Abstract
PCR mapping of staphylococcal cassette chromosome mec type IVa and adjacent mobile elements in 94 methicillin-resistant Staphylococcus aureus (MRSA) strains identified two primary structures (A and B) that could be further classified into two (A1 and A2) and five (B1 to B5) variants, primarily based on structural differences in the orfX-J3 region. While spa type t008 (USA300) invariably contained the A variants, other spa types belonging to clonal complex 8 and unrelated lineages generally contained B variants. These findings have important implications for the typing and identification of MRSA strains containing B variants.
TEXT
One of the most widespread community-associated (CA) methicillin-resistant Staphylococcus aureus (MRSA) clones is USA300 (14, 16). This clone has become the most common cause of CA MRSA infections in North America (15) and has recently spread to western Europe, including Denmark (14). USA300 isolates are characterized by a common pulsed-field gel electrophoresis (PFGE) pattern, generally belong to spa type t008 and multilocus sequence type 8 (ST8, clonal complex 8), and harbor staphylococcal cassette chromosome mec (SCCmec) type IVa. In addition, USA300 isolates typically contain the arginine catabolic mobile element (ACME) and the lukS-PV and lukF-PV genes encoding Panton-Valentine leukocidin (PVL) (5, 14, 16).
Genotypic differences were recently shown among Danish health care-associated MRSA isolates belonging to clonal complex 8 (CC8) and resembling USA300 by PFGE. In particular, t024 isolates were PVL negative and inconsistently carried ACME, whereas all t008 isolates carried both elements (9). A study evaluating the sensitivity and specificity of a commercial PCR-based kit for MRSA detection failed to identify 85% of Danish t024 MRSA isolates due to a structural variation in the orfX J3 region of SCCmec IVa (1). Whole-genome sequencing of two t024 isolates identified a novel J3 region containing two unique clusters of open reading frames (ORFs). One of the isolates contained two additional direct repeats between J3 and orfX. These direct repeats delimited a region containing an SCC element followed by an ACME II variant (SCCmec IVa variant B1, Fig. 1) (2). Based on these findings, we decided to further investigate the J3 region and adjacent structures of SCCmec IVa in a large collection of MRSA isolates.
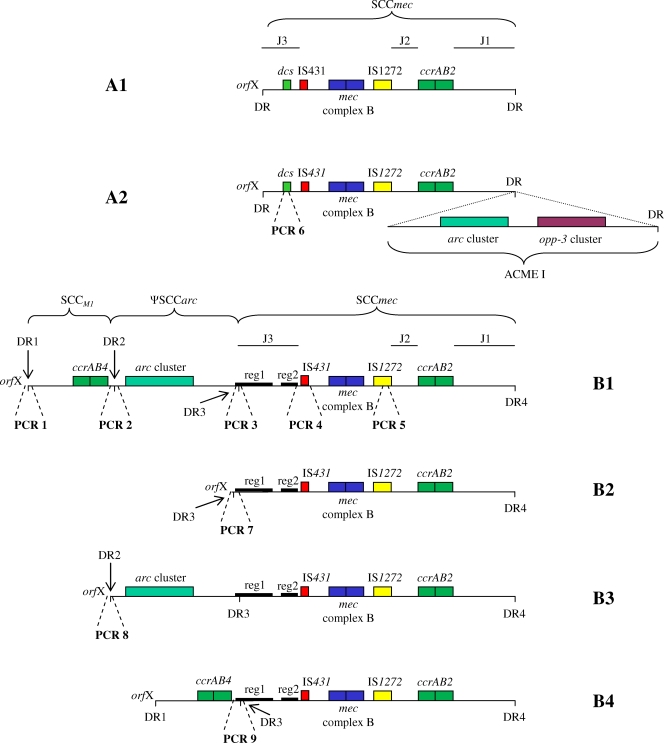
Fig. 1.
Schematic drawing of SCCmec composite elements detected in the present study. “A” structures have prototype SCCmec IVa (11), and “B” structures have SCCmec IVa with the novel J3 region characterized by two unique clusters of ORFs (reg1 and reg2) (2). A1 (SCCmec IVa), A2 (SCCmec IVa with ACME I downstream), B1 (SCCM1, ΨSCCarc, SCCmecIVa [2B]; J1, subtype 1-specific ORFs; J3, subtype 3-specific ORFs for M1), and B2 (SCCmec IVa [2B]; J1, subtype 1-specific ORFs; J3, subtype 3-specific ORFs for M1) have been previously described (2, 5, 11) and fully sequenced (GenBank accession numbers NC_003923, NC_007793, HM030720, and HM030721). B3 and B4 are proposed structures of new SCCmec IVa composite elements based on PCR mapping and partial sequencing. Approximate targets of PCRs 1 to 9 are shown by broken lines.
MRSA strains isolated at Hvidovre Hospital are routinely spa typed, SCCmec typed, and screened by PCR for detection of ACME-specific (arcA and opp3AB) and PVL-specific (lukS-PV and lukF-PV) genes (4, 5, 6, 8, 10, 12). For this study, we selected 94 MRSA isolates obtained between 2003 and 2010 from patients with health care-associated or CA MRSA in the greater Copenhagen area. All isolates contained SCCmec IVa and belonged to spa types where arcA had been detected. A representative number of isolates was selected on the basis of the relative isolation frequency of each spa type. When available, arcA-negative isolates were included for each spa type. Ninety-one isolates had spa types belonging to CC8, primarily t024 (n = 50) and t008 (n = 25). Three arcA-positive isolates had spa types related to other MRSA lineages, i.e., CC1, CC5, and CC30. Most isolates were epidemiologically unrelated, except for 36 isolates from persons living or working at nine nursing homes with multiple cases of MRSA. These 36 isolates were included based on the same criteria as other isolates but with the purpose of studying within-outbreak variability. They represented approximately one-third of all MRSA isolates and half of all spa types obtained from these nursing homes between 2003 and 2010.
We investigated all isolates by PCR mapping using previously described primers targeting the SCCmec IVa prototype (3, 7, 13) and novel primers targeting the new orfX-J3 elements identified by our previous work (2) (Table 1; Fig. 1). PCRs were performed with a final volume of 20 μl using the DreamTaq Green PCR Master Mix (Fermentas, Helsingborg, Sweden) and purified DNA from each isolate. The cycling conditions for PCRs 1, 3 to 6, and 9 were initial denaturation at 94°C for 4 min, followed by 25 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 60 s and a 5-min final elongation step at 72°C. The cycling conditions for PCRs 2, 7, and 8 were identical, except that annealing temperatures of 55°C (PCRs 2 and 7) and 54°C (PCR 8) were used.
Table 1.
Primers used in the study
| PCR no. and primer | Sequence (5′–3′) | Amplicon size (bp) | Reference |
|---|---|---|---|
| 1 | |||
| cR1 | AAGAATTGAACCAACGCATGA | 7 | |
| insert orf | CTTTGATAAGCCATTCATTC | 326 | This study |
| 2 | |||
| DR2/IR2_F2 | GATTTTCCCAACCGCACC | ||
| DR2/IR2_R2 | ACCTTTATTCGTTACGCCC | 975 | This study |
| 3 | |||
| DR3/IR3_F2 | ATATAGGTGAGAGTATGATAAGTGAG | ||
| ins-orf-M299 | GCTATCAATCTGTCCTCAAA | 325 | This study |
| 4 | |||
| Insert_IS431 | GTAAATGAACTGCAGCTATTG | ||
| IS431F | CTTCTGGTTGGAATGTGTGG | 939 | This study |
| 5 | |||
| IS1272F1 | GCCACTCATAACATATGGAA | 3 | |
| IS1272R1 | CATCCGAGTGAAACCCAAA | 415 | |
| 6 | |||
| DCS F2 | CATCCTATGATAGCTTGGTC | 13 | |
| DCS R1 | CTAAATCATAGCCATGACCG | 342 | |
| 7 | |||
| cR1 | AAGAATTGAACCAACGCATGA | 7 | |
| ins-orf-M299 | GCTATCAATCTGTCCTCAAA | 395 | This study |
| 8 | |||
| cR1 | AAGAATTGAACCAACGCATGA | 7 | |
| DR2/IR2_R2 | ACCTTTATTCGTTACGCCC | 606 | This study |
| 9 | |||
| DR2/IR2_F2 | ACCTTTATTCGTTACGCCC | ||
| ins-orf-M299 | GCTATCAATCTGTCCTCAAA | 764 | This study |
We identified two main SCCmec IVa structures, A and B (Table 2; Fig. 1). Structure A had the dcs region typical of the SCCmec IVa prototype, and structure B had the novel J3 region comprising two unique clusters of ORFs (2). This variation in J3 has important implications for MRSA diagnostics, since only J3 of structure A is targeted by one of the commercially available PCR kits for MRSA detection (1). Variants of A (A1 and A2) and B (B1 to B5) were defined for the purpose of this study based on the observed structural variations in the regions immediately down- and upstream of SCCmec, respectively (Fig. 1). B5 had the novel J3 region, but the organization of orfX-J3 could not be discerned based on PCR results (Table 2). Elements from J3 were not detected in five isolates, which were therefore considered nontypeable (NT1 and NT2, Table 2). Further analysis of SCCmec composite elements in B5, NT1, and NT2 was attempted by standard- and long-range PCRs using various combinations of primers from Table 1. However, the structures could not be deduced, possibly due to mutations in primer binding sites or because of the presence of novel variants. Further studies sequencing the entire SCCmec composite elements of isolates representing NT1, NT2, and B5 are warranted.
Table 2.
Description of the two SCCmec IVa structures (A and B) detected by PCR mappinga
| SCCmec IVa variant | Strain used as referenceb |
spa type(s) (no. of strains) |
ccr allotype | ACME gened |
PVL | Profile according to PCRs 1 to 9 | ||
|---|---|---|---|---|---|---|---|---|
| CC8 | Other CCc | arcA | opp3AB | |||||
| A1 | MW2 | t008 (2), t064 (2) | ccrAB2 | 0 | 0 | 1/0e | 0 0 0 0 1 1 0 0 0 | |
| A2 | FPR3757 | t008 (23), t068 (1), t121 (1), t211 (1), t3980 (1), t5098 (1) | ccrAB2 | 1 | 1 | 1/0e | 0 0 0 0 1 1 0 0 0 | |
| B1 | M1 | t024 (17), t430 (3), t351 (1), t842 (1), t2555 (1) | t012 (1) | ccrAB2 and ccrAB4 | 1 | 0 | 0 | 1 1 1 1 1 0 0 0 0 |
| B2 | M299 | t024 (11) | ccrAB2 | 0 | 0 | 0 | 0 0 0 1 1 0 1 0 0 | |
| B3 | M15 | t024 (16), t430 (1), t846 (1) | t127 (1) | ccrAB2 | 1 | 0 | 0 | 0 0 1 1 1 0 0 1 0 |
| B4 | M1298 | t024 (2) | ccrAB2 and ccrAB4 | 0 | 0 | 0 | 1 0 0 1 1 0 0 0 1 | |
| B5 | M289 | t979 (1) | ccrAB2 | 1 | 0 | 0 | 0 0 1 1 1 0 0 0 0 | |
| NT1f | M620 | t024 (3) | ccrAB2 | 0 | 0 | 0 | 1 0 0 0 1 0 0 0 0 | |
| NT2f | M220 | t024 (1), t064 (1) | ccrAB2 | 0 | 0 | 0 | 0 0 0 0 1 0 0 0 0 | |
A1 and A2 correspond to the prototype SCCmec IVa cassette and the ACME I-positive variant associated with USA300, respectively. B1 and B2 were described recently (2), and B3, B4, and B5 are novel variants identified in this study.
MW2 and FPR3757, isolated in Japan and in the United States, respectively, were kindly provided by T. Ito. The remaining standards were Danish clinical MRSA isolates from Hvidovre Hospital, Hvidovre, Denmark.
spa types not associated with CC8: t012 (CC30), t127 (CC1), and t979 (CC5).
Possession of both arcA and opp3AB suggests the presence of ACME I, which is typical of the S. aureus USA300 lineage. Possession of only arcA suggests the presence of the recently described ACME II variant ΨSCCarc (2).
50% and 83% of the isolates with variants A1 and A2 were PVL positive, respectively.
Isolates without any apparent loci in J3 were considered nontypeable (NT1 and NT2).
The SCCmec IVa cassette originally discovered by Ma et al. (11) was identified in four isolates (A1 variant), and the same cassette followed by ACME I (A2 variant), which is characteristic of USA300 (5), was detected in 28 isolates (Fig. 1). The majority (92%) of the t008 isolates were assigned to variant A2 (Table 2; Fig. 1), thus supporting the notion that spa type t008 is strongly associated with USA300 (17). Most of the other isolates possessed B variants. In addition to the two variants recently described by our group (B1 and B2), two closely related new variants (B3 and B4) were described for the first time in this study. B variants were detected in 92% of the t024 isolates but not in t008 isolates (Table 2). Interestingly, B1 and B3 were detected in both CC8 and non-CC8 isolates (Table 2), suggesting either horizontal transfer of these large composite elements or convergent evolution across S. aureus lineages.
B3 differed from B1 only by the lack of ccrAB4. We hypothesized that this difference was due to an excision or insertion of SCCM1 between DR1 and DR2 (Fig. 1). Sequencing of the 606-bp amplicon generated from this region (PCR 8) confirmed this hypothesis, since DR2 was adjacent to orfX in the B3 variant (Fig. 1). Similarly, B4 differed from B1 only by the lack of ΨSCCarc. Sequencing of the 764-bp amplicon generated from this region (PCR 9) proved that excision or insertion of ΨSCCarc had taken place between DR2 and DR3 (Fig. 1). These results are in favor of our previously described hypothesis that B2 derived from B1 by excision of ΨSCCarc and SCCM1 (2). The results also indicate that possession of more than the usual two direct repeat regions flanking SCCmec may lead to a greater variety of recombination events and consequently more variants. Further experiments are needed to elucidate which factors influence mobilization of ΨSCCarc and SCCM1.
Analysis of the 36 isolates originating from nine nursing homes with multiple cases of MRSA showed that in some facilities a single clone occurred over several years, whereas in others different spa types and SCCmec IVa variants developed over the years. In nursing home 1, the B1 variant of SCCmec IVa was detected in three spa types belonging to CC8 (Table 3). This may be suggestive of evolutionary changes in region X of spa or horizontal transfer of composite SCCmec elements between closely related lineages. Detection of both variants B1 and B3 in t024 isolates within three nursing homes (1, 3, and 4, Table 3) may suggest that B3 was generated in one of these places by deletion of SCCM1, followed by transmission in local hospitals or through staff or residents moving between these nursing homes. A less likely explanation would be that the same excision or insertion event happened in all three nursing homes. Overall, a clear pattern of local SCCmec evolution could not be discerned, since strain isolation dates did not suggest gradual replacement of one variant by another.
Table 3.
Distribution of novel SCCmec IVa variants in MRSA CC8 isolated from nine nursing homes
| Nursing home | spa type (no. of strains) | SCCmec IVa variant(s) | Time of isolation |
|---|---|---|---|
| 1 | t024 (5) | B1, B3 | 2003–2008 |
| t430 (3) | B1 | 2004–2006 | |
| t351 (1) | B1 | 2004 | |
| t846 (1) | B3 | 2009 | |
| 2 | t024 (4) | B1 | 2003–2009 |
| t842 (1) | B1 | 2005 | |
| 3 | t024 (5) | B1, B3 | 2003–2009 |
| 4 | t024 (5) | B1, B2, B3, NT1 | 2004–2007 |
| 5 | t024 (2) | B4 | 2007–2009 |
| 6 | t024 (2) | B3 | 2004 |
| 7 | t024 (1) | B1 | 2005 |
| t2555 (1) | B1 | 2007 | |
| 8 | t024 (3) | B1 | 2004–2006 |
| 9 | t024 (2) | B1, NT1 | 2006–2008 |
The present study reveals a wide dispersal of novel SCCmec IVa structures in CC8 and other MRSA lineages in the greater Copenhagen area. The B variants probably evolved from a common ancestral structure through ccr-mediated excision of the mobile elements between J3 and orfX. The variability in the structure of this region has important implications for the typing and identification of MRSA, since strains carrying B variants of SCCmec IVa cannot not be detected by current commercial PCR kits targeting the orfX J3 region. Our PCR mapping approach targeting the unique regions in J3 and upstream of SCCmec can be used for the subtyping of CC8 isolates diverging from the typical USA300 clone by lacking the opp3 gene cluster, being PVL negative, and having spa types other than t008.
Acknowledgments
This work was funded in part by a grant from Fondation Idella (Vaduz, Liechtenstein).
Footnotes
Published ahead of print on 6 June 2011.
REFERENCES
- 1. Bartels M. D., et al. 2009. A common variant of staphylococcal cassette chromosome mec type IVa in isolates from Copenhagen, Denmark, is not detected by the BD GeneOhm methicillin-resistant Staphylococcus aureus assay. J. Clin. Microbiol. 47:1524–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartels M. D., Hansen L. H., Boye K., Sørensen S. J., Westh H. 2011. An unexpected location of the arginine catabolic mobile element (ACME) in a USA300-related MRSA strain. PLoS One 6:e16193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boye K., Bartels M. D., Andersen I. S., Moller J. A., Westh H. 2007. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I-V. Clin. Microbiol. Infect. 13:725–727 [DOI] [PubMed] [Google Scholar]
- 4. Diep B. A., et al. 2008. The arginine catabolic metabolic element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 197:1523–1530 [DOI] [PubMed] [Google Scholar]
- 5. Diep B. A., et al. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739 [DOI] [PubMed] [Google Scholar]
- 6. Harmsen D., et al. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ito T., Katayama Y., Hiramatsu K. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kondo Y., et al. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Larsen A. R., et al. 2009. Two distinct clones of methicillin-resistant Staphylococcus aureus (MRSA) with the same USA300 pulsed-field gel electrophoresis profile: a potential pitfall for identification of USA300 community-associated MRSA. J. Clin. Microbiol. 47:3765–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Larsen A. R., Stegger M., Sorum M. 2008. spa typing directly from a mecA, spa and pvl multiplex PCR assay—a cost-effective improvement for methicillin-resistant Staphylococcus aureus surveillance. Clin. Microbiol. Infect. 14:611–614 [DOI] [PubMed] [Google Scholar]
- 11. Ma X. X., et al. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Milheiriço C., Oliveira D. C., de Lencastre H. 2007. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J. Antimicrob. Chemother. 60:42–48 [DOI] [PubMed] [Google Scholar]
- 13. Oliveira D. C., de Lencastre H. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Otter J. A., French G. L. 2010. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Europe. Lancet Infect. Dis. 10:227–239 [DOI] [PubMed] [Google Scholar]
- 15. Tenover F. C., Goering R. V. 2009. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J. Antimicrob. Chemother. 64:441–446 [DOI] [PubMed] [Google Scholar]
- 16. Vandenesch F., et al. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamamoto T., et al. 2010. Community-acquired methicillin-resistant Staphylococcus aureus: community transmission, pathogenesis, and drug resistance. J. Infect. Chemother. 16:225–254 [DOI] [PMC free article] [PubMed] [Google Scholar]