Abstract
Therapeutic options for tuberculosis (TB) are limited and notoriously ineffective despite the wide variety of potent antibiotics available for treating other bacterial infections. We investigated an approach that enables an expansion of TB therapeutic strategies by using synergistic combinations of drugs. To achieve this, we devised a high-throughput synergy screen (HTSS) of chemical libraries having known pharmaceutical properties, including thousands that are clinically approved. Spectinomycin was used to test the concept that clinically available antibiotics with limited efficacy against Mycobacterium tuberculosis might be used for TB treatment when coadministered with a synergistic partner compound used as a sensitizer. Screens using Mycobacterium smegmatis revealed many compounds in our libraries that acted synergistically with spectinomycin. Among them, several families of antimicrobial compounds, including macrolides and azoles, were also synergistic against M. tuberculosis in vitro and in a macrophage model of M. tuberculosis infection. Strikingly, each sensitizer identified for synergy with spectinomycin uniquely enhanced the activities of other clinically used antibiotics, revealing a remarkable number of unexplored synergistic drug combinations. HTSS also revealed a novel activity for bromperidol, a butyrophenone used as an antipsychotic drug, which was discovered to be bactericidal and greatly enhanced the activities of several antibiotics and drug combinations against M. tuberculosis. Our results suggest that many compounds in the currently available pharmacopoeia could be readily mobilized for TB treatment, including disease caused by multi- and extensively drug-resistant strains for which there are no effective therapies.
INTRODUCTION
Tuberculosis (TB) is a major cause of morbidity and mortality worldwide. The World Health Organization estimates that there are 9.4 million new TB cases and 1.7 million deaths annually (43). The current TB regimen recommended for drug-susceptible (DS) disease is lengthy (at least 6 months), with a cure rate of 95% under optimal conditions (26). Coinfection with HIV and the emergence of resistant strains has reaffirmed TB as a global public health threat (43). Multidrug-resistant Mycobacterium tuberculosis (MDR-TB) strains are resistant to rifampin and isoniazid, the two first-line TB drugs; extensively drug-resistant M. tuberculosis (XDR-TB) strains have, in addition, acquired resistance to any fluoroquinolone and to any one of the three injectable second-line anti-TB drugs (amikacin, kanamycin, or capreomycin) (23). Effective MDR-TB therapy is more toxic to patients than conventional treatment for DS-TB, costly, prolonged (lasting up to 2 years), and uncertain (cure rates typically range from 50% to 70%) (26). All of these problems are even more acute for XDR-TB patients (11). While several new compounds are under investigation (26), alternative therapies are urgently needed both to shorten the duration of the current TB treatment and to treat MDR- and XDR-TB.
Development of new therapies for bacterial infections has traditionally focused on empirical screening for single compounds that inhibit growth. As identification of active new compounds using this approach became less fruitful, pharmaceutical companies embarked on massive target-based high-throughput screening campaigns. This approach proved to be a lengthy process, with unsustainably low yields and profits (35). An analysis of 68 approved drugs showed that it takes an average of 14 years and $800 million (U.S. dollars) to bring a single drug to the market (17). One economical solution would be to identify new uses for existing drugs (“repurposing”) (9, 12), either alone or in combination therapies. Because approved drugs have known pharmacokinetic and safety profiles, any newly identified application can be more rapidly evaluated in phase II clinical trials, thereby decreasing the average time for FDA approval from 14 years to about 5 years (12). This avenue would be of particular importance for the development of new TB therapies since it would allow predictions of suitability in a combinatorial regimen and for use with antiretroviral drugs. In addition, physicians can elect to use drugs that are clinically approved for other indications if available treatments are ineffective (9), as is the case for XDR-TB.
A major obstacle to finding a more effective treatment for TB is the intrinsic resistance of M. tuberculosis to most clinically approved antibiotics. Most of these antibiotics, such as spectinomycin, have little or no activity against M. tuberculosis. Like other aminocyclitols (streptomycin, kanamycin, or gentamicin), spectinomycin inhibits protein synthesis by binding to the 30S ribosomal subunit (41) but does not have ototoxic effects (32). While it has poor in vitro activity against most bacteria, including M. tuberculosis, it is used to treat patients infected with Neisseria gonorrhoeae who cannot tolerate first-line treatments. We used spectinomycin to test the concept that clinically available drugs with limited efficacy against M. tuberculosis might be used for TB treatment when coadministered with a synergistic partner compound.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and chemicals.
Strains and plasmids used in this study are listed in Table S1 in the supplemental material. Mycobacterium smegmatis mc2155 was cultivated at 37°C in Middlebrook 7H9 broth supplemented with 10% (vol/vol) albumin-dextrose-catalase (ADC) (Difco), 0.2% glycerol, and 0.05% (vol/vol) tyloxapol or on Middlebrook 7H10 (Difco) agar plates supplemented with 10% (vol/vol) oleic acid albumin-dextrose-catalase (Difco) and 0.2% glycerol. M. smegmatis experiments were performed in NE medium (glucose, 10 g/liter; yeast extract, 2 g/liter; Casamino Acids, 2 g/liter; and Lab-Lemco powder [Oxoid], 1 g/liter) (30). M. tuberculosis H37Rv was cultivated at 37°C in Middlebrook 7H9 broth supplemented with 10% (vol/vol) ADC (Difco), 0.2% glycerol, and 0.05% (vol/vol) tyloxapol or on Middlebrook 7H10 (Difco) agar plates supplemented with 10% (vol/vol) oleic acid albumin-dextrose-catalase (Difco) and 0.2% glycerol. Experiments were performed in Middlebrook 7H9 broth supplemented with 0.2% glycerol and 10% (vol/vol) ADC (Difco). Chemical libraries (Prestwick, Sigma, Microsource, and BioMol) were provided by the Canadian Chemical Biology Network (CCBN). Compounds in the chemical libraries were provided at a concentration of 5 mM in 100 μl of dimethyl sulfoxide (DMSO) in 96-well plates and were stored at −20°C.
HTSS.
A qualitative high-throughput synergy screen (HTSS) on solid medium was developed to find compounds that enhanced the activity of spectinomycin against M. smegmatis. M. smegmatis cells were grown to stationary phase, diluted 10-fold, treated for 1 min in a 2510 Branson bath sonicator to disrupt cell aggregates, diluted to a concentration of 105 cells/ml in 22 ml of NE medium containing 0.5% agar, and uniformly poured over 45 ml of NE-1.5% agar in OmniTrays (Nunc). When appropriate, spectinomycin was added to the agar at subinhibitory concentrations that were previously determined by 2-fold serial dilutions of the drug under the same HTSS growing conditions. Chemicals were transferred from 96-well plates onto cell lawns using a BioRobotics TAS1 pinning robot with a pin diameter of 0.7 mm, which transferred approximately 340 nl (1.7 nmol of each compound). A pin density of 16 compounds (4 × 4) per 96-well position corresponded to a total of 1,536 compounds per OmniTray. When necessary, active hits on these high-density plates were identified by spotting localized groups of compounds at a lower density (deconvolution; 96 compounds per OmniTray). Controls were included to ensure that DMSO had no effect on bacterial growth. The OmniTrays were incubated at 37°C, and growth inhibition zones were measured after 60 h. HTSS was carried out in duplicate from independent bacterial cultures.
Drug susceptibility tests.
Resazurin or MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] assays (29) were used to determine viability and hence the sensitivity of M. smegmatis and M. tuberculosis strains to the drugs studied. Briefly, 2-fold antibiotic serial dilutions were inoculated with a suspension of mycobacteria in microtiter plates at 37°C. The plates were incubated for 3 and 8 days for M. smegmatis and M. tuberculosis, respectively. After the addition of resazurin, the plates were further incubated for 1 day in the case of M. smegmatis and 2 to 3 additional days for M. tuberculosis. A change from blue to pink indicated growth of bacteria, and the MIC was defined as the lowest concentration of drug that prevented this color change. The MTT assay was carried out essentially as described above, except that the MTT reagent was added to the cells, followed by incubation at 37°C overnight. The formation of the formazan precipitate indicated bacterial growth. We used an established technique (46) to differentiate the bacteriostatic or bactericidal effect of a single drug or drug combination against M. tuberculosis. Briefly, the protocol for MIC determination was followed. In addition, before resazurin or MTT was added, 5 μl from every well was transferred to 200 μl of a drug-free medium and processed as described above. Isoniazid, a bactericidal drug, was used as a positive control. A compound or drug combination was considered bactericidal if the ratio between the minimal bactericidal concentration (MBC) and the MIC was equal to or smaller than 2 (MBC/MIC ≤ 2). All drug sensitivity tests were carried out in at least three independent experiments.
Checkerboard synergy assay.
The fractional inhibitory concentration index (FICI) was determined in a 96-well plate format using the resazurin or MTT assay as described above. The fractional inhibitory concentration for each compound was calculated as follows (38): FICA = (MIC of compound A in the presence of compound B)/(MIC of compound A alone), where FICA is the fractional inhibitory concentration of compound A. Similarly, the FIC for compound B was calculated. The FICI was calculated as FICA plus FICB. Synergy was defined by FICI values of ≤0.5, antagonism by FICI values of >4.0, and no interaction by FICI values from 0.5 to 4.0 (33). A graphical representation of a typical checkerboard assay is shown in Fig. S1 in the supplemental material.
Ex vivo checkerboard synergy assay.
An ex vivo checkerboard assay was developed to study the synergistic interactions between pairwise combinations of selected compounds against M. tuberculosis H37Rv in a macrophage model of infection. Frozen stocks of macrophage THP1 cells (ATCC TIB-202) were thawed in RPMI medium with 1% glutamine, 10% fetal bovine serum (FBS), 10,000 units/ml penicillin G (sodium salt), and 10 mg/ml streptomycin sulfate in 0.85% saline. Cells were passaged only 5 times and maintained without antibiotics. Four thousand cells per well were allowed to differentiate in RPMI medium containing 1% glutamine, 80 ng/ml phorbol myristate acetate (PMA), and 10% FBS without antibiotics (incomplete medium) and left to adhere to the culture dish for 24 h. For toxicity evaluation, black, clear-bottom 96-well microtiter plates were used. After cell differentiation, medium was removed from the wells and 100 μl of phenol red-free RPMI medium containing 10% FBS medium was added. Twofold serial dilutions of drugs were added in 100 μl of medium. Cells were incubated in the presence of the drugs for 72 h, after which 20 μl of resazurin (0.01%) was added to each well. After 4 h of incubation, fluorescence (λexc = 535 nm; λem = 590 nm, where λexc is the excitation wavelength and λem is the emission wavelength) was read to determine the viability of the macrophages; the 50% inhibitory concentration (IC50) was calculated relative to that for untreated cells. Compounds with a MIC smaller than the macrophage toxicity concentration were analyzed further in ex vivo checkerboard assays. Macrophages were differentiated as described above in white polystyrene 96-well plates and washed three times with 100 μl RPMI medium. Before infection, M. tuberculosis H37Rv harboring the firefly luciferase lux gene cloned into the pMV361 integrative vector (M. tuberculosis lux) (39) was opsonized. For this, cultures at an optical density at 600 nm (OD600) of 0.1 (∼3 × 107 cells) were washed three times with 7H9 salts medium containing 0.05% tyloxapol. Bacteria were resuspended in 450 μl of RPMI medium (with no supplement) containing 50 μl of human serum and incubated for 20 min at 37°C. Cells were harvested and resuspended in an appropriate volume of RPMI medium. Opsonized M. tuberculosis lux was used to infect macrophages at a multiplicity of infection (MOI) of 10 for 3 h at 37°C. Infected macrophages were gently washed three times with drug-free RPMI medium in order to remove noninternalized bacteria and incubated for 24 h at 37°C before media were aspirated and the test compounds were added. Test compounds were prepared in a checkerboard format in an independent 96-well plate as described above. After 96 h, compound-containing medium was aspirated and 50 μl of Bright-Glo luciferase assay reagent (Promega) added to the wells. M. tuberculosis lux viability was measured during a 1-s exposure with a PerkinElmer Tropix luminometer 15 min after addition of the luciferase assay reagent. Wells containing drug-free medium with and without infected macrophages established maximum and minimal light production, respectively. A 90% reduction in light production was considered growth inhibition. The macrophage checkerboard data were processed as described above. Every combination was assayed in duplicate in at least two independent experiments.
RESULTS
HTSS revealed chemically diverse compounds that enhanced the activity of spectinomycin against M. smegmatis.
The nonpathogenic fast-growing bacterium M. smegmatis was used as a surrogate model for M. tuberculosis to facilitate screening of chemical libraries and accelerate the discovery process. Our screening program included intermediate steps of validation and, ultimately, testing of the efficacy of the combination directly against M. tuberculosis in vitro and ex vivo (Fig. 1). Chemical libraries used in the screening contained 4,900 compounds (including about 250 recognized as antibiotics), many with known pharmacologic properties (Table 1). Compounds were screened for activity against M. smegmatis in the presence or absence of subinhibitory concentrations (one-quarter the MIC [1/4×MIC]) of spectinomycin (Fig. 2). This qualitative HTSS was carried out by pinning compounds onto lawns of bacterial cells growing on solid agar media. Hits were defined as those whose activity increased the zone of inhibition by at least 1 mm in the presence of spectinomycin compared to the zone of inhibition on control plates containing no spectinomycin. Fifty-one compounds that increased spectinomycin activity (sensitizers) were identified, representing an overall hit rate of 1.4%. To validate HTSS results, the checkerboard assay was used to measure the synergistic interaction of 23 randomly selected hits with spectinomycin (Table 2). Synergy was defined by equal or >4-fold reductions in the MICs of both compounds in combination, compared to the MICs of the compounds alone (FICI ≤ 0.5). Synergistic activities for 78% of the hits (18/23 compounds) were confirmed in liquid cultures (see Table S2 in the supplemental material). Many compounds having synergy with spectinomycin, including macrolides, targeted the ribosome (10). However, antibacterials with other targets, including those that inhibit DNA gyrase, redox systems, or P450 monooxygenases, were identified (10). Other compounds that enhanced the inhibitory activity of spectinomycin were not known antibiotics but have other pharmaceutically defined activities for unrelated indications. These compounds included bromperidol (antipsychotic), telmisartan (hypertension control), fiduxosin (adrenoceptor antagonist), or clioquinol (antifungal) (Table 2). Compounds affecting redox balance were also active, including shikonin, a compound that reacts with and greatly reduces total thiols, protein thiols, and glutathione levels (19); menadione, a vitamin K precursor that can act as a redox cycler; dequalinium (20), an inhibitor of MshC enzyme, catalyzing a step in the biosynthesis of mycothiol, the primary thiol-reducing agent in Mycobacterium; and cadmium, a nonspecific disruptor of oxidative balance inside the cell (21). To further investigate the possible link between redox balance and spectinomycin activity, the thiol-specific alkylating agent monobromobimane (mBBr) was also tested and found to act in synergy with spectinomycin (Table 2).
Fig. 1.

Combinatorial drug discovery program.
Table 1.
Chemical libraries
| Library | No. of compoundsa | Comment(s) |
|---|---|---|
| Prestwickb | 1,120 | Off-patent compounds; 85% are FDA approved |
| Sigmac | 1,280 | Pharmacologically active compounds |
| Microsourced | 2,000 | Known drugs and exptl bioactives |
| BioMole | 500 | Purified natural products |
Libraries contained 3,600 unique compounds from a total of 4,900 chemicals.
Prestwick Collection (Prestwick Chemical).
Sigma LOPAC library (Sigma-Aldrich).
SPECTRUM Collection (MicroSource Discovery Systems, Inc.).
BIOMOL Library (BIOMOL International).
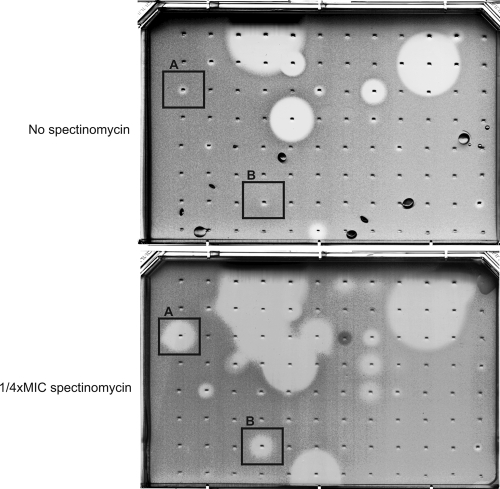
Fig. 2.
High-throughput synergy screening (HTSS). Compounds spotted against M. smegmatis in an agar-based HTSS plate in the absence or in the presence of subinhibitory concentrations of spectinomycin (1/4×MIC). Compounds whose zones of inhibition were larger in the presence of spectinomycin than in plates without spectinomycin (i.e., example A or B) were selected for further studies. Up to 1,536 compounds (4 × 4 × 96) could be tested per plate; however, groups of active compounds on such high-density plates often needed to be tested by spotting at a lower density (deconvolution; 96 compounds per OmniTray), as illustrated in the figure.
Table 2.
Degree of interaction of pairwise spectinomycin combinations against mycobacterial species
| Compound | Drug class | Cellular target | FICI range for:a |
||
|---|---|---|---|---|---|
| M. smegmatis | M. tuberculosis | M. tuberculosis THP1 | |||
| Telmisartan | Benzoates | Angiotensin II receptor (type AT1) | 0.37 | 0.625 | |
| Ethambutol | Ethylenediamines | Cell wall | ND | 1–3 | |
| Vancomycin | Glycopeptide | Cell wall | 2.03 | ||
| Isoniazid | Isonicotinic acids | Cell wall | 0.37–0.5 | 1 | |
| Bromperidol | Butyrophenones | D2 dopamine receptor | 0.09–0.16 | 0.06–0.19 | 0.19–0.25 |
| Haloperidol | Butyrophenones | D2 dopamine receptor | 0.19 | ||
| Spiperone | Butyrophenones | D2 dopamine receptor | 0.25 | ||
| Trifluoperidol | Butyrophenones | D2 dopamine receptor | 0.25 | ||
| Novobiocin | Coumarins | DNA gyrase | 0.5 | ||
| Moxifloxacin | Fluoroquinolones | DNA gyrase | 2.25 | ||
| Econazole | Azoles | P450 | 0.5 | 0.5 | |
| Isoconazole | Azoles | P450 | 0.37–0.5 | 0.28–0.5 | |
| Ketoconazole | Azoles | P450 | 0.19–0.5 | 0.16–0.5 | 0.5 |
| Miconazole | Azoles | P450 | 0.5 | 0.5 | |
| Sertaconazole | Azoles | P450 | 0.5 | 0.5 | |
| Terconazole | Azoles | P450 | 0.12–0.25 | 0.5 | |
| Monobromobimane | Bicyclo compounds | Redox balance | 0.5 | ||
| Menadione | Naphthoquinones | Redox balance | 0.37–0.5 | ||
| Plumbagin | Naphthoquinones | Redox balance | 0.52–0.75 | ||
| Shikonin | Naphthoquinones | Redox balance | 0.37 | ||
| Dequalinium | Quaternary ammonium cations | Redox balance | 0.31–0.37 | ||
| Cadmium acetate | Heavy metals | Redox balance | 0.37–0.5 | ||
| Kanamycin | Aminoglycosides | Ribosome | 2.03 | ||
| Streptomycin | Aminoglycosides | Ribosome | ND | 4.25 | |
| Thiostrepton | Cyclic peptides | Ribosome | 0.5–0.75 | ||
| Azithromycin | Macrolides | Ribosome | ND | <0.51 | 0.25–0.5 |
| Clarithromycin | Macrolides | Ribosome | 0.06 | 0.31–0.5 | 1 |
| Roxythromycin | Macrolides | Ribosome | 0.31–0.5 | ||
| Telithromycin | Macrolides | Ribosome | 0.16–0.19 | ||
| Fusidic acid | Sterols | Ribosome | 0.25 | ||
| Minocycline | Tetracyclines | Ribosome | 0.54 | ||
| Oxytetracycline | Tetracyclines | Ribosome | 0.62 | ||
| Tetracycline | Tetracyclines | Ribosome | 1 | ||
| Rifampin | Rifamycins | RNA polymerase | 0.37 | 0.22–0.5 | 0.37–0.75 |
| Clioquinol | Hydroxyquinolines | Unknown | 0.37–0.5 | 1 | |
| Solasodine | Steroidal alkaloids | Unknown | 0.12–0.25 | 0.375 | |
| Fiduxosin | Pyrimidinones | α1A- and α1D-adrenoceptors | 0.19–0.37 | 0.62–0.75 | |
The FICI range represents the lowest and highest FICI value observed for at least two independent experiments. Synergy is defined as a FICI of ≤0.5 and antagonism as a FICI of >4. ND, not determined (azithromycin, ethambutol, and streptomycin were included at a later stage of the study).
Chemical-structure clustering was performed to analyze the chemical diversity of the HTSS hits (Fig. 3). It is known that structurally related molecules often exhibit similar biological effects. The Tanimoto similarity coefficient is one of the most appropriate distance metrics for topology-based chemical similarity studies. Cutoff values indicating significant similarities are between 0.85 and 0.70; values of 0.5 to 0.6 indicate lack of similarity (27). Although hits were identified by a shared biological effect, i.e., synergy with spectinomycin, most chemical structures had similarity coefficients lower than 0.7, with two main dissimilar groups showing a value of 0.55.
Fig. 3.
Chemical structure clustering of the compounds identified by HTSS as enhancers of spectinomycin activity against M. smegmatis. A total of 48 chemical structures were clustered using the Chemical Structure Clustering tool available at PubChem (http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?p=clustering). Cutoff values for similarity are between 0.85 and 0.70. A value of 0.5 to 0.6 or lower indicates lack of similarity.
Collectively, these data indicated that synergistic modes of action cannot simply be explained by inhibition of common cellular targets and highlights the importance of the intracellular oxidative and thiol balance in the bacterial cell as a modulator of the antibacterial activity of spectinomycin in mycobacteria.
Synergistic drug interactions are widely distributed.
Combinatorial drug sensitivity assays were performed to determine whether HTSS-selected compounds acted in synergy only with spectinomycin or might have more-extensive repertoires of interactions (Fig. 4). Seven spectinomycin sensitizers were tested for their abilities to sensitize M. smegmatis to 10 other clinically available and commonly used antibiotics. This study revealed that more than one-third of the 70 drug combinations tested were synergistic. Furthermore, every compound that acted in synergy with spectinomycin generated its own specific and distinctive sensitivity pattern, a drug-specific “barcode.”
Fig. 4.
Spectinomycin sensitizers have their own characteristic pattern of synergy with other antibiotics. Seven compounds that were synergistically active with spectinomycin in M. smegmatis (sensitizers, horizontal axis) were assayed for their abilities to induce sensitivity to an array of 11 representative antibiotics (vertical axis). A sensitivity “barcode” was generated for each compound. Sensitivity was determined in the presence or absence of a 1/4×MIC concentration of the sensitizers. Solid lines indicate a ≥4-fold increase in sensitivity to one of the 11 antibiotics in the presence of the sensitizer.
HTSS hits identified using M. smegmatis were active against M. tuberculosis.
HTSS hits were selected for further analysis based on their pharmacological potential. For example, although cadmium acted in synergy with spectinomycin, it was discarded because of its toxicity. Three major families of compounds having synergistic activities with spectinomycin in M. smegmatis were also synergistic in M. tuberculosis in liquid cultures (FICI ≤ 0.5): macrolides, azoles, and butyrophenones (Table 2). Representatives of these three families, azithromycin, ketoconazole, and bromperidol, had FICIs ranging from 0.19 to 0.5 when tested in combination with spectinomycin against M. tuberculosis lux within macrophages, a well-established luciferase-based method to evaluate antimycobacterial drug activity (7). These values reflect a 4- to 16-fold reduction in the MIC for each of the two drugs (synergistic MIC [MICsyn]) compared to the MIC of each drug alone. Although not identified by HTSS, the interaction of spectinomycin with the first-line drugs rifampin, isoniazid, and ethambutol, as well as streptomycin, was also analyzed in M. tuberculosis. There was strong synergy between spectinomycin and rifampin (Table 2), whereas no interaction was observed with isoniazid or ethambutol; streptomycin had an antagonistic profile. In summary, three families of compounds (macrolides, azoles, and butyrophenones) were identified as having activities synergistic with spectinomycin against M. tuberculosis in vitro and ex vivo (within macrophages).
Bromperidol is bactericidal against M. tuberculosis.
Clarithromycin, ketoconazole, bromperidol, rifampin, and isoniazid (a bactericidal positive control), alone and in combination with spectinomycin, were tested in vitro to determine whether they were bacteriostatic or bactericidal against M. tuberculosis (Table 3). If a drug had bactericidal activity, culture growth would not resume after dilution into fresh medium. When the MBC/MIC ratio for the individual drugs was determined, only isoniazid and bromperidol had a value of 1, indicating a bactericidal (MBC/MIC ≤ 2) rather than a bacteriostatic effect. Interestingly, when the bacteriostatic antibiotic spectinomycin was assayed in combination with a fixed concentration (1/4×MIC) of bromperidol, the MICsyn and MBCsyn of spectinomycin were identical, suggesting that the bactericidal character of bromperidol was dominant in the combination. This effect was not observed when spectinomycin was assayed in combination with ketoconazole, clarithromycin, or rifampin. These results revealed bromperidol as a potential new bactericidal chemical entity for TB therapy.
Table 3.
Bactericidal activities of compounds alone and in combination with spectinomycin
| Compound | Result for M. tuberculosis H37Rv witha: |
|||||
|---|---|---|---|---|---|---|
| Compound aloneb |
Compound in combination with spectinomycinc |
|||||
| MIC | MBC | MBC/MIC | MICsyn | MBCsyn | MBC/MIC | |
| Spectinomycin | 80 | 320–640 | 4–8 | NA | NA | |
| Bromperidol | 64 | 64 | 1 | 5 | 5–10 | 1–2 |
| Clarithromycin | 32–64 | >128 | ≥4–8 | 10–20 | 320 | 16–32 |
| Isoniazid | 0.03 | 0.03 | 1 | 80 | 320–640 | 4–8 |
| Ketoconazole | 16–32 | 64–128 | 4 | 5–10 | 160 | 16–32 |
| Rifampin | 0.06 | 0.25–0.5 | 4–8 | 10–20 | 160 | 8–16 |
MIC was determined after 8 days of incubation in the presence of the compound(s). To calculate the MBC, 5 μl was transferred to 200 μl of drug-free medium and incubated for 8 additional days. NA, not applicable.
MIC and MBC (mg/liter) for every compound tested individually.
Spectinomycin synergistic MIC (MICsyn) and synergistic MBC (MBCsyn) (mg/liter) in the presence of a 1/4×MIC of the compound, as follows: bromperidol, 16 mg/liter; clarithromycin, 16 mg/liter; isoniazid, 0.007 mg/liter; ketoconazole, 8 mg/liter; and rifampin, 0.015 mg/liter.
Bromperidol enhanced the inhibitory activities of a variety of antibiotics against M. tuberculosis in vitro and ex vivo.
In addition to spectinomycin, bromperidol enhanced the activities of clarithromycin, clofazimine, and rifampin against M. smegmatis (Fig. 4). Similarly, the sensitivity of M. tuberculosis to a set of 13 antibiotics with different structures and cellular targets was analyzed in the presence or absence of subinhibitory concentrations of bromperidol (1/4× to 1/8×MIC) (Table 4). In these experiments, bromperidol increased the potency (at least 4-fold) of clarithromycin, clofazimine, econazole, novobiocin, rifampin, spectinomycin, and streptomycin against M. tuberculosis in vitro. Bromperidol was equally active in an M. tuberculosis-macrophage model of infection. Importantly, the MIC of bromperidol against M. tuberculosis ex vivo was 8- to 16-fold lower than its MIC in vitro (Table 4), indicating that bromperidol accumulated inside macrophages. These results led us to pursue the idea that bromperidol or structural analogs might be a promising active adjuvant in combinatorial TB therapy.
Table 4.
Combinatorial sensitivity assay for bromperidol against M. tuberculosis in vitro and ex vivo
| Compound | MIC for M. tuberculosis (mg/liter)a |
|||||
|---|---|---|---|---|---|---|
|
In vitrob |
Fold increased |
Ex vivoc |
Fold increase | |||
| − BPDL | + BPDL | − BPDL | + BPDL | |||
| Bromperidol | 64–128 | ND | 8–16 | ND | ||
| Azithromycin | ND | ND | 8–16 | 1–2 | 8 | |
| Bacitracin | >512 | >512 | 1 | ND | ND | |
| Clarithromycin | 64 | 4 | 8 | 0.5–1 | 0.125 | 4–8 |
| Clofazimine | 0.125–0.25 | 0.015–0.03 | 4–8 | 0.6 | 0.15 | 4 |
| Econazole | 32 | 4–8 | 4–8 | ND | ND | |
| Isoniazid | 0.025 | 0.025 | 1 | ND | ND | |
| Novobiocin | 32–64 | 8–16 | 4 | ND | ND | |
| p-Aminosalicylate | 0.06 | 0.06 | 1 | ND | ND | |
| Rifampin | 0.125 | 0.03 | 4 | 0.125 | 0.03 | 4 |
| Spectinomycin | 80–160 | 2.5 | 64 | 80–160 | 5 | 16–32 |
| Streptomycin | 1 | 0.25 | 4 | ND | ND | |
| Tetracycline | 8 | 8 | 1 | ND | ND | |
| Vancomycin | >256 | >256 | 1 | ND | ND | |
MICs were assayed in a range of 2-fold dilutions of antibiotics in the presence or absence of bromperidol (BPDL) at 1/4×MIC. ND, not determined.
The MIC was determined in 7H9 liquid medium.
The MIC was determined in M. tuberculosis-infected THP1 macrophages.
Fold increase in sensitivity in the presence of bromperidol.
Trio of synergistically active drugs.
Since pairwise combinations of bromperidol, rifampin, and spectinomycin were all synergistic in M. tuberculosis (Tables 2 and 4), possible second-order multiplicative effects within the triple drug combination were investigated. The MIC of spectinomycin, rifampin, or bromperidol alone, the MICsyn of each pairwise combination, and the MICsyn of both spectinomycin and rifampin at a fixed concentration of bromperidol (1/4× to 1/8×MIC) were determined (Table 5). The introduction of bromperidol multiplied the antibacterial activities of the drugs. Compared to the MIC of the antibiotic alone, the MICsyn of spectinomycin in combination with bromperidol was 16-fold lower, and in combination with rifampin, it was 4- to 8-fold lower. When the three drugs were tested together, the activity of spectinomycin was increased 128-fold (16 × 8 = 128). A similar multiplicative effect was observed for rifampin (Table 5). These results support the concept that using synergistic drug combinations of two, three, or even more drugs to treat TB could greatly increase the activities of the individual drugs, thereby increasing their efficacy or reducing their toxicity (drug dosage).
Table 5.
Bromperidol enhancement of pairwise synergistic combinations against M. tuberculosis
| Drug(s)b | MIC (mg/liter) for M. tuberculosis H37Rv ofa: |
|||||
|---|---|---|---|---|---|---|
| Spectinomycin | Rifampin | Bromperidol | ||||
| Drug alone | 80 | 0.025 | 64–128 | |||
| SPT + RIF | 10–20 | (4–8) | 0.06 | (4) | NA | |
| SPT + BPDL | 5 | (16) | NA | 8 | (8–16) | |
| RIF + BPDL | NA | 0.06 | (4) | 16 | (4–8) | |
| SPT + RIF + BPDL | 0.6 | (128) | 0.015–0.03 | (8–16) | 16b | (4–8) |
MICs of the antibiotics alone or within different combinations. When in combination, values represent the lowest MIC for each antibiotic. Values in parentheses indicate fold reduction in the MIC compared to that of the antibiotic alone. Bromperidol was included in the combination at a fixed concentration of 16 mg/liter (4- to 8-fold below the MIC). NA, not applicable.
BPDL, bromperidol; RIF, rifampin; SPT, spectinomycin.
DISCUSSION
While hundreds of new antibiotics were introduced during the last 50 years, few are sufficiently active for clinical use against M. tuberculosis. The concept of using combinations of drugs to treat bacterial diseases was first developed to treat TB in the 1950s because single-drug therapy rapidly led to resistance (11). It has since been routinely employed against M. tuberculosis as well as other bacterial pathogens (13). However, there have been no systematic studies of how these drugs might affect each other's activities, and little attention has been paid to the possible efficacy of synergistic interactions involving antibiotics not clinically effective against TB. Combinatorial therapy has identified synergistic combinations against fungal infections (24, 46) and is the foundation for treating HIV using formulations of 3 to 4 drugs (43). This principle is also the basis for several effective antibacterial drug combinations, including beta-lactam antibiotics/beta-lactamase inhibitors, sulfanilamide-trimethoprim, and pristinamycin A and B (13, 41). More recently, the idea has reemerged for TB therapy (22). Compared to standard drug discovery programs, the strategy of using clinically approved compounds would allow the timely introduction of existing drugs as new antibacterial therapies. The use of innovative combination therapies is recognized by the FDA for its therapeutic potential (42). Our studies build on an FDA initiative, implemented by the Critical Path Institute, which supports clinical trials to test TB drug candidates in combination regimens. In summary, HTSS promises to be more efficient in terms of both capital investment and the amount of time before a drug can be used in clinics. However, to our knowledge, a systematic screen to identify new synergistic drug pair combinations for bacterial therapy has not been described in the literature.
In our approach, HTSS was used to systematically screen libraries containing compounds with known pharmacodynamic and pharmacokinetic properties to improve spectinomycin activity. The novelty of our approach was the implementation of a high-throughput screen on solid media that provided a clear, reproducible, and reliable signal for synergistic hits. As many as 1,536 compounds could be pinned at the same concentration in a 4 × 4 × 96 spot density, and changes in the zone of inhibition could be readily monitored (Fig. 2). The fact that such a variety of compounds enhanced the activity of spectinomycin implied that hits might involve different cellular mechanisms. A systematic study of drug interactions showed that compounds inactivating the same cellular function typically act more efficiently together (45). This could explain the synergy observed between spectinomycin and macrolides, both of which target ribosome function. However, the diversity of chemical structures and targets associated with compounds that shared the same synergistic partner made this model inapplicable to many other combinations (Fig. 3). The simplest interpretation is that each structurally different sensitizer induced specific alterations in the cell (envelope) structure or cellular physiology, with a common downstream effect (25) that resulted in synergy with spectinomycin. Interestingly, several spectinomycin sensitizers are known to alter the oxidative state within the cell. Genetic screens, conceptually similar to our compound screens, have been done to identify mutations that sensitize bacteria to specific drugs. These studies demonstrated that intrinsic drug resistance relies on contributions from many genetic loci having diverse physiological functions (3, 31, 37). The complexity of factors that determine intrinsic drug resistance is further documented by our “barcode” assays (Fig. 4). In these assays, every compound that acted synergistically with spectinomycin provoked its own specific and distinct sensitivity pattern, indicating a compound-specific mode of antibacterial action. Considering this, it is difficult to imagine a common cellular target for all compounds that act synergistically with spectinomycin. Furthermore, the fact that 24 of the 70 random combinations surveyed were synergistically active in M. smegmatis suggests a large, unexplored pool of possible synergistic drug combinations for TB treatment. We are currently carrying out systematic synergy screens to unravel these interactions for other clinically relevant antibiotics.
Several spectinomycin combinations of biological interest as well as potential clinical applications were analyzed in detail (Table 2). In vitro and ex vivo macrophage assays confirmed the synergy of several of these combinations against M. tuberculosis. We focused on three major families of compounds that acted in synergy with spectinomycin: macrolides, azoles, and butyrophenones. Macrolides are first-line ribosome-targeting drugs for treating Mycobacterium avium infection. Clarithromycin is considered an oral reserve drug with uncertain antituberculosis activity, and azithromycin is a close analog with better pharmacological properties (11). Antifungal azoles have recently been proposed as potential anti-TB drugs (34); they inhibit cytochrome P450 monooxygenases in both fungi and mycobacteria (28). In the case of butyrophenones, our studies are the first to describe their antibacterial activity; studies are now under way to identify their mode of action.
Here we demonstrated that several butyrophenones acted in synergy with spectinomycin against M. smegmatis (Table 2) and that a representative compound, bromperidol, was bactericidal and effective against M. tuberculosis in a macrophage model of infection. In macrophages, the MIC of bromperidol was about 10 times lower than in vitro (Table 4), suggesting that it accumulates within macrophages, similar to what has been reported for macrolides (6). Importantly, bromperidol enhanced the activities of a broad spectrum of other antibiotics against M. tuberculosis, including rifampin, streptomycin, clofazimine, and clarithromycin, all anti-TB drugs (Table 4). Bromperidol represents a new bactericidal chemical compound class (butyrophenone) having broadly synergistic activity with diverse antibiotics against M. tuberculosis. Butyrophenones have high affinity for dopamine D2 receptors, and they are used to treat various psychiatric disorders (8, 44). The similarities of the butyrophenones with the phenothiazines suggest their potential use as adjuvants in TB therapy. Both are antidopaminergic antipsychotic drugs with potential use in TB therapy. Although two major phenothiazines, chlorpromazine and its less toxic derivative thioridazine, have poor in vitro activity against M. tuberculosis, like bromperidol, they accumulate inside macrophages and lungs. Thioridazine reaches concentrations in pulmonary tissue that are 100-fold in excess of levels found in plasma (5). In fact, thioridazine had activity in a mouse model of multidrug-resistant tuberculosis, and it has been successfully used to cure XDR-TB patients within a therapeutic drug regimen (1, 4, 40). Similarly, butyrophenones (including bromperidol) accumulate largely in the lung tissues of animal models (16). Furthermore, lower dosages over longer periods may allow additional accumulation in lung macrophages, as suggested by our ex vivo experiments.
We found that all possible pairwise combinations of bromperidol, spectinomycin, and rifampin were synergistic (Table 5), leading to our hypothesis that the synergistic activity of the triplet combination might have multiplicative effects. In fact, the spectinomycin-rifampin synergistic combination displayed a dramatic reduction in the MICsyn when bromperidol was added to the combination (Table 5). To illustrate the magnitude of the synergistic changes observed in the triple combination, spectinomycin's MIC decreased about 128-fold and rifampin's MIC decreased 8- to 16-fold in the presence of subinhibitory concentrations of bromperidol. These results and the implications that they have for other unexplored drug combinations could have important consequences in TB therapy.
There are two important benefits of synergistic drug combinations. First, efficacy can be increased without augmenting the toxicity of individual compounds; alternatively, toxic side effects can be decreased without compromising efficacy. For example, in the case of TB treatment, rifampin is not used at its most effective concentrations because of its associated toxicity. However, optimal dosages have never been clearly established (15). Within the proposed triple synergistic combination, the same rifampin therapeutic dose could be used with a 16-fold increase in efficacy. Since rifampin is a dose-dependent, rather than a time-dependent, antibiotic (18), optimal synergistic concentrations need to be maintained for a minimal period of time. This makes it an especially appealing candidate for use within synergistic combinations. In the case of spectinomycin, routinely given by intramuscular injection to achieve therapeutic concentrations in serum (about 100 mg/liter 1 h after a single 2-g dose [36]), the 128-fold increase in its effectiveness within the triple-combination MICsyn (0.6 mg/liter) would potentially allow oral administration, a fundamental requirement for TB drugs to treat off-site patients. While little is known about the oral bioavailability of spectinomycin in humans, extensive studies have been performed in animals (14). After oral administration of spectinomycin to dogs (14) and chickens (2), a maximal concentration of 22 mg/liter and 13 mg/liter, respectively, was achieved; these values are well above the MICsyn obtained in our in vitro studies. Finally, when rats and dogs were given spectinomycin orally, no significant side effects were observed after 90 days (14). This suggests that spectinomycin could be orally administered for prolonged periods.
In this work, we described the use of HTSS in mycobacteria to identify synergistic combinations that enhanced the activity of spectinomycin, an antibiotic that is not clinically effective in treating TB on its own. We demonstrated that clinically approved drugs, such as macrolides, azoles, or butyrophenones, could be used to enhance the activities of not only spectinomycin but also other non-clinically effective antibiotics against M. tuberculosis. We are now using HTSS to identify synergistic combinations that improve the activities of existing anti-TB drugs. In drug combination therapy, similar pharmacokinetic properties for each drug in the combination would be ideal, avoiding periods of “monotherapy” and loss of synergy. Therefore, it is important to test the activities of synergistic combinations in mouse models. Because drugs described in this study are available in today's pharmacopoeia, such synergistic combinations could be readily evaluated in phase II clinical trials. This may be of critical importance for TB patients, especially those infected with MDR- or XDR-TB strains, for which there are no effective therapies. Finally, our “barcode” assays among randomly tested drugs revealed an understudied array of synergistic drug interactions in Mycobacterium and underline the need for similar studies of other bacterial pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Gaye Sweet for lab management and critical reading of the manuscript, Heidi Hare for technical assistance, Clara Westwell-Roper for participation in initial studies, Vivian Miao and Julian Davies for critical reading of the manuscript, and the Canadian Chemical Biology Network for providing the chemical libraries used in this study. We thank the British Columbia Centre for Disease Control for providing access to a containment level 3 facility.
This work was supported by Canadian Institute of Health Research grants MOP-82745 (C.J.T. and Y.A.-G.) and MOP-82855 (C.J.T.), the Centre for Drug Research and Development, and the British Columbia Lung Association. S.R.-G. held a postdoctoral grant from the Fundación Alfonso Martín Escudero (Spain).
S.R-G. designed research, performed experiments, analyzed data, and wrote the manuscript; C.N. performed in vitro and ex vivo M. tuberculosis experiments; H.A. performed and analyzed HTSS; J.D.C. performed in vitro M. tuberculosis experiments; X.Z. performed ex vivo M. tuberculosis experiments; T.P. designed research; Y.A.-G. designed research; M.R. designed research and analyzed HTSS data; and C.J.T. designed research, analyzed data, and wrote the manuscript.
Footnotes
Supplemental material for this article may be found at http://aac.asmusa.org/.
Published ahead of print on 16 May 2011.
REFERENCES
- 1. Abbate E., et al. 2007. Extensively resistant tuberculosis (XDR-TB) in Argentina: epidemiology, bacteriology, therapy and outcome. Rev. Argentina Med. Respir. 1:19–25 [Google Scholar]
- 2. Abu-Basha E. A., Gehring R., Albwa'neh S. J. 2007. Pharmacokinetics and bioavailability of spectinomycin after i.v., i.m., s.c. and oral administration in broiler chickens. J. Vet. Pharmacol. Ther. 30:139–144 [DOI] [PubMed] [Google Scholar]
- 3. Alvarez-Ortega C., Wiegand I., Olivares J., Hancock R. E., Martinez J. L. 2010. Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to beta-lactam antibiotics. Antimicrob. Agents Chemother. 54:4159–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amaral L., Boeree M. J., Gillespie S. H., Udwadia Z. F., van Soolingen D. 2010. Thioridazine cures extensively drug-resistant tuberculosis (XDR-TB) and the need for global trials is now! Int. J. Antimicrob. Agents 35:524–526 [DOI] [PubMed] [Google Scholar]
- 5. Amaral L., Kristiansen J. E., Viveiros M., Atouguia J. 2001. Activity of phenothiazines against antibiotic-resistant Mycobacterium tuberculosis: a review supporting further studies that may elucidate the potential use of thioridazine as anti-tuberculosis therapy. J. Antimicrob. Chemother. 47:505–511 [DOI] [PubMed] [Google Scholar]
- 6. Anonymous 2008. Handbook of anti-tuberculosis agents. Tuberculosis (Edinb.) 88:85–170 [DOI] [PubMed] [Google Scholar]
- 7. Arain T. M., Resconi A. E., Singh D. C., Stover C. K. 1996. Reporter gene technology to assess activity of antimycobacterial agents in macrophages. Antimicrob. Agents Chemother. 40:1542–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benfield P., Ward A., Clark B. G., Jue S. G. 1988. Bromperidol. A preliminary review of its pharmacodynamic and pharmacokinetic properties, and therapeutic efficacy in psychoses. Drugs 35:670–684 [DOI] [PubMed] [Google Scholar]
- 9. Boguski M. S., Mandl K. D., Sukhatme V. P. 2009. Drug discovery. Repurposing with a difference. Science 324:1394–1395 [DOI] [PubMed] [Google Scholar]
- 10. Bryskier A., Bergogne-Berezin E. 2005. Antimicrobial agents: antibacterials and antifungals. ASM Press, Washington, DC [Google Scholar]
- 11. Chiang C. Y., Centis R., Migliori G. B. 2010. Drug-resistant tuberculosis: past, present, future. Respirology 15:413–432 [DOI] [PubMed] [Google Scholar]
- 12. Chong C. R., Sullivan D. J., Jr 2007. New uses for old drugs. Nature 448:645–646 [DOI] [PubMed] [Google Scholar]
- 13. Cottarel G., Wierzbowski J. 2007. Combination drugs, an emerging option for antibacterial therapy. Trends Biotechnol. 25:547–555 [DOI] [PubMed] [Google Scholar]
- 14. Cuerpo L., Livingston R. C. 1994. Spectinomycin. In Residues of some veterinary drugs in animals and foods. Monographs prepared by the Forty-second Meeting of the Joint FAO/WHO Expert Committee on Food Additives FAO Food Nutr. Pap. 41:1-86. [PubMed] [Google Scholar]
- 15. Diacon A. H., et al. 2007. Early bactericidal activity of high-dose rifampin in patients with pulmonary tuberculosis evidenced by positive sputum smears. Antimicrob. Agents Chemother. 51:2994–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Digenis G. A., et al. 1981. Tissue distribution studies of [18F]haloperidol, [18F]-beta-(4-fluorobenzoyl)propionic acid, and [82Br]bromperidol by external scintigraphy. J. Pharm. Sci. 70:985–989 [DOI] [PubMed] [Google Scholar]
- 17. DiMasi J. A., Hansen R. W., Grabowski H. G. 2003. The price of innovation: new estimates of drug development costs. J. Health Econ. 22:151–185 [DOI] [PubMed] [Google Scholar]
- 18. Donald P. R., Diacon A. H. 2008. The early bactericidal activity of anti-tuberculosis drugs: a literature review. Tuberculosis (Edinb.) 88(Suppl. 1):S75–S83 [DOI] [PubMed] [Google Scholar]
- 19. Gao D., Hiromura M., Yasui H., Sakurai H. 2002. Direct reaction between shikonin and thiols induces apoptosis in HL60 cells. Biol. Pharm. Bull. 25:827–832 [DOI] [PubMed] [Google Scholar]
- 20. Gutierrez-Lugo M. T., Baker H., Shiloach J., Boshoff H., Bewley C. A. 2009. Dequalinium, a new inhibitor of Mycobacterium tuberculosis mycothiol ligase identified by high-throughput screening. J. Biomol. Screen. 14:643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Helbig K., Grosse C., Nies D. H. 2008. Cadmium toxicity in glutathione mutants of Escherichia coli. J. Bacteriol. 190:5439–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hugonnet J. E., Tremblay L. W., Boshoff H. I., Barry C. E., III, Blanchard J. S. 2009. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science 323:1215–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jassal M., Bishai W. R. 2009. Extensively drug-resistant tuberculosis. Lancet Infect. Dis. 9:19–30 [DOI] [PubMed] [Google Scholar]
- 24. Johnson M. D., MacDougall C., Ostrosky-Zeichner L., Perfect J. R., Rex J. H. 2004. Combination antifungal therapy. Antimicrob. Agents Chemother. 48:693–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kohanski M. A., Dwyer D. J., Hayete B., Lawrence C. A., Collins J. J. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]
- 26. Koul A., Arnoult E., Lounis N., Guillemont J., Andries K. 2011. The challenge of new drug discovery for tuberculosis. Nature 469:483–490 [DOI] [PubMed] [Google Scholar]
- 27. Manley P. W., et al. 2010. Structural resemblances and comparisons of the relative pharmacological properties of imatinib and nilotinib. Bioorg. Med. Chem. 18:6977–6986 [DOI] [PubMed] [Google Scholar]
- 28. McLean K. J., et al. 2002. Azole antifungals are potent inhibitors of cytochrome P450 mono-oxygenases and bacterial growth in mycobacteria and streptomycetes. Microbiology 148:2937–2949 [DOI] [PubMed] [Google Scholar]
- 29. Montoro E., et al. 2005. Comparative evaluation of the nitrate reduction assay, the MTT test, and the resazurin microtitre assay for drug susceptibility testing of clinical isolates of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 55:500–505 [DOI] [PubMed] [Google Scholar]
- 30. Murakami T., et al. 1986. The bialaphos biosynthetic genes of Streptomyces hygroscopicus: molecular cloning of the gene cluster. Mol. Gen. Genet. 205:42–50 [Google Scholar]
- 31. Nguyen L., Thompson C. J. 2006. Foundations of antibiotic resistance in bacterial physiology: the mycobacterial paradigm. Trends Microbiol. 14:304–312 [DOI] [PubMed] [Google Scholar]
- 32. Novak E., Gray J. E., Pfeifer R. T. 1974. Animal and human tolerance of high-dose intramuscular therapy with spectinomycin. J. Infect. Dis. 130:50–55 [DOI] [PubMed] [Google Scholar]
- 33. Odds F. C. 2003. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 52:1. [DOI] [PubMed] [Google Scholar]
- 34. Ouellet H., Johnston J. B., Ortiz de Montellano P. R. 2010. The Mycobacterium tuberculosis cytochrome P450 system. Arch. Biochem. Biophys. 493:82–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Payne D. J., Gwynn M. N., Holmes D. J., Pompliano D. L. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29–40 [DOI] [PubMed] [Google Scholar]
- 36. Pharmacia & Upjohn Company 2003. Trobicin (spectinomycin) for injectable suspension prescribing information. Pharmacia & Upjohn Company, Kalamazoo, MI [Google Scholar]
- 37. Poole K. 2007. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 39:162–176 [DOI] [PubMed] [Google Scholar]
- 38. Ramon-Garcia S., Martin C., Ainsa J. A., De Rossi E. 2005. Characterization of tetracycline resistance mediated by the efflux pump Tap from Mycobacterium fortuitum. J. Antimicrob. Chemother. 57:252–259 [DOI] [PubMed] [Google Scholar]
- 39. Sun J., et al. 2009. A broad-range of recombination cloning vectors in mycobacteria. Plasmid 62:158–165 [DOI] [PubMed] [Google Scholar]
- 40. van Soolingen D., et al. 2010. The antipsychotic thioridazine shows promising therapeutic activity in a mouse model of multidrug-resistant tuberculosis. PLoS One 5:e12640 doi:10.1371/journal.pone.0012640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walsh C. 2003. Antibiotics: actions, origins, resistance. ASM Press, Washington, DC [Google Scholar]
- 42. Woodcock J., Griffin J. P., Behrman R. E. 2011. Development of novel combination therapies. N. Engl. J. Med. 364:985–987 [DOI] [PubMed] [Google Scholar]
- 43. World Health Organization 2010. Report: global tuberculosis control World Health Organization, Geneva, Switzerland [Google Scholar]
- 44. Yasui-Furukori N., et al. 2002. Therapeutic effects of bromperidol on the five dimensions of schizophrenic symptoms. Prog. Neuropsychopharmacol. Biol. Psychiatry 26:53–57 [DOI] [PubMed] [Google Scholar]
- 45. Yeh P., Tschumi A. I., Kishony R. 2006. Functional classification of drugs by properties of their pairwise interactions. Nat. Genet. 38:489–494 [DOI] [PubMed] [Google Scholar]
- 46. Zhang L., et al. 2007. High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections. Proc. Natl. Acad. Sci. U. S. A. 104:4606–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.