Abstract
One method that bacteria employ to reduce their susceptibility to antibiotics is the formation of biofilms. We developed a robust 6-well plate biofilm assay to evaluate early-stage discovery compounds against methicillin-resistant Staphylococcus aureus (MRSA). Tissue culture-treated 6-well plates were selected for this assay because they facilitate the adherence of MRSA and enable accurate determination of the number of CFU in each well. The MRSA biofilms formed in this assay exhibit increased tolerances to clinically used antibiotics. Using this biofilm assay, we identified a novel potentiator of gentamicin against MRSA biofilms. The combination of gentamicin and pentadecenyl tetrazole is superior to clinically used MRSA antibiotics against these MRSA biofilms. This novel combination also exhibits synergistic effects on MRSA planktonic cells. This plant-derived compound reveals promise for its effectiveness and warrants further lead optimization as an antibiotic and aminoglycoside potentiator.
INTRODUCTION
Antibiotic resistance among pathogenic bacteria is increasing, creating the need for new antibacterial agents with different modes of action. Among these pathogenic bacteria, Staphylococcus aureus has developed remarkable antibiotic resistance, causing health care practitioners to appeal for increased research efforts to find new treatments. Methicillin-resistant S. aureus (MRSA) has been found to be the most common identifiable cause of infections in skin and soft tissue in several U.S. cities (17), and occurrences are on the rise (11).
The difficulty of treating MRSA may be exacerbated by its ability to form biofilms in the body. Biofilms in general account for more than 65% of bacterial infections (13). These infections are costly to treat, often requiring multiple rounds of antibiotics. As a disease, bacterial biofilm infections have an extensive effect on the human body, including skin, bone, ear, sinus, urinary tract, heart, and lung (3, 7, 20, 24, 25). Biofilms form on indwelling catheters and other surgically implanted medical devices, necessitating additional surgery to remove or replace the device. Biofilms are especially problematic because they exhibit increased resistance to antibiotics; using a variety of mechanisms, bacteria in biofilms can be 10 to 1,000 times more resistant than their planktonic counterparts (16). In order to continue to provide effective antibacterial treatments, it will be advantageous to develop new antibiotics or new agents that can act synergistically with existing antibiotics to prolong their usefulness.
To better understand how to attack biofilms, researchers are continually trying to develop new methods to study biofilms and evaluate new antibiofilm treatments in the laboratory. Assays have been developed to study biofilms in flow cells, on agar plates, at liquid-air interfaces, and on the surfaces of wells in microtiter plates (8, 9, 13, 23). Numerous variations of these assays have been established to answer different questions about different species of bacteria. To facilitate our search for new antimicrobial compounds active against MRSA in biofilms, we chose to develop a method that grows biofilms in 6-well plates instead of the more widely used 96-well plates. Using 6-well plates makes it possible to scrape and remove a biofilm and quantify the viable CFU in each well. Therefore, this method enables a side-by-side comparison of the effectiveness of antibiotics and investigational compounds used alone or in combination at reducing viable CFU from established MRSA biofilms. Most importantly, experiments performed weeks apart or in different laboratories can be compared based upon reduction of CFU instead of the more qualitative reduction of absorbances produced using conventional staining techniques. Furthermore, the resulting biofilm produced using the method described herein exhibits increased tolerances to clinically used MRSA antibiotics compared to their corresponding planktonic MICs. This increased tolerance to antibiotics demonstrates that these MRSA biofilms exhibit this hallmark phenotype of biofilms.
Here, we describe a reliable method for growing and quantifying MRSA biofilms. We further describe the use of this procedure in evaluating a new antibacterial compound, SEQ-914 [(Z)-5-(pentadec-8-enyl)-2H-tetrazole], and comparing it to clinically used MRSA antibiotics.
(Portions of this work were previously shown at ASM Biofilm Conference 2009.)
MATERIALS AND METHODS
Bacterial strains and general methods.
Cation-adjusted Mueller-Hinton broth (CAMHB) and Trypticase soy broth (TSB) were obtained from BD Biosciences (San Jose, CA). GNT (glucose plus NaCl plus TSB) was prepared by supplementing TSB with 0.5% glucose and 3% NaCl. Colony counts were determined on TSB agar plates. Overnight broth cultures were inoculated in TSB from frozen stocks stored at −70°C in TSB plus 20% glycerol.
The isolates of MRSA and methicillin-susceptible S. aureus (MSSA) are described in Table 1. Two laboratory strains were obtained from the American Type Culture Collection (ATCC). Clinical isolates were obtained from David Hunstad at St. Louis Children's Hospital (SLCH). The strains were first isolated from patient specimens on 5% sheep blood agar plates, and their relevance and genotypes have been described (15). Briefly, Panton-Valentine leukocidin (PVL) has pathogenic importance with regulation of the accessory gene regulator (Agr) (12, 27). Agr regulates protein expression during the transition from exponential to stationary phase in the quorum sensing system (19). An enzyme gene that codes for the bacteriocin biosynthesis pathway (bsaB) was also reported, along with the arginine catabolic mobile element (ACME), which was originally identified in a USA300 isolate of S. aureus.
Table 1.
S. aureus strains used in this studya
| Strain | Characteristic(s) | Source |
|---|---|---|
| ATCC 25923 | MSSA clinical isolate | ATCC |
| ATCC BAA-40 | MRSA clinical isolate; nasal passage | ATCC |
| Superficial | ||
| MRSA-105 | MRSA; PVL+ ACME+bsaB+ Agr1 | SLCH |
| MRSA-106 | MRSA; PVL+ ACME+bsaB+ Agr1 | SLCH |
| MRSA-107 | MRSA; PVL+ ACME+bsaB+ Agr1 | SLCH |
| MRSA-108 | MRSA; PVL+ ACME−bsaA+ Agr1 | SLCH |
| MSSA-109 | MSSA; PVL+ ACME−bsaB+ Agr1 | SLCH |
| MRSA-111 | MRSA; PVL+ ACME+bsaB+ Agr1 | SLCH |
| MRSA-148 | MRSA; PVL+ ACME+bsaB+ Agr1 | SLCH |
| MRSA-158 | MRSA; PVL+ ACME+bsaB+ Agr1 | SLCH |
| MSSA-175 | MSSA; PVL+ ACME−bsaB+ Agr1 | SLCH |
| MRSA-295 | MRSA; PVL− ACME−bsaB+ Agr1 | SLCH |
| Invasive | ||
| MRSA-186 | MRSA; PVL+ ACME−bsaA+ Agr1 | SLCH |
| MRSA-194 | MRSA; PVL+ ACME+bsaB+ Agr1 | SLCH |
Strains used in this study included methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-susceptible Staphylococcus aureus (MSSA). Genotype information available for each clinical isolate included Panton-Valentine leukocidin (PVL), arginine catabolic mobile element (ACME), biosynthesis pathway (bsaB), and accessory gene regulator (Agr). Sources: American Type Culture Collection (ATCC) and St. Louis Children's Hospital (SLCH).
Antibacterial agents.
Gentamicin (GEN) and trimethoprim-sulfamethoxazole (SXT) were obtained from Sigma-Aldrich (St. Louis, MO), vancomycin (VAN) was purchased from Gold Biotechnology (St. Louis, MO), and daptomycin (DAP) and linezolid (LZD) were from Tecoland (Edison, NJ).
MIC determination.
Standardized planktonic antibiotic MICs were determined by the broth microdilution method outlined by the CLSI (5), using CAMHB medium. The initial inoculum contained 106 CFU/ml. The MIC was defined as the lowest concentration that prevented visible growth of the bacteria after overnight incubation at 37°C. The optical density at 600 nm (OD600) of each well was measured with a Versamax plate reader (Molecular Devices, Sunnyvale, CA). MICs were obtained for GEN and SEQ-914 in 14 isolates (see Table 3). MRSA-295 was used for VAN, LZD, SXT, and DAP (Table 2).
Table 3.
MICs of gentamicin and SEQ-914 against MSSA and MRSA isolates in CAMHB
| Isolate | MIC (μg/ml)a |
|
|---|---|---|
| Gentamicin | SEQ-914 | |
| MSSA ATCC 25923 | 0.5 | 32 |
| MRSA ATCC BAA-40 | >400 | 16 |
| MRSA-105 | 1 | 16 |
| MRSA-106 | 4 | 32 |
| MRSA-107 | 1 | 32 |
| MRSA-108 | 1 | 32 |
| MRSA-109 | 1 | 32 |
| MRSA-111 | 1 | 32 |
| MRSA-148 | 1 | 32 |
| MRSA-158 | 2 | 32 |
| MRSA-175 | 4 | 32 |
| MRSA-186 | 4 | 64 |
| MRSA-194 | 2 | 32 |
| MRSA-295 | 2 | 32 |
Determined by broth microdilution from 0.03125 to 400 μg/ml.
Table 2.
Antibiofilm activity against MRSA-295
| Treatment groupa | MIC (μg/ml)b | Concn (μg/ml)c | Log10 no. of CFU/biofilm (mean ± SD)d | Log reduction |
|---|---|---|---|---|
| Inoculum | NA | 8.24 ± 0.16 | ||
| Control untreated | NA | 9.32 ± 0.07 | ||
| DAP | 2 | 8 | 7.92 ± 0.16 | 0.32 |
| 16 | 5.38 ± 0.28 | 2.85 | ||
| LZD | 2 | >64 | 8.51 ± 0.07 | −0.27 |
| GEN | 2 | 4 | 7.97 ± 0.14 | 0.27 |
| 8 | 6.11 ± 0.21 | 2.13 | ||
| SXT | 1 | >64 | 8.83 ± 0.01 | −0.59 |
| VAN | 1 | 4 | 9.07 ± 0.09 | −0.82 |
| 8 | 6.02 ± 0.30 | 2.22 |
Antibiotics currently on the market in our biofilm assay: daptomycin (DAP), linezolid (LZD), gentamicin (GEN), trimethoprim-sulfamethoxazole (SXT), and vancomycin (VAN).
MIC was determined by broth microdilution.
Biofilm assay. NA, not applicable.
A minimum of two independent experiments were performed, with three wells per experiment. Data are reported as averages (n = 6 as a minimum) for these experiments.
Time-kill assay.
The time dependence of bacterial killing was measured by incubating an initial inoculum of MRSA-295 between 3 × 106 and 4 × 107 CFU/ml with varied concentrations of test compound. Assays were performed with 5 ml of CAMHB medium in triplicate 14-ml round-bottom plastic tubes. Tubes were harvested after 0, 2, 5, or 24 h of static incubation at 37°C, and numbers of CFU were determined using a standard titer method on TSB agar plates. Time-kill assays were performed at least three times, and results are averages for the three.
Biofilm assay development.
Twelve different kinds of commercially treated 96-well plates, including tissue culture plate (92096; TPP, Trasadingen, Switzerland), Gelatin (543689; BD Scientific, Franklin Lakes, NJ), poly-d-lysine (356461; BD Scientific), collagen I rat tail (356701; BD Scientific) and soft u-bottom (353911; BD Scientific), Immulon IB (6505; Thermo Scientific, Hudson, NH), delta tissue (167008; Thermo Scientific), Immulon 4 HBX (3855; Thermo Scientific), polystyrene u-bottom (3378; Corning, Corning, NY) and flat (3370; Corning), tissue culture round (3358; Corning) and flat (3596; Corning), were tested for adhesion capacity against the 14 isolates of MRSA and MSSA. Overnight cultures were diluted 1:100 into fresh TSB plus 0.5% glucose or GNT and grown with shaking at 37°C. The resulting log-phase culture was then used to inoculate the same medium to a final density of 1.0 × 107 CFU/ml. This culture (200 μl per well) was placed into the test plates and incubated overnight at 37°C. In order to visualize the biofilm and qualitatively measure its adhesion capability, a modified Kolter assay was used (21, 22). Plates were gently rinsed in three tubs of deionized water, three times per tub, and were then allowed to dry for 10 min. The biofilms were stained with 200 μl of 0.1% crystal violet (Sigma-Aldrich) for 10 min and then rinsed as described above and dried inverted for 10 min. The crystal violet was then dissolved in 200 μl of 33% acetic acid, 100 μl of the resulting solution was transferred to a flat-bottom assay plate, and the OD600 was measured.
Biofilm assay.
Based on the biofilm assay development described above, we adopted the following scaled-up method for assessing the effect of chemical agents on MRSA biofilms. Previous research has demonstrated that GNT increases adherence of Gram-positive bacteria (1, 10). Therefore, we compared CAMHB and GNT in the six-well plate biofilm assay. Our data demonstrated that CAMHB produced adherence of MRSA equivalent to that observed in the well plates containing GNT. We subsequently chose to continue our research with CAMHB, the accepted medium for testing antimicrobial activity in MRSA (5). Overnight MRSA-295 cultures were diluted 1:100 in CAMHB to achieve an OD600 of 0.3. They were immediately diluted 1:1,000. Cultures (5 ml/well) were then placed in tissue culture-treated 6-well plates (3516; Costar). Static biofilms were allowed to form at 37°C for 5 h, resulting in a cell density of 1 × 108 CFU per biofilm. Half of the liquid culture was then removed and replaced with an equal volume of fresh medium plus test compound. Treatments were done in triplicate, repeated at least one time, and represented by averages in data. After 20 h of static incubation at 37°C, the biofilm was quantitated as follows. The medium was aspirated, and the wells were rinsed with 5 ml fresh TSB to remove loosely adhered MRSA. The biofilms were then scraped with a cell lifter into 5 ml fresh TSB and enumerated for bacterial growth.
Synthesis of SEQ-914.
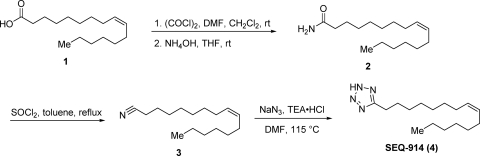
SEQ-914 was synthesized from palmitoleic acid (compound 1) as shown in Fig. 1. To a solution of compound 1 (2.0 g; 7.9 mmol) in dimethylformamide (DMF) (1.0 ml) and CH2Cl2 (30 ml) at room temperature was added a solution of oxalyl chloride (2.0 M in CH2CH2; 7.8 ml; 15.6 mmol) under nitrogen. The solution was stirred for 1.5 h and then concentrated under reduced pressure. The residue was taken up in tetrahydrofuran (THF) (30 ml), and concentrated ammonium hydroxide (21 ml; 314 mmol) was added. After stirring overnight, the mixture was diluted in CH2Cl2 (50 ml) and the organic layer was separated. The aqueous layer was extracted with CH2Cl2 (50 ml), and the combined organic layers were dried over sodium sulfate. The drying agent was removed by filtration, and the filtrate was concentrated to dryness under reduced pressure to provide (Z)-hexadec-9-enamide (compound 2; 1.9 g; 95%) as an oil: 1H NMR (300 MHz, CD3OD) δ 5.40 to 5.28 (m, 2H), 2.18 (t, J = 7.5 Hz, 2H), 2.05 to 1.98 (m, 4H), 1.63 to 1.57 (m, 2H), 1.45 to 1.25 (m, 16H), 0.90 (t, J = 6.6 Hz, 3H).
Fig. 1.
Synthesis of SEQ-914.
A solution of compound 2 (1.9 g; 7.5 mmol), thionyl chloride (2.7 ml; 37.4 mmol), and toluene (75 ml) was heated at reflux for 1.5 h. The reaction mixture was cooled and the solvent removed under reduced pressure. The residue was purified by flash chromatography (silica; 0 to 50% ethyl acetate [EtOAc] in hexanes) to provide (Z)-hexadec-9-enenitrile (compound 3; 2.1 g; 100%) as an oil: 1H NMR (300 MHz, CDCl3) δ 5.37 to 5.32 (m, 2H), 2.33 (t, J = 7.1 Hz, 2H), 2.10 to 1.95 (m, 4H), 1.68 to 1.61 (m, 2H), 1.55 to 1.20 (m, 16H), 0.91 to 0.86 (m, 3H).
A mixture of compound 3 (250 mg; 1.06 mmol), sodium azide (345 mg; 5.3 mmol), triethylamine hydrochloride (876 mg; 6.36 mmol), and DMF (10 ml) was heated to 115°C under nitrogen. The mixture was heated for 40 h and then cooled to room temperature. The solvent was removed under reduced pressure, and the residue was purified by flash chromatography (silica; 20 to 100% EtOAc in hexanes) to provide SEQ-914 (135 mg; 45%) as a white solid. The structure was verified using two-dimensional nuclear magnetic resonance (2D-NMR). Mp 28 to 30°C; 1H NMR (300 MHz, CDCl3) δ 5.34 to 5.29 (m, 2H), 3.15 to 3.10 (t, J = 7.7 Hz, 2H), 2.00 to 1.84 (m, 6H), 1.43 to 1.07 (m, 16H), 0.88 to 0.84 (m, 3H); 13C NMR (75 MHz, CDCl3) δ 156.9, 130.1, 129.8, 31.9, 29.9, 29.4, 29.1, 28.9, 27.7, 27.2, 27.1, 23.4, 22.8, 22.6, 14.3, 14.0; ESI MS m/z 277 [M-H]−.
RESULTS
MRSA biofilm assay.
In order to develop a robust plate-based assay for evaluating the effects of antibiotics and biofilm inhibitors on MRSA biofilms, we performed preliminary experiments examining the adhesion of 14 isolates of MSSA and MRSA to different commercially available plastic 96-well plates in two different media. Some of the tested plates were coated with collagen, gelatin, or lysine, and some were tissue culture or immunoassay treated. Of the strains examined, the greatest adhesion was observed for clinical isolates MRSA-106, -108, and -295 (data not shown). For future experiments, MRSA-295 was randomly selected out of these three isolates for consistency. Of the plates tested, Costar tissue culture-treated polystyrene plates allowed the most adhesion (data not shown; determined by staining). Subsequent experiments were carried out with the six-well version of this plate in order to increase the biofilm surface area to allow more accurate biofilm quantification. For these scaled-up assays, a relatively large inoculum (approximately 1 × 108 CFU/ml) was used to allow the bacteria to quickly reach stationary phase. Biofilms were quantified by scraping and measuring CFU, as described in Materials and Methods.
When establishing a biofilm model, it is essential to demonstrate that the experimental system produces increased tolerances to clinically used antibiotics compared to their corresponding planktonic MICs. We therefore tested the susceptibility of our experimental biofilms to several antibiotics currently used against MRSA (Table 2). Six-well plates were inoculated with MRSA-295 in CAMHB, and the bacteria were allowed to adhere for 5 h. The resulting biofilms typically then contained 1 × 108 CFU. Previous experiments demonstrated that a 5-h biofilm generates results equivalent to those generated by a 24-h biofilm (data not shown). Half the medium was then removed and replaced with medium containing antibiotic. Remaining bacteria in the biofilm were quantitated after 20 h. For comparison, MICs were determined for planktonic MRSA-295 using the broth microdilution assay. The data in Table 2 demonstrate that this MRSA biofilm model indeed results in reduced antibiotic susceptibilities compared to the level for planktonic cells. For example, daptomycin has an MIC of 2 μg/ml for planktonic bacteria, and 16 μg/ml is required to show activity in the biofilm assay. Other antibiotics, including linezolid and trimethoprim-sulfamethoxazole, were inactive in the biofilm assay even at 64 μg/ml.
Evaluation of SEQ-914 against planktonic bacteria.
In our effort to identify new agents for treatment of MRSA, we have examined the effects of novel natural and synthetic compounds against planktonic and biofilm-associated MRSA. The results for one of these compounds, SEQ-914 (Fig. 1), are described here. MIC values for SEQ-914 and gentamicin against the strains in Table 1 were determined and are listed in Table 3. SEQ-914 displayed low activity while the level for gentamicin was equivalent to that reported in the current literature (14).
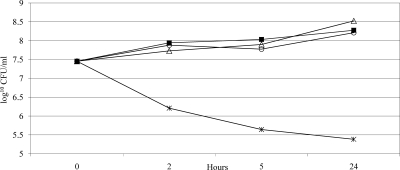
The time dependence of bacterial inhibition was then determined for SEQ-914 and gentamicin against MRSA-295, which was the isolate chosen for the biofilm assay. Planktonic cultures were grown in CAMHB and treated with various concentrations of compound. CFU were then enumerated at the 2-, 5-, and 24-h time points (Fig. 2). In this assay, 8 μg/ml SEQ-914 demonstrates a 2-log reduction at 2 or 5 h but no effect at 24 h (data not shown). Additionally, higher concentrations (16 or 32 μg/ml) show growth reduction at all time points and a 2-log reduction at 24 h with 32 μg/ml. Gentamicin demonstrated a 2-log reduction at the MIC of 2 μg/ml.
Fig. 2.
Time-kill kinetics of MRSA-295 in CAMHB. Viability was enumerated at the indicated time points by serial dilution plates. Each point represents the mean of results from duplicate experiments, with three wells per experiment. ■, untreated control; ▵, 2 μg/ml SEQ-914 (MIC, 32 μg/ml); ○, 0.5 μg/ml GEN (MIC, 2 μg/ml); *, 2 μg/ml SEQ-914 plus 0.5 μg/ml GEN.
To determine whether SEQ-914 and gentamicin show synergy against planktonic MRSA, the time-kill experiment described above was repeated using combinations of the two compounds. Synergy is defined as a ≥2-log decrease in number of CFU/ml between a combination and its most active constituent after a given time, with the number of surviving organisms in the presence of the combination at ≥2-log CFU/ml below the level for the starting inoculum. As shown in Fig. 2, combining 0.5 μg/ml gentamicin with 2 μg/ml SEQ-914 (one-fourth and 1/16 of their respective MIC values) results in a 2-log reduction of growth. The combination thus shows synergistic effects.
Evaluation of SEQ-914 against biofilms.
The ability of SEQ-914 to inhibit MRSA-295 biofilms was evaluated in the six-well plate assay described above; the results are shown in Table 4. Unlike the other antibiotics examined, SEQ-914 reduces biofilm bacteria at its MIC for planktonic bacteria.
Table 4.
Synergy of gentamicin and SEQ-914 against MRSA-295 biofilms in CAMHB
| Treatment group | Concn (μg/ml)a | Log10 no. of CFU/biofilm (mean ± SD)b | Log reduction |
|---|---|---|---|
| Pretreatmentc | 8.24 ± 0.29 | ||
| Untreated control | 9.34 ± 0.07 | ||
| GEN | 2 | 9.03 ± 0.07 | −0.79 |
| 4 | 7.99 ± 0.08 | 0.25 | |
| 8 | 6.11 ± 0.21 | 2.14 | |
| 16 | 3.87 ± 0.64 | 4.37 | |
| SEQ-914 | 16 | 8.57 ± 0.06 | −0.33 |
| 32 | 5.86 ± 0.53 | 2.39 | |
| GEN–SEQ-914 | 0.5; 8d | 5.79 ± 1.01 | 2.45 |
| 1; 4 | 7.50 ± 0.17 | 0.75 | |
| 1; 8 | 4.44 ± 0.69 | 3.81 | |
| 2; 2 | 7.15 ± 0.48 | 1.10 | |
| 2; 4 | 6.70 ± 0.28 | 1.55 | |
| 2; 8 | 3.59 ± 0.42 | 4.66 |
Biofilm assay concentration.
A minimum of two independent experiments were performed, with three wells per experiment. Data are reported as averages (n = 6 as a minimum) for these experiments.
This is the biofilm at 5 h prior to treatment.
GEN and SEQ-914 concentrations, respectively.
To determine whether SEQ-914 and gentamicin exhibit synergy against biofilms as well as planktonic bacteria, the compounds were tested in the biofilm assay using CAMHB medium at varied concentrations, alone and in combination (Table 4). The combination of 2 or 4 μg/ml SEQ-914 with 2 μg/ml of gentamicin results in a 1-log reduction of CFU/biofilm. At 8 μg/ml, SEQ-914 has a synergistic effect with 0.5, 1, and 2 μg/ml of gentamicin, with log reductions of CFU/biofilm of 2.45, 3.81, and 4.66, respectively.
DISCUSSION
In this study, we have described a reproducible assay for screening potential antimicrobial compounds against MRSA biofilms. Although many different experimental systems exist for studying biofilms, microtiter plate-based assays allow for the rapid assessment of large numbers of compounds and conditions. One drawback of studying S. aureus biofilms in microtiter plates is the high well-to-well variability of staining. During the development of this method, we compared the crystal violet staining of untreated and treated biofilms to scraping and determining numbers of CFU. Repeatedly, the staining of biofilms with crystal violet did not correlate with the determined numbers of CFU (data not shown). Therefore, we choose to determine numbers of CFU for each experiment because this is the accepted methodology for bactericidal experiments (5). Vacuum aspiration of medium is used to ensure that all unattached bacterial cells are removed. The biofilm is then rinsed one time, and the rinse medium is removed using the same method prior to the scraping and suspending of the biofilm in medium. Different types of plates were screened to choose the best adhesion properties along with multiple isolates of MRSA. Our conclusion is that the Costar tissue culture 6-well plates gave the best results. This plate type has previously demonstrated good adhesion properties (10). We also tested different bacterial isolates and had best results from three, MRSA-106, MRSA-108, and MRSA-295. From these, we randomly selected MRSA-295 to continue our studies. This model produces significant attachment of MRSA to the plates, requiring the biofilms to be physically scraped off the well surface. We have found that this method enables us to compare results generated weeks apart.
We are routinely using the biofilm assay described herein to evaluate potential new antimicrobial agents. One of these, SEQ-914, shows significant antibiofilm activity at its MIC for planktonic bacteria. This is in contrast to the known antibiotics we examined, which affected biofilms only at concentrations above the MIC. This difference may suggest a new mechanism of action for SEQ-914. Currently, there are limited numbers of compounds in early stages of development against nonmultiplying bacteria such as dormant bacteria or those in biofilms (6, 20).
Based on an earlier analogue of SEQ-914 and its activity with the aminoglycoside antibiotic gentamicin, we initiated combination treatment with this antibiotic. Other antibiotics are currently being screened. The in vitro synergy of SEQ-914 with the aminoglycoside antibiotic gentamicin in both biofilms and planktonic bacteria suggests that this compound could lower the dose of gentamicin required for treatment in vivo. Aminoglycosides are effective against Gram-positive bacterial infections but exhibit dose-limiting toxicities; for this reason, other antibiotics are usually favored in treatment regimens. However, gentamicin has prevailed over other antibiotic treatments in vitro (2, 4), and a recent in vivo study suggests that gentamicin is more effective than linezolid, vancomycin, or ciprofloxacin against S. aureus and MRSA (6). Thus, a compound that lowers the required dose of gentamicin could reduce the toxic effects of an otherwise effective treatment.
Future work will focus on the mechanism of SEQ-914 and its analogues. Our time-kill and biofilm experiments demonstrate that SEQ-914 and gentamicin act synergistically. One hypothesis to explain this effect is that gentamicin's penetration into intact bacterial cells is limited, rendering it less effective against nutrient-limited and anaerobic regions of the S. aureus biofilm. The combination of gentamicin and SEQ-914 may increase penetration of gentamicin into bacterial cells, resulting in a decrease of CFU. Compounds structurally similar to SEQ-914 have recently been identified and shown to inhibit Pseudomonas aeruginosa biofilms (18, 24).
The synthesis of SEQ-914 was inspired by a structurally complex compound isolated from a plant. The data described herein suggest that further lead optimization is warranted. Recently, researchers at Takeda reported the identification of a potent development candidate for type 2 diabetes inspired by docosahexaenoic acid (26), which consists of a linear carbon chain structurally comparable to palmitoleic acid. This example offers compelling evidence that the optimization of SEQ-914 or other biologically active fatty acids into more potent and druglike development candidates is feasible.
ACKNOWLEDGMENTS
We thank David Hunstad and the St. Louis Children's Hospital clinical bacteriology laboratory for supplying us with our clinical isolates and the molecular typing of the strains.
All of the authors are employed with Sequoia Sciences or AMRI.
Footnotes
Published ahead of print on 6 June 2011.
REFERENCES
- 1. Beenken K. E., Blevins J. S., Smeltzer M. S. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bookstaver P. B., Williamson J. C., Tucker B. K., Raad I. I., Sherertz R. J. 2009. Activity of novel antibiotic lock solutions in a model against isolates of catheter-related bloodstream infections. Ann. Pharmacother. 43:210–219 [DOI] [PubMed] [Google Scholar]
- 3. Brady R. A., Leid J. G., Calhoun J. H., Costerton J. W., Shirtliff M. E. 2008. Osteomyelitis and the role of biofilms in chronic infection. FEMS Immunol. Med. Microbiol. 52:13–22 [DOI] [PubMed] [Google Scholar]
- 4. Ceri H., et al. 1999. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. CLSI 2008. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 8th ed., vol. 29 (2). Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Coates A. R., Hu Y. 2007. Novel approaches to developing new antibiotics for bacterial infections. Br. J. Pharmacol. 152:1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grundmann H., Aires-de-Sousa M., Boyce J., Tiemersma E. 2006. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368:874–885 [DOI] [PubMed] [Google Scholar]
- 8. Guvener Z. T., McCarter L. L. 2003. Multiple regulators control capsular polysaccharide production in Vibrio parahaemolyticus. J. Bacteriol. 185:5431–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kearns D. B., Chu F., Branda S. S., Kolter R., Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 55:739–749 [DOI] [PubMed] [Google Scholar]
- 10. Kennedy C. A., O'Gara J. P. 2004. Contribution of culture media and chemical properties of polystyrene tissue culture plates to biofilm development by Staphylococcus aureus. J. Med. Microbiol. 53:1171–1173 [DOI] [PubMed] [Google Scholar]
- 11. Klevens R. M., et al. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771 [DOI] [PubMed] [Google Scholar]
- 12. Labandeira-Rey M., et al. 2007. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science 315:1130–1133 [DOI] [PubMed] [Google Scholar]
- 13. Lemon K. P., Earl A. M., Vlamakis H. C., Aguilar C., Kolter R. 2008. Biofilm development with an emphasis on Bacillus subtilis. Curr. Top. Microbiol. Immunol. 322:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin G., Pankuch G. A., Ednie L. M., Appelbaum P. C. 2010. Antistaphylococcal activities of telavancin tested alone and in combination by time-kill assay. Antimicrob. Agents Chemother. 54:2201–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Loughman J. A., Fritz S. A., Storch G. A., Hunstad D. A. 2009. Virulence gene expression in human community-acquired Staphylococcus aureus infection. J. Infect. Dis. 199:294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mah T. F., O'Toole G. A. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39 [DOI] [PubMed] [Google Scholar]
- 17. Moran G. J., et al. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355:666–674 [DOI] [PubMed] [Google Scholar]
- 18. Muh U., et al. 2006. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob. Agents Chemother. 50:3674–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Novick R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429–1449 [DOI] [PubMed] [Google Scholar]
- 20. Ooi N., et al. 2010. XF-70 and XF-73, novel antibacterial agents active against slow-growing and non-dividing cultures of Staphylococcus aureus including biofilms. J. Antimicrob. Chemother. 65:72–78 [DOI] [PubMed] [Google Scholar]
- 21. O'Toole G. A., Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 22. O'Toole G. A., et al. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91–109 [DOI] [PubMed] [Google Scholar]
- 23. Pratt L. A., Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285–293 [DOI] [PubMed] [Google Scholar]
- 24. Richards J. J., et al. 2009. Amide isosteres of oroidin: assessment of antibiofilm activity and C. elegans toxicity. J. Med. Chem. 52:4582–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rosen D. A., Hooton T. M., Stamm W. E., Humphrey P. A., Hultgren S. J. 2007. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 4:e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sasaki S., et al. 2011. Design, synthesis, and biological activity of potent and orally available G protein-coupled receptor 40 agonists. J. Med. Chem. 54:1365–1378 [DOI] [PubMed] [Google Scholar]
- 27. Ventura C. L., et al. 2010. Identification of a novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PLoS One 5:e11634. [DOI] [PMC free article] [PubMed] [Google Scholar]