Ni-NTA-agarose (Qiagen) equilibrated with buffer A and then incubated 3 h at room temperature with agitation. Agarose was precipitated by centrifugation and washed twice with buffer B (10 mM Tris-Cl [pH 8.0], 8 M urea, 0.1 M NaH2PO4) and three times with 1:4-diluted buffer B. The precipitates were resuspended in 100 μl of 2× Laemmli buffer (4% sodium dodecyl sulfate [SDS], 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromphenol blue, 0.125 M Tris-HCl [pH 6.8]) with 200 mM imidazole, heated at 95°C for 10 min, and subjected to Western blotting.
Coimmunoprecipitation.
Cells were treated with trypsin, spun down, washed twice with PBS, and lysed for 30 min in IP lysis buffer (30 mM Tris [pH 7.5], 150 mM NaCl, 10% glycerol, 1% Triton X-100, 10 mM NaF, 100 μM orthovanadate, 200 μM phenylmethylsulfonyl fluoride [PMSF]) supplemented with protease inhibitor cocktail (Biosciences). The cell lysate was centrifuged at 13,000 × g at 4°C. The supernatant was precleared with protein A/G agarose (Roche) and incubated with primary antibodies overnight at 4°C with agitation. The lysates were then incubated with protein A/G-agarose for 2 h, washed twice with IP lysis buffer with 500 mM NaCl and twice with IP lysis buffer, and heated in 2× SDS sample buffer at 95°C for 5 min.
Size exclusion chromatography.
Cells were lysed in IP lysis buffer (30 mM Tris [pH 7.5], 150 mM NaCl, 10% glycerol, 1% Triton X-100, 10 mM NaF, 100 μM orthovanadate, 200 μM PMSF) supplemented with protease inhibitor cocktail (Biosciences). Cell lysates were cleared by centrifugation at 4°C at 13,000 × g) and filtered through a 0.45-μm-pore-size filter. The supernatant (3 mg of total protein for each sample) was loaded onto a Superdex 200 10/300 GL column equilibrated with washing buffer (30 mM Tris [pH 7.5], 150 mM NaCl), subsequently eluted with washing buffer at flow speed of 0.5 ml/min, and finally collected in 0.5-ml volumes.
Acetone precipitation.
Four times the sample volume of cold (−20°C) acetone was added to each sample, mixed, and incubated for 120 min at −20°C. The proteins were precipitated for 15 min at 4°C at 13,000 × g. The pellets were dried for 10 min at room temperature and boiled in 2× SDS sample buffer at 95°C for 5 min.
Caspase activity assay.
Caspase activity was determined by using a Caspase-Glo 8 assay kit (Promega catalog no. G8200), which uses the respective luminogenic caspase substrates, according to the manufacturer's instructions. The luciferase activities were read by a luminescence reader (Spectramax M5; Molecular Devices).
Immunofluorescence.
Cells were fixed in 4% paraformaldehyde in PBS for 25 min at room temperature. After two washes with PBS, the cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min. The cells were blocked in 5% goat serum in PBS for 1 h at room temperature. After a brief wash with PBS with 0.1% Tween 20 (PBST), the cells were incubated with primary antibodies in PBST with 5% goat serum overnight at 4°C. The cells were washed four times with PBST, followed by incubation with fluorophore-conjugated secondary antibodies in PBST with 5% goat serum for 1 h at room temperature. The cells were then washed three times with PBST and incubated with 1 μg of DAPI/ml in PBS for 5 min. After two washes with PBS, the cells were mounted with Immu-Mount (Thermo Scientific). The slides were observed and imaged using a Zeiss LSM 510 META NLO two-photon laser scanning confocal microscope or a Zeiss inverted Axiovert 200M deconvolution microscope. Active caspase-8 was detected with a caspase-8 detection kit (Calbiochem catalog no. QIA114) according to the recommended protocol.
Trypsinization of HM and LM proteins.
A total of 5 × 106 cells were resuspended in hypotonic buffer A (250 mM sucrose, 20 mM HEPES [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA) on ice for 30 min. The cells were disrupted by passage through 26-gauge needles 30 times and then through 30-gauge needles 30 times. The cell lysates were centrifuged at 750 × g for 10 min at 4°C. The supernatants were collected and centrifuged at 750 × g for 10 min at 4°C again. The supernatants were divided into two parts. To one part trypsin was added to a final concentration of 100 μg/ml, followed by incubation for 30 min on ice. Trypsin inhibitor (60 μg/ml) was added to stop the proteolysis reaction. All of the supernatants with or without trypsin treatment were centrifuged at 10,000 × g for 30 min at 4°C. The pellets were recovered as a heavy-membrane (HM) fraction. The supernatants were recovered and centrifuged at 100,000 × g for 1 h at 4°C. These pellets were recovered as a light-membrane (LM) fraction. All of the fractions were resuspended in 2× SDS sample buffer before being subjected to Western blotting.
Measurement of cell death.
For cell viability, cells (including the detached ones) were collected and resuspended in culture medium with propidium iodide (PI) at 1 μg/ml. Cell viability was determined by PI exclusion using a FACSCalibur (BD Biosciences). Alternatively, trypan blue staining was used to determine cell viability. Portions (0.5 ml) of the cells (∼105 cells per ml) were mixed with 0.1 ml of 0.4% trypan blue and incubated for 5 min at room temperature. The cells were counted under a phase-contrast light microscope.
Statistics.
Data from cell death assays are presented as means ± the standard errors of the mean (SEM). A Student t test was used to compare the differences between two groups. Significance was judged when P < 0.05.
Image processing and densitometry measurement.
Images taken from deconvolution and confocal microscopes were viewed and processed by AxioVision LE and Zeiss LSM image browser, respectively. Images were processed in Adobe Photoshop to enhance the brightness and contrast. Densitometry of the immunoblot bands was determined by using ImageJ software unless indicated otherwise.
RESULTS
Inhibition of protein degradation leads to caspase-8 activation and apoptosis.
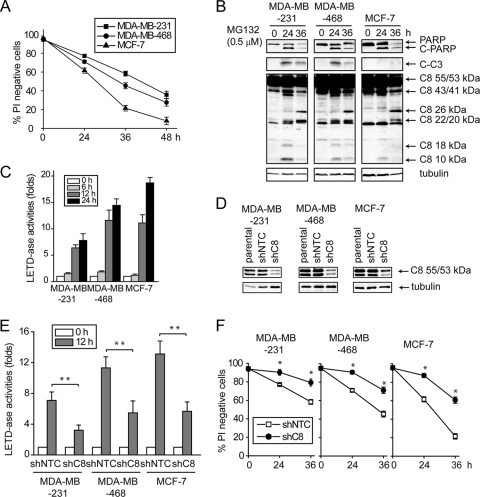
To study the mechanism underlying proteotoxicity, we treated human breast cancer cell lines MDA-MB231, MDA-MB-468, and MCF7 with the proteasome inhibitor MG132. MG132 induced cell death and apoptotic cleavage of poly(ADP-ribose) polymerase (PARP), caspase-3, and caspase-8 in MDA-MB-231 and MDA-MB-468 cells (Fig. 1A and B). Similar apoptotic features were observed in MCF-7 cell line, although it is caspase-3 deficient (Fig. 1B). The cleavage of caspase-8 accompanies its activation determined using a Caspase-Glo 8 luminogenic assay kit (Fig. 1C). To determine whether the activation of caspase-8 contributes to cell death induced by MG132, caspase-8 was knocked down using shRNA (Fig. 1D). Partial knockdown of caspase-8 led to a significant lower activity (Fig. 1E) and conferred protection, albeit incomplete, against MG132 (Fig. 1F). These results raise the possibility that activation of caspase-8 may be involved in proteotoxicity caused by proteasome inhibition.
Fig. 1.
MG132 induces caspase-8-dependent apoptosis in human breast cancer cells. Human breast cancer cells—MDA-MB-231, MDA-MB-468, and MCF-7—were treated with MG132 (0.5 μM). At the indicated time points, cells were collected. (A) Cell death was measured by PI exclusion. The averages of four independent assays ± the SEM are shown. (B) Cell lysates were examined by immunoblotting with the indicated antibodies. (C) The activities of caspase-8 were assayed by using the Caspase-Glo 8 luminogenic assay kit. The fold increase of caspase-8 activity relative to that of the respective untreated cells is shown. The average of four independent assays ± the SEM is shown. (D to F) Cells were infected with lentiviral nontargeting control (shNTC) and caspase-8 (shC8) shRNAs. Cells with caspase-8 knockdown were treated with MG132 (0.5 μM). (D) Knockdown of the caspases was verified by immunoblotting. (E) Caspase-8 activity was measured using the Caspase-Glo 8 luminogenic assay kit. The fold increase in the caspase-8 activity relative to that of the respective untreated cells is shown. The averages of four independent assays ± the SEM are shown. **, P < 0.01. (F) Cell death was measured by PI exclusion. The averages of four independent assays ± the SEM are shown. *, P < 0.05.
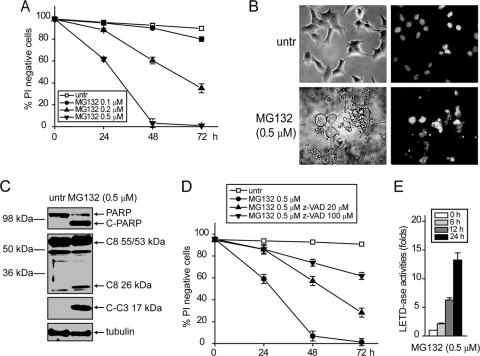
These results suggest that the activation of the initiator caspase-8 plays a role in MG132-induced apoptosis. To further study the molecular mechanisms underlying MG132-induced caspase-8 activation, we used bax−/− bak−/− BMK cells in which the mitochondrial apoptotic pathway is inhibited (15). It has been previously shown that proteasome inhibitors can induce cell death in bax−/− bak−/− cells (37); hence, we reasoned this genetically modified system is ideal for studying the molecular mechanism of activation of initiator caspases without interference from mitochondrial apoptosis and has strong relevance to cancer cells with compromised mitochondrial apoptosis. Indeed, MG132 induced cell death in bax−/− bak−/− BMK cells in a dose- and time-dependent manner (Fig. 2A). This MG132-induced cell death in bax−/− bak−/− BMK cells appeared to be caspase dependent, judging by nuclear fragmentation and chromatin condensation (Fig. 2B) and apoptotic cleavage of caspase-3 and PARP (Fig. 2C) and the fact that z-VAD protected cells from MG132 treatment (Fig. 2D). Importantly, proteolytic cleavage of caspase-8 was also observed (Fig. 2C). The Caspase-Glo 8 luminogenic assay showed that MG132 treatment induced activation of caspase-8 (Fig. 2E). Caspase-8 partial knockdown conferred significant protection against MG132 (see Fig. 4E to G). These results indicate that proteasome inhibition can induce apoptosis, which may involve caspases, in cells with defective mitochondrial apoptosis.
Fig. 2.
MG132 induces caspase-dependent apoptosis in cells deficient in mitochondrial apoptosis. bax−/− bak−/− BMK cells were treated with indicated concentration of MG132. (A) At the indicated time points cells were collected and cell death measured by PI exclusion. The averages of three independent assays ± the SEM are shown. (B) Cells untreated or treated with MG132 for 24 h were stained with Hoechst 33342 and observed with a fluorescence microscope. Representative images are shown. Note the nuclear condensation and fragmentation in MG132-treated cells. (C) Cells untreated or treated with MG132 for 24 h were lysed and probed for indicated proteins by immunoblotting. (D) The pan-caspase inhibitor z-VAD was added to cell culture. Note that z-VAD protected cells from MG132 treatment. The averages of three independent assays ± the SEM are shown. (E) Upon treatment of MG132 at indicated time points, caspase-8 activity was assayed using the Caspase-Glo 8 luminogenic assay kit. The averages of three independent assays plus the SEM are shown.
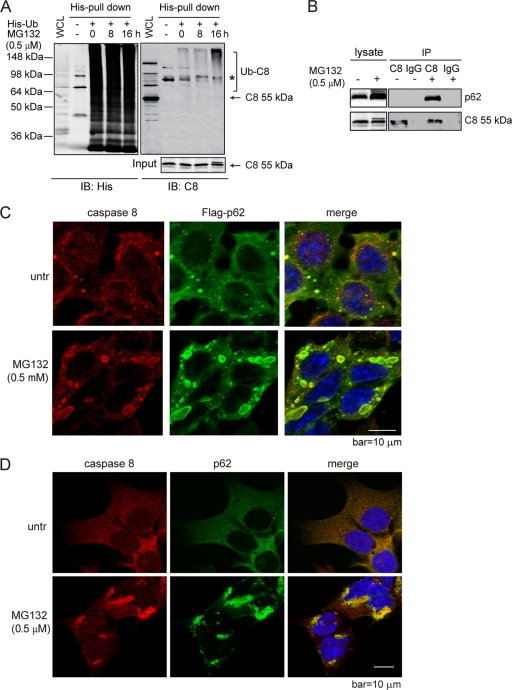
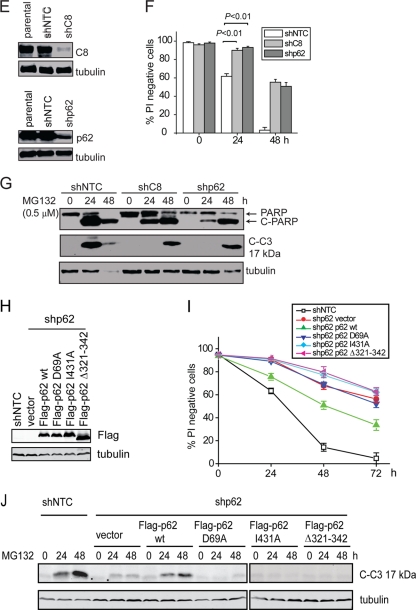
Fig. 4.
Proteasome inhibition promotes apoptosis by enhancing the interaction of caspase-8 and SQSTM1/p62. (A) bax−/− bak−/− BMK cells were transiently transfected with plasmid encoding His-tagged ubiquitin (His-Ub). At 24 h posttransfection, cells were treated with MG132 for the indicated periods of time and subjected to His pulldown. The pulldown samples were probed with an anti-His and an anti-caspase-8 antibody. The asterisk indicates a nonspecific band. The samples subjected to His pulldown were probed with an anti-caspase-8 antibody. (B) bax−/− bak−/− BMK cells were left untreated or were treated with MG132. Cell lysates were subjected to IP with rat IgG or an anti-caspase-8 antibody. IP samples and cell lysates were probed for caspase-8 or p62. (C) bax−/− bak−/− BMK cells stably expressing Flag-p62 were left untreated or were treated with MG132 for 18 h. Immunofluorescence analysis was performed using anti-Flag (green) and anti-caspase-8 (red) antibodies. DAPI was used to visualize the nucleus. (D) bax−/− bak−/− BMK cells were left untreated or were treated with MG132 for 18 h. Immunofluorescence was performed using anti-p62 (green) and anti-caspase-8 (red) antibodies. DAPI was used to visualize the nucleus. (E) bax−/− bak−/− BMK cells were stably infected with lentivirus encoding nontarget control shRNA (shNTC), caspase-8 shRNA (shC8), or p62 shRNA (shp62). The expression of p62 or caspase-8 was examined by immunoblotting. (F and G) Cells with different shRNAs were treated with MG132 (0.5 μM). Cell death was determined by PI exclusion (G) or immunoblotting (G). Note that the lower amount of cleaved PARP and C3 in shNTC at 48 h is due to excessive cell death. The averages of four independent assays plus the SEM is shown for the cell death curve. (H to J) Characterization of the p62 dependence of MG132-induced apoptosis. (H) Wild-type p62 and mutants deficient in dimerization (D69A), ubiquitin binding (I431A), and LC3-interaction (Δ321-342) were expressed in bax−/− bak−/− BMK cells stably expressing shp62. (I) Cells were treated with MG132 (0.5 μM) for the indicated periods of time, and cell death was determined by PI exclusion. The averages of four experiments ± the SEM are shown. (J) Cell lysates were probed for cleaved caspase-3 (C-C3) and tubulin.
Proteasome inhibition leads to increased caspase-8 oligomerization and activation.
The results presented above suggest that caspase-8, an initiator caspase, may be involved in MG132-induced apoptosis (Fig. 1 and 2). Since we observed caspase-8 activation in both apoptosis-competent human cancer lines (Fig. 1) and in apoptosis-deficient bax−/− bak−/− BMK cells (Fig. 2), we continued to explore the mechanism underlying caspase-8 activation using the bax−/− bak−/− cells to minimize the interference from mitochondrial apoptosis in the following studies.
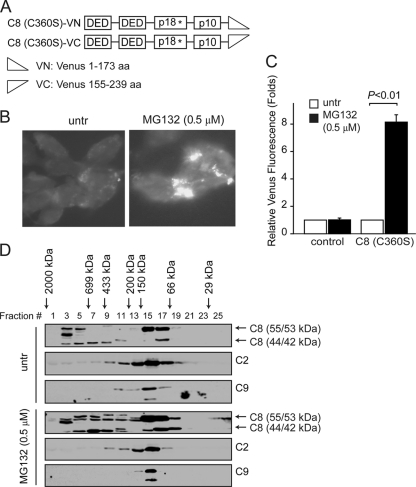
In response to the activation of death receptors upon death ligand engagement, caspase-8 is activated and self-cleaved by increased oligomerization through its recruitment to the death-inducing signaling complex (DISC) on the plasma membrane (5, 9, 31, 43, 58). Since caspase-8 cleavage and activation were induced by MG132 treatment (Fig. 1 and 2), we explored whether MG132 can induce oligomerization of caspase-8 using the bimolecular fluorescence complementation (BiFC) assay (25). To avoid cell death induced by overexpression of caspase-8, we used the catalytically inactive C360S mutant of the full-length caspase-8 (10). The C8 (C360S) mutant was fused to the split Venus proteins, Venus N1-173 and Venus C155-239 (Fig. 3A), and transfected into bax−/− bak−/− BMK cells. MG132 treatment significantly induced BiFC of C8 (C360S) (Fig. 3B and C). To determine whether MG132 induces oligomerization of caspase-8 at endogenous levels, we performed size exclusion chromatography in bax−/− bak−/− BMK cells. Upon MG132 treatment, increased amounts of both the 55/53-kDa and 44/42-kDa species of caspase-8 were detected in high-molecular-mass fractions (Fig. 3D). Oligomerization-induced activation has been shown as a general mechanism for the activation of caspases. In contrast to caspase-8, caspase-2 and caspase-9, two other initiator caspases that can be induced to oligomerize (50, 59), did not display apparent oligomerization upon MG132 treatment (Fig. 3D). Together, these results indicate that inhibition of proteasomal degradation can specifically induce oligomerization and aggregation of caspase-8.
Fig. 3.
MG132 induces caspase-8 oligomerization and aggregation. (A) Schematic representation of the full-length and truncated caspase-8 BiFC constructs. (B) bax−/− bak−/− BMK cells were transiently transfected with plasmids encoding C8 (C360S)-VN and C8 (C360S)-VC for 18 h, followed by treatment with MG132 for an additional 18 h. Representative fluorescence images were taken. (C) Cells untransfected, or transfected with the Venus pairs of C8 (C360S) were left untreated or were treated with MG132 for 18 h. The percentages of Venus green cells were analyzed by flow cytometry. The number of Venus-positive cells in MG132-treated samples was normalized against that of the untreated samples. The average of four experiments plus the SEM are shown. (D) Lysates from untreated or MG132-treated bax−/− bak−/− BMK cells were resolved by size exclusion chromatography on a Superdex 200 column. Eluate was collected in 0.5-ml fractions, concentrated by acetone precipitation, and probed for indicated proteins. The molecular masses of the protein standards used to indicate the approximate sizes of the fractions are indicated in kilodaltons.
Proteasome inhibition results in increased caspase-8 ubiquitination and interaction with SQSTM1/p62.
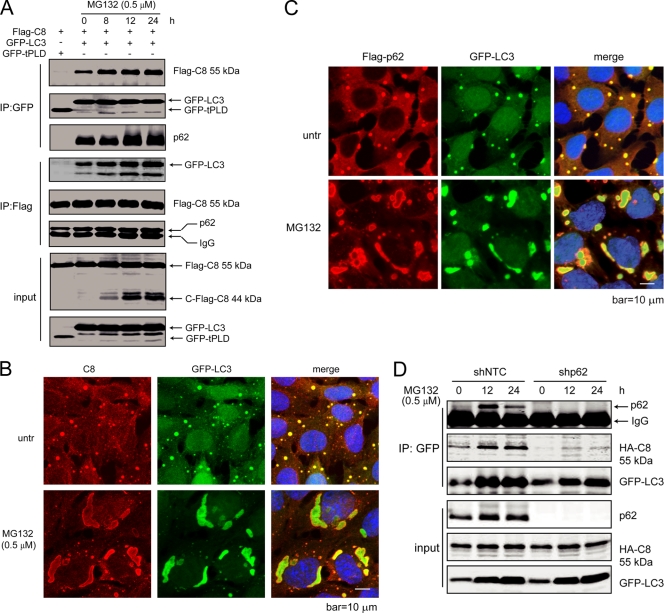
It was recently reported that upon the engagement of death ligand to the death receptor, caspase-8 is ubiquitinated by the E3 ubiquitin ligase cullin-3, which facilitates its interaction with the ubiquitin-binding protein Sequestosome-1 (SQSTM1/p62) and aggregation at the DISC (26). Since proteasome inhibition generally leads to the accumulation of ubiquitinated proteins, we reasoned that MG132-induced caspase-8 aggregation and activation may also involve its ubiquitination and subsequent interaction with p62. Indeed, a His-tagged ubiquitin (His-Ub) pulldown assay showed that MG132 treatment led to increased levels of ubiquitinated caspase-8 (Fig. 4A), and both the caspase-8/p62 interaction and colocalization were enhanced upon MG132 treatment at both overexpression (Fig. 4B and C) and endogenous (Fig. 4D) levels. Knockdown of p62 led to protection against MG132-induced cell death, as shown by reduced plasma membrane permeability (Fig. 4E and F) and delayed apoptotic cleavage of caspase-3 and PARP (Fig. 4G). The partial protection may be due to the incomplete knockdown of p62 expression (Fig. 4E) and/or activation of an alternative cytotoxic effect due to loss of p62. Although restored expression of wild-type p62 in the p62 knockdown cells significantly enhanced MG132-induced apoptosis, the dimerization-deficient mutant (D69A) and the ubiquitin binding-deficient mutant (I431A) of p62 (8) failed to restore apoptosis (Fig. 4H to J). These results indicate that proteasome inhibition-induced apoptosis is dependent on caspase-8 ubiquitination and its interaction with p62.
LC3 is required for p62-mediated caspase-8 aggregation and activation.
p62 forms complexes with a number of proteins and acts as a central player in regulating NF-κB activation (48) or facilitating apoptosis at the DISC (26). Another function of p62 is to facilitate the packing and delivery of misfolded or aggregated proteins for disposal through the autophagolysosomal degradation pathway (8). Indeed, a well-known binding partner of p62 is LC3 (33, 41, 49). LC3 is enriched on intracellular membranes during autophagy (28), which can be induced by proteasome inhibition (40). This raised the possibility that the caspase-8 aggregation and activation upon MG132 treatment involve the interaction between p62 and LC3. To test this, we first examined whether caspase-8, p62, and LC3 interact with each other. Flag-tagged caspase-8 and GFP-tagged LC3 were coexpressed in HEK293T cells. Upon MG132 treatment, GFP IP pulled down increased amounts of Flag-caspase-8 and p62, and Flag IP pulled down increased amounts of GFP-LC3 and p62 (Fig. 5A). Consistent with this result, it was observed that subcellular colocalization of caspase-8, p62, and LC3 was also enhanced upon MG132 treatment (Fig. 4C, 4B, 5B, and 5C). In addition, knockdown of p62 significantly diminished caspase-8 and LC3 association, indicating that the interaction between caspase-8 and LC3 requires p62 (Fig. 5D).
Fig. 5.
LC3 forms a complex with p62 and caspase-8, which is enhanced by proteasome inhibition. (A) Caspase-8, p62, and LC3 form a complex, which is enhanced by proteasome inhibitor treatment. HEK293 cells were transfected with plasmids encoding Flag-caspase-8, together with GFP-LC3 or a GFP control (GFP-tPLD). At 24 h posttransfection, cells with GFP-LC3 were treated with MG132 for the indicated periods of time. Cell lysates were subjected to IP with GFP or Flag antibodies and probed for the indicated proteins. (B) LC3 colocalizes with caspase-8. bax−/− bak−/− BMK cells stably expressing Flag-p62 and GFP-LC3, untreated or treated with MG132 for 18 h, were stained with an anti-caspase-8 antibody. DAPI was used to visualize the nucleus. (C) LC3 colocalizes with p62. Flag-p62 was transfected into bax−/− bak−/− BMK cells expressing GFP-LC3. Cells were left untreated or were treated with MG132 (0.5 μM) for 18 h. Immunofluorescence analysis was performed using an anti-Flag antibody. DAPI was used to visualize the nucleus. (D) p62 knockdown abolishes caspase-8 and LC3 interaction. Lentivirus encoding control shRNA (shNTC) or p62 shRNA (shp62) were used to infect bax−/− bak−/− BMK cells stably expressing HA-caspase-8 (C360S) and GFP-LC3. Cells were treated with MG132 for indicated periods of time. Cell lysates were subjected to IP with GFP and probed for indicated proteins.
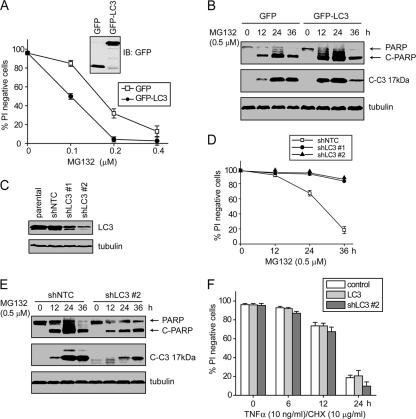
These results suggest the theory that LC3 serves as a molecular hub for caspase-8 aggregation and activation, which would predict that the levels of LC3 can dictate cell sensitivity to proteotoxicity. To test this, GFP or GFP-LC3 was expressed in bax−/− bak−/− BMK cells (Fig. 6A). Expression of GFP-LC3 accelerated MG132-induced apoptosis as shown by PI exclusion, caspase-3 activation, and PARP cleavage (Fig. 6A and B). Conversely, LC3 partial knockdown conferred marked protection against MG132-induced apoptosis (Fig. 6C to E). Importantly, this LC3-mediated apoptosis in response to MG132 is independent of the death receptor pathway, as either the accelerated apoptosis upon LC3 overexpression, or the protection conferred by LC3 knockdown, was not observed when cells were treated with TNF-α (Fig. 6F). Together, these results indicate that proteasomal inhibition facilitates the formation of a caspase-8/p62/LC3 complex and subsequent apoptosis.
Fig. 6.
LC3 promotes MG132-induced apoptosis. (A) GFP or GFP-LC3 was expressed in bax−/− bak−/− BMK cells. Cells were treated with indicated concentration of MG132, and cell death was determined by PI exclusion. The averages of four independent assays + the SEM are shown. (B) Cells expressing GFP or GFP-LC3 were treated with MG132 and probed for the indicated proteins. Note that the apoptotic cleavage of caspase-3 and PARP was more pronounced in GFP-LC3 cells. Note that the lower amount of cleaved PARP and caspase-3 at 36 h is due to excessive cell death. (C) bax−/− bak−/− BMK cells were infected with lentivirus encoding control shRNA (shNTC) or LC3 shRNA (shLC3). Two individual cell clones with LC3 knockdown are shown. (D and E) shNTC and the two shLC3 cell lines were treated with MG132. (D) Cell death was measured by PI exclusion. The averages of four independent assays + the SEM are shown. (E) Cell lysates from the shNTC and shLC3 #2 were probed for indicated proteins. Note the reduced apoptosis in shLC3 cells. (F) The parental, LC3-overexpressing, and shLC3 #2 bax−/− bak−/− BMK cells were treated with TNF-α plus cycloheximide for indicated periods of time. Cell death was determined by PI exclusion. The averages of four independent assays plus SEM are shown.
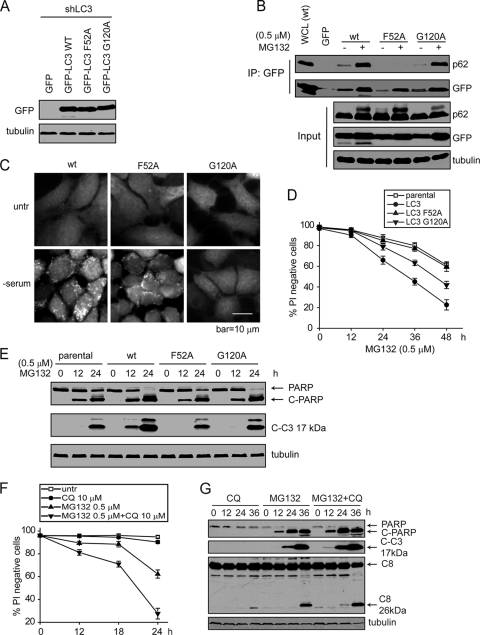
To further test the theory that membrane localized LC3 can act as a molecular hub to recruit p62 and caspase-8 and to mediate subsequent apoptosis, we used the LC3 F52A mutant that is defective in p62 interaction (49), and the G120A mutant that is defective in its lipidation, a prerequisite for LC3 membrane localization (29). To compare their abilities to restore cell sensitivity to apoptosis, we stably expressed GFP-tagged LC3 wt, LC3 F52A, or LC3 G120A in the bax−/− bak−/− BMK cells containing LC3 stable knockdown (Fig. 7A). As expected, LC3 F52A failed to interact with p62 (Fig. 7B), and LC3 G120A failed to form membrane-associated punctate structures upon serum starvation (Fig. 7C). While the wild-type LC3 led to enhanced cell death, both the F52A and the G120A mutants failed to restore cell death as efficiently as did the wild-type LC3 (Fig. 7D and E). Consistent with the importance of LC3/p62 interaction, the p62 Δ321-342 mutant that is defective in LC3 interaction (8) failed to restore MG132-induced apoptosis in p62 knockdown cells (Fig. 4H to J). Consistent with the theory that the accumulation of LC3 can lead to apoptosis, the pharmacological inhibitor of autolysosomal turnover, chloroquine, also enhanced MG132-induced apoptosis (Fig. 7F and G). Taken together, these results indicate that the LC3/p62 interaction and LC3 membrane localization are essential for apoptosis induced by the inhibition of protein degradation.
Fig. 7.
The proapoptotic activity of LC3 requires its p62 interaction and membrane localization. (A) bax−/− bak−/− BMK cells with stable LC3 knockdown were infected with retrovirus encoding GFP-LC3wt, GFP-LC3 F52A, or GFP-LC3 G120A. (B) Cells were left untreated or were treated with MG132. Cell lysates were subjected to IP with an anti-GFP antibody, and the IP samples were probed for GFP and p62. Note that while MG132 induced p62 interaction with LC3wt and LC3 G120A, the LC3 F52A mutant failed to interact with p62. (C) Cells were cultured in regular medium or serum-free medium for 18 h. Note the GFP-LC3 puncta formation in the LC3wt and the LC3 F52A mutant but not in the LC3 G120A mutant. (D and E) Cells were treated with MG132 for the indicated periods of time. Cell death was determined by PI exclusion. The averages of four independent assays and the SEM are shown (D) or immunoblotting for apoptotic cleavage of caspase-3 and PARP (E). (F and G) Chloroquine (CQ) enhances MG132-induced apoptosis. bax−/− bak−/− BMK cells were treated with the indicated conditions of MG132 and/or chloroquine. (F) Cell death was measured by PI exclusion. The averages of four independent assays plus the SEM are shown. (G) PARP, caspase-3, and caspase-8 cleavage was examined by immunoblotting.
Caspase-8 is recruited to and activated at intracellular membranes.
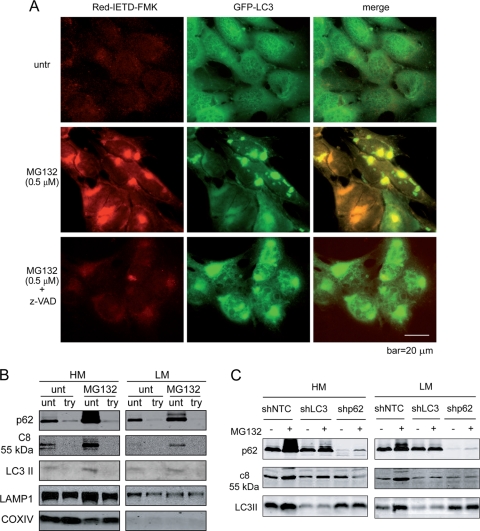
While the results presented above indicate that caspase-8 forms a protein complex with p62 and LC3, the question remains as to whether this is a mechanism for caspase-8 activation, or whether it is merely a mechanism for a cell to dispose caspase-8 by the autophagolysosomal pathway via the p62/LC3 cargo. To address this, we first determined whether caspase-8 recruited to the LC3/p62 complex is enzymatically active. Indeed, a fluorescence-based caspase-8 specific substrate showed that the z-VAD-inhibitable activated caspase-8, induced by MG132, colocalized with LC3 (Fig. 8A). To further characterize caspase-8 localization on intracellular membranes, we separated the heavy membrane (HM; consisting of mitochondria and their associated membranes) and the light membrane (LM; consisting predominantly of the ER, lysosomes, and the Golgi apparatus) fractions. Caspase-8, p62, and LC3 were detected in both heavy- and light-membrane fractions (Fig. 8B). The LC3 localization on various membranes is consistent with reports showing various membrane sources of autophagosomes, including that from the ER, mitochondria, and the plasma membrane (6, 20, 22, 45). Importantly, trypsin treatment led to a significant decrease of membrane-associated caspase-8, p62, and LC3 but not of COX IV and LAMP1 (Fig. 8B), indicating that a significant amount of caspase-8, p62, and LC3 are on the cytosolic side of the intracellular membranes rather than in the lumen of the autophagolysosomes. Knockdown of either LC3 or p62 significantly decreased MG132-induced membrane association of caspase-8 (Fig. 8C). Taken together, these results indicate that upon the inhibition of protein degradation, caspase-8 is recruited to intracellular membranes via p62 and LC3 and is activated to induce apoptosis by its enhanced oligomerization.
Fig. 8.
Proteasome inhibition leads to caspase-8 aggregation and activation on the cytosolic side of intracellular membranes. (A) bax−/− bak−/− BMK cells stably expressing GFP-LC3 were left untreated (one well) or were treated with MG132 (two wells). At 16 h after treatment, one well of MG132-treated cells was treated with z-VAD for 2 h. All three wells were incubated with Red-IETD-FMK for 1 h. The cells were observed under a Zeiss inverted Axiovert 200M microscope. Note that MG132 induced caspase-8 activity that colocalized with GFP-LC3. (B) bax−/− bak−/− BMK cells were left untreated or were treated with MG132 (0.5 μM) for 18 h. Cells were homogenized and left untreated or were treated with trypsin (100 μg/ml) and fractionated. Heavy-membrane (HM) and light-membrane (LM) fractions were subjected to immunoblotting for the indicated proteins. (C) MG132-induced caspase-8 membrane recruitment is dependent on both p62 and LC3. bax−/− bak−/− BMK cells expressing control shRNA (shNTC), shLC3, or shp62 were left untreated or were treated with MG132 (0.5 μM) for 18 h. The cells were fractionated. The heavy-membrane (HM) and light-membrane (LM) fractions were probed with the indicated antibodies. Note that in the LC3 and p62 knockdown cells, MG132-induced increased membrane-localized caspase-8 was diminished.
DISCUSSION
In this study, we found a novel mechanism to explain proteotoxicity. Upon the inhibition of protein degradation pathways, cells upregulate the autophagic response to facilitate autophagolysosomal protein turnover, which is mediated by the lipidation and membrane localization of LC3. However, severely damaged proteasomal or autophagolysosomal pathways may “clog” each other (16, 34). This leads to the enrichment of LC3 on intracellular membranes that can serve as a molecular hub to recruit the ubiquitin-binding protein SQSTM1/p62 and one of its binding partners, caspase-8. The induced oligomerization and activation of caspase-8 subsequently initiate the downstream apoptosis cascade. Although in the present study we mainly focused on the use of the proteasome inhibitor MG132, which has been shown to induce caspase-8 activation in an osteosarcoma line (56), we believe this caspase-8-mediated proteotoxicity has a broad implication under a wide range of conditions where cellular protein homeostasis is irreversibly disrupted. A separate study in our laboratory has also shown that elevated expression of the endogenous serine/cysteine protease inhibitor SerpinB3b led to the blockade of lysosomal degradation, caspase-8 aggregation and activation, and apoptosis in response to ER stress (53).
Our finding demonstrates a novel mechanism for caspase-8 activation and amplification. It is well established that caspase-8 aggregation and activation take place at the DISC and plays an essential role in death receptor signaling (47). While evidence exists showing that cleaved caspase-8 can also function on mitochondrial membranes through its interaction with cardiolipin (19), our finding is the first to show that caspase-8 oligomerization and activation can be induced intracellularly. We found that this intracellular caspase-8 aggregation correlates with its ubiquitination and interaction with p62. While we show that caspase-8 ubiquitination is enhanced upon proteasome inhibition, our finding is reminiscent of a recent report that at the DISC, caspase-8 is ubiquitinated by cullin-3 and interacts with p62, which facilitates its aggregation and activation (26). Together with a recent report that cullin-3 is a LC3-interacting protein (7), our finding suggests the possibility that in addition to the death receptors, caspase-8 may also be ubiquitinated at intracellular membranes in a LC3-dependent manner. This provides a mechanism to amplify caspase-8-dependent apoptosis downstream of death receptors and can help to explain the observation that proteasome inhibitors can potentiate TRAIL-induced cell death (27) and suggests that the use of proteasome or lysosome inhibitors may be considered to sensitize tumors that are resistant to death receptor-mediated apoptosis.
Our study demonstrates a proapoptotic role of LC3, a well-known molecule that is involved in autophagy (28). Upon the initiation of autophagy, LC3 conjugation to phosphatidylethanolamine at its C-terminal glycine and its subsequent membrane localization promote the subsequent dynamic rearrangement of intracellular membranes, leading to the formation of autophagosomes (28). LC3 directly interacts with p62 via its N-terminal domain, which is essential for the cargo function of p62 to transport polyubiquitylated protein aggregates to autophagosomes (49). In the present study, we found that this LC3/p62 interaction is required for MG132-induced caspase-8 aggregation and activation on the cytosolic side of intracellular membranes. This suggests that LC3/p62-mediated protein complex formation and membrane localization are not merely a garbage disposal mechanism. It can serve as a safeguard mechanism by which, when sensing a stress resulting from a massive amount of misfolded proteins that cannot be resolved by autophagy flux, cells commit to die by apoptosis to maintain organismal homeostasis. Indeed, the proapoptotic function of LC3 has been recently reported (14, 30). A number of cancers have been reported to have elevated levels of LC3 (39, 57). Our finding that LC3 levels can influence cell sensitivity to proteotoxicity suggests that the LC3 level may be a predictor for the outcome of treatment using proteasome inhibitors.
Most malignancies have developed various mechanisms to evade apoptosis through the upregulation of antiapoptotic proteins such as the Bcl-2 family of proteins or the mutation of proapoptotic molecules such as p53 (2, 21). A main goal in cancer treatment is to selectively kill these apoptosis-deficient cancer cells. Our finding here that bax−/− bak−/− cells, in which the mitochondrial apoptosis pathway is heavily impaired, are still sensitive to proteotoxicity indicates that the cell death mediated by LC3, p62, and caspase-8 may be effectively induced in many types of cancer cells regardless of their various survival signaling mechanisms. Elevated levels of protein translation, turnover, and metabolism render cancer cells more susceptible to proteotoxicity, providing a therapeutic opportunity to eradicate cancers based on their fundamental biochemical properties.
ACKNOWLEDGMENTS
We thank Eileen White for the bax−/− bak−/− BMK cells, Erich Mackow for the polyHis-Ub construct, Michael Frohman and Huiyan Huang for the Venus and linker sequences, William Lennarz and Gang Zhao for assistance on the size exclusion chromatography, and Juei-Suei Chen for molecular cloning. We thank Susan Van Horn and Guowei Tian (Stony Brook Microscopy Imaging Center) for assistance on transmission electron microscopy and fluorescence microscopy, respectively.
E.U. is supported by the NIH training grant T32CA009176. This study was supported by NIH (CA098092 and CA129536), Susan Komen for the Cure (KG081538), and the Carol Baldwin Breast Cancer Research Foundation (W.-X.Z.).
The authors declare that they have no conflicts of interest.
Footnotes
Published ahead of print on 31 May 2011.
REFERENCES
- 1. Adams J. 2004. The proteasome: a suitable antineoplastic target. Nat. Rev. Cancer 4:349–360 [DOI] [PubMed] [Google Scholar]
- 2. Adams J. M., Cory S. 2007. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 26:1324–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allen C., et al. 2008. Bortezomib-induced apoptosis with limited clinical response is accompanied by inhibition of canonical but not alternative nuclear factor-κB subunits in head and neck cancer. Clin. Cancer Res. 14:4175–4185 [DOI] [PubMed] [Google Scholar]
- 4. Amaravadi R. K., Thompson C. B. 2007. The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin. Cancer Res. 13:7271–7279 [DOI] [PubMed] [Google Scholar]
- 5. Ashkenazi A., Dixit V. M. 1999. Apoptosis control by death and decoy receptors. Curr. Opin. Cell Biol. 11:255–260 [DOI] [PubMed] [Google Scholar]
- 6. Axe E. L., et al. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Behrends C., Sowa M. E., Gygi S. P., Harper J. W. 2010. Network organization of the human autophagy system. Nature 466:68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bjorkoy G., et al. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boatright K. M., Salvesen G. S. 2003. Mechanisms of caspase activation. Curr. Opin. Cell Biol. 15:725–731 [DOI] [PubMed] [Google Scholar]
- 10. Boldin M. P., Goncharov T. M., Goltsev Y. V., Wallach D. 1996. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell 85:803–815 [DOI] [PubMed] [Google Scholar]
- 11. Buchberger A., Bukau B., Sommer T. 2010. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol. Cell 40:238–252 [DOI] [PubMed] [Google Scholar]
- 12. Carew J. S., Nawrocki S. T., Giles F. J., Cleveland J. L. 2008. Targeting autophagy: a novel anticancer strategy with therapeutic implications for imatinib resistance. Biologics 2:201–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen K. F., et al. 2009. Bortezomib overcomes tumor necrosis factor-related apoptosis-inducing ligand resistance in hepatocellular carcinoma cells in part through the inhibition of the phosphatidylinositol 3-kinase/Akt pathway. J. Biol. Chem. 284:11121–11133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen Z. H., et al. 2010. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc. Natl. Acad. Sci. U. S. A. 107:18880–18885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Degenhardt K., Sundararajan R., Lindsten T., Thompson C., White E. 2002. Bax and Bak independently promote cytochrome c release from mitochondria. J. Biol. Chem. 277:14127–14134 [DOI] [PubMed] [Google Scholar]
- 16. Ding W. X., Yin X. M. 2008. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy 4:141–150 [DOI] [PubMed] [Google Scholar]
- 17. Dou Z., et al. 2010. The class IA phosphatidylinositol 3-kinase p110-β subunit is a positive regulator of autophagy. J. Cell Biol. 191:827–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fehrenbacher N., Jaattela M. 2005. Lysosomes as targets for cancer therapy. Cancer Res. 65:2993–2995 [DOI] [PubMed] [Google Scholar]
- 19. Gonzalvez F., et al. 2008. Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J. Cell Biol. 183:681–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hailey D. W., et al. 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hainaut P., Hollstein M. 2000. p53 and human cancer: the first ten thousand mutations. Adv. Cancer Res. 77:81–137 [DOI] [PubMed] [Google Scholar]
- 22. Hayashi-Nishino M., et al. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437 [DOI] [PubMed] [Google Scholar]
- 23. Hideshima T., et al. 2003. Molecular mechanisms mediating antimyeloma activity of proteasome inhibitor PS-341. Blood 101:1530–1534 [DOI] [PubMed] [Google Scholar]
- 24. Hightower L. E. 1991. Heat shock, stress proteins, chaperones, and proteotoxicity. Cell 66:191–197 [DOI] [PubMed] [Google Scholar]
- 25. Huang H., Choi S. Y., Frohman M. A. 2010. A quantitative assay for mitochondrial fusion using Renilla luciferase complementation. Mitochondrion 10:559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jin Z., et al. 2009. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 137:721–735 [DOI] [PubMed] [Google Scholar]
- 27. Johnson T. R., et al. 2003. The proteasome inhibitor PS-341 overcomes TRAIL resistance in Bax and caspase 9-negative or Bcl-xL overexpressing cells. Oncogene 22:4953–4963 [DOI] [PubMed] [Google Scholar]
- 28. Kabeya Y., et al. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kabeya Y., et al. 2004. LC3, GABARAP, and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117(Pt. 13):2805–2812 [DOI] [PubMed] [Google Scholar]
- 30. Kar R., Singha P. K., Venkatachalam M. A., Saikumar P. 2009. A novel role for MAP1 LC3 in nonautophagic cytoplasmic vacuolation death of cancer cells. Oncogene 28:2556–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kawadler H., Gantz M. A., Riley J. L., Yang X. 2008. The paracaspase MALT1 controls caspase-8 activation during lymphocyte proliferation. Mol. Cell 31:415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kirkin V., McEwan D. G., Novak I., Dikic I. 2009. A role for ubiquitin in selective autophagy. Mol. Cell 34:259–269 [DOI] [PubMed] [Google Scholar]
- 33. Komatsu M., et al. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131:1149–1163 [DOI] [PubMed] [Google Scholar]
- 34. Korolchuk V. I., Mansilla A., Menzies F. M., Rubinsztein D. C. 2009. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol. Cell 33:517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Korolchuk V. I., Menzies F. M., Rubinsztein D. C. 2010. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 584:1393–1398 [DOI] [PubMed] [Google Scholar]
- 36. Loehberg C. R., et al. 2007. Ataxia telangiectasia-mutated and p53 are potential mediators of chloroquine-induced resistance to mammary carcinogenesis. Cancer Res. 67:12026–12033 [DOI] [PubMed] [Google Scholar]
- 37. Lomonosova E., Ryerse J., Chinnadurai G. 2009. BAX/BAK-independent mitoptosis during cell death induced by proteasome inhibition? Mol. Cancer Res. 7:1268–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mathew R., Karantza-Wadsworth V., White E. 2007. Role of autophagy in cancer. Nat. Rev. Cancer 7:961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Namgoong G. M., et al. 2010. The prolyl isomerase Pin1 induces LC-3 expression and mediates tamoxifen resistance in breast cancer. J. Biol. Chem. 285:23829–23841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pandey U. B., et al. 2007. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447:859–863 [DOI] [PubMed] [Google Scholar]
- 41. Pankiv S., et al. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 [DOI] [PubMed] [Google Scholar]
- 42. Podar K., et al. 2008. A pivotal role for Mcl-1 in Bortezomib-induced apoptosis. Oncogene 27:721–731 [DOI] [PubMed] [Google Scholar]
- 43. Pop C., Fitzgerald P., Green D. R., Salvesen G. S. 2007. Role of proteolysis in caspase-8 activation and stabilization. Biochemistry 46:4398–4407 [DOI] [PubMed] [Google Scholar]
- 44. Puthalakath H., et al. 2007. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 129:1337–1349 [DOI] [PubMed] [Google Scholar]
- 45. Ravikumar B., Moreau K., Jahreiss L., Puri C., Rubinsztein D. C. 2010. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 12:747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reimertz C., Kogel D., Rami A., Chittenden T., Prehn J. H. 2003. Gene expression during ER stress-induced apoptosis in neurons: induction of the BH3-only protein Bbc3/PUMA and activation of the mitochondrial apoptosis pathway. J. Cell Biol. 162:587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Salvesen G. S., Dixit V. M. 1999. Caspase activation: the induced-proximity model. Proc. Natl. Acad. Sci. U. S. A. 96:10964–10967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sanz L., Diaz-Meco M. T., Nakano H., Moscat J. 2000. The atypical PKC-interacting protein p62 channels NF-κB activation by the IL-1-TRAF6 pathway. EMBO J. 19:1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shvets E., Fass E., Scherz-Shouval R., Elazar Z. 2008. The N terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J. Cell Sci. 121(Pt. 16):2685–2695 [DOI] [PubMed] [Google Scholar]
- 50. Tinel A., Tschopp J. 2004. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304:843–846 [DOI] [PubMed] [Google Scholar]
- 51. Uddin S., et al. 2008. Bortezomib (Velcade) induces p27Kip1 expression through S-phase kinase protein 2 degradation in colorectal cancer. Cancer Res. 68:3379–3388 [DOI] [PubMed] [Google Scholar]
- 52. Ullman E., et al. 2008. Autophagy promotes necrosis in apoptosis-deficient cells in response to ER stress. Cell Death Differ. 15:422–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ullman E., Pan J.-A., Zong W.-X. 2011. Squamous cell carcinoma antigen 1 promotes caspase-8-mediated apoptosis in response to endoplasmic reticulum stress while inhibiting necrosis induced by lysosomal injury. Mol. Cell. Biol. 31:2902–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Voorhees P. M., Orlowski R. Z. 2006. The proteasome and proteasome inhibitors in cancer therapy. Annu. Rev. Pharmacol. Toxicol. 46:189–213 [DOI] [PubMed] [Google Scholar]
- 55. Wu W. K., et al. 2010. Proteasome inhibition: a new therapeutic strategy to cancer treatment. Cancer Lett. 293:15–22 [DOI] [PubMed] [Google Scholar]
- 56. Yan X. B., et al. 2007. Caspase-8-dependent osteosarcoma cell apoptosis induced by proteasome inhibitor MG132. Cell Biol. Int. 31:1136–1143 [DOI] [PubMed] [Google Scholar]
- 57. Yoshioka A., et al. 2008. LC3, an autophagosome marker, is highly expressed in gastrointestinal cancers. Int. J. Oncol. 33:461–468 [PubMed] [Google Scholar]
- 58. Yu J. W., Jeffrey P. D., Shi Y. 2009. Mechanism of procaspase-8 activation by c-FLIPL. Proc. Natl. Acad. Sci. U. S. A. 106:8169–8174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zou H., Li Y., Liu X., Wang X. 1999. An APAF-1 cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 274:11549–11556 [DOI] [PubMed] [Google Scholar]