Abstract
Echinocandins, including caspofungin (CSP) and micafungin (MCF), are highly active versus Candida glabrata (MIC of ≤0.06 μg/ml). True resistance (MIC of ≥1 μg/ml) is a rare event and strictly associated with mutations in β-1,3-glucan synthase gene FKS1 or FKS2. In contrast, we show here that mutants exhibiting reduced susceptibility to CSP (CRS; MICs of 0.12 to 0.5 μg/ml) are readily selected in vitro and, paradoxically, demonstrate increased susceptibility to MCF (MIS) ranging from 4- to 32-fold. CRS-MIS mutants were generated from all 10 C. glabrata strains tested and were tentatively identified within a collection of clinical isolates. Intriguingly, sequencing and gene disruption demonstrated that CRS-MIS is Fks independent.
TEXT
The echinocandins caspofungin (CSP; FDA approved in 2001), micafungin (MCF; FDA approved in 2005), and anidulafungin (ANF; FDA approved in 2006) are semisynthetic lipopeptide inhibitors of β-1,3-glucan synthase and hence fungal cell wall synthesis (3, 11). They are highly active versus most Candida species, including C. albicans and C. glabrata (9, 12). The latter activity is particularly important, since resistance to widely used azole antifungals often complicates treatment of C. glabrata infections. Consequently, echinocandins have been elevated recently to first-line agents for treatment of invasive C. glabrata infection (10). While acquired echinocandin resistance remains rare, it appears to be increasing in C. glabrata in response to increasing clinical use (5, 12, 14, 18). Resistance to date in clinical isolates has been strictly associated with mutations in the putative β-1,3-glucan synthases Fks1 and Fks2 and is typically characterized by CSP, MCF, and ANF cross-resistance (1, 2, 6, 8, 13, 14, 18). Here, we describe initial laboratory studies examining the potential for echinocandin resistance in C. glabrata and the mechanisms behind it.
Experimental procedures.
The medium was YPD (1% yeast extract, 2% peptone, 2% dextrose) or, where indicated, RPMI (RPMI 1640, 2% dextrose, 0.165 M MOPS [morpholinepropanesulfonic acid] [pH 7], supplemented as needed with 50 μg/ml uridine). CSP (Merck, Rahway, NJ), MCF (Astellas, Deerfield, IL), and ANF (Pfizer, New York, NY) stocks were prepared in dimethyl sulfoxide; the final solvent concentration was <0.5% in all cases. Colonies were selected on CSP-containing YPD (or RPMI) agar overlaid with 1 × 106 to 1 × 107 cells with incubation at 35°C for 3 to 4 days, followed by isolation on drug-free plates with incubation for 2 days. The MIC (defined as ≥80% inhibition relative to a drug-free control) was determined by broth microdilution as described previously (8, 16) with incubation in YPD (or RPMI) at 35°C for 24 h. Strain 66032u (ura3 auxotroph) has been described previously (16); additional strains were obtained from diverse sources (Table 1). The fks1Δ, fks2Δ, and fks3Δ disruptants were generated in mutant 66032u-C2 and its wild-type (WT) 66032u parent by the PRODIGE method with a URA3 marker (for primers, see Table S1 in the supplemental material), as previously described (4, 7). FKS1, FKS2, and FKS3 coding sequences were amplified and directly sequenced using gene-specific primers (see Table S1), as described previously (8).
Table 1.
CSP and MCF susceptibilities of C. glabrata WT strains and their CRS-MIS mutants determined by broth microdilution in YPD
| Strain | Source or reference | WT CSP/MCF MIC in μg/ml | Selection medium (CSP concn in μg/ml)a | Mutant CSP/MCF MIC in μg/ml (CRS-MIS differential) |
||
|---|---|---|---|---|---|---|
| C1 | C2 | C3 | ||||
| 66032u | 16 | 0.03/0.008 | YPD (0.2) | 0.5/0.00025 (512) | 0.5/0.0005 (256) | 0.5/0.001 (128) |
| RPMI (0.25) | 0.5/0.00025 (512) | 1/0.002 (128) | ||||
| 380 | 15 | 0.016/0.016 | YPD (0.25) | 0.12/0.002 (64) | 0.12/0.004 (32) | 0.12/0.004 (32) |
| RPMI (0.25) | 0.5/0.004 (128) | |||||
| 945 | 15 | 0.008/0.008 | YPD (0.2) | 0.25/0.001 (256) | 0.12/0.0005 (256) | 0.06/0.0005 (128) |
| 2807990 | 17 | 0.03/0.008 | YPD (0.25, 0.3) | 0.5/0.001 (128) | 0.5/0.001 (128) | |
| 2806145 | 17 | 0.06/0.008 | YPD (0.3) | 0.12/0.004 (4) | 012/0.002 (8) | |
| 34-023-157 | 9 | 0.03/0.008 | YPD (0.25) | 0.25/0.002 (32) | 0.12/0.001 (32) | 0.125/0.001 (32) |
| 34-023-038.02 | 9 | 0.06/0.03 | YPD (0.25) | 0.12/0.004 (16) | 0.12/0.004 (16) | 0.125/0.004 (16) |
| 33-94R-0024-119 | 9 | 0.03/0.03 | YPD (0.25) | 0.12/0.008 (16) | ||
| 38326 | ATCCb | 0.03/0.03 | YPD (0.25) | 0.5/0.004 (128) | 0.5/0.004 (128) | 0.5/0.004 (128) |
| 2001 | ATCC | 0.008/0.016 | YPD (0.2) | 0.12/0.004 (64) | 0.03/0.004 (16) | 0.03/0.008 (8) |
Mutants were selected on YPD or RPMI agar with the indicated concentration of CSP. Following selection, 3 to 22 (median of 6) colonies were tested for each strain, yielding 1 to 6 (median of 3) CRS-MIS mutants.
ATCC, American Type Culture Collection, Manassas, VA.
Mutant selection and characterization.
In initial studies, we attempted to isolate CSP-resistant mutants (MIC of ≥1 μg/ml) of C. glabrata 66032u (CSP MIC of 0.03 μg/ml) by plating 3.8 × 107 total cells on 1 or 2 μg/ml CSP-containing YPD agar. This yielded only one mutant (frequency of ≈3 × 10−8), which exhibited cross-resistance to CSP, MCF, and ANF (MICs in YPD of 8, >16, and 8 μg/ml, respectively) and the commonly described Fks1 hot spot 1 mutation, S629P.
In contrast, plating on 0.2 μg/ml CSP-containing YPD yielded mutants exhibiting reduced susceptibility to CSP (CRS; MICs in YPD of 0.12 to 0.5 μg/ml) at relatively high frequency (2 × 10−5). Surprisingly, 5 of the 8 CRS mutants of strain 66032u that were isolated and tested exhibited 4- to 32-fold increased susceptibility to MCF (MIS); representative MIC data are presented in Table 1. The remaining three mutants exhibited minimal (≤2-fold) change in susceptibility to both CSP and MCF, and susceptibility to ANF was minimally changed for all eight mutants (not shown).
To explore the generality of this paradoxical CRS-MIS phenotype, mutants of 9 additional C. glabrata strains were similarly selected on YPD plates containing 0.2 to 0.3 μg/ml CSP. CRS-MIS mutants were obtained for all strains, again at relatively high frequency (range, 2 × 10−6 to 2 × 10−5). CSP and MCF MICs for representative mutants (labeled C1, C2, and C3) are presented in Table 1. The magnitude of the phenotype, expressed as CRS-MIS differential (CSP fold increase × MCF fold decrease), ranged from 4 to 512, with a median of 32, and appeared to be strain dependent. Again, for all mutants, ANF MICs were ≤2-fold changed (not shown).
With respect to medium, CRS-MIS mutants were also readily obtained following selection on RPMI agar rather than YPD for both strains tested (Table 1). However, for both YPD- and RPMI-selected mutants, the CRS-MIS differential was consistently higher when assayed in YPD than when assayed in RPMI, primarily due to the higher activity of CSP in the former compared to the latter (Table 1 and see Table S2 in the supplemental material). Specifically, the CRS-MIS differential in RPMI ranged from 2-fold to 32-fold, with a median of 8-fold, i.e., 4-fold less than the differential obtained in YPD. This diminishing of the CRS-MIS phenotype in RPMI may explain why it has not been previously noted.
Using mutant 66032u-C2 (Table 1) as a model, we examined stability and reversion. Its CRS-MIS phenotype was unchanged after six passages on drug-free YPD. However, following selection on 0.004 μg/ml MCF, partial revertants of both the CRS and MIS phenotypes were isolated at low frequency (3 × 10−7) (data not shown). Several attempts were made to isolate mutants with reduced susceptibility to MCF that exhibited increased susceptibility to CSP (i.e., the inverse of CRS-MIS), but this phenotype was never observed. Similar attempts with ANF also failed.
Mechanism.
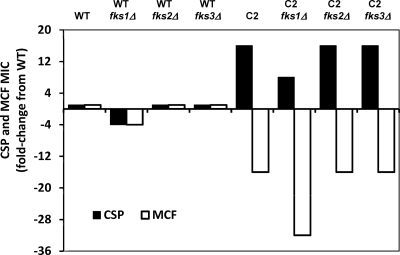
The mechanism behind CRS-MIS is of considerable interest given its paradoxical effects, relatively high frequency, and potential implications for echinocandin therapy (see below). In light of the central roles of FKS genes in true echinocandin resistance (1, 2, 6, 8, 13, 14, 18), we tested their roles in the CRS-MIS phenotype, initially by constructing fks1Δ, fks2Δ, and fks3Δ disruptants of both mutant 66032u-C2 and its WT 66032u parent. The CRS-MIS phenotype (256-fold differential) of 66032u-C2 was retained in all three disruptants (Fig. 1). (The CSP and MCF MICs decreased 2-fold in the fks1Δ disruptant of 66032u-C2, but they similarly decreased in the fks1Δ disruptant of its WT parent.)
Fig. 1.
The CRS-MIS phenotype (256-fold differential) of mutant 66032u-C2 is unaltered in fks1Δ, fks2Δ, and fks3Δ disruptants.
Next, the complete 5.5- to 5.7-kbp coding regions of FKS1, FKS2, and FKS3 were amplified from 66032u-C2 and directly sequenced. No mutation was identified relative to parent 66032u. These data strongly suggest that the CRS-MIS mechanism is Fks-independent.
Clinical implications.
It will be important to determine if CRS-MIS mutants, readily generated in vitro, also arise in vivo during CSP therapy of C. glabrata infection. Initial support for this was provided by the identification of three potential CRS-MIS mutants (strains 4719, 4743, and 4771) within a large collection of bloodstream isolates with characterized echinocandin MICs (1). Relative to representative wild-type C. glabrata strains (Table 1) (median CSP and MCF MICs of 0.03 and 0.008 to 0.016 μg/ml, respectively), these three bloodstream isolates (CSP and MCF MICs of 0.12 to 0.25 and 0.0005 to 0.001 μg/ml) exhibited 64- to 256-fold CRS-MIS differentials in YPD medium. For one of these isolates (4771), complete FKS1 and FKS2 coding regions were sequenced, and no amino acid substitutions were identified relative to WT Fks1 (XP_446406) and Fks2 (XP_448401). In the absence of their WT parents, confirmation that these clinical isolates are truly CRS-MIS mutants must await elucidation of the CRS-MIS mechanism. Alternatively, the generation of CRS-MIS mutants in vivo can be tested by characterization of paired isolates obtained before and after CSP therapy.
If CRS-MIS mutants do indeed arise during CSP therapy of C. glabrata infection, there are two important implications. First, these mutants could represent a CSP-tolerant population from which truly resistant clones with Fks mutations arise. Second, the CRS-MIS phenotype implies that sequential therapy—CSP followed by MCF—may be more effective than conventional therapy with a single echinocandin.
Nucleotide sequence accession numbers.
The sequences of the coding regions of FKS1, FKS2, and FKS3 have been deposited in GenBank under accession no. HQ845283, HQ845284, and HQ845285, respectively.
Supplementary Material
Acknowledgments
We thank J. Rex, J. Sobel, and J.-P. Vermitsky for generously providing strains.
These studies were supported by National Institutes of Health grant AI075272 and Pfizer Investigator-Initiated Research grant WS428829.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 31 May 2011.
REFERENCES
- 1. Castanheira M., et al. 2010. Low prevalence of fks1 hot spot 1 mutations in a worldwide collection of Candida strains. Antimicrob. Agents Chemother. 54:2655–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cleary J. D., Garcia-Effron G., Chapman S. W., Perlin D. S. 2008. Reduced Candida glabrata susceptibility secondary to an FKS1 mutation developed during candidemia treatment. Antimicrob. Agents Chemother. 52:2263–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Douglas C. M. 2001. Fungal β(1,3)-D-glucan synthesis. Med. Mycol. 39(Suppl. 1):55–66 [DOI] [PubMed] [Google Scholar]
- 4. Edlind T. D., et al. 2005. Promoter-dependent disruption of genes: simple, rapid, and specific PCR-based method with application to three different yeast. Curr. Genet. 48:117–125 [DOI] [PubMed] [Google Scholar]
- 5. Garcia-Effron G., et al. 2010. Novel FKS mutations associated with echinocandin resistance in Candida species. Antimicrob. Agents Chemother. 54:2225–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garcia-Effron G., Lee S., Park S., Cleary J. D., Perlin D. S. 2009. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-β-d-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob. Agents Chemother. 53:3690–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Katiyar S., Johnson M. E., Edlind T. D. 2010. Role of Fks1, Fks2, and Fks3 in Candida glabrata susceptibility and resistance to echinocandins, abstr. F-1691, p. 131 Abstr. 110th Gen. Meet. Am. Soc. Microbiol American Society for Microbiology, Washington, DC [Google Scholar]
- 8. Katiyar S., Pfaller M., Edlind T. 2006. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 50:2892–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ostrosky-Zeichner L., et al. 2003. Antifungal susceptibility survey of 2,000 bloodstream Candida isolates in the United States. Antimicrob. Agents Chemother. 47:3149–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pappas P. G., et al. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 48:503–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perlin D. S. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist. Update 10:121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pfaller M., et al. 2011. Use of epidemiological cutoff values to examine 9-year trends in susceptibility of Candida species to anidulafungin, caspofungin, and micafungin. J. Clin. Microbiol. 49:624–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pfaller M. A., et al. 2011. Clinical breakpoints for the echinocandins and Candida revisited: integration of molecular, clinical, and microbiological data to arrive at species-specific interpretive criteria. Drug Resist. Updates. 14:164–176 [DOI] [PubMed] [Google Scholar]
- 14. Pfeiffer C. D., et al. 2010. Breakthrough invasive candidiasis in patients on micafungin. J. Clin. Microbiol. 48:2373–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sobel J. D., et al. 2003. Fluconazole susceptibility of vaginal isolates obtained from women with complicated Candida vaginitis: clinical implications. Antimicrob. Agents Chemother. 47:34–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vermitsky J.-P., et al. 2006. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol. Microbiol. 61:704–722 [DOI] [PubMed] [Google Scholar]
- 17. Vermitsky J. P., et al. 2008. Survey of vaginal-flora Candida species isolates from women of different age groups by use of species-specific PCR detection. J. Clin. Microbiol. 46:1501–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zimbeck A. J., et al. 2010. FKS mutations and elevated echinocandin MIC values among Candida glabrata isolates from U.S. population-based surveillance. Antimicrob. Agents Chemother. 54:5042–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.