Abstract
This work reports, for the first time, the presence of New Delhi metallo-β-lactamase 1 (NDM-1) in Pseudomonas aeruginosa. Moreover, this is the first report of the NDM-1 presence in the Balkan region. Cosmid gene libraries of carbapenem-nonsusceptible Pseudomonas aeruginosa clinical isolates MMA83 and MMA533 were screened for the presence of metallo-β-lactamases. Accordingly, both MMA83 and MMA533 carried the blaNDM-1 gene. Pulsed-field gel electrophoresis (PFGE) analysis indicated that strains MMA83 and MMA533 belonged to different clonal groups. Five additional isolates from different patients clonally related to either MMA83 or MMA533 were found to be NDM-1 positive.
TEXT
Enterobacteriaceae carrying blaNDM-1 have been found in India, Pakistan, Bangladesh, Singapore, the United States, Canada, Australia, and 13 European countries (3, 4, 5, 6, 8, 10, 13). The majority of European metallo-β-lactamase 1 (NDM-1) cases had a history of recent travel and hospital admission in India or Balkan countries (10). These findings imply the importance of NDM-1 screening in Balkan region countries like Serbia, which are being reported as potential regions of NDM-1 endemicity.
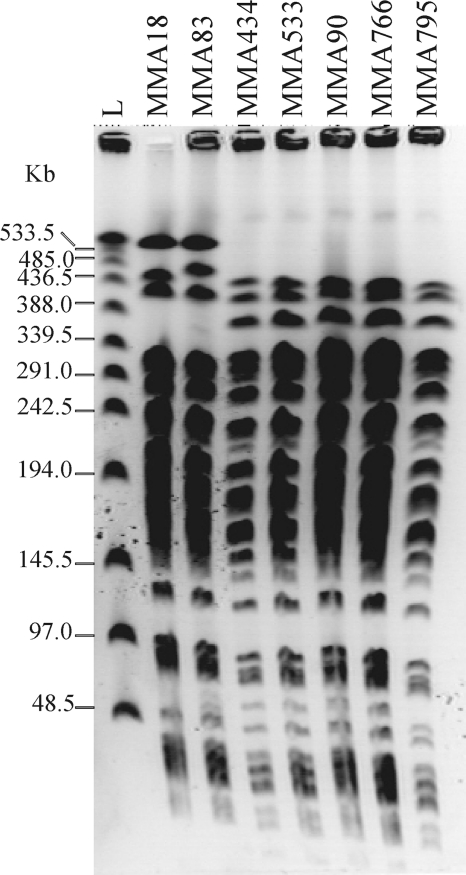
During the routine analyses of carbapenemase-producing Gram-negative bacteria in the Military Medical Academy (MMA; Belgrade, Serbia) in 2010, based on Etest (AB Biodisk, Solna, Sweden) and the combined disk method with imipenem-EDTA, seven Pseudomonas aeruginosa isolates were selected as potential metallo-β-lactamase (MBL) producers. Pulsed-field gel electrophoresis (PFGE; SpeI macrorestriction) analysis (11) revealed two major genotype groups; thus, representative strains for both genotypes (MMA83 and MMA533) were used in further experiments (Fig. 1). MMA83 and MMA533 were isolated from two patients that underwent invasive surgical interventions, with no epidemiological connection determined between them. Patient 1 (carrying MMA83), a 61-year-old Serbian woman, was admitted to the MMA in March 2010 as an emergency case diagnosed with stercoral fistulas and intra-abdominal abscess in the left lumbal retroperitoneal region. During her hospitalization, multiple Gram-negative bacteria were isolated, including NDM-1-producing P. aeruginosa isolated from urine. The patient died 1 month after admission. Patient 2 (carrying MMA533), a 63-year-old Serbian women with intestinal carcinoma, received an explorative laparotomy as an emergency case in the MMA on March 2010. Multiple Gram-negative and Gram-positive bacteria were isolated, including P. aeruginosa (NDM-1 producer MMA533), Acinetobacter spp., and Staphylococcus aureus (methicillin resistant) obtained from a wound. The patient died 2 months after admission. It is difficult to establish whether MMA83 and MMA533 were colonizers or the cause of the infection.
Fig. 1.
PFGE profiles of P. aeruginosa isolates carrying the blaNDM-1 gene (MMA18, MMA83, MMA434, MMA533, MMA90, MMA766, and MMA795). L, λ concatemers (New England Biolabs).
Antimicrobial susceptibility tests for MMA83 and MMA533 were done by use of the disk diffusion method on Mueller-Hinton agar (Bio-Rad Laboratories, Marnes-la-Coquette, France). Susceptibility was determined using breakpoints recommended by the Clinical and Laboratory Standards Institute (2011) (1). MICs of selected antimicrobials were determined by Etest (AB Biodisk, Solna, Sweden). Both strains were resistant to imipenem and meropenem (Table 1). Strains were positive for MBL production in the imipenem-EDTA, ceftazidime-EDTA, and cefepime-EDTA combined disk tests and in the MBL Etest. PCRs with primers specific for the blaIMP (5′-GAAGGYGTTTATGTTCATAC-3′ and 5′-GTAMGTTTCAAGAGTGATGC-3′), blaSPM (5′-CTGCTTGGATTCATGGGCGC-3′ and 5′-CCTTTTCCGCGACCTTGATC-3′), blaGIM (5′-TCGACACACCTTGGTCTG-3′ and 5′-AACTTCCAACTTTGCCAT-3′), blaSIM (5′-TACAAGGGATTCGGCATCC-3′ and 5′-TAATGGCCTGTTCCCATG-3′), and blaVIM (5′-GTTTGGTCGCATATCGCAAC-3′ and 5′-AATGCGCAGCACCAGGATAG-3′) genes were negative (2, 7). In order to reveal the genetic determinant of MBL production, cosmid gene libraries of MMA83 and MMA533 were constructed with Gigapack III Gold packaging extract (Stratagene, Amsterdam, Netherlands). Total DNA from strains was partially digested with PstI and cloned into the pLAFR3 cosmid (9). Cosmid libraries were screened for the presence of β-lactamases by plating on tetracycline (20 μg/ml)- and ampicillin (100 μg/ml)-containing plates. Selected cosmids pLAFR83 and pLAFR533 from MMA83 and MMA533, respectively, conferring resistance to beta-lactams, were digested with EcoRI and BamHI. Restriction patterns of the pLAFR83 and pLAFR533 cosmids differed substantially. Plasmid libraries of DNA fragments from these cosmids were generated in pBluescript vector (the gene for resistance to ampicillin was replaced with a neomycin resistance gene from pUC4K) by using EcoRI and BamHI. Constructs pBS83E, pBS83B, pBS533E, and pBS533B carrying determinants for beta-lactam resistance were selected. Sequencing revealed the presence of identical blaNDM-1 genes (100% identity) on 1,059-bp EcoRI fragments in both cosmids. Both nucleotide and amino acid sequences of the blaNDM-1 genes and their corresponding proteins, respectively, from P. aeruginosa MMA83 and MMA533 isolates analyzed in this work shared 100% of identity with the blaNDM-1 genes and their proteins from Klebsiella pneumoniae KP-05-506 and Escherichia coli 271. The presence of NDM-1 was confirmed in other selected P. aeruginosa strains (MMA18, MMA434, MMA90, MMA766, and MMA795) by using primers specific for blaNDM-1 (14). Referring to the previous data on NDM-1 distribution among bacterial genera (4, 10, 13), we report, for the first time, the presence of NDM-1 in Pseudomonaceae. Also, this is the first report of blaNDM-1-positive isolates detected in the Balkan region, which were previously suspected as sources of travel-associated NDM-1 infections, along with the Indian subcontinent (10). Moreover, 6 out of 55 travel-associated NDM-1 cases reported in European countries had a link to the Balkan region, presumably to Serbia (including Kosovo), Montenegro, and Bosnia and Herzegovina (10). However, it should be emphasized that patients from these surveillances were not screened for the presence of NDM-1 prior to departure from resident countries; thus, it is difficult to determine whether the Balkan region is the primary source of NDM-1 in those studies. According to our results and indications from previous reports, one could assume that NDM-1 is present in the Balkan region, including Serbia, and maybe in higher frequency than in the rest of Europe, but experimental data supporting this speculation are required.
Table 1.
Antibiotic susceptibility of P. aeruginosa strains MMA83 and MMA533
| Antibiotic | MIC (μg/ml) |
|
|---|---|---|
| MMA83 | MMA533 | |
| Meropenem | >32 | >32 |
| Imipenem | >32 | >32 |
| Cefepime | >256 | >256 |
| Aztreonam | 6 | 3 |
| Piperacillin-tazobactam | 96 | 24 |
| Gentamicin | >256 | >256 |
| Amikacin | >256 | >256 |
| Netilmicin | >256 | >256 |
| Ciprofloxacin | >32 | >32 |
Both of the NDM-1 cases described in this paper had comorbidity after the patients had undergone invasive procedures, indicating that the severities of illness were similar to those in Enterobacteriaceae-associated NDM-1 infections. Prior to the detection of NDM-1-positive P. aeruginosa strains, none of the patients from our study had travel history with the Indian subcontinent or any European country. Accordingly, either these patients experienced cross-transmission with NDM-1 cases that were travel associated (but not reported) or NDM-1 is an endemic in Serbia or in the entire Balkan region as previously suggested (10). Since the two NDM-1-positive P. aeruginosa strains analyzed in our study belong to different clonal groups, we can speculate that these strains came from two different sources of infection. Considering that NDM-1 genes in Pseudomonas are identical to those from Enterobacteriaceae, further experiments are required to determine transmissible elements and transfer mechanisms existing in both groups of bacteria. DNA-DNA hybridization revealed that the blaNDM-1 gene probe hybridized to a ∼55-kb band in MMA83 and MMA533 SpeI macrorestriction (data not shown).
The ability of NDM-1 to spread not only among Enterobacteriaceae but also among other bacterial families, like Pseudomonaceae, implies the possibility for numerous new NDM-1 cases to be detected in the near future. If we look at the chronology of VIM-1 spreading, a rapid emergence of new NDM-1 cases might be expected (12).
Nucleotide sequence accession numbers.
The nucleotide sequences of the blaNDM-1 genes from MMA83 and MMA533 are available in GenBank/EMBL under accession numbers FR820590 and FR820591, respectively.
Acknowledgments
This work was supported by grant 173019 from the Ministry of Education and Science of the Republic of Serbia.
Footnotes
These authors equally contributed to this work.
Published ahead of print on 6 June 2011.
REFERENCES
- 1. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. M100–S21, replaces M100-S20 and M100-S20-U Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 2. Ellington M., Kistler J., Livermore D., Woodford N. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J. Antimicrob. Chemother. 59:321–322 [DOI] [PubMed] [Google Scholar]
- 3. Koh T. H., et al. 2010. Global spread of New Delhi metallo-beta-lactamase 1. Lancet Infect. 10:828. [DOI] [PubMed] [Google Scholar]
- 4. Kumarasamy K. K., et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mulvey M. R., Grant J. M., Plewes K., Roscoe D., Boyd D. A. 2011. New Delhi metallo-beta-lactamase in Klebsiella pneumoniae and Escherichia coli, Canada. Emerg. Infect. Dis. 17:103–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Muir A., Weinbren M. J. 2010. New Delhi metallo-beta-lactamase: a cautionary tale. J. Hosp. Infect. 75:239–240 [DOI] [PubMed] [Google Scholar]
- 7. Pitout J. D. D., et al. 2005. Detection of Pseudomonas aeruginosa producing metallo-β-lactamases in a large centralized laboratory. J. Clin. Microbiol. 43:3129–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poirel L., Lagrutta E., Taylor P., Pham J., Nordmann P. 2010. Emergence of metallo-beta-lactamase NDM-1 producing multiresistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 54:4914–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Staskawicz B., Dahlbeck D., Keen N., Napoli C. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789–5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Struelens M. J., et al. 2010. New Delhi metallo-beta-lactamase 1-producing Enterobacteriaceae: emergence and response in Europe. Euro Surveill. 15:pii19716 [DOI] [PubMed] [Google Scholar]
- 11. Tenover C. F., et al. 1995. Interpreting chromosomal DNA restriction patterns produced by Pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Toleman M. A., Rolston K., Jones R. N., Walsh T. R. 2004. blaVIM-7, an evolutionarily distinct metallo-β-lactamase gene in a P. aeruginosa isolate from the United States. Antimicrob. Agents Chemother. 48:329–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yong D., Toleman M. A., Giske C. G., Walsh T. R. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zarfel G., et al. 2011. Emergence of New Delhi metallo-β-lactamase, Austria. Emerg. Infect. Dis. 17:129–130 [DOI] [PMC free article] [PubMed] [Google Scholar]