Abstract
Vancomycin-intermediate Staphylococcus aureus (VISA) is generated from vancomycin-susceptible Staphylococcus aureus by multiple spontaneous mutations. We previously reported that sequential acquisition of mutations in the two-component regulatory systems vraSR and graRS was responsible for the VISA phenotype of strain Mu50. Here we report on the identification of a novel set of regulator mutations, a deletion mutation in two-component regulatory system walRK (synonyms, vicRK and yycFG), and a truncating mutation in a proteolytic regulatory gene, clpP, responsible for the raised vancomycin resistance in a laboratory-derived VISA strain, LR5P1-V3. The contributory effect of the two mutations to vancomycin resistance was confirmed by introducing the walK and clpP mutations into the vancomycin-susceptible parent strain N315LR5P1 by a gene replacement procedure. The vancomycin MIC of N315LR5P1 was raised from 1 to 2 mg/liter by the introduction of the walK or clpP mutation, but it was raised to 4 mg/liter by the introduction of both the walK and clpP mutations. The vancomycin MIC value of the double mutant was equivalent to that of strain LR5P1-V3. Like VISA clinical strains, LR5P1-V3 and the double mutant strain LR5P1walK*clpP* exhibited a thickened cell wall, slow growth, and decreased autolytic activity. Transcriptional profiles of the mutants with gene replacements demonstrated that introduction of both the walK and clpP mutations could alter expression of dozens or hundreds of genes, including those involved in cell envelope and cellular processes, intermediary metabolism, and information pathway. A mutation prevalence study performed on 39 worldwide clinical VISA strains showed that 61.5, 7.7, 10.3, and 20.5% of VISA strains harbored mutations in walRK, clpP, graRS, and vraSR, respectively. The mutation of walRK was most frequently carried by VISA strains. Together, these results suggested that the mutations of walK and clpP identified in LR5P1-V3 constitute a new combination of genetic events causing vancomycin resistance in Staphylococcus aureus.
INTRODUCTION
Staphylococcus aureus has become one of the most frequent causes of a wide range of both hospital- and community-acquired infections that range from superficial skin and soft tissue infections to life-threatening toxic shock, pneumonia, endocarditis, and septicemia. The spectacular adaptive capacity of this pathogen has resulted in the worldwide emergence and spread of clonal strains that have acquired resistance to the majority of currently available antimicrobial agents, including vancomycin. Vancomycin used to be the first-line antibiotic for the therapy of infections due to methicillin-resistant S. aureus (MRSA). However, increased use of vancomycin led to the emergence of vancomycin-intermediate S. aureus (VISA) in 1996 (33).
The genetic basis for vancomycin resistance in VISA remains largely unknown. Ever since the emergence of VISA in 1996, a number of investigations have been carried out to discover the molecular mechanism involved in the generation of VISA. More than a dozen genes have been reported to be associated with glycopeptide resistance, including pbpB (58), pbpD (25, 60), sigB (2, 13, 46, 52), ddh (4, 49), tcaA (44), and walRK (37). We have also reported on 12 other genes (mgrA, msrA2, msrR, malR, lysC, graA, graB, graC, graD, graE, murZ, and rsbU) whose overexpression raised vancomycin resistance in S. aureus (13). Most of these reports have provided evidence for the correlation between gene transcriptional levels with altered glycopeptide susceptibilities of tested cells. Several sets of up- and downregulated genes associated with glycopeptide resistance, including global regulators, were also reported (13, 41, 51, 63). However, none of these reports identified the mutations responsible for the altered vancomycin susceptibility.
Recently, a number of mutations were found in some VISA clinical strains, such as Mu50 (42, 54, 55), JH9 (53), and JKD6008 (35, 36), by comparison of their genomes with those of their clinically relevant glycopeptide-susceptible parent strains. However, except for the mutations in vraSR and graRS, the role of the mutations found in these strains has not been established. The vraS I5N mutation has been proven to confer heterogeneous vancomycin resistance when it was introduced into a vancomycin-susceptible MRSA strain (38).
The graR N197S mutation was shown to convert the heterogeneous resistant VISA (hVISA) phenotype (MIC ≤ 2 mg/liter) of strain Mu3 into the VISA phenotype (MIC ≥ 4 mg/liter) when it was introduced into Mu3 (54). Furthermore, the plasmid-mediated overexpression of graR N197S but not that of intact graR could raise the vancomycin MIC of vancomycin-susceptible strain N315 (13). In addition, when we sequentially introduced the vraS and graR mutations into VSSA strain Mu50Ω (MICs of both vancomycin and teicoplanin = 0.5 mg/liter), a mutant expressing a VISA phenotype indistinguishable from that of Mu50 (vancomycin MIC = 6 mg/liter, teicoplanin MIC = 12 mg/liter) was obtained (16). These results clearly demonstrated that point mutations in the two regulatory genes of vraS and graR were sufficient, at least in a strain having the Mu50 genetic background, for expression of the VISA phenotype. The contribution of the graRS regulator mutation to raising vancomycin resistance in another VISA clinical strain has also been confirmed by Howden et al. by introducing the graS T136I mutation found in VISA strain JKD6008 into a vancomycin-susceptible S. aureus strain (36). It was noticed, however, that the vraSR and graRS mutations are not frequently found in VISA clinical strains (36, 39). This implies that there are genetic mechanisms other than vraSR-graRS mutations that achieve a VISA phenotype. In this study, we demonstrated that another set of regulator mutations comprises an alternative pathway to the VISA phenotype, i.e., walK, encoding a histidine kinase of walRK two-component regulatory system (TCRS), and clpP, encoding a proteolytic regulatory protein. We also found that walK mutations were carried by as many as 24 of 39 (61.5%) VISA clinical strains isolated from various countries.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
N315LR5P1 is a laboratory MRSA strain in which mecI gene function has been inactivated by a 62-bp deletion of nucleotide positions 130 to 191 and its penicillinase (PCase) plasmid has been eliminated (1, 30). It is a derivative of pre-MRSA strain N315, whose mecI gene is intact and methicillin resistance is not fully expressed (43). N315LR5P1 represents the genotype of hospital-acquired MRSA (HA-MRSA) in Japan whose mecI gene function is inactivated by mutations. The absence of the PCase plasmid from this strain makes it more pertinent as a model to trace the development of vancomycin-intermediate resistance in Japanese HA-MRSA strains, since the hVISA and VISA representative strains Mu3 and Mu50, respectively, lacked the PCase plasmid. N315LR5P1 was classified as sequence type 5-staphylococcal cassette chromosome mec type IIa, and its agr type is II. LR5P1-V3 is an N315LR5P1-derived VISA strain generated by serial passage of N315LR5P1 (vancomycin and teicoplanin MICs = 1 mg/liter) on brain heart infusion (BHI) plates containing vancomycin with gradually increasing concentrations of 1 to 4 mg/liter as described previously (65). All the strains were grown in BHI broth (Difco, Detroit, MI) at 37°C with aeration (shaking at 20 rpm without a CO2 supply), if not otherwise indicated. For each experiment, an overnight culture was diluted 100-fold in prewarmed fresh BHI broth and further incubated with aeration to ensure exponential growth condition before sampling. The cell growth was monitored by measuring the optical density of the culture at 578 nm (OD578) with a spectrophotometer (Phamacia LKB Biotechnology, Inc., Uppsala, Sweden).
Antimicrobial susceptibility testing.
The MICs for selected antimicrobials were determined for the constructed mutants and parent strains using Etest strips (AB Biodisk, Sweden). A sterile cotton swab was immersed in a 0.5 McFarland standard of tested bacterial culture and streaked on Mueller-Hinton (MH) agar and BHI agar plates. Antimicrobial strips were then applied 10 min after bacterial inoculation. Plates were then incubated at 37°C, and the MICs were read after 24 to 48 h of incubation. MICs of vancomycin and teicoplanin were also determined by both broth microdilution and agar dilution methods for vancomycin concentrations of 0.25, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 4.5, and 5.0 mg/liter and for teicoplanin at concentrations up to 16 mg/liter in 1-mg/liter increments. For daptomycin MIC determination, the test medium was supplemented with 50 mg/liter Ca2+ according to Clinical and Laboratory Standards Institute (CLSI) criteria (8).
Analysis of teicoplanin- and vancomycin-resistant subpopulations (population analysis).
To know how many subpopulations of each strain in a fixed number of cells (usually about 107 colonies) are resistant to various given concentrations of vancomycin and teicoplanin, population analysis was carried out as described previously (32). Briefly, overnight cultures of tested strains in BHI broth were adjusted to an OD578 of 0.3 (about 1.0 × 108 CFU). A series of 10-fold dilutions of these cell suspensions was then prepared, and 0.1 ml of each suspension was spread on BHI agar plates containing various concentrations of teicoplanin or vancomycin. Plates were incubated at 37°C for 48 h, and the number of cell colonies was counted. The number of resistant cells in 0.1 ml of the starting cell suspension was plotted against each corresponding teicoplanin or vancomycin concentration.
Whole-genome-sequence comparison of N315LR5P1 and LR5P1-V3.
Whole-genome-sequence comparison of VISA strain LR5P1-V3 and its vancomycin-susceptible parent strain, N315LR5P1, was performed using the array-based service (comparative genome sequencing [CGS]) provided by Roche NimbleGen, Inc. (Madison, WI), as described previously (17). Briefly, LR5P1-V3 (test) and N315LR5P1 (control) genome DNA samples were separately cleaved to form pools of low-molecular-weight fragments and labeled with the fluorescent dyes Cy3 and Cy5, respectively. The labeled samples were then competitively hybridized to two NimbleGen CGS whole-genome tiling arrays, which were generated with the S. aureus N315 genome sequence as a reference. The resulting hybridization signals were analyzed using NimbleScan (version 2.5) software, and the signal ratios of control versus test samples for all probes were plotted against the N315 genomic position. The locations of probes along the genome which have a significant ratio shift between test and control probes for both strands represent regions of possible sequence differences, including single nucleotide polymorphisms, deletions, sites of insertion, endpoints of inversion, or translocation. These locations were checked by PCR and resequencing of both test and control genomic DNA.
Generation of gene replacement mutants where an intended chromosome mutation(s) was introduced.
To introduce chromosomal mutations identified in a drug-resistant mutant into its drug-susceptible parent strain, pKOR1-mediated gene replacement was performed as described previously (16, 54). Briefly, the gene in frame containing the intended nucleotide sequence was amplified from sequence donor strain LR5P1-V3 containing walK or clpP mutations using primers that contain attB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-) and attB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGG-) sites on the respective up- and downstream sequences. This fragment was then cloned into pKOR1. Following cloning and ccdB selection in Escherichia coli, the constructed plasmid was then introduced into strain N315LR5P1 by electroporation using a GenePulser Xcell apparatus (Bio-Rad Laboratories, Inc., Hercules, CA) with the parameters described previously (43). Overnight culture of plasmid-carrying clones at 43°C selects for single-crossover mutants that carry both mutated and wild-type nucleotides. Single-crossover mutants were then cultured in drug-free broth to facilitate plasmid excision and subjected to anhydrotetracycline induction, whereby only non-plasmid-carrying mutants could survive. To check for successful introduction of the mutations, the resulting mutants were checked for the target sequences via sequencing. The colonies carrying the expected mutation that survived were set up as mutants, and those without the expected mutation were used as reverted controls as described previously (16, 17). By using this strategy, the chromosomal mutations, a deletion point mutation of walK, and a truncating mutation of clpP identified in drug-resistant strain LR5P1-V3 were introduced into the chromosome of drug-susceptible parent strain N315LR5P1.
Growth curve and autolysis assay.
Growth curve and autolysis experiments were carried out in succession. Overnight cultures of tested strains were diluted 1/1,000 in 10 ml fresh BHI broth and grown at 37°C with shaking at 25 rpm in a photorecording incubator (TN-2612; Advantec, Tokyo, Japan). The OD was monitored automatically every 2 min, and cells were grown to an OD600 of 1.2. Following this, the cells were placed on ice for 10 min and pelleted by centrifugation at 7,500 rpm for 5 min for autolytic activity determination. The cell pellets were then washed twice with chilled 0.05 M Tris-Cl buffer (pH 7.2) and suspended in 10 ml Tris-Cl (0.05 M, pH 7.2) containing 0.05% Triton X-100. The suspensions were then incubated in the same photorecording incubator at 37°C with continuous shaking at 25 rpm. The decrease of the OD was monitored, with recording of the OD values every 2 min for 24 h. For growth curve and doubling time determinations, the values of OD versus time of each strain were plotted. Doubling times were calculated as follows: [(t2 − t1) × log 2]/(log OD600 at t2 − log OD600 at t1), where t1 is the first sampling time and t2 is the second.
Nucleotide sequencing and mutation determination.
Chromosomal DNAs of 39 VISA strains (14), including 23 VISA strains from the network on antimicrobial resistance in S. aureus (NARSA; http://www.narsa.net/content/default.jsp), were extracted using phenol-chloroform purification methods after digestion of cells with 2.0 μg/ml lysostaphin (Wako, Japan), and the nucleotide sequences of vraSR, graRS, walRK, and clpP (from 150 bp upstream to 100 bp downstream of the open reading frames [ORFs]) were determined using an ABI 3730 DNA analyzer (Applied Biosystems).
Electron microscope evaluation of cell wall thickness.
Preparation of cells of the tested S. aureus strains for transmission electron microscopy and examination of the cells by transmission electron microscopy were performed as described previously (15). Morphometric evaluation of cell wall thickness was performed using photographic images at a final magnification of ×30,000. Thirty cells of each tested strain with nearly equatorial cut surfaces were measured for the evaluation of cell wall thickness, and results were expressed as mean values ± standard deviations (SDs).
RNA preparation and microarray analysis.
The preparation of microarray chips with the whole ORF of the S. aureus N315 chromosome and transcriptional profile analysis of the constructed mutants versus controls were carried out as described previously (13). Bacterial culture, RNA extraction, cDNA labeling, hybridization, and data analysis for microarray analysis were carried out according to protocols described previously (13, 17).
Statistical analysis of data.
The statistical significance of the data were evaluated with Student's t test.
Microarray data accession number.
Transcriptional profiles of walK and/or clpP mutant-related strains may be found in CIBEX under accession number CBX144.
RESULTS AND DISCUSSION
Whole-genome-sequence comparison between N315LR5P1 and LR5P1-V3.
To study the genetic mechanism of vancomycin resistance in S. aureus, we generated an in vitro VISA strain, LR5P1-V3, by stepwise vancomycin selection. The strain was derived from N315LR5P1, a teicoplanin- and vancomycin-susceptible laboratory MRSA strain (1, 30). The raised vancomycin MICs of LR5P1-V3 were 4 mg/liter in MH medium and 8 mg/liter in BHI medium, compared to 1 mg/liter each for N315LR5P1 (Table 1). Besides decreased susceptibility to vancomycin, the strain showed typical VISA phenotypes; i.e., thickened cell wall, slow growth, and decreased autolytic activity (14) (see Fig. 3 and 4). Sequencing of the vraSR and graRS genes of LR5P1-V3 revealed no mutation, indicating that LR5P1-V3 has a genetic mechanism distinct from that of Mu50 for the raised vancomycin resistance. This prompted us to carry out whole-genome-sequence comparison between strains LR5P1-V3 and N315LR5P1 to identify genes responsible for the raised vancomycin resistance.
Table 1.
Antibiotic susceptibilities and doubling times of S. aureus strains used in this study
| Strain | MIC (mg/liter)a |
Doubling time (min) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BHI medium |
MH medium |
|||||||||||||
| VCM | TEIC | VCM | TEIC | LZD | DPC | IPM | BA | TC | MINO | CP | MUP | LVFX | ||
| N315LR5P1 | 1.5 (1, 1) | 1.5 (1, 1) | 0.75 (1, 1) | 0.75 (1, 1) | 1.5 | 0.5 | >32 | 48 | 0.38 | 0.25 | 8 | 0.38 | 0.19 | 35.62 |
| LR5P1-V3 | 4 (8, 8) | 12 (8, 8) | 3 (4, 4) | 4 (4, 8) | 0.75 | 1 | >32 | 96 | 0.064 | 0.032 | 6 | 0.125 | 0.19 | 44.16 |
| LR5P1walK | 1.5 (1, 1) | 2 (1, 1) | 0.75 (1, 1) | 0.75 (1, 1) | 1.5 | 0.38 | >32 | 64 | 0.38 | 0.25 | 8 | 0.38 | 0.19 | 35.90 |
| LR5P1walK* | 3 (4, 4) | 6 (4, 4) | 2 (2, 2) | 3 (2, 2) | 1.5 | 1 | >32 | 64 | 0.38 | 0.25 | 8 | 0.38 | 0.19 | 37.20 |
| LR5P1clpP | 1.5 (1, 1) | 1.5 (1, 1) | 0.75 (1, 1) | 0.75 (1, 1) | 1.5 | 0.25 | >32 | 48 | 0.38 | 0.25 | 8 | 0.38 | 0.19 | 36.30 |
| LR5P1clpP* | 2 (2, 2) | 3 (4, 4) | 1.5 (2, 2) | 2 (2, 2) | 0.75 | 0.5 | >32 | 48 | 0.094 | 0.032 | 6 | 0.125 | 0.19 | 43.23 |
| LR5P1walK*clpP | 2 (1, 1) | 3 (1, 1) | 1 (1, 1) | 2 (1, 1) | 1 | 0.5 | >32 | 64 | 0.25 | 0.25 | 8 | 0.38 | 0.19 | 38.25 |
| LR5P1walK*clpP* | 4 (4, 4) | 12 (8, 8) | 2.5 (4, 4) | 4 (4, 4) | 0.75 | 1 | >32 | 64 | 0.047 | 0.032 | 6 | 0.125 | 0.19 | 45.29 |
MICs were determined by Etest (Sysmex bioMérieux Co., Ltd.), broth microdilution, and agar dilution methods. The results were read after 24 h incubation at 37°C. Since some strains had slow growth, vancomycin and teicoplanin MICs were read after 48 h incubation to improve accuracy. The data in parentheses indicate MICs determined by broth microdilution, agar dilution. Abbreviations: VCM, vancomycin; TEIC, teicoplanin; LZD, linezolid; DPC, daptomycin; IPM, imipenem; BA, bacitracin; TC, tetracycline; MINO, minocycline; CP, chloramphenicol; MUP, mupirocin; LVFX, levofloxacin.
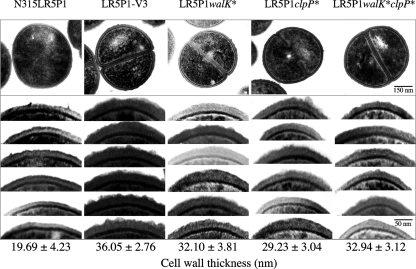
Fig. 3.
Transmission electron microscopy of the gene replacement mutants and their control strains. Transmission electron microscopy was carried out on gene replacement mutants LR5P1walK* and LR5P1clpP*, which were generated from N315LR5P1 by substitution of its walK and clpP genes with those of LR5P1-V3, respectively. The LR5P1walK*clpP* mutant was generated from LR5P1walK* by substitution of its clpP gene with that of LR5P1-V3. The values given under each picture are the means and standard deviations of each strain's cell wall thickness. Note that all gene replacement cells had thick cell walls compared to those of the parent strain. Magnifications, ×30,000.
Fig. 4.
Growth curves (A) and autolytic activities (B) of gene replacement mutants and their control strains. Note that strains with the clpP* mutation (LR5P1-V3, LR5P1clpP*, and LR5P1walK*clpP*) had significantly slower growth than the parent strain, and strains with dual deletion mutations of clpP* and walK* (LR5P1-V3 and LR5P1walK*clpP*) had autolytic activities remarkably different from the activity of the parent strain. Data shown are means of duplicate determinations.
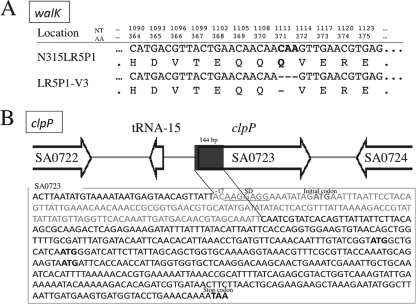
We employed the NimbleGen tiling array to compare the whole-genome sequences of the two strains as reported previously (17). Two mutations in separate genes were identified in LR5P1-V3 compared to the sequence in parent strain N315LR5P1. The first, a deletion mutation, was found in two-component regulatory system walRK, where 3 nucleotides (CAA) from positions 1111 to 1113 were deleted from the walK gene, which encodes a histidine kinase, WalK. The mutation, designated walK*, which did not cause a shift in the reading frame, resulted in the loss of a single amino acid, glutamine (Q), from the intact WalK protein (Fig. 1A). The other mutation was a deletion of clpP, where a 144-bp DNA fragment located at base positions −17 to 127 of clpP was deleted. Amino acid sequence alignment with the sequence in the conserved domain database (45) was carried out and found that the functional core motif was located at the N terminus of ClpP. Since the deletion contained the translation initiation codon ATG and a part of the promoter region of clpP and some possible initial codons existed, we predict that the gene function probably remained but was inactivated (Fig. 1B). The mutation was designated clpP*.
Fig. 1.
VISA strain LR5P1-V3 harbors two mutations on its chromosome. Whole-genome-sequence comparison of VISA LR5P1-V3 and its vancomycin-susceptible parent strain, N315LR5P1, revealed two mutations: a deletion mutation in walK and a truncating mutation in clpP of the LR5P1-V3 chromosome. (A) walK deletion mutation. Three nucleotides (NT; CAA, positions 1111 to 1113) of walK were deleted, resulting in an amino acid (AA) deletion of Q371. (B) clpP truncating mutation. A total of 144 nucleotides, located at positions −17 to 127 of clpP, were deleted in LR5P1-V3, which results in a clpP truncation. Putative initial codons for translation are in boldface.
Evaluation of effects of walK and clpP mutations on glycopeptide resistance.
To evaluate the roles of the walK and clpP mutations in raising LR5P1-V3 vancomycin resistance, the mutations were introduced one by one into the chromosome of parent strain N315LR5P1 by a gene replacement method (16, 54). The obtained mutants were then studied for phenotypic changes, including susceptibility to vancomycin and teicoplanin.
(i) Effect of walK mutation.
We first introduced the walK deletion mutation, walK*, into the N315LR5P1 chromosome, obtaining the strain with the gene replacement, LR5P1walK*, and a revertant strain, LR5P1walK. The latter strain had the intact walK gene that was recovered from the gene replacement procedure (see Materials and Methods). Revertants were obtained in every gene replacement experiment and used as isogenic control strains throughout this study. Antibiotic susceptibilities for the constructed mutants were evaluated by determination of the MICs of various antibiotics, including vancomycin and teicoplanin. In order to accurately evaluate the susceptibility changes, MIC determinations for glycopeptides were carried out by Etest, broth microdilution, and agar dilution methods. A significant increase in vancomycin and teicoplanin MICs was detected between parent strain N315LR5P1 and mutant strain LR5P1walK* in both BHI medium and MH medium, while no such difference was observed between the parent and revertant strain LR5P1walK (Table 1). However, the level of glycopeptide resistance of LR5P1walK* still fell short of that of LR5P1-V3, indicating that the walK* mutation alone was not enough to express the same level of glycopeptide resistance as that of LR5P1-V3.
(ii) Effect of clpP mutation.
Since the walK* mutation was not sufficient for N315LR5P1 to attain a level of glycopeptide resistance similar to that of LR5P1-V3, we suspected that the clpP mutation detected in our genome comparison study is important in contributing toward the glycopeptide resistance detected in LR5P1-V3. The clpP mutation was first tested for an independent effect by introducing clpP* into N315LR5P1, obtaining strain LR5P1clpP*. Antibiotic susceptibility tests showed that the mutant had increased vancomycin and teicoplanin MICs compared to those of parent strain N315LR5P1 and revertant strain LR5P1clpP (Table 1). However, this increase in glycopeptide resistance was smaller than that as a result of the effect of the single walK mutation (Table 1). In other words, on its own, the clpP* mutation does raise glycopeptide resistance, but the other mutation was clearly needed for N315LR5P1 to attain the level of glycopeptide resistance of LR5P1-V3.
(iii) Combination effect of walK* and clpP* on glycopeptide resistance.
With the results of the experiments described above, we speculated that both the walK* and clpP* mutations are required for achieving LR5P1-V3 glycopeptide resistance. To test this hypothesis, the clpP* mutation was additionally introduced into the chromosome of LR5P1walK*, obtaining the double mutant strain LR5P1walK*clpP*. As expected, the glycopeptide MIC values of the double mutant strain attained levels of glycopeptide resistance comparable to that of LR5P1-V3, though interestingly, revertant strain LR5P1walK*clpP had a reduced level of glycopeptide resistance compared to that of LR5P1walK* (Table 1). LR5P1-V3 possessed a characteristic resistance pattern with a much higher teicoplanin MIC (12 mg/liter) than vancomycin MIC (4 mg/liter) in BHI medium (Table 1). This feature of resistance was also retained by the double mutant strain (Table 1).
Though LR5P1-V3 recorded an MIC of 4 mg/liter for vancomycin, which is the criterion of VISA (9), its population analysis profile (PAP) showed a profile which is typical of heterogeneous-type resistance (Fig. 2A). The single mutant strains LR5P1walK* and LR5P1clpP* had PAPs that were between those of N315LR5P1 and LR5P1-V3. LR5P1walK* possessed much larger resistant subpopulations than LR5P1clpP* for each concentration of vancomycin and teicoplanin. In agreement with the MIC results, the PAP of double mutant strain LR5P1walK*clpP* was similar to that of LR5P1-V3. The PAP of LR5P1walK*clpP* against teicoplanin was almost identical to that of LR5P1-V3, even at the higher teicoplanin concentrations of 8 to 12 mg/liter (Fig. 2B). Taken together, the glycopeptide resistance of LR5P1-V3 was shown to be caused by the combined effects of the two mutations, walK* and clpP*.
Fig. 2.
Analysis of vancomycin- and teicoplanin-resistant subpopulations of gene replacement mutants and their isogenic controls. The data presented are representative of data from three experiments. The numbers of colonies on plates containing various concentrations of vancomycin (A) and teicoplanin (B) were counted after 48 h of incubation at 37°C.
(iv) Truncation mutation of clpP confers susceptibility to protein synthesis inhibitor antibiotics.
Table 1 shows that not only the susceptibilities to glycopeptides but also those to other antibiotics were affected by the introduction of the mutations. Daptomycin and bacitracin MICs were slightly increased in LR5P1-V3. This seemed to be associated with the introduction of walK* but not of clpP*, as far as this can be judged from the data presented in Table 1. More significant changes were observed with protein synthesis inhibitor antibiotics. MICs of linezolid, tetracycline, minocycline, chloramphenicol, and mupirocin for the mutants were significantly decreased with the introduction of the clpP* mutation (Table 1). The reason for these changes, which were probably due to the loss of clpP gene function, is currently unknown. However, a possible explanation for the increased susceptibility to protein synthesis inhibitor antibiotics is that a loss or decrease of ClpP function results in an impaired survival ability of the clpP* mutants through the anaerobic respiratory pathway. ClpP is essential for the growth and survival of S. aureus through the anaerobic respiratory pathway (48), and full expression of the anaerobic respiratory pathway contributes to resistance to protein synthesis inhibitor antibiotics (50).
walK* and clpP* mutations caused cell wall thickening.
A thickened cell wall has been reported to be a common phenotypic feature of clinical VISA strains (11, 14, 31). The feature is closely associated with the peptidoglycan-clogging theory that explains vancomycin resistance by the difficulty and delay of passage of vancomycin molecules across the thickened peptidoglycan layers (12). We considered that either one of the mutations, walK* or clpP*, would cause thickening of the host cell wall. This hypothesis was tested by transmission electron microscopy of the mutant strains (Fig. 3). The glycopeptide-susceptible parent strain N315LR5P1 had a cell wall thickness of 19.69 ± 4.23 nm. Introduction of the walK* mutation in N315LR5P1 caused the cell wall to be 1.63 times thickener. On the other hand, introduction of the clpP* mutation caused the cell wall to be 1.48 times thicker (Fig. 3), while the revertant mutants had the same cell wall thickness as the parent strain (data not show). The double mutant LR5P1walK*clpP* had a cell wall 1.67 times thicker than that of N315LR5P1. The cell wall of the double mutant strain was the thickest among the strains with gene replacements. However, the cell wall of LR5P1-V3 was still thicker than that of the double mutant strain (36.05 versus 32.94 nm). The reason for this discrepancy is unknown at the moment.
Slow growth and decreased autolysis of the walK and clpP mutants.
Next, the growth rates of the mutant strains were compared with the growth rate of the parent strain. The doubling time and growth curve of mutant strain LR5P1walK* were similar to those of parent strain N315LR5P1 (Table 1; Fig. 4A). On the other hand, introduction of the clpP* mutation into N315LR5P1, whether singly or in combination with the walK* mutation, significantly prolonged the doubling time of the cell, as shown in Table 1. Therefore, the slow-growth phenotype of LR5P1-V3 was found to be due to the effect of the clpP* mutation. Slow growth has repeatedly been reported in VISA strains and is considered one of the important features of the VISA phenotype (10, 14, 18, 27, 59). Nevertheless, the contribution of slow growth to S. aureus glycopeptide resistance remains controversial. We recently studied a laboratory-derived hVISA strain with decreased susceptibility to both vancomycin and daptomycin (6). The strain had mutations in rplV and rplC (which code for 50S ribosomal proteins L22 and L3, respectively), which were considered to be the reason for the remarkably prolonged growth rate of the strain. However, this slow growth was not associated with decreased susceptibility to vancomycin and daptomycin (17). Therefore, the slow growth of LR5P1-V3 is not necessarily a factor contributing to the raised glycopeptide resistance. In this study, the slow growth and raised glycopeptide resistance were inseparable features of the clpP* mutation; however, the direct contribution of slow growth to vancomycin resistance could still not be evaluated comprehensively.
Decreased autolytic activity is one of the features of VISA (3, 29). Our present study also demonstrated a correlation between the levels of autolytic activity and glycopeptide resistance (Fig. 4B). Glycopeptide-susceptible parent strain N315LR5P1 and LR5P1walK had the highest autolytic activity, while LR5P1-V3 had the lowest. The mutants LR5P1walK* and LR5P1clpP* had autolytic activities slightly lower than those of N315LR5P1 and LR5P1walK. The double mutant strain LR5P1walK*clpP* exhibited a significantly reduced autolytic activity comparable to that of LR5P1-V3.
Microarray transcriptional analysis.
In the above experiments, the walK* and clpP* mutations identified in the laboratory VISA LR5P1-V3 strain were shown to contribute to vancomycin resistance; however, the impact of these mutations on the regulatory function toward vancomycin resistance remains to be clarified. On the basis of this consideration, we carried out genome-scale transcriptional profiling, comparing the mutants from gene replacement or drug selection with their parent strains. Microarray analysis was carried out for a total of four combinations of strains: LR5P1walK* versus N315LR5P1, LR5P1clpP* versus N315LR5P1, LR5P1walK*clpP* versus N315LR5P1, and LR5P1-V3 versus N315LR5P1. All pairs consisted of the vancomycin-resistant mutant and its vancomycin-susceptible parent strain. Analyses of the first two pairs revealed the effect of the individual walK* and clpP* mutations, while analyses of the last two elucidate the combination effect of the walK* and clpP* mutations. All transcriptional profiles and comparison data are available on the CIBEX site under accession number CBX144. Table 2 shows the extracted representative data. The results revealed significant genome-wide differences in expression patterns between the mutants and parent strains. As shown in Table 2, significant differences in gene expression of the four vancomycin-resistant mutants and vancomycin-susceptible parent strain N315LR5P1 were repression of the metabolic pathways of galactose (lac operon), succinate dehydrogenase, the efeUMN operon, and aminoacyl-tRNA biosynthesis and enhancement of the metabolic pathways of urease formation, the cydAB operon, the agr locus, and many ABC transporters. The enhanced expression of the sirABC, ahp, clpC, and opuC operons was commonly seen in mutants with clpP* (Table 2). A calculation of genes with 2-fold alterations in the transcriptional level showed that, compared to the sequence of parent strain N315LR5P1, there were a total of 39 and 28 genes down- and upregulated, respectively, in mutant strain LR5P1walK*, into which walK* was introduced (see Tables S1 and S2 at http://www.jj.em-net.ne.jp/∼longzhu/data/pub1/table_s1-s4.pdf), while there were 56 and 141 genes (Table S3 and S4) down- and upregulated, respectively, in mutant LR5P1clpP*, into which the clpP* mutation was introduced, revealing a broad regulatory impact of these mutations on bacterial physiology (see Tables S1 to S4).
Table 2.
Representatives of genes differentially expressed in mutants with walK* and/or clpP* compared to that in parent strain N315LR5P1
| Function and ORF | Gene | Product | Ratio of signal intensitya |
|||
|---|---|---|---|---|---|---|
| LR5P1walK*/N315LR5P1 | LR5P1clpP*/N315LR5P1 | LR5P1walK*clpP*/N315LR5P1 | LR5P1-V3/N315LR5P1 | |||
| Galactose metabolism | ||||||
| SA1991 | lacG | 6-Phospho-beta-galactosidase | 0.44 | 0.24 | 0.20 | 0.20 |
| SA1992 | lacE | PTSb system lactose-specific IIBC component | 0.32 | 0.14 | 0.11 | 0.10 |
| SA1993 | lacF | PTS system, lactose-specific IIA component | 0.35 | 0.17 | 0.13 | 0.11 |
| SA1994 | lacD | Tagatose-1,6-diphosphate aldolase | 0.42 | 0.21 | 0.18 | 0.15 |
| SA1995 | lacC | Tagatose-6-phosphate kinase | 0.36 | 0.18 | 0.14 | 0.12 |
| SA1996 | lacB | Galactose-6-phosphate isomerase LacB subunit | 0.41 | 0.19 | 0.17 | 0.14 |
| SA1997 | lacA | Galactose-6-phosphate isomerase LacA subunit | 0.56 | 0.24 | 0.26 | 0.18 |
| SA2288 | gtaB | Cell envelope, UTP-glucose-1-phosphate uridylyltransferase | 0.62 | 0.63 | 0.66 | 0.85 |
| efeUMN operon | ||||||
| SA0331 | efeM | Putative lipoprotein | 0.25 | 0.26 | 0.43 | 0.35 |
| SA0332 | efeN | Dyp-type peroxidase family protein | 0.23 | 0.27 | 0.43 | 0.35 |
| SA0333 | efeU | Ferrous iron transport permease EfeU | 0.24 | 0.28 | 0.37 | 0.34 |
| Succinate dehydrogenase | ||||||
| SA0963 | pycA | Pyruvate carboxylase | 0.65 | 0.41 | 0.43 | 0.39 |
| SA0994 | sdhC | Succinate dehydrogenase, cytochrome b-558 subunit | 1.18 | 0.49 | 0.45 | 0.49 |
| SA0995 | sdhA | Succinate dehydrogenase flavoprotein subunit | 1.01 | 0.38 | 0.35 | 0.40 |
| SA0996 | sdhB | Succinate dehydrogenase iron-sulfur subunit 8 | 0.97 | 0.44 | 0.42 | 0.47 |
| Aminoacyl-tRNA biosynthesis | ||||||
| SA0009 | serS | Seryl-tRNA synthetase | 0.93 | 1.07 | 1.06 | 0.94 |
| SA0448 | metS | Methionyl-tRNA synthetase | 0.69 | 0.73 | 0.78 | 0.74 |
| SA0475 | lysS | Lysyl-tRNA synthetase | 1.10 | 1.12 | 1.11 | 1.00 |
| SA0486 | gltX | Glutamyl-tRNA synthetase | 1.37 | 1.71 | 1.81 | 1.55 |
| SA0488 | cysS | Cysteinyl-tRNA synthetase | 0.94 | 0.85 | 0.96 | 0.91 |
| SA0564 | argS | Arginyl-tRNA synthetase | 0.55 | 0.67 | 0.66 | 0.66 |
| SA0855 | trpS | Tryptophanyl-tRNA synthetase | 0.57 | 0.66 | 0.69 | 0.65 |
| SA0985 | pheS | Phenylalanyl-tRNA synthetase subunit alpha | 0.81 | 0.67 | 0.70 | 0.70 |
| SA0986 | pheT | Phenylalanyl-tRNA synthetase subunit beta | 0.95 | 0.74 | 0.77 | 0.80 |
| SA1036 | ileS | Isoleucyl-tRNA synthetase | 0.71 | 0.69 | 0.73 | 0.78 |
| SA1106 | proS | Prolyl-tRNA synthetase | 1.18 | 0.95 | 0.96 | 0.94 |
| SA1287 | asnC | Asparaginyl-tRNA synthetase | 0.79 | 0.78 | 0.93 | 0.79 |
| SA1446 | alaS | Alanyl-tRNA synthetase | 0.81 | 0.65 | 0.70 | 0.64 |
| SA1456 | aspS | Aspartyl-tRNA synthetase | 0.94 | 0.86 | 0.89 | 0.77 |
| SA1457 | hisS | Histidyl-tRNA synthetase | 0.81 | 0.80 | 0.76 | 0.73 |
| SA1488 | valS | Valyl-tRNA synthetase | 0.82 | 0.64 | 0.65 | 0.62 |
| SA1506 | thrS | Threonyl-tRNA synthetase | 0.72 | 0.63 | 0.64 | 0.63 |
| SA1550 | tyrS | Tyrosyl-tRNA synthetase | 0.59 | 0.62 | 0.72 | 0.56 |
| SA1563 | ytpR | Phenylalanyl-tRNA synthetase protein | 0.86 | 0.81 | 0.84 | 0.70 |
| SA1579 | leuS | Leucyl-tRNA synthetase | 0.73 | 0.64 | 0.68 | 0.64 |
| SA1715 | gatB | Aspartyl-/glutamyl-tRNA amidotransferase subunit B | 1.14 | 0.92 | 0.96 | 0.77 |
| SA1716 | gatA | Aspartyl-/glutamyl-tRNA amidotransferase subunit A | 0.89 | 0.77 | 0.76 | 0.64 |
| SA1717 | gatC | Aspartyl-/glutamyl-tRNA amidotransferase subunit C | 0.87 | 0.76 | 0.77 | 0.61 |
| Urease formation | ||||||
| SA2081 | Urea transporter | 1.32 | 2.11 | 2.51 | 3.11 | |
| SA2082 | ureA | Amino acid biosynthesis (urease subunit gamma) | 1.45 | 2.70 | 2.93 | 4.38 |
| SA2083 | ureB | Amino acid biosynthesis (urease subunit beta) | 1.49 | 2.74 | 2.95 | 4.23 |
| SA2084 | ureC | Urease subunit alpha | 1.44 | 2.59 | 2.78 | 4.19 |
| SA2085 | ureE | Urease accessory protein UreE | 1.49 | 2.59 | 2.77 | 3.99 |
| SA2086 | ureF | Urease accessory protein UreF | 1.50 | 2.50 | 2.73 | 4.06 |
| SA2087 | ureG | Urease accessory protein UreG | 1.46 | 2.39 | 2.52 | 3.48 |
| SA2088 | ureD | Urease accessory protein UreD | 1.52 | 2.85 | 2.97 | 4.29 |
| cydAB operon | ||||||
| SA0937 | cydA | Cytochrome d ubiquinol oxidase subunit I protein | 2.37 | 3.61 | 3.25 | 3.35 |
| SA0938 | cydB | Cytochrome d ubiquinol oxidase subunit II protein | 2.39 | 3.62 | 3.42 | 3.52 |
| SA0939 | ktrC | TrkA potassium uptake family protein | 1.73 | 2.23 | 2.15 | 2.14 |
| agr locus | ||||||
| SA1842 | agrB | Accessory gene regulator protein B | 1.66 | 2.52 | 1.98 | 1.44 |
| SAS066 | agrD | Accessory gene regulator protein D | 1.50 | 2.23 | 1.77 | 1.31 |
| SA1843 | agrC | Accessory gene regulator protein C | 1.61 | 2.34 | 1.83 | 1.30 |
| SA1844 | agrA | Accessory gene regulator protein A | 1.55 | 2.12 | 1.81 | 1.24 |
| sirABC operon | ||||||
| SA0110 | sirB | Siderophore compound ABC transporter permease protein | 1.04 | 2.55 | 2.02 | 1.85 |
| SA0111 | sirA | Iron compound ABC transporter | 1.09 | 4.31 | 3.61 | 3.00 |
| SA0112 | sbnA | Pyridoxal-5′-phosphate-dependent enzyme, beta subunit | 1.05 | 3.03 | 2.76 | 2.23 |
| SA0113 | ocd | Ornithine cyclodeaminase protein | 1.36 | 3.42 | 3.16 | 2.53 |
| SA0114 | sbnC | IucA/IucC family siderophore biosynthesis protein | 1.24 | 2.77 | 2.68 | 2.12 |
| ahp operon and nfrA | ||||||
| SA0365 | ahpF | Alkyl hydroperoxide reductase subunit F | 0.92 | 3.91 | 4.62 | 4.66 |
| SA0366 | ahpC | Alkyl hydroperoxide reductase subunit C | 0.89 | 3.24 | 3.58 | 3.51 |
| SA0367 | nfrA | NADPH-dependent oxidoreductase | 1.15 | 2.46 | 2.61 | 2.32 |
| clpC operon | ||||||
| SA0480 | ctsR | Transcriptional regulator of class III stress genes | 0.57 | 4.24 | 4.41 | 4.42 |
| SA0481 | mcsA | UvrB/UvrC motif-containing protein | 0.53 | 3.98 | 4.31 | 4.43 |
| SA0482 | mcsB | ATP guanido phosphotransferase | 0.53 | 4.19 | 4.52 | 4.53 |
| SA0483 | clpC | Putative stress response-related Clp ATPase | 0.94 | 1.07 | 1.11 | 1.00 |
| ABC transporters | ||||||
| SA0109 | sirC | Siderophore compound ABC transporter permease protein SirC | 0.89 | 1.78 | 1.56 | 1.40 |
| SA0110 | sirB | Siderophore compound ABC transporter permease protein SirB | 1.04 | 2.55 | 2.02 | 1.85 |
| SA0111 | sirA | Iron compound ABC transporter | 1.09 | 4.31 | 3.61 | 3.00 |
| SA0209 | yvfM | Maltose/maltodextrin transport permease protein | 1.47 | 1.77 | 1.45 | 1.23 |
| SA0229 | appAA | Nickel ABC transporter nickel-binding protein | 1.73 | 2.80 | 2.79 | 2.02 |
| SA0639 | cydC | ABC transporter ATP-binding protein | 1.50 | 1.98 | 1.89 | 1.85 |
| SA0640 | cydD | ABC transporter ATP-binding protein | 1.77 | 2.05 | 2.05 | 1.98 |
| SA0677 | opuBA | Glycine betaine ABC transporter ATP-binding protein | 1.28 | 1.85 | 1.54 | 1.74 |
| SA0891 | Hypothetical protein, similar to ferrichrome ABC transporter | 1.16 | 1.79 | 1.67 | 1.17 | |
| SA1213 | opp-2C | Putative oligopeptide transporter membrane permease | 1.33 | 1.80 | 1.87 | 1.97 |
| SA1214 | opp-2B | Putative oligopeptide transporter membrane permease | 1.48 | 2.28 | 2.02 | 2.32 |
| SA1979 | yhfQ | Iron ABC transporter binding protein, | 0.82 | 1.67 | 1.94 | 2.35 |
| SA2234 | opuCD | Glycine betaine/carnitine/choline ABC transporter membrane | 1.02 | 2.14 | 2.02 | 2.40 |
| SA2235 | opuCC | Glycine betaine/carnitine/choline ABC transporter binding | 1.04 | 2.17 | 2.28 | 2.54 |
| SA2236 | opuCB | Glycine betaine/carnitine/choline ABC transporter opuCB | 1.08 | 2.22 | 2.22 | 2.52 |
| SA2237 | opuCA | Glycine betaine/carnitine/choline ABC transporter ATP-bindi | 1.21 | 1.86 | 2.16 | 2.24 |
| clpP gene SA0723 | clpP | ATP-dependent Clp protease proteolytic subunit | 1.04 | 0.44 | 0.45 | 0.44 |
| walRK system | ||||||
| SA0017 | walR | Two-component response regulator | 0.86 | 1.34 | 1.29 | 1.14 |
| SA0018 | walK | Two-component sensor histidine kinase | 0.87 | 1.26 | 1.25 | 1.10 |
Ratios of signal intensity of strains with mutations to that of the isogenic parent strain.
PTS, phosphotransferase.
S. aureus is one of the organisms known to exclusively use enzymes of the d-tagatose-6-phosphate pathway (encoded by the lac operon) to metabolize lactose and d-galactose (47). In the lac operon, the lacABCD genes comprise the d-tagatose-6-phosphate pathway and are cotranscribed with the lacFEG genes, which specify proteins for transport and metabolism of lactose and galactose in S. aureus (47). Although the impact of the downregulated lac operon on the vancomycin-resistant phenotype still remains to be clarified, the downregulated lac operon in vancomycin-resistant strains was also observed in our previous study that was carried out with an hVISA strain with an rpoB mutation-mediated heteroresistance (17), suggesting that decreased lactose/galactose metabolism is associated with vancomycin resistance in S. aureus. The mutual biological relationship of the vancomycin-resistant phenotype with the genes or operons that were differentially expressed in vancomycin-resistant strains remains to be studied.
Impact of walRK mutation on vancomycin-resistant phenotypes.
walRK (vicRK) was originally identified as yycFG in Bacillus subtilis (23, 26) to be an essential two-component system for the viability of cell growth (23, 26). Recently, Dubrac et al. stressed the importance of the system in cell wall metabolism in S. aureus and proposed renaming the system walRK (20). Two reports have described the correlation of the walRK system with the phenotype of vancomycin intermediate resistance in S. aureus. Jansen et al. reported that walRK was highly upregulated due to an insertion mutation in the walRK promoter in a VISA clinical isolate (37). Mwangi et al. found a mutation in the yycH gene in a clinical VISA strain (53). The yycH product is reported to downregulate the walRK system in B. subtilis (62). Thus, both studies have suggested that the increment of vancomycin resistance was mediated by activation of the walRK system. Nevertheless, in our study, we did not find any significant change (i.e., a more or less than 2-fold change) in the expression of walRK in any of our resistant mutants (Table 2; see Table S1 and Table S2 at http://www.jj.em-net.ne.jp/∼longzhu/data/pub1/table_s1-s4.pdf). The causes of raised resistance due to the walK* mutation remain unknown. From the microarray data, we could neither estimate the biological function of walK* nor confirm the activation or deactivation of WalRK due to introduction of walK*, since there was only one gene (SA0710) downregulated by walK* introduction out of nine genes that have been reported to be directly regulated by the walRK system (21, 22). Recently, Delaune et al. reported on the effect of walRK on cell morphology, showing that walRK depletion could raise the cell wall thickness of S. aureus (19). As the mutant into which walK* was introduced, LR5P1walK*, had a thickened cell wall, it might be reasonable to consider that walK* might hamper the function of the walRK system, but its regulatory pathway toward cell wall thickening remains to be studied.
Impact of clpP mutation on vancomycin-resistant phenotypes.
ClpP is a proteolytic subunit of the ATP-dependent Clp protease, which consists of an ATPase specificity factor (ClpA or ClpX in E. coli; ClpX, ClpC, or ClpE in Bacillus subtilis) and a proteolytic domain (ClpP) (28). The global regulatory impact of ClpP on the virulence, stress response, and physiology of S. aureus was well documented by Michel et al. by transcriptional profile analysis using a clpP deletion mutant (48). Under normal growth conditions, the clpP deletion mutant showed a growth defect affecting the expression of many regulatory genes, such as agr, sigB, sarT, and arlRS. walRK was also reported to be upregulated by clpP deletion. Deletion of clpP in NTCT8325 caused accelerated autolysis (48). However, our findings with N315LR5P1 were contradictory to the previous ones. Introduction of mutated clpP, either singly or with the walK mutation, decreased autolysis of N315LR5P1 (Fig. 4). This signifies that the physiological role of clpP may differ from strain to strain, depending on their genetic backgrounds, and this is supported by our microarray data. Comparing the transcriptional profile of LR5P1clpP* with that of the NTCT8325 mutant with the clpP deletion that was published by Michel et al (48), we found that only 44 out of 137 genes that were differentially expressed according to the data of Michel et al. were also similarly up- or downregulated in LR5P1clpP* (data not shown).
Microarray analysis of mutant LR5P1clpP* into which clpP* was introduced showed quite a number of differentially expressed genes compared to the number in the walK* mutants (see Tables S3 and S4 at http://www.jj.em-net.ne.jp/∼longzhu/data/pub1/table_s1-s4.pdf). Among the genes which were differentially expressed include those involved in cell envelope and cellular processes, intermediary metabolism, and various regulatory functions, e.g., sarA, sarX, norR, malR, the agr locus, yuaC, ctsR, ccrA, sarH1, lytR, ydcH, and sarV, revealing a strong regulatory impact of ClpP on the expression of genes. Regarding the role of regulation of clpP* in raising vancomycin resistance, the explanations for differentially expressed genes in the clpP* mutants are speculative at this point; however, we suspect that the clpP* mutation alters metabolic pathways of the whole cell by reducing protein biosynthesis and accumulation of cell wall material, directing the cell toward vancomycin resistance. Consistent with this hypothesis, our microarray data showed lowered expression of almost all of aminoacyl-tRNA biosynthesis genes and enhancement of many ABC transporters, including the opuC operon, which is related to cell wall and membrane envelope biogenesis (Table 2). tagE, encoding a poly(glycerol-phosphate) alpha-glucosyltransferase that is one of the important enzymes in the pathway of teichoic acid biosynthesis, and some operons related to the aerobic respiratory chain, such as cydAB in all clpP* mutants, were also highly expressed (Table 2; see Table S4 at http://www.jj.em-net.ne.jp/∼longzhu/data/pub1/table_s1-s4.pdf). The strong influence of the truncated clpP mutation on transcription of regulators observed in this study suggests that ClpP proteolytic activity may serve as an important mechanism to control gene expression in S. aureus.
Prevalence of vraSR, graRS, clpP, and walRK mutations among VISA clinical strains.
To test if walK and clpP mutations are naturally occurring genetic events, we carried out a study of the prevalence of walRK and clpP mutations among a worldwide collection of clinical VISA strains. A total of 39 clinical VISA strains isolated from various countries were subjected to nucleotide sequencing of vraSR, graRS, clpP, and walRK. The mutations of the first two operons are causative of the Mu50 VISA phenotype (16, 54). Eight VISA strains, including Mu50, were found to harbor mutations in vraS, while the graR mutation was identified in only four strains, i.e., Mu50, HIP5836, HIP10267, and HIP13057 (Table 3). The clpP mutations were rare and found only in strains 99/3759-V, HIP07920, and HIP09313. All clpP mutations were accompanied by single amino acid substitutions and not by huge deletions, as seen in LR5P1-V3. On the other hand, walK mutations were found in as many as 61.5% of the VISA strains (Table 3). None of the four tested genes was mutated in 9 of the 39 VISA strains (23.1%). Therefore, there might still be alternative genetic pathways for generation of the VISA phenotype in nature.
Table 3.
Mutations of the genes associated with vancomycin resistance in clinical VISA strainsa
| Strain | NARSA no. | Vancomycin MIC (mg/liter) | Isolation information |
Amino acid substitution(s)d |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yr | Country | Reference or source | WalK | WalR | ClpP | GraS | GraR | VraS | VraR | |||
| N315b | 1 | 1982 | Japan | 1 | ||||||||
| Mu3c | 2 | 1996 | Japan | 32 | I5N | |||||||
| Mu50 | 8 | 1996 | Japan | 33 | N197S | I5N | ||||||
| MI (HIP5827) | 8 | 1997 | USA | 61 | V494L | |||||||
| NJ (HIP5836) | 8 | 1997 | USA | 61 | I28T, I341V | S79F | A260V | |||||
| PC (HIP06297) | 8 | 1998 | USA | 60 | A567D | |||||||
| IL | 8 | 2001 | USA | 5 | D496N | |||||||
| AMC11094 | 8 | 1997 | South Korea | 40 | A113V | |||||||
| 99/3759-V | 8 | 1999 | UK | 34 | V156Q | M1V | ||||||
| 99/3700-W | 8 | 1999 | UK | 34 | R222K, V366M, A468T | |||||||
| LIM2 | 8 | 1995 | France | 57 | ||||||||
| 98141 | 8 | 1998 | France | 7 | ||||||||
| 28160 | 8 | 1998 | South Africa | 24 | ||||||||
| BR 1 | 8 | 1998 | Brazil | 56 | R222K, A468T | |||||||
| BR 2 | 8 | 1998 | Brazil | 56 | R222K, A468T | |||||||
| BR 3 | 8 | 1998 | Brazil | 56 | R222K, A468T | |||||||
| BR 4 | 8 | 1998 | Brazil | 56 | R222K, A468T | |||||||
| BR 5-1 | 8 | 1998 | Brazil | 56 | R222K, A468T | |||||||
| SA MER-S6 | NRS12 | 8 | 1999 | France | NARSA | |||||||
| HIP06854 | NRS18 | 4 | 1998 | USA | NARSA | T492K | ||||||
| HIP07920 | NRS21 | 4 | 1998 | USA | NARSA | H83R | ||||||
| HIP07930 | NRS22 | 4 | 1999 | USA | NARSA | G9V | ||||||
| HIP08926 | NRS23 | 4 | 2000 | USA | NARSA | R222I, T492K | ||||||
| HIP09143 | NRS24 | 4 | 2000 | USA | NARSA | |||||||
| HIP09313 | NRS26 | 4 | 2000 | USA | NARSA | L10F, S437T | R152H | P327S | ||||
| HIP09433 | NRS27 | 4 | 2000 | USA | NARSA | V1454G | G9V | |||||
| HIP09662 | NRS28 | 4 | 2000 | USA | NARSA | Ins.433N, 434De | ||||||
| HIP09735 | NRS29 | 4 | 2000 | USA | NARSA | A468T | ||||||
| HIP09740 | NRS51 | 6 | 2000 | USA | NARSA | V380I | ||||||
| HIP09737 | NRS52 | 4 | 2000 | USA | NARSA | G275V | ||||||
| LY-1999-01 | NRS63 | 4 | 1998 | Oman | NARSA | N48K, R222K, A468T | ||||||
| LY-1999-03 | NRS65 | 4 | 1998 | Oman | NARSA | N48K, R222K, A468T | ||||||
| HIP10540 | NRS73 | 8 | 2000 | USA | NARSA | |||||||
| HIP10267 | NRS74 | 4 | 2000 | USA | NARSA | T11A | ||||||
| C2000001227 | NRS76 | 8 | 2000 | USA | NARSA | A243T | A314V | |||||
| NRS118 | NRS118 | 4 | 2002 | USA | NARSA | F330S | ||||||
| NRS126 | NRS126 | 4 | 2000 | USA | NARSA | |||||||
| P1V44 | NRS272 | 16 | 1999 | Belgium | NARSA | |||||||
| HIP12864 | NRS402 | 8 | 2003 | USA | NARSA | |||||||
| HIP13057 | NRS403 | 8 | 2004 | USA | NARSA | R282C | E15K | |||||
| HIP13036 | NRS404 | 8 | 2004 | USA | NARSA | T104A | ||||||
Sequence determination was carried out with both corresponding forward and reverse primers. Mutations found in 10 vancomycin-susceptible S. aureus strains whose whole genomes were sequenced (including N315, COL, JH1, MRSA252, MSSA472, NCTC8325, Newman, RF122, MW2, and TW20) are not described in this table.
Vancomycin-susceptible MRSA strain used as a sequence control.
hVISA strain.
Amino acid substitutions are given with reference to the amino acid sequence of N315 as follows: the first letter of the N315 sequence, the amino acid sequence position, and the last letter of the altered amino acid sequence.
Two amino acids were inserted at positions 433 and 434.
In conclusion, we proved that the combination of the walK and clpP mutations confers glycopeptide resistance on S. aureus. Very recently, we have reported on the involvement of the mutation of rpoB, encoding the RNA polymerase β subunit, in the mechanism of glycopeptide resistance in some VISA strains (17, 64). Therefore, besides the set of vraS-graR and rpoB mutations, the novel set of mutations in walK-clpP represents an alternative genetic pathway through which the VISA phenotype is achieved in S. aureus. Investigation of the biological significance of these mutations is ongoing with the hope of obtaining an understanding of the entire scheme by which S. aureus achieves resistance to glycopeptide antibiotics.
ACKNOWLEDGMENTS
We thank Mitutaka Yoshida, Laboratory of Electron Microscopy, for his help with electron microscopy.
This work was supported by a grant-in-aid (S0991013) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) for the Foundation of Strategic Research Projects in Private Universities and partially by a Grant-in-Aid for Scientific Research (18590438) to L. Cui from the Ministry of Education, Science, Sports, Culture and Technology of Japan.
Footnotes
Published ahead of print on 31 May 2011.
REFERENCES
- 1. Asada K., et al. 1995. Evolution and resistance expression of MRSA. Evaluation of beta-lactam antibiotics against a set of isogenic strains with different types of phenotypic expression. Acta Biochim. Pol. 42:517–524 [PubMed] [Google Scholar]
- 2. Bischoff M., et al. 2001. Involvement of multiple genetic loci in Staphylococcus aureus teicoplanin resistance. FEMS Microbiol. Lett. 194:77–82 [DOI] [PubMed] [Google Scholar]
- 3. Boyle-Vavra S., Challapalli M., Daum R. S. 2003. Resistance to autolysis in vancomycin-selected Staphylococcus aureus isolates precedes vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 47:2036–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyle-Vavra S., de Jonge B. L., Ebert C. C., Daum R. S. 1997. Cloning of the Staphylococcus aureus ddh gene encoding NAD+-dependent d-lactate dehydrogenase and insertional inactivation in a glycopeptide-resistant isolate. J. Bacteriol. 179:6756–6763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyle-Vavra S., Labischinski H., Ebert C. C., Ehlert K., Daum R. S. 2001. A spectrum of changes occurs in peptidoglycan composition of glycopeptide-intermediate clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 45:280–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Camargo I. L., Neoh H. M., Cui L., Hiramatsu K. 2008. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob. Agents Chemother. 52:4289–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chesneau O., Morvan A., Solh N. E. 2000. Retrospective screening for heterogeneous vancomycin resistance in diverse Staphylococcus aureus clones disseminated in French hospitals. J. Antimicrob. Chemother. 45:887–890 [DOI] [PubMed] [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed., vol. 29, no. 2 M07-A8 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9. Clinical and Laboratory Standards Institute 2009. Performance standards for antimicrobial susceptibility testing; nineteenth informational supplement, M100-S19, vol. 29, No. 3 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 10. Cosgrove S. E., Carroll K. C., Perl T. M. 2004. Staphylococcus aureus with reduced susceptibility to vancomycin. Clin. Infect. Dis. 39:539–545 [DOI] [PubMed] [Google Scholar]
- 11. Cui L., Hiramatsu K. 2003. Vancomycin-resistant Staphylococcus aureus, p. 187–212 In Fluit A. C., Schmitz F. J. (ed.), MRSA: current perspectives. Caister Academic Press, Norfolk, England [Google Scholar]
- 12. Cui L., et al. 2006. Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cui L., Lian J., Neoh H., Ethel R., Hiramatsu K. 2005. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3404–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cui L., et al. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cui L., Murakami H., Kuwahara-Arai K., Hanaki H., Hiramatsu K. 2000. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 44:2276–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cui L., Neoh H. M., Shoji M., Hiramatsu K. 2009. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 53:1231–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cui L., Neoh H. M., Shoji M., Hiramatsu K. 2010. An RpoB mutation confers dual hetero-resistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 54:5222–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cui L., Tominaga E., Neoh H. M., Hiramatsu K. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:1079–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Delaune A., et al. 2011. Peptidoglycan crosslinking relaxation plays an important role in Staphylococcus aureus WalKR-dependent cell viability. PLoS One 6:e17054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dubrac S., Bisicchia P., Devine K. M., Msadek T. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70:1307–1322 [DOI] [PubMed] [Google Scholar]
- 21. Dubrac S., Boneca I. G., Poupel O., Msadek T. 2007. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 189:8257–8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dubrac S., Msadek T. 2004. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 186:1175–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fabret C., Feher V. A., Hoch J. A. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181:1975–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ferraz V., et al. 2000. Vancomycin-resistant Staphylococcus aureus occurs in South Africa. S. Afr. Med. J. 90:1108–1109 [PubMed] [Google Scholar]
- 25. Finan J. E., Archer G. L., Pucci M. J., Climo M. W. 2001. Role of penicillin-binding protein 4 in expression of vancomycin resistance among clinical isolates of oxacillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:3070–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fukuchi K., et al. 2000. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146(Pt 7):1573–1583 [DOI] [PubMed] [Google Scholar]
- 27. Gardete S., Aires-De-Sousa M., Faustino A., Ludovice A. M., de Lencastre H. 2008. Identification of the first vancomycin intermediate-resistant Staphylococcus aureus (VISA) isolate from a hospital in Portugal. Microb. Drug Resist. 14:1–6 [DOI] [PubMed] [Google Scholar]
- 28. Gottesman S., Maurizi M. R. 1992. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol. Rev. 56:592–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hanaki H., et al. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199–209 [DOI] [PubMed] [Google Scholar]
- 30. Hiramatsu K. 1995. Molecular evolution of MRSA. Microbiol. Immunol. 39:531–543 [DOI] [PubMed] [Google Scholar]
- 31. Hiramatsu K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147–155 [DOI] [PubMed] [Google Scholar]
- 32. Hiramatsu K., et al. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670–1673 [DOI] [PubMed] [Google Scholar]
- 33. Hiramatsu K., et al. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135–136 [DOI] [PubMed] [Google Scholar]
- 34. Hood J., Edwards G. F. S., Cosgrove B., Curran E., Morrison D., Gemmell C. G. 2000. Vancomycin-intermediate Staphylococcus aureus at a Scottish hospital. J. Infect. 40:A11 [Google Scholar]
- 35. Howden B. P., et al. 2010. Complete genome sequence of Staphylococcus aureus strain JKD6008, an ST239 clone of methicillin-resistant Staphylococcus aureus with intermediate-level vancomycin resistance. J. Bacteriol. 192:5848–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Howden B. P., et al. 2008. Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob. Agents Chemother. 52:3755–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jansen A., et al. 2007. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int. J. Med. Microbiol. 297:205–215 [DOI] [PubMed] [Google Scholar]
- 38. Katayama Y., Murakami-Kuroda H., Cui L., Hiramatsu K. 2009. Selection of heterogeneous vancomycin-intermediate Staphylococcus aureus by imipenem. Antimicrob. Agents Chemother. 53:3190–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kato Y., Suzuki T., Ida T., Maebashi K. 2009. Genetic changes associated with glycopeptide resistance in Staphylococcus aureus: predominance of amino acid substitutions in YvqF/VraSR. J. Antimicrob. Chemother. 65:37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim M. N., Pai C. H., Woo J. H., Ryu J. S., Hiramatsu K. 2000. Vancomycin-intermediate Staphylococcus aureus in Korea. J. Clin. Microbiol. 38:3879–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kuroda M., Kuwahara-Arai K., Hiramatsu K. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485–490 [DOI] [PubMed] [Google Scholar]
- 42. Kuroda M., et al. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240 [DOI] [PubMed] [Google Scholar]
- 43. Kuwahara-Arai K., Kondo N., Hori S., Tateda-Suzuki E., Hiramatsu K. 1996. Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP 2′ production. Antimicrob. Agents Chemother. 40:2680–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maki H., McCallum N., Bischoff M., Wada A., Berger-Bachi B. 2004. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:1953–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marchler-Bauer A., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McAleese F., et al. 2006. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate- S. aureus-type resistance to vancomycin. J. Bacteriol. 188:1120–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miallau L., Hunter W. N., McSweeney S. M., Leonard G. A. 2007. Structures of Staphylococcus aureus d-tagatose-6-phosphate kinase implicate domain motions in specificity and mechanism. J. Biol. Chem. 282:19948–19957 [DOI] [PubMed] [Google Scholar]
- 48. Michel A., et al. 2006. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188:5783–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Milewski W. M., Boyle-Vavra S., Moreira B., Ebert C. C., Daum R. S. 1996. Overproduction of a 37-kilodalton cytoplasmic protein homologous to NAD+-linked d-lactate dehydrogenase associated with vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:166–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mingeot-Leclercq M. P., Glupczynski Y., Tulkens P. M. 1999. Aminoglycosides: activity and resistance. Antimicrob. Agents Chemother. 43:727–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mongodin E., et al. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Morikawa K., et al. 2001. Overexpression of sigma factor, sigma(B), urges Staphylococcus aureus to thicken the cell wall and to resist beta-lactams. Biochem. Biophys. Res. Commun. 288:385–389 [DOI] [PubMed] [Google Scholar]
- 53. Mwangi M. M., et al. 2007. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 104:9451–9456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Neoh H. M., et al. 2008. Mutated response regulator graR is responsible for phenotypic conversion of Staphylococcus aureus from heterogeneous vancomycin-intermediate resistance to vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 52:45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ohta T., et al. 2004. Nucleotide substitutions in Staphylococcus aureus strains, Mu50, Mu3, and N315. DNA Res. 11:51–56 [DOI] [PubMed] [Google Scholar]
- 56. Oliveira G. A., et al. 2001. Isolation in Brazil of nosocomial Staphylococcus aureus with reduced susceptibility to vancomycin. Infect. Control Hosp. Epidemiol. 22:443–448 [DOI] [PubMed] [Google Scholar]
- 57. Ploy M. C., Grelaud C., Martin C., de Lumley L., Denis F. 1998. First clinical isolate of vancomycin-intermediate Staphylococcus aureus in a French hospital. Lancet 351:1212. [DOI] [PubMed] [Google Scholar]
- 58. Shlaes D. M., et al. 1993. Teicoplanin-resistant Staphylococcus aureus expresses a novel membrane protein and increases expression of penicillin-binding protein 2 complex. Antimicrob. Agents Chemother. 37:2432–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sieradzki K., Leski T., Dick J., Borio L., Tomasz A. 2003. Evolution of a vancomycin-intermediate Staphylococcus aureus strain in vivo: multiple changes in the antibiotic resistance phenotypes of a single lineage of methicillin-resistant S. aureus under the impact of antibiotics administered for chemotherapy. J. Clin. Microbiol. 41:1687–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sieradzki K., Pinho M. G., Tomasz A. 1999. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J. Biol. Chem. 274:18942–18946 [DOI] [PubMed] [Google Scholar]
- 61. Sieradzki K., Roberts R. B., Haber S. W., Tomasz A. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340:517–523 [DOI] [PubMed] [Google Scholar]
- 62. Szurmant H., Nelson K., Kim E. J., Perego M., Hoch J. A. 2005. YycH regulates the activity of the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 187:5419–5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Utaida S., et al. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719–2732 [DOI] [PubMed] [Google Scholar]
- 64. Watanabe Y., Cui L., Katayama Y., Kozue K., Hiramatsu K. 27 April 2011. Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J. Clin. Microbiol. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Watanabe Y., Neoh H. M., Cui L., Hiramatsu K. 2008. Improved antimicrobial activity of linezolid against vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 52:4207–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]