Abstract
The emergence of multidrug resistance among Acinetobacter baumannii is leading to an increasing dependence on the use of polymyxins as last-hope antibiotics. Here, we utilized genetic and biochemical methods to define the involvement of the pmrCAB operon in polymyxin resistance in this organism. Sequence analysis of 16 polymyxin B-resistant strains, including 6 spontaneous mutants derived from strain ATCC 17978 and 10 clinical isolates from diverse sources, revealed that they had independent mutations in the pmrB gene, encoding a sensor kinase, or in the response regulator PmrA. Knockout of the pmrB gene in two mutants and two clinical isolates led to a decrease in the polymyxin B susceptibility of these strains, which could be restored with the cloned pmrAB genes from the mutants but not from the wild type. Reverse transcription-quantitative PCR (RT-qPCR) analysis also showed a correlation between the expression of pmrC and polymyxin B resistance. Characterization of lipid A species from the mutant strains, by thin-layer chromatography and mass spectrometry, indicated that the addition of phosphoethanolamine to lipid A correlated with resistance. This addition is performed in Salmonella enterica serovar Typhimurium by the product of the pmrC gene, which is a homolog of the pmrC gene from Acinetobacter. Knockout of this gene in the mutant R2 [pmrB(T235I)] reversed resistance as well as phosphoethanolamine modification of lipid A. These results demonstrate that specific alterations in the sequence of the pmrCAB operon are responsible for resistance to polymyxins in A. baumannii.
INTRODUCTION
Acinetobacter baumannii has emerged worldwide as an important nosocomial and opportunistic pathogen, particularly in intensive care units (25). It has been implicated in ventilator-associated pneumonia, bacteremia, wound infections, and secondary meningitis. Its natural resistance to desiccation, disinfectants, and antibiotics plays a key role in environmental survival (39) and subsequently in outbreaks in health care settings (25).
Treatment of these infections is often difficult, as clinical isolates of A. baumannii are generally resistant to multiple antibiotics (25). Carbapenems have remained the first-choice option for serious infections caused by multidrug-resistant (MDR) A. baumannii (5). However, the ability of this organism to develop resistance has led to the emergence of extremely drug-resistant (XDR) isolates displaying susceptibility only to the polymyxins (8). Reduced susceptibility to tigecycline, the first glycylcycline, in A. baumannii isolates has also been reported (25).
As a result, polymyxins are being prescribed as the last remaining therapeutic option (3, 18). It has been demonstrated in a prospective study that intravenous colistin is safe and effective (7), and there are lesser issues with polymyxin toxicity than expected based on early experiences (18). Nevertheless, the intensive use and the lack of an optimal dosing creates a potential for colistin resistance (12, 17, 21, 31), and indeed, some cases have already been reported (3, 16, 26, 34), including a nosocomial outbreak of colistin-resistant A. baumannii (37).
The mode of action of polycationic polymyxins involves the initial interaction with polyanionic lipopolysaccharide (LPS) found within the outer membrane, displacing divalent cations that normally cross bridge adjacent LPS molecules required for membrane stabilization (10). The resultant localized disruption of the outer membrane enables self-promoted uptake of polycationic peptides across the outer membrane. In Salmonella enterica serovar Typhimurium, Escherichia coli, and Pseudomonas aeruginosa, resistance can occur through modification of the lipid anchor of LPS known as lipid A (2, 4, 9, 11, 13, 32). For example, the addition of the cationic sugar l-4-aminoarabinose to the lipid A phosphate groups of LPS alters the dependency on divalent cation cross bridging for stabilization of the outer membrane and consequently blocking self-promoted uptake (9). This process is regulated by two-component regulatory systems, including PhoPQ, PmrAB, and ParRS, which respond to signals such as antimicrobial peptides, low pH, and low Mg2+ (2, 4, 9). Other lipid A modifications, including alteration of the number of acyl chains and addition of phosphoethanolamine, can also occur (13, 32). Colistin resistance in P. aeruginosa has been shown to occur in both cystic fibrosis and other patients, and in the latter, the involvement of the PhoPQ and PmrAB two-component regulatory systems has been described (29).
Recently, two papers addressed the mechanism of colistin resistance in A. baumannii (1, 22). Adams et al. (1) identified a homolog of the pmrCAB operon in A. baumannii and demonstrated that resistant mutants trained by growth on 1 μg/ml of colistin contained mutations in the pmrB gene with one mutant having an additional pmrA mutation. However, this study was somewhat limited by the fact that no genetic complementation studies were performed to link these changes to resistance, and indeed, MAC103, a revertant mutant isolated by multiple passages on colistin-free medium, had the same mutations as the original mutant, MAC101, despite a wild-type (WT) MIC. Subsequently, Moffatt et al. (22) indicated that the basis for polymyxin resistance in A. baumannii was mutations in the first 3 genes of the lipid A biosynthesis pathway, namely, lpxACD, and that this led to complete loss of LPS production and supersusceptibility to other antibiotics.
Here, we utilized genetic methods, including knockouts of pmrB and complementation of spontaneous and clinical resistant mutants, to define the critical role of the pmrCAB system in A. baumannii polymyxin resistance. Furthermore, we provide evidence that certain strains have lipid A modifications that occur in a fashion consistent with a blockage of self-promoted uptake of polymyxin across the outer membrane.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and antimicrobial agents.
The bacterial strains and plasmids used in this study are described in Table 1. A collection of 10 polymyxin B-resistant A. baumannii clinical isolates from 5 different countries (isolated from 2001 to 2008 through the SENTRY surveillance system or the University of Seville) was also included; susceptibility to a variety of antibiotics and epidemiological data are shown in Table 2 and, overall, suggested that these strains were independent isolates. All clinical isolates were identified at the species level by amplified ribosomal rRNA gene restriction analysis (ARDRA) (38). Cultures were routinely grown in Luria-Bertani (LB) broth or cation-adjusted Müller-Hinton (CAMH) broth (BBL-Becton Dickinson). The antibiotics and concentrations used for selection in A. baumannii were as follows: carbenicillin, 750 μg/ml, and kanamycin, 50 μg/ml. The antibiotics and concentrations used for selection in E. coli were as follows: ampicillin, 100 μg/ml, and kanamycin, 50 μg/ml. All antibiotics were purchased from Sigma. Polymyxin B and colistin MICs were measured by the broth microdilution method according to the Clinical and Laboratory Standards Institute (CLSI), using CAMH broth. P. aeruginosa ATCC 27853 was included as a quality control strain. An MIC breakpoint of ≥4 μg/ml was used to define resistance to polymyxin B and colistin in clinical isolates of A. baumannii according to CLSI M100-S20 (2010).
Table 1.
Strains, plasmids, and primers used to make the deletion mutants
| Strain, plasmid, or primer | Genotype, relevant phenotype,a or sequence | Source or reference |
|---|---|---|
| A. baumannii strains | ||
| ATCC 17978 | WT | ATCC |
| R2, R3, R5, R6, R7, and R9 | Polymyxin-resistant mutants spontaneously derived on gradient plates from ATCC 17978 | This study (Table 3) |
| E. coli strains | ||
| TOPO10 | F−mcrA D(mrr-hsdRMS-mcrBC) φ80lacZ M15 D lacX74 recA1 araD 139 D (ara-leu)7697 galU galK rpsL (strR) endA1 nupG | Invitrogen |
| DH5α | Invitrogen | |
| W3110 | Wild type; F− λ− | E. coli Genetic Stock Center (Yale) |
| WD101 | W3110 constitutive pmrA mutant (polymyxin resistant) | 13 |
| Plasmids | ||
| pCR-Blunt II-TOPO | PCR cloning vector; Kmr | Invitrogen |
| pUCK4 | Source of Kmr kanamycin cassette gene | Pharmacia |
| pEX18 Amp | Suicide plasmid; Ampr; sacB | 14 |
| pWH1266 | E. coli-Acinetobacter shuttle plasmid; Ampr CBr Tetr | 15 |
| pABN | pWH1266 harboring pmrAB from the WT; CBr | This study |
| pAB25 | pWH1266 harboring pmrAB from R2; CBr | This study |
| pAB10 | pWH1266 harboring pmrAB from R5; CBr | This study |
| Primers | ||
| AB pmrA NF | CAGCAGTTTCTGTGCCATGT | This study |
| KanPmrA NR | TGAGACACAACGTGGCTTTCCCATCCATCATAGGCAATCC | This study |
| KanPmrA CF | ATTACGCTGACTTGACGGGAACCGTTTGGGGCAATCATA | This study |
| AB pmrA CR | TTTTGCATAGCCAAGTTGACC | This study |
| nest-A-F | CGGGTATGCCACGTGTAGAT | This study |
| nest-A-R | AATGCAGTCACAGGTGTTCG | This study |
| AB pmrB NF | GGTCATCAACCTCAACTGG | This study |
| KanPmrB NR | TGAGACACAACGTGGCTTTCGGGTGCTCAGCTGTTCTTTC | This study |
| KanPmrB CF | ATTACGCTGACTTGACGGGATTTACATGAAACAAGAGCGTGA | This study |
| AB pmrB CR | TTAAAGCGACCATTGGCATT | This study |
| nestB-F | GGTGGAATGGGTCAATAACG | This study |
| nestB-R | GAGTTCCGTGGTGCAGACTT | This study |
| C0F | TGATAAATTACGTGCGACTGC | This study |
| KC1R1 | TGAGACACAACGTGGCTTTCTTCAGCATCGTGAGTGACTACA | This study |
| KC2F2 | ATTACGCTGACTTGACGGGAGTGCCCATGTAAACTAAAGCG | This study |
| YnewR | TTGCCAATGCTAAAAGCTGA | This study |
| C1F | GCGTATTCGCCATGCTTATT | This study |
| C2R | GCTTCACGAACCCTCTCTTG | This study |
Antibiotic resistance phenotypes: Kmr, kanamycin resistant, Ampr, ampicillin resistant for E. coli; CBr, carbenicillin resistant for A. baumannii; Tetr, tetracycline resistant.
Table 2.
Sources of specimens, origins, and antibiotic profiles for 10 polymyxin B-resistant clinical isolates of A. baumannii
| Isolate | Clinical sourcea | Origin | MIC (μg/ml) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COLb | PXBb | IMPc | MEMc | FEPc | CAZc | P/Tc | A/Sc | AMKc | GENc | TOBc | CIPc | T/Sc | DOXc | MINc | TIGc | |||
| C2 | BAL (XDR)d | Spain | 64 | 16 | >8 | >8 | >16 | >16 | >64 | >16 | >32 | >8 | 16 | >2 | >2 | >8 | >8 | 1 |
| C4 | Blood (XDR)d | U.S. | 32 | 64 | >8 | >8 | >16 | >16 | >64 | >16 | >32 | >8 | >16 | >4 | >2 | >8 | >8 | 0.5 |
| C5 | Pleural fluid | U.S. | 16 | 32 | 8 | >8 | 16 | >16 | >64 | 8 | 8 | >8 | 1 | >4 | >2 | 4 | 2 | 2 |
| C8 | Blood | U.S. | 32 | 16 | 4 | 8 | 8 | >16 | 64 | 4 | ≤4 | ≤2 | 0.5 | >4 | 1 | 2 | 2 | 2 |
| C11 | BAL | Israel | 8 | 16 | 0.5 | 4 | >16 | >16 | 64 | 8 | >32 | >8 | >16 | >4 | >2 | ≤1 | 1 | 1 |
| C12 | Blood (MDR)d | U.S. | 16 | 16 | >8 | >8 | 16 | >16 | >64 | 32 | 16 | >8 | >16 | >4 | >2 | 2 | 2 | 1 |
| C13 | BAL | U.S. | 16 | 16 | 4 | 8 | >16 | >16 | >64 | >16 | 8 | >8 | 16 | >4 | >2 | 2 | 1 | ND |
| C14 | Wound | Brazil | 32 | 8 | 0.5 | 1 | 8 | >16 | >64 | 8 | 2 | ≤2 | 0.5 | >4 | ≤0.5 | 0.25 | 0.25 | ND |
| C15 | Blood | France | 128 | 64 | 2 | 1 | 8 | >16 | ≤0.5 | 4 | 32 | >8 | >16 | >4 | ≤0.5 | >8 | 4 | ND |
| C87 | VCT (MDR)d | Spain | 64 | 16 | >8 | >8 | >16 | >16 | >64 | <8 | >32 | >8 | >16 | >4 | >2 | ND | 0.5 | 0.5 |
BAL, bronchoalveolar lavage; VCT, ventricular catheter tip.
Colistin (COL) and polymyxin B (PXB) MICs were determined by the broth microdilution method according to the Clinical and Laboratory Standards Institute (CLSI; 2009).
MICs were determined by the broth microdilution M07-A8 method (CLSI; 2009) using commercially prepared and validated panels (TREK Diagnostic Systems, OH) in CAMH broth to the following antibiotics: IMP, imipenem; MER, meropenem; FEP, cefepime; CAZ, ceftazidime; P/T, piperacillin-tazobactam; A/S, ampicillin-sulbactam; AMK, amikacin; GEN, gentamicin; TOB, tobramycin; CIP, ciprofloxacin; T/S, trimethoprim-sulfamethoxazole; DOX, doxycycline; MIN, minocycline; and TIG, tigecycline. ND, not done.
The clinical isolates were classified according to their susceptibility to the antimicrobials listed above (excluding colistin/polymyxin B and tigecycline) as follows: multidrug resistant (MDR), resistant to at least 3 antimicrobial families (including carbapenems) and displaying variable susceptibility to the remaining agents, and extremely drug resistant (XDR), resistant to all the antibiotics listed (8). Susceptibility interpretations were performed according to the CLSI (2010) criteria, when available. Due to the lack of CLSI breakpoints for tigecycline in Acinetobacter, no susceptibility could be inferred. Antibiotic profiles were categorized as MDR or XDR.
In vitro generation of polymyxin B-resistant mutants.
Late-log-phase WT LB cultures were plated on gradient (4 to 50 μg/ml) polymyxin B-LB agar plates and then incubated at room temperature for 72 to 96 h (23). Colonies were confirmed by six serial passages on polymyxin (4 μg/ml) LB plates and subsequently by six serial passages on drug-free LB plates, resulting in a polymyxin B resistance phenotype.
Gene knockouts by homologous recombination.
Splicing PCR was performed as described previously (24), with slight modifications, to generate recombinant DNA fragments consisting of a kanamycin cassette (obtained by PCR from pUCK4 plasmid; Pharmacia) flanked by ∼1-kb chromosomal regions upstream and downstream of the gene to be deleted. Forward and reverse primers for this study (Table 1) were designed from the ATCC 17978 genome sequence (30) using Primer3 (28). Phusion DNA polymerase (Fynnzimes, Finland) was used as a high-fidelity DNA polymerase. The final PCR constructs were cloned into the TOPO blunt-ended vector and excised using KpnI and PstI restriction enzymes, and the excised fragment was cloned into the cognate sites of the pEX18Amp plasmid (a gene replacement suicide vector for Acinetobacter spp.) (14). The recombinant knockout plasmids were introduced into parental strains by electroporation, and transformants were selected for kanamycin resistance and carbenicillin susceptibility, resulting from a double crossover recombination event. The final structure of cloned and knockout genes in all replacement vectors and knockout mutant strains, including the flanking regions, was confirmed by PCR analysis and sequencing. For generation of knockout A. baumannii clinical isolates, strains C8 and C14 were selected because their antibiotic profile was suitable for enabling mutant selection.
Sequencing of pmrCAB in clinical isolates and WT-derived mutant strains.
Genomic DNA was isolated using a DNeasy blood and tissue kit (Qiagen, Inc., Canada) according to the manufacturer's instructions. The pmrCAB genes were amplified and sequenced. A set of PCR primers was designed to encompass the entire operon and flanking regions (∼4.5 kb). Sequencing of both strands was performed using Applied Biosystems BigDye Terminator version 3.1 chemistry and run on an Applied Biosystems Prism 377 automated sequencer.
Construction of the pmrAB-complemented strains.
To complement the disrupted pmrB mutants, the contiguous pmrAB genes were cloned together with 252 bp of upstream DNA and 89 bp of downstream DNA. Fragments were PCR amplified from ATCC 17978, and mutants R2 and R5, cloned into the TOPO vector, were excised using BamHI and subsequently cloned into the cognate sites of the expression vector pWH1266 (15), resulting in plasmids pABN, pAB25, and pAB10, respectively.
RT-qPCR.
The A. baumannii WT strain ATCC 17978 or derivatives and clinical isolates were grown to the mid-logarithmic phase of growth in CAMH broth, and cells were harvested at an optical density at 600 nm (OD600) between 0.5 and 0.6. Total RNA was extracted using a Qiagen Easy minikit and RNeasy minicolumns. DNase treatment of RNA samples, cDNA synthesis, and reverse transcription-quantitative PCR (RT-qPCR) were carried out as described previously (4). cDNA was diluted 1:100, and 2.5 μl was used as a template for each reaction, using 1× SYBR green PCR master mix (Applied Biosystems, Foster City, CA) and an ABI Prism 7000 instrument (Applied Biosystems). Internal forward and reverse primers for each gene were designed using PrimerExpress (Applied Biosystems). Experiments were repeated with three independent cultures, each tested in duplicate. Comparison to the 16S ribosomal gene allowed calculation of the fold change by the threshold cycle (CT) method (19).
TLC analysis of lipid A species from 32Pi-labeled cells.
Strains of E. coli and A. baumannii were labeled using 2.5 μCi/ml 32Pi at a starting OD600 of ∼0.05 and harvested at an OD600 of ∼1.0. Cultures were grown at 37°C in LB broth. 32P-labeled lipid A species were isolated and separated by thin-layer chromatography (TLC) using published protocols (13). TLC plates were exposed overnight to a phosphorimager screen and visualized using a Bio-Rad personal molecular imager (PMI) equipped with Quantity One software.
Analysis of lipid A by mass spectrometry.
Bacterial strains were grown at 37°C in LB broth until each culture reached an OD600 of ∼1.0. Lipid A was purified as described previously and analyzed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry in the negative-ion-reflectron mode (11). The matrix used was a saturated solution of 6-aza-2-thiothymine in 50% acetonitrile and 5% tribasic ammonium citrate (20:1, vol/vol). A 0.6-μl volume of matrix solution was deposited on the sample plate, followed by 0.4 μl of sample dissolved in chloroform-methanol (4:1, vol/vol) (11).
Nucleotide sequence accession numbers.
The nucleotide sequences of pmrCAB from polymyxin B-resistant spontaneous laboratory WT-derived mutants and clinical isolates of A. baumannii have been submitted to the GenBank nucleotide sequence databases under the accession numbers JF766938 to JF766953.
RESULTS
Polymyxin-resistant spontaneous mutants.
To isolate polymyxin-resistant mutants of the wild-type A. baumannii strain ATCC 17978, the organism was plated onto polymyxin B gradient plates. After 72 to 96 h, nine colonies grew at concentrations that inhibited the parent strain. After 6 serial passages, only six, R2, R3, R5, R6, R7, and R9, demonstrated stable resistance, forming colonies that were able to grow on polymyxin B plates; all of these continued to display polymyxin resistance after 6 serial passages on drug-free plates. The resulting polymyxin B MICs were between 2 and 8 μg/ml, compared to the parental MIC of 0.5 μg/ml (Table 3). No significant changes were observed in susceptibility to any other tested antibiotic, including imipenem, meropenem, cefepime, ceftazidime, piperacillin-tazobactam, ampicillin-sulbactam, amikacin, gentamicin, tobramycin, ciprofloxacin, trimethoprim-sulfamethoxazole, doxycycline, minocycline, tigecycline, or different cationic peptides (indolicidin, LL-37, hLF1-11, HH2, E6, Bac2A, and MX-226).
Table 3.
Amino acid changes or mutations in the pmrCAB polymyxin B-resistant WT-derived strains (compared to their parental WT strain ATCC 17978) and pmrCAB variants of polymyxin B-resistant clinical isolates (compared to a consensus sequence)a
| Strain | PXBb MIC (μg/ml) | Amino acid change(s) inc: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
pmrC (549 aa) |
pmrA (224 aa) |
pmrB (444 aa) |
|||||||
| aa 1–236 | Sulfatase (aa 237–532) | Rec (aa 5–116) | aa 117–131 | aa 1–215 | HisK (aa 216–276) | aa 277–330 | HATPaseC (aa 331–419) | ||
| WT | 0.5 | ||||||||
| R9 | 2 | R231L | |||||||
| R3 | 4 | G315D | |||||||
| R5 | 4 | M12I | |||||||
| R6 | 4 | R263P | Q277H | ||||||
| R7 | 4 | G315D | |||||||
| R2 | 8 | T235I | |||||||
| C14 | 8 | T7I, A211V | Δ160 | ||||||
| C2 | 16 | D64V, L208F | |||||||
| C8 | 16 | L208F | |||||||
| C11 | 16 | H499R* | Δ32–35 | P360Q* | |||||
| C13 | 16 | P170Q | |||||||
| C12 | 16 | N256I | |||||||
| C87 | 16 | F90L | S119T | P233S | P360Q* | ||||
| C5 | 32 | R263C | P377L | ||||||
| C4 | 64 | A226V | |||||||
| C15 | 64 | A80V, P170L | |||||||
Sequences were compared to a consensus sequence made by alignment of the pmrCAB operons from the genome sequences of the following strains: ATCC 17978 (Yale University); AYE (Genoscope, France); ACICU (National Research Centre, Italy); AB0057, AB307-0294, and AB900 (Case Western Reserve University); 1656-2 (Kyungpook National University/GenoTech Corporate, South Korea), and TCDC-AB0715 (Taiwan Center for Disease Control, Taiwan). Accordingly, the identified single nucleotide polymorphisms (SNPs; naturally variant between the sequenced isolates) pmrC(F166L), pmrC(A370S), pmrC(K531T), and pmrB(A444V) occurred in each of the clinical isolates C2, C4, C5, C8, C12, C13, and C15 but not in C11, C14, or C87. In addition, the SNP pmrC(H499R) (marked with an asterisk) was found only in clinical isolate C11. The SNP pmrB(P360Q) (marked with an asterisk) occurred in clinical isolates C11 and C87 but none of the other strains. aa, amino acids.
Polymyxin B (PXB) MICs were assessed by the broth microdilution method according to the CLSI.
The predicted domains according to the NCBI domain predictor (www.ncbi.nlm.nhi.gov/protein) are indicated as follows: sulfatase, sulfatase domain; Rec, signal receiver domain; HisK, histidine kinase (dimerization/phosphoacceptor) domain; and HATPaseC, histidine-kinase-like ATPase. Only domains or regions displaying mutations or variants are shown. The amino acid (aa) positions corresponding to these domains are displayed in brackets.
Sequence analysis of the polymyxin resistance operon pmrCAB in mutants.
Homology searches revealed an operon (loci A1S_2752 to -2750) with homology to the pmrCAB operon of Salmonella with the predicted genes having moderate 42, 45, and 28% identity (60, 61, and 47% similarity) to the corresponding genes of Salmonella, STM4293 (pmrC), -4291 (pmrA), and -4290 (pmrB), respectively. The genes in Salmonella encode a lipid A phosphoethanolamine transferase (pmrC), response regulator (pmrA), and histidine kinase (pmrB), with the last two being components of a two-component signal transduction system. No other two-component regulators with homology to that of known polymyxin resistance regulatory genes were observed in A. baumannii, which was in good agreement with the conclusions of Adams et al. (1). We were unable to identify strong homologs of any other polymyxin resistance genes of Pseudomonas or Salmonella, including the arnBCADTEF (lipid A arabinosaminylation) operon, pagP, pagL, or lpxR genes. The pmrCAB operons from the above-described polymyxin-resistant mutants of strain ATCC 17978 were sequenced to reveal potential differences (Table 3).
No changes in the pmrC gene were observed in any mutant. One mutant, R5, had a mutation in the predicted receiver domain of the response regulator gene pmrA, while the remaining 5 mutants had mutations in the histidine kinase gene pmrB in either the predicted histidine kinase domain (R2 and R9 with mutations in residues 235 and 231, respectively) or residue 315 (R3 and R7) or two separate mutations, one of which was in the histidine kinase domain (R6).
Knockout mutants exhibit different MICs for polymyxin B.
To confirm that the described mutations were responsible for resistance, pmrB was knocked out by insertion of a kanamycin cassette into the pmrB gene of 2 selected mutants, R2 and R5 (Table 4). In each case, MICs for polymyxin B were decreased by 8- to 16-fold. In contrast, knockout of pmrB (or pmrA or pmrC; data not shown) in the wild-type background caused no significant change (≤2-fold) in polymyxin B MICs, indicating that this operon contributes modestly at most to intrinsic resistance. Knockout of pmrC, the first gene in the operon led to complete suppression of polymyxin resistance in the mutant R2::pmrC (MIC = 0.25 μg/ml), although, despite multiple attempts, we were unable to perform the same knockout in the mutant R5. Knockout mutants exhibited no significant changes in susceptibility to other antibiotics (data not shown).
Table 4.
Polymyxin B MIC changes in pmrB::Km knockout mutants
| Parental strain | Polymyxin B MIC (μg/ml) |
|
|---|---|---|
| Parent | pmrB::Km insertion mutant | |
| WT | 0.5 | 0.25 |
| R2 | 8 | 0.5 |
| R5 | 4 | 0.5 |
| C8 | 16 | 0.25 |
| C14 | 8 | 2 |
Clinical isolates.
Ten clinical polymyxin B-resistant isolates were obtained from the SENTRY surveillance program and University of Seville. Each was resistant to multiple antibiotics (Table 2), with 4 strains being designated multidrug resistant (C12 and C87) or extremely drug resistant (C2 and C4). Polymyxin MICs varied from 8 to 64 μg/ml, and colistin MICs ranged from 8 to 128 μg/ml. To define the potential presence of variants leading to resistance, the entire pmrCAB operon from each isolate was sequenced. Considering these were independent clinical isolates from a variety of clinical situations and countries, the gene sequences were fairly conserved. Regarding pmrC, two isolates displayed variants in a region of unknown function (T7I and A211V in C14 and F90L in C87). All clinical isolates had different variants of pmrB, with 4 of them having alterations in the kinase domain. Excluding the potential single nucleotide polymorphisms (SNPs) identified in the pmrB gene (Table 3), we found that three isolates (C2, C5, and C15) had two variants while C11 and C14 had small in-frame deletions. One strain, C87, had an additional variant of pmrA, although not within the receiver domain.
To test the involvement of the two-component system PmrAB in the resistant phenotype of these strains, the pmrB gene was knocked out by insertion of a kanamycin cassette into the pmrB genes of 2 clinical isolates, C8 and C14 (Table 4). This deletion caused 64-fold and 4-fold increases in susceptibility to polymyxin B, respectively.
Complementation of pmrAB mutations.
To further demonstrate that pmrB played a role in the polymyxin-resistant phenotype, we attempted to complement the susceptibility phenotype created through the knockout of pmrB in the mutant R2 (Table 5). As expected, the cloned pmrAB genes from the wild-type strain did not increase the level of polymyxin resistance but the pmrAB genes from the R2 mutant [carrying the pmrB(T235I) mutation] were able to reconstitute resistance. Similarly, the pmrAB genes from the mutant R5 [carrying the pmrA(M12I) mutation] were able to reconstitute resistance, indicating that alterations in either component of this regulatory system, PmrA or PmrB, can lead to resistance.
Table 5.
Polymyxin B susceptibility and lipid A profile lane in mutant R2 and complemented A. baumannii strains
| Inserted mutation | Complementing gene | Polymyxin B MIC (μg/ml) | Lipid A profile (lane in Fig. 1) |
|---|---|---|---|
| None (mutant R2) | None | 8 | 2 |
| pmrB::Km | None | 0.5 | 3 |
| pmrB::Km | pWH1266 (empty vector) | 0.5 | |
| pmrB::Km | pmrABWT | 0.25 | |
| pmrB::Km | pmrABR2 | 4 | 4 |
| pmrB::Km | pmrABR5 | 4 | 5 |
Analysis of pmrCAB transcription in different strains.
We hypothesized that the polymyxin-resistant phenotype would be associated with increased expression of the pmrCAB operon, in particular that of pmrC, which encodes the protein that adds phosphoethanolamine to lipid A. For this reason, we studied the transcription levels of this operon in different strains compared to the level for ATCC 17978. These strains included the resistant mutants and clinical isolates R2, R5, C8, and C14, the pmrB deletion mutants corresponding to R2, R5, C8 and C14, and the complemented R2 pmrB deletion mutant carrying the pmrAB genes from the wild type or from the R2-resistant strain (Table 6).
Table 6.
RT-qPCR analysis of the expression of pmrCAB in different strains compared to the level for the laboratory wild-type strain ATCC 17978a
| Strain | Mean fold change ± SD |
||
|---|---|---|---|
| pmrC | pmrA | pmrB | |
| R2 | 224.8 ± 140.6 | 51.7 ± 32.7 | 30.5 ± 18.5 |
| R2 pmrB::Km | 1.43 ± 0.09 | 1.3 ± 0.4 | * |
| R2 pmrB::Km + pmrABWT | 1.0 ± 0.3 | 49.5 ± 8.1 | 189.9 ± 17.0 |
| R2 pmrB::Km + pmrABR2 | 567.0 ± 137.0 | 58.8 ± 6.2 | 77.0 ± 9.2 |
| R5 | 165.2 ± 27.6 | 18.5 ± 0.4 | 10.6 ± 0.3 |
| R5 pmrB::Km | 2.0 ± 1.2 | 0.9 ± 0.2 | * |
| C8 | 292.1 ± 18.3 | 4.3 ± 0.05 | 2.7 ± 0.3 |
| C8 pmrB::Km | 6.8 ± 0.6 | 1.7 ± 0.08 | * |
| C14 | 26.5 ± 3.3 | 4.1 ± 0.7 | 3.2 ± 0.5 |
| C14 pmrB::Km | 16.2 ± 1.8 | 2.9 ± 1.7 | 0.9 ± 0.1 |
The values represent three independent samples, with each assessment being repeated in duplicate. *, no expression of pmrB could be detected in these strains.
In general, we observed a direct correlation between the greater expression of the pmrC gene and resistance to polymyxin B. Thus, the five resistant strains tested, which included the clinical isolates C8 and C14 together with the laboratory isolated mutants R2 and R5, showed increases in the transcription of pmrC from 26- to 292-fold, compared to the level for our laboratory wild-type strain ATCC 17978.
Upon deletion of pmrB, the spontaneous resistant mutants R2 and R5 recovered transcription levels and polymyxin B MICs that were comparable to those of their parent strain. The results of the analysis of the pmrB deletion mutants derived from the clinical isolates C8 and C14 were slightly more complex. Thus, deletion of pmrB in strain C8 resulted in a loss of the resistance phenotype and a significant 43-fold decrease in the transcription of pmrC, although the expression of pmrC was still ∼6-fold higher than that of ATCC 17978. For isolate C14, for which the deletion mutant demonstrated only a partial restoration of polymyxin B susceptibility, RT-qPCR analysis revealed that this mutant did not completely lose the expression of the deleted gene pmrB. This is consistent with the interpretation that there might be a more complex regulatory mechanism or a second copy of the operon in this strain.
Finally, we also evaluated the expression of the genes in the pmrCAB operon in two complemented pmrB deletion mutants derived from strain R2, carrying in trans copies of the pmrAB genes from either the parent ATCC 17978 (pmrABWT) or the R2-resistant mutant (pmrABR2). In both cases, a high level of overexpression of pmrA and pmrB can be observed. However, only the strain carrying the mutant copy of pmrB (pmrABR2) leads to an overexpression of pmrC and to polymyxin B resistance. In contrast, overexpression of the wild-type copy of these genes does not result in increased expression of the chromosomally located gene pmrC, nor does it confer increased resistance.
Polymyxin-resistant strain mutants have a modified lipid A profile.
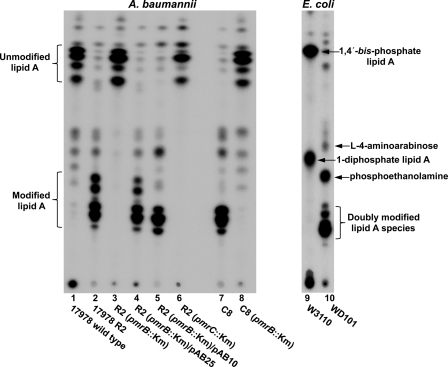
To examine whether modification of lipid A played a role in conferring polymyxin resistance in A. baumannii, cultures were radiolabeled with 32Pi and the purified lipid A species was isolated and separated using TLC (Fig. 1). As controls, lipid A of a polymyxin-resistant E. coli K-12 strain (WD101) and its polymyxin-sensitive parent strain (W3110) were included in the analysis. PmrA is constitutively active in WD101, resulting in modification of lipid A phosphate groups with phosphoethanolamine and l-4-aminoarabinose (13, 32). E. coli K-12 produced two hexa-acylated lipid A species, 1,4′-bis-phosphate lipid A and 1-diphosphate lipid A, respectively (Fig. 1, lane 9). As previously reported for WD101, modification of both phosphate groups with phosphoethanolamine and l-4-aminoarabinose resulted in slower-migrating species (lane 10) (13, 32).
Fig. 1.
Analysis of 32P-labeled lipid A species isolated from A. baumannii and E. coli strains. 32P-labeled lipid A species isolated from the indicated bacterial strains were separated by TLC and visualized by phosphorimaging analysis. As shown previously, polymyxin-resistant E. coli (WD101) produced lipid A species modified with l-4-aminoarabinose and phosphoethanolamine, including doubly modified lipid A species (13, 32). Polymyxin B-resistant strains of A. baumannii produced slower migrating, more hydrophilic lipid A species (Rf values of 0.17 to 0.29), compared to the lipid A of polymyxin B-sensitive strains (Rf values of 0.63 to 0.70). The major 32P-labeled lipid A species are indicated with arrows.
Wild-type A. baumannii ATCC 17978 produced lipid A species migrating with Rf values of 0.63 to 0.70 (Fig. 1, lane 1), whereas the lipid A species from strain R2 were more hydrophilic in nature, with Rf values of 0.17 to 0.29 (lane 2), suggesting modification of lipid A. Deletion of pmrB in strain R2 resulted in the loss of this modification in lipid A migration (lane 3), while the complementation with the pmrAB genes from R2 (plasmid pAB25) restored lipid A modification (lane 4). A similar result was obtained using plasmid pAB10 expressing pmrAB genes from the polymyxin-resistant strain R5 (lane 5). An insertion in the pmrC gene, encoding a putative A. baumannii phosphoethanolamine transferase, also resulted in the loss of lipid A modification. Finally, clinical isolate C8 had a modified lipid A profile (lane 7), and deletion of pmrB in this background resulted in loss of lipid A modification. Taken together, these results demonstrate that polymyxin-resistant strains of A. baumannii displayed modified forms of lipid A on their surface and that the appearance of these modified forms was consistent with the polymyxin B MICs of the respective strains.
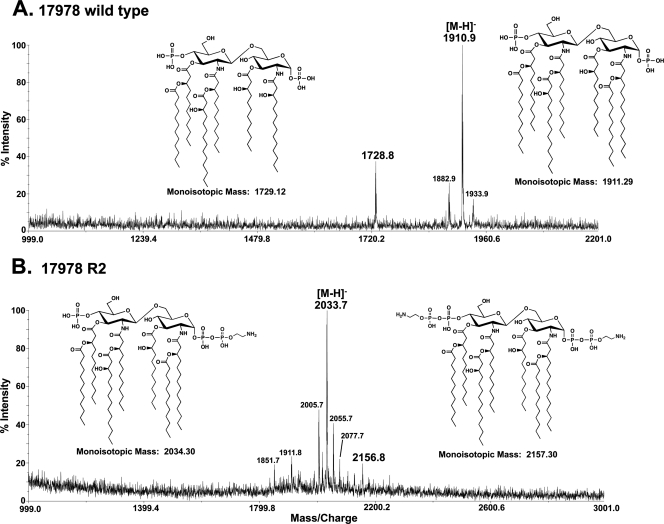
Purified lipid A from A. baumannii was also subjected to analysis by MALDI-TOF mass spectrometry in the negative-ion mode (Fig. 2). The wild-type spectrum showed a predominant peak at an m/z of 1,910.9, consistent with a [M-H]− ion of a hepta-acylated, bis-phosphorylated lipid A species (predicted [M-H]− at an m/z of 1,910.29). A second predominant peak at an m/z of 1,728.8 was consistent with a [M-H]− ion of a hexa-acylated, bis-phosphorylated lipid A species (predicted [M-H]− at an m/z of 1,728.12). The predicted lipid A structures are shown in Fig. 2A and are in agreement with a previously reported mass spectrometry analysis of wild-type A. baumannii lipid A (20). As expected, mass spectrometry analysis of the polymyxin-resistant R2 variant (Fig. 2B) revealed modified lipid A structures. The predominant peak at an m/z of 2,033.7 was consistent with the [M-H]− ion of hepta-acylated, bis-phosphorylated lipid A modified with a phosphoethanolamine residue (predicted [M-H]− at an m/z of 2,033.30). A minor peak at an m/z of 2,156.8 was also present and was consistent with a lipid A species bearing two phosphoethanolamine residues (predicted [M-H]− at an m/z of 2,156.30). The predicted lipid A structures of strain R2 are shown in Fig. 2B. Similar spectra were obtained upon analysis of lipid A isolated from strain C8 (data not shown). Additional peaks of lower intensity are explained in the figure legend.
Fig. 2.
Mass spectrometry of lipid A isolated from wild-type and polymyxin-resistant A. baumannii. Lipid A was isolated from wild-type strain ATCC 17978 and wild-type-derived R2, followed by MALDI-TOF mass spectrometry in the negative-ion mode. (A) Wild-type 17978 produced peaks at m/z values of 1,910.9 and 1,728.8, corresponding to bis-phosphorylated hepta- and hexa-acylated lipid A species, respectively. The minor peak at an m/z of 1,933.9 represents a sodium adduct of the parent ion. The peak at an m/z of 1,882.9 arises from lipid A with an acyl chain reduced by 2 carbons in length compared to that of the major ion. (B) Strain 17978 R2 produced peaks at m/z values of 2,033.7 and 2,156.8, corresponding to bis-phosphorylated hepta-acylated lipid A decorated with one and two phosphoethanolamine groups, respectively. Minor peaks at m/z values of 2,055.7 and 2,077.7 represent sodium adducts of the major parent ion. The minor peak at an m/z of 2,005.7 arises from lipid A with an acyl chain reduced by 2 carbons in length compared to that of the major ion. The peak at an m/z of 1,851.7 is bis-phosphorylated, hexa-acylated lipid A with a single phosphoethanolamine residue.
DISCUSSION
As polymyxin is becoming a drug of last hope for the often multidrug-resistant species A. baumannii, understanding polymyxin resistance is of great interest. Here, we have confirmed, using genetic means, the involvement of an operon that was suggested by Adams et al. (1) to control polymyxin resistance in A. baumannii and that demonstrates moderate homology to the pmrCAB operon of Salmonella Typhimurium. Our results indicate that the two-component regulator PmrAB regulates polymyxin resistance. Spontaneous mutants isolated from the wild-type A. baumannii strain ATCC 17978 had 4- to 16-fold changes in polymyxin B MIC and mutations in either pmrB (4 different mutations in 5 strains) or pmrA (1 mutation isolated). These results are in part consistent with those found by Adams et al. (1) and also in polymyxin-resistant E. coli and Salmonella Typhimurium, where polymyxin resistance mutations were found to be located within the receiver domain of the PmrA regulator (6, 27, 33). In addition, mutations have been identified in pmrB in polymyxin-resistant P. aeruginosa (23).
The effects of these mutations on polymyxin resistance in 2 selected mutants could be eliminated by knocking out the pmrB gene by insertion of a kanamycin cassette and then restored by complementation with the mutated pmrAB genes from two spontaneous mutants but not with the wild-type genes. Transcriptional analysis showed that the resistant mutants had an overexpression of the pmrCAB operon that was abrogated when pmrB was deleted. Adams et al. (1) had already reported that the level of pmrA expression was higher in colistin-resistant mutants. Furthermore, our data indicate that pmrAB overexpression is not sufficient to result in increased polymyxin resistance, unless there is also an overexpression of pmrC, which encodes the lipid A modification enzyme phosphoethanolamine transferase. These data indicate that the mutations have a dominant effect (gain of function) on the activity of the pmrCAB operon, leading to constitutive expression of the regulon controlled by these genes. Interestingly, we also tested these mutants and complemented strains for susceptibility to 6 diverse antimicrobial peptides and demonstrated minimal changes in susceptibility (≤2-fold; data not shown), consistent with our data for Pseudomonas (4), indicating that resistance to polymyxin and antimicrobial peptides is separately determined in A. baumannii.
Results for clinical isolates taken from the SENTRY surveillance program and the University of Seville were also consistent with these data. Although these mutants were obtained from 5 different countries, and had diverse other resistances, they demonstrated moderate overall conservation of sequence in the pmrAB and pmrC genes, but each of these had sequence alterations in the pmrB gene as well as apparent SNPs (P360Q and A444V). Only one of these strains had an additional variant in the pmrA gene, although it was not located within the receiver domain. Four common SNPs, pmrC(F166L), pmrC(A370S), pmrC(K531T), and pmrB(A444V), which occur in a subset of the sequenced A. baumannii strains, were also found within the pmrC and pmrB genes in 7 clinical isolates, C2, C4, C5, C8, C12, C13, and C15, but not C11, C14, or C87. An apparent SNP, pmrC(H499R), was found only in isolate C11. For the two clinical isolates investigated in greater detail, C14 and C8, the knockout of pmrB caused a decrease in resistance. However, this appeared to be only partial (4-fold) in the case of C14, which might be explained, based on expression analysis carried out with these strains, by additional regulatory elements or the existence of a second copy of the pmrC gene. The presence of additional copies of some of the genes in the pmrCAB operon might also be a factor in some clinical strains, as additional open reading frames (ORFs) are annotated as lipid A phosphoethanolamine transferases in the genomes of MDR clinical isolates such as strains 1656-2 and TCDC-AB0715. However, further studies, including genome sequencing, would be necessary to establish the likelihood of any these possibilities.
In other Gram-negative bacteria, two-component regulatory systems control polymyxin resistance by altering the lipid A species of LPS, consequently decreasing self-promoted uptake of this polycationic antibiotic. These modifications can include the addition of arabinosamine or phosphoethanolamine to the phosphate residues of lipid A or the alteration in acylation to change the charge or fluidity of the lipid A, respectively, and consequently decrease self-promoted uptake across the outer membrane. Here, it appeared that the pmrAB operon also controlled lipid A modification, specifically, in the case of strain ATCC 17978, the addition of phosphoethanolamine. These data differ from the observations of Moffatt et al. (22), who indicated that polymyxin resistance in A. baumannii is associated with a loss of LPS. Our results clearly show that loss of LPS is not the only mechanism of resistance to polymyxins in this microorganism, as we clearly demonstrated the presence of LPS/lipid A in the mutants studied, as well as the importance of modified LPS in these strains as a result of alterations in PmrAB. Another difference from the Moffatt et al. study is that we did not observe the 32- to 256-fold increase in susceptibility to other classes of antibiotics. Furthermore, the observation of heteroresistance to polymyxin in A. baumannii is entirely consistent with differential (epigenetic) regulation of pmrAB, as observed for skipped well resistance in Pseudomonas (29).
In Salmonella, the pmrC gene is involved in adding phosphoethanolamine to lipid A. Knockout of the pmrC gene in the A. baumannii spontaneous mutant R2 led to increased susceptibility to polymyxin B and reversed the modification in lipid A profiles observed in thin-layer-chromatography analyses. However, the situation was not as clear for the two clinical isolates investigated, C8 and C14, which could be related to additional regulatory elements or the presence of additional copies of pmrC or the pmrCAB operon, as suggested above. This could also indicate that polymyxin resistance might be more complicated in clinical isolates and that there might be other pmrAB-dependent and/or -independent mechanisms in these isolates.
Overall, we have identified here an important mechanism of resistance for the last-hope antibiotics polymyxins. Although the sequenced A. baumannii strains lack homologs of the other regulatory and effector mechanisms involved in polymyxin resistance in other bacteria, which reduces the number of potential target genes that upon mutation lead to polymyxin resistance in the laboratory or the clinic, this bacterium can become resistant through a diversity of independent mutations in the pmrCAB operon. Given that we found no obvious overlap in resistance between polymyxin and cationic antimicrobial peptides, it will be interesting to see if new modified polymyxin derivatives demonstrate cross-resistance (35, 36), providing a potential new avenue to therapeutic success.
ACKNOWLEDGMENTS
This work was supported by grants from the Canadian Cystic Fibrosis Foundation and the Canadian Institutes of Health Research. R.E.W.H. holds a Canada Research Chair. L.A.A. was the recipient of La Caixa Postgraduate Research Award (Barcelona, Spain). M.S.T., C.M.H., and J.V.H. are supported by National Institutes of Health (NIH) grants AI064184 and AI76322.
We are grateful to R. N. Jones and R. Mendes (JMI Labs) and J. Aznar (University of Seville, Spain) for kindly providing antibiotic-resistant isolates.
Footnotes
Published ahead of print on 6 June 2011.
REFERENCES
- 1. Adams M. D., et al. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53:3628–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bader M. W., et al. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472 [DOI] [PubMed] [Google Scholar]
- 3. Falagas M. E., et al. 2005. Outcome of infections due to pandrug-resistant (PDR) Gram-negative bacteria. BMC Infect. Dis. 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fernandez L., et al. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob. Agents Chemother. 54:3372–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fishbain J., Peleg A. Y. 2010. Treatment of Acinetobacter infections. Clin. Infect. Dis. 51:79–84 [DOI] [PubMed] [Google Scholar]
- 6. Froelich J. M., Tran K., Wall D. 2006. A pmrA constitutive mutant sensitizes Escherichia coli to deoxycholic acid. J. Bacteriol. 188:1180–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garnacho-Montero J., et al. 2003. Treatment of multidrug-resistant Acinetobacter baumannii ventilator-associated pneumonia (VAP) with intravenous colistin: a comparison with imipenem-susceptible VAP. Clin. Infect. Dis. 36:1111–1118 [DOI] [PubMed] [Google Scholar]
- 8. Giske C. G., Monnet D. L., Cars O., Carmeli Y. 2008. Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob. Agents Chemother. 52:813–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gooderham W. J., Hancock R. E. W. 2009. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 33:279–294 [DOI] [PubMed] [Google Scholar]
- 10. Hancock R. E. W., Chapple D. S. 1999. Peptide antibiotics. Antimicrob. Agents Chemother. 43:1317–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hankins J. V., Trent M. S. 2009. Secondary acylation of Vibrio cholerae lipopolysaccharide requires phosphorylation of KDO. J. Biol. Chem. 284:25804–25812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hawley J. S., Murray C. K., Jorgensen J. H. 2008. Colistin heteroresistance in Acinetobacter and its association with previous colistin therapy. Antimicrob. Agents Chemother. 52:351–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Herrera C. M., Hankins J. V., Trent M. S. 2010. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol. Microbiol. 76:1444–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 15. Hunger M., Schmucker R., Kishan V., Hillen W. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45–51 [DOI] [PubMed] [Google Scholar]
- 16. Ko K. S., et al. 2007. High rates of resistance to colistin and polymyxin B in subgroups of Acinetobacter baumannii isolates from Korea. J. Antimicrob. Chemother. 60:1163–1167 [DOI] [PubMed] [Google Scholar]
- 17. Li J., et al. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:2946–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li J., et al. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 6:589–601 [DOI] [PubMed] [Google Scholar]
- 19. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 20. March C., et al. 2010. Dissection of host cell signal transduction during Acinetobacter baumannii-triggered inflammatory response. PLoS One 5:e10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matthaiou D. K., et al. 2008. Risk factors associated with the isolation of colistin-resistant gram-negative bacteria: a matched case-control study. Crit. Care Med. 36:807–811 [DOI] [PubMed] [Google Scholar]
- 22. Moffatt J. H., et al. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide. Antimicrob. Agents Chemother. 54:4971–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moskowitz S. M., Ernst R. K., Miller S. I. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186:575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murphy K. C., Campellone K. G., Poteete A. R. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321–330 [DOI] [PubMed] [Google Scholar]
- 25. Peleg A. Y., Seifert H., Paterson D. L. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reis A. O., Luz D. A., Tognim M. C., Sader H. S., Gales A. C. 2003. Polymyxin-resistant Acinetobacter spp. isolates on dry surfaces. Emerg. Infect. Dis. 9:1025–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roland K. L., Martin L. E., Esther C. R., Spitznagel J. K. 1993. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J. Bacteriol. 175:4154–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rozen S., Skaletsky H. J. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365–386 In Krawetz S., Misener S. (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 29. Schurek K. N., et al. 2009. Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:4345–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith M. G., et al. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21:601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tan C. H., Li J., Nation R. L. 2007. Activity of colistin against heteroresistant Acinetobacter baumannii and emergence of resistance in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob. Agents Chemother. 51:3413–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tran A. X., et al. 2005. Resistance to the antimicrobial peptide polymyxin requires myristoylation of Escherichia coli and Salmonella typhimurium lipid A. J. Biol. Chem. 280:28186–28194 [DOI] [PubMed] [Google Scholar]
- 33. Trent M. S., et al. 2001. Accumulation of a polyisoprene-linked amino sugar in polymyxin-resistant Salmonella typhimurium and Escherichia coli: structural characterization and transfer to lipid A in the periplasm. J. Biol. Chem. 276:43132–43144 [DOI] [PubMed] [Google Scholar]
- 34. Urban C., et al. 2001. Polymyxin B-resistant Acinetobacter baumannii clinical isolate susceptible to recombinant BPI and cecropin P1. Antimicrob. Agents Chemother. 45:994–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vaara M., et al. 2008. Novel polymyxin derivatives carrying only three positive charges are effective antibacterial agents. Antimicrob. Agents Chemother. 52:3229–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vaara M., et al. 2010. A novel polymyxin derivative that lacks the fatty acid tail and carries only three positive charges has strong synergism with agents excluded by the intact outer membrane. Antimicrob. Agents Chemother. 54:3341–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Valencia R., et al. 2009. Nosocomial outbreak of infection with pan-drug-resistant Acinetobacter baumannii in a tertiary care university hospital. Infect. Control Hosp. Epidemiol. 30:257–263 [DOI] [PubMed] [Google Scholar]
- 38. Vaneechoutte M., et al. 1995. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J. Clin. Microbiol. 33:11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wendt C., Dietze B., Dietz E., Rüden H. 1997. Survival of Acinetobacter baumannii on dry surfaces. J. Clin. Microbiol. 35:1394–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]