Abstract
Significant increases in STM3031, STM1530, and AcrD protein levels and significant decreases in OmpC and OmpD protein levels are present when the ceftriaxone-resistant Salmonella enterica serovar Typhimurium R200 strain is compared with the ceftriaxone-susceptible strain 01-4. AcrD is known to be involved in drug export, and STM3031 seems to play a key role in ceftriaxone resistance. Here, we examine the roles of STM1530, OmpC, and OmpD in ceftriaxone resistance. An ompD gene deletion mutant showed 4-fold higher ceftriaxone resistance than 01-4. An ompC gene deletion mutant showed 4-fold higher cephalothin and erythromycin resistance than 01-4, but there was no effect on ceftriaxone resistance. However, a stm1530 deletion mutant did show >64-fold lower ceftriaxone resistance than R200. Moreover, the STM3031 protein was significantly decreased in R200(Δstm1530) compared to R200. STM3031 expression has been shown to be influenced by the two-component system regulator gene baeR. CpxR seems to modulate BaeR. A cpxA-cpxR gene deletion mutant showed >2,048-fold lower ceftriaxone resistance than R200. The outer membrane protein profile of R200(ΔcpxAR) showed significant decreases in STM3031 and STM1530 compared to R200, while OmpD had returned to the level found in 01-4. Furthermore, the stm3031, stm1530, and ompD mRNA levels were correlated with their protein expression levels in these strains, while decreases in the mRNA levels of the efflux pump acrB, acrD, and acrF genes were found in R200(ΔcpxAR). Findings similar to those for R200(ΔcpxAR) were found for R200(ΔbaeSR). These results, together with those for STM3031 and the fact that STM1530 is an outer membrane protein, suggest that STM1530 and OmpD are influenced by the CpxAR and BaeSR two-component systems and that this contributes to S. enterica serovar Typhimurium ceftriaxone resistance.
INTRODUCTION
Salmonella enterica serovar Typhimurium is a bacterial pathogen that causes gastroenteritis in humans. Resistance to multiple antimicrobial agents is common among S. enterica serovar Typhimurium strains. Emergence of resistance to a variety of antibiotics in Salmonella has been found in many areas of the world and is a potentially serious public health problem. Although extended-spectrum cephalosporins and fluoroquinolones have proved effective, resistance to these agents has been reported (20, 28, 36). The mechanism of resistance to extended-spectrum cephalosporins is mainly via the acquisition of multidrug-resistant plasmids that encode a variety of extended-spectrum and AmpC-type beta-lactamases (1, 9, 13, 23). In general, there have been fewer reports that have found a reduction in permeability to a β-lactam drug or an increase in drug efflux to be the resistance mechanism in Salmonella (3, 10, 12).
The bacterial envelope is the front line for most external stress conditions that may alter envelope components and thus induce an extracytoplasmic stress response. In Escherichia coli, a number of response pathways are induced in response to envelope stress (5, 31). These systems are involved in the biogenesis, maintenance, and repair of the bacterial envelope and thus contribute to cell surface integrity. In addition, they modulate key bacterial life functions, such as motility, colony formation, and biofilm formation (4, 11). These are found in nearly every sequenced bacterial genome. In response to environmental change, the signal is transduced from a sensor histidine kinase to a cognate response regulator through phosphoryl transfer (19, 35). Although studies have shown that two-component signal transduction systems do contribute to antibiotic resistance, only a few reports have shown that CpxAR or BaeSR is associated with antibiotic resistance (14, 15).
It has been shown that outer membrane proteins participated in antibiotic resistance either by decreasing permeability or by increasing export of drug. A reduction in the permeation of antibiotics is generally related to a decrease in porin expression or an alteration in the porin structure (27). For example, a lack of porin OmpK35 is found in most extended-spectrum beta-lactamase-expressing Klebsiella pneumoniae clinical isolates (7). Enhanced export of a drug is usually related to an increase in transporter efflux pump activity. For example, the efflux pump AcrAB-TolC, which belongs to the RND family, is known to play an important role in multidrug resistance in E. coli and Salmonella (3, 10, 12, 21). As part of this efflux pump, TolC is an outer membrane protein and has been suggested to cooperate with a number of transporters, such as AcrD and MdtABC, to expel drugs from bacteria (2, 24, 26). In a previous study, we showed that significant increases in the levels of outer membrane protein STM3031 and putative outer membrane protein STM1530 were present in the ceftriaxone-resistant strain R200 (18). Of these proteins, STM3031 has been suggested to play a key role in ceftriaxone resistance, possibly by reducing permeability via a decreased OmpD level and by enhancing export via an increase in AcrD efflux pump activity (18). Furthermore, the expression of this protein is influenced by baeR, the regulator gene of a two-component system of S. enterica serovar Typhimurium (17). In E. coli, CpxR is believed to act as a modulator of other extracytoplasmic response proteins, especially BaeR and PspF, rather than as a stand-alone regulator (5). In addition, bioinformatics analysis predicts that CpxR is likely to be a transcription factor that affects stm3031 gene expression (data not shown). Thus, in this study, we investigated the role of OmpD and STM1530 in ceftriaxone resistance; furthermore, we also examined whether the expression of these two proteins is influenced by the CpxAR and/or the BaeSR two-component signal transduction systems of S. enterica serovar Typhimurium.
MATERIALS AND METHODS
Bacterial strains and plasmids.
S. enterica serovar Typhimurium strain 01-4 is a ceftriaxone-susceptible strain that was isolated in Taiwan. S. enterica serovar Typhimurium strain R200 is a ceftriaxone-resistant strain that was generated from parental strain 01-4. S. enterica serovar Typhimurium strain, R200(Δstm3031) was generated from strain R200. All these strains have been described previously (18). Strains R200(Δstm1530), R200(ΔcpxAR), and R200(ΔbaeSR) were generated from strain R200. Strains 01-4(ΔompC) and 01-4(ΔompD) were generated from strain 01-4. All the above deletion mutants were created using the one-step inactivation of chromosomal genes method that is described below. The plasmid pGEM-T Easy (Promega) was used as the cloning vector. The plasmids pKD46 and pKD4 was used to generate strains R200(ΔcpxAR), R200(ΔbaeSR), R200(Δstm1530), 01-4(ΔompC), and 01-4(ΔompD). E. coli and S. enterica serovar Typhimurium were routinely grown in Luria broth (LB) and tryptic soy broth (TSB) at 37°C for 18 h, respectively. Bacterial cultures were supplemented, when necessary, with an appropriate amount of antibiotic.
Antibiotics for susceptibility testing.
The MICs for the various antibiotics were determined by the agar dilution method as recommended by the Clinical and Laboratory Standards Institute (25). The antibiotics used for the agar dilution method were tetracycline (TET), streptomycin (STR), gentamicin (GEN), chloramphenicol (CHL), novobiocin (NOV), erythromycin (ERY), nalidixic acid (NAL), rifampin (RIF), cephalothin (CEF), cefamandole (FAM), and ampicillin (AMP) (Sigma-Aldrich Corp., St. Louis, MO). An Etest strip was used, according to the manufacturer's recommendations (AB Biodisk, Solna, Sweden), to measure the resistance to ceftriaxone (CRO) of strains 01-4 and R200, as well as their isogenic strains.
Construction of various gene deletion mutants.
The procedures for DNA purification, restriction, ligation, agarose gel electrophoresis, and transformation of E. coli and S. enterica serovar Typhimurium were carried out as described previously (17). Each specific gene deletion mutant was constructed by the one-step inactivation of chromosomal genes method of Datsenko and Wanner (6). To construct the S. enterica serovar Typhimurium R200(Δstm1530) mutant, a Red recombinase expression plasmid, pKD46-apra, was transformed into strain R200 by electroporation. For the construction of plasmid pKD46-apra, the apramycin resistance gene was amplified by PCR using plasmid pVK173T as template DNA and primer set ApaLI-apra-F and AhdI-apra-R. The amplified product was cloned into plasmid pGEM-T Easy to form pGEM-apra. The 1,200-bp ApaLI-AhdI fragment containing an apramycin resistance gene from pGEM-apra was directionally inserted into pKD46 that had been cut with ApaLI/AhdI to form pKD46-apra. Then, pKD46-apra was introduced into S. enterica serovar Typhimurium strain R200 cells by electroporation, and bacteria were selected on LB agar plates containing apramycin. A 1,579-bp DNA fragment containing the first 40 bp of the 5′ end of the stm1530 gene, a kanamycin resistance gene, and the last 40 bp of the 3′ end of the stm1530 gene was amplified by PCR using pKD4 as template DNA and primer set stm1530-kan-F/stm1530-kan-R. R200 cells containing pKD46-apra were induced by 0.2% arabinose; then the 1,579-bp PCR product was introduced by electroporation, and bacteria were selected on a LB agar plate with kanamycin. The R200(Δstm1530) mutant was verified by PCR using two sets of primers, primer set 1, stm1530-F/stm1530-R, and primer set 2, NcoI-stm1530-F/XbaI-stm1530-R. The construction of the R200(ΔcpxAR) and R200(ΔbaeSR) mutants was similar to that of the R200(Δstm1530) mutant, except for the primers used to generate the gene deletion mutants and for verification of mutants. The primer set used to generate R200(ΔcpxAR) was cpxAR-kan-F/cpxAR-kan-R, and the primer sets used for verification were primer set 1, NcoI-cpxAR-F/XbaI-cpxAR-R, and primer set 2, NcoI-cpxR-F/XbaI-cpxR-R. The primer set used for generation of R200(ΔbaeSR) was baeSR-kan-F/baeSR-kan-R, and the primer sets used for verification were primer set 1, NcoI-baeSR-F/XbaI-baeSR-R, and primer set 2, NcoI-baeR-F/XbaI-baeR-R. The construction of the 01-4(ΔompC) and 01-4(ΔompD) mutants was similar to that of the R200(Δstm1530) mutant, except that the plasmid pKD46-apra was introduced into S. enterica serovar Typhimurium strain 01-4. The primer set used for generation of 01-4(ΔompC) was ompC-kan-F/ompC-kan-R, and the primer sets used for verification were primer set 1, kan-F/kan-R, and primer set 2, NcoI-ompC-F/XbaI-ompC-R. The primer set used for generation of 01-4(ΔompD) was ompD-kan-F/ompD-kan-R, and the primer sets used for verification were primer set 1, kan-F/kan-R, and primer set 2, NcoI-ompD-F/XbaI-ompD-R. All primers are listed in Table 1.
Table 1.
Sequences of primers designed for this study
| Function and gene | Primer | Sequence (5′ → 3′)a | Reference or source |
|---|---|---|---|
| Gene deletion from R200 | |||
| Δstm1530 | stm1530-kan-F | ATGAAAAAAGTAGTGGTGTTATCTGCGGTAGCCGCAGCCGATTGTGTAGGCTGGAGCTGC | This study |
| stm1530-kan-R | TCAAAACTCGTAGACTAAGCCGAGCGCCACGGTGTTGTCTATGGGAATTAGCCATGGTCC | This study | |
| stm1530-F | CGTCTCGGTTTTGCTGGTTTGG | 18 | |
| stm1530-R | GCCGTCATTTTTACCCTGATACTGC | 18 | |
| NcoI-stm1530-F | CCATGGCGATGAAAAAAGTAGTGGTGTTATCTG | This study | |
| XbaI-stm1530-R | TCTAGAGCAAACTCGTAGACTAAGCCG | This study | |
| ΔcpxAR | cpxAR-kan-F | ATGAATAAAATCCTGTTAGTTGATGATGACGAGAGCTGAGTGTAGGCTGGAGCTGCTTC | This study |
| cpxAR-kan-R | TTAGGTTCGCTTGTACAGCGGTAGCCACAGCGTAAGCCGCGACATGGGAATTAGCCATGGTCC | This study | |
| NcoI-cpxAR-F | CCATGGCGATGAATAAAATCCTGTTAGTTGATGAT | This study | |
| XbaI-cpxAR-R | TCTAGAGCGGTTCGCTTGTACAGCGGTA | This study | |
| NcoI-cpxR-F | CCATGGCGATGAATAAAATCCTGTTAGTTGATGAT | This study | |
| XbaI-cpxR-R | TCTAGAGCTGAAGCGGAAACCATCAGAT | This study | |
| ΔbaeSR | baeSR-kan-F | ATGAAAGTCTGGCGGCCCGGAATCACCGGCAAGCTGTTTCGTGTAGGCTGGAGCTGCTTC | This study |
| baeSR-kan-R | TACCAGGCGACACGCATCCGCTTCCCAGCGATATCCCACCGACATGGGAATTAGCCATGGTCC | This study | |
| NcoI-baeSR-F | CCATGGCGATGAAAGTCTGGCGGCCCGG | This study | |
| XbaI-baeSR-R | TCTAGAGCTACCAGGCGACACGCATCC | This study | |
| NcoI-baeR-F | CCATGGCGATGAATAAAATCCTGTTAGTTGATGAT | This study | |
| XbaI-baeR-R | TCTAGAGCTACCAGGCGACACGCATCC | This study | |
| Gene deletion from 01-4 | |||
| ΔompC | ompC-kan-F | ATGAAAGTTAAAGTACTGTCCCTCCTGGTACCAGCTCTGCGTGTAGGCTGGAGCTGCTTC | This study |
| ompC-kan-R | TTAGAACTGGTAAACCAGACCCAGCGCTACGATGTCGTCGGACATGGGAATTAGCCATGGTCC | This study | |
| kan-F | GACAGGATGAGGATCGTTTC | This study | |
| kan-R | CCTTCTATCGCCTTCTTGAC | This study | |
| NcoI-ompC-F | CCATGGCGATGAAAGTTAAAGTACTGTCCCTC | This study | |
| XbaI-ompC-R | TCTAGAGCGAACTGGTAAACCAGACCCAG | This study | |
| ΔompD | ompD-kan-F | ATGAAACTTAAGTTAGTGGCAGTGGCAGTGACTTCCCTGTTGGCAGCAGGCGTTGGTGTAGGCTGGAGCTGCTTC | This study |
| ompD-kan-R | TTAGAACTGGTAGTTCAGACCAACAGCAACGATGTTGTCGGTAGACACTTTAGCCATGGGAATTAGCCATGGTCC | This study | |
| kan-F | Sequence described above | This study | |
| kan-R | Sequence described above | This study | |
| NcoI-ompD-F | CCATGGCGAAACTTAAGTTAGTGGCAGTGG | This study | |
| XbaI-ompD-R | TCTAGAGCGAACTGGTAGTTCAGACCAAC | This study | |
| Apramycin gene for pKD46 | ApaLI-apra-F | GTGCACATCCCCGGGGACCTGCA | This study |
| AhdI-apra-R | GACTCCCCGTCGTGTTGCCCCAGCAATCAGC | This study | |
| stm1530 gene expression | NcoI-stm1530-F | Sequence described above | This study |
| XbaI-stm1530-R | Sequence described above | This study | |
| RT-PCR | |||
| stm3031 | stm3031-F | TGCAAGCAGGGAGTAATAACGGGT | |
| stm3031-R | TCACTTGGATACGCCCAGTCCCAT | ||
| stm1530 | stm1530-F | Sequence described above | |
| stm1530-R | Sequence described above | ||
| acrA | acrA-F | TATCGCTGAACTGACGGTGA | |
| acrA-R | ACCAATACGACCGCTAATCG | ||
| acrB | acrB-F | GTTCGACCTAACGGTCTGGA | |
| acrB-R | AACATGCGGTATTTCGCTTC | ||
| acrD | acrD-F | TCCGGCCAAATTGAATAGTT | |
| acrD-R | TCGGAACCGTCCTGATTAAC | 18 | |
| acrF | acrF-F | TATCTGGCTGGATGCGAATCTGCT | |
| acrF-R | ACTTTGCCGAACTCTTCCGGATCT | ||
| tolC | tolC-F | CACCCAGTACGACGATAGCA | |
| tolC-R | CTGCTGATGGAGGCGTTAAT | ||
| ompC | ompC-F | TCGCAGCCTGCTGAACCAGAAC | |
| ompC-R | ACGGGTTGCGTTATAGGTCTGAG | ||
| ompD | ompD-F | GCAACCGTACTGAAAGCCAGGG | |
| ompD-R | GCCAAAGAAGTCAGTGTTACGGT | ||
| vacJ | vacJ-F | TTTCCTGCAGGGCGATCCTTATCA | |
| vacJ-R | CTGCCGTAGAACGGAAGTTGCATA | ||
| psd | psd-F | AAACGTGCCGTTGCGGAACTTATC | |
| psd-R | ATTTCTTTGTTCGTCCGCTGCGTG | ||
| 16S RNA | 16S RNA-F | TAACCGCAGCAATTGACGTTACCC | |
| 16S RNA-R | TCTACAAGACTCAAGCCTGCCAGT |
Restriction enzyme sites are underlined.
Preparation of outer membrane proteins and SDS-PAGE.
Outer membrane fraction preparation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for protein analysis were performed as described previously (17, 18). The detection of STM3031 protein or STM1530 protein was by Western blotting as described previously (18).
RT-PCR.
Total RNA was isolated from 1 × 109 cells of S. enterica serovar Typhimurium using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. The concentration and the quality of the RNA in each sample were determined by measuring their absorbance at 260 nm. All RNAs were adjusted to a concentration of 100 ng μl−1. The detection of S. enterica serovar Typhimurium 16S rRNA was employed as an internal control. The reverse transcriptase PCR (RT-PCR) protocol was carried out with the SuperScript first-strand synthesis system (Invitrogen). The primers used for the RT-PCR analysis are listed in Table 1.
Overexpression and purification of STM1530 protein and antibody production.
A DNA fragment containing the stm1530 gene was amplified by PCR from the R200 strain using primers NcoI-stm1530-F and XbaI-stm1530-R (Table 1). The PCR product was ligated into the pGEM-T Easy vector to construct pGEM-1530. The NcoI/XbaI DNA fragment containing the stm1530 gene was then subcloned into similarly digested pBAD/Myc-His and transformed into E. coli JM109. The resulting E. coli strain harboring pBAD/Myc-His-1530 was cultured in 100 ml LB and then induced with arabinose; after this the protein was identified by SDS-PAGE. The recombinant protein 6×His-STM1530 was then purified by affinity chromatography using nickel-nitrilotriacetic acid resin (Qiagen) and SDS-PAGE. The purified recombinant 6×His-STM1530 protein was finally sent to LTK BioLaboratories (Taiwan) and used to immunize a rabbit in order to generate polyclonal antibodies against the STM1530 protein.
Fractionation into outer membrane, periplasm, and inner membrane fractions.
Using the various Salmonella strains, the separation of the bacterial cells into cytoplasmic, periplasmic, inner membrane, and outer membrane fractions was carried out as previously described (18).
Western blotting.
The procedure for detection of STM3031 and STM1530 by Western blotting was as previously described (18). The primary polyclonal rabbit antibodies, anti-STM3031 and anti-STM1530, were diluted 1/625,000 and 1/20,000, respectively.
RESULTS
Construction of the 01-4(ΔompC) and 01-4(ΔompD) mutants and determination of their susceptibility to various antibiotics.
On the basis of the findings that there were decreased levels of OmpC and OmpD in strain R200, we investigated whether OmpC and/or OmpD were involved in resistance to ceftriaxone. The ompC gene and ompD gene deletion mutants, 01-4(ΔompC) and 01-4(ΔompD), were independently generated from the 01-4 strain, and each strain's MICs for a number of different antibiotics were then determined. As shown in Table 2, strain 01-4(ΔompD) showed a 4-fold elevation in resistance to ceftriaxone, cefamandole, and cephalothin compared to the ceftriaxone-susceptible strain, 01-4 (0.047 μg ml−1 versus 0.19 μg ml−1, 2 μg ml−1 versus 8 μg ml−1, and 4 μg ml−1 versus 16 μg ml−1, respectively). For the same strain, there was also a 2- to 4-fold elevation in resistance to streptomycin, erythromycin, and chloramphenicol. However, there was no effect on resistance to ampicillin, tetracycline, gentamicin, novobiocin, nalidixic acid, and rifampin. For strain 01-4(ΔompC), there was no effect on resistance to any of the tested antibiotics, except for a 4-fold elevation in resistance to cephalothin and erythromycin. Thus, these results suggest that the decrease in the level of the putative outer membrane porin precursor OmpD is involved in ceftriaxone resistance and may also participate in resistance to cephalothin and cefamandole.
Table 2.
MICs for Salmonella enterica serovar Typhimurium strains 01-4, 01-4(ΔompC), and 01-4(ΔompD)
| Strain | MIC (μg ml−1) of the following druga: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CEF | FAM | CROb | AMP | TET | STR | GEM | NOV | ERY | NAL | RIF | CHL | |
| 01-4 | 4 | 2 | 0.047 | >512 | 8 | 128 | 1 | >512 | 128 | 4 | 16 | 256 |
| 01-4(ΔompC)c | 16 | 2 | 0.047 | >512 | 8 | 128 | 1 | >512 | 512 | 4 | 16 | 256 |
| 01-4(ΔompD)d | 16 | 8 | 0.19 | >512 | 8 | 256 | 1 | >512 | 512 | 4 | 16 | 512 |
CEF, cephalothin; FAM, cefamandole; CRO, ceftriaxone; AMP, ampicillin; TET, tetracycline; STR, streptomycin; GEM, gentamicin; NOV, novobiocin; ERY, erythromycin; NAL, nalidixic acid; RIF, rifampin; CHL, chloramphenicol.
The MIC was determined by Etest.
An ompC gene deletion mutant strain derived from 01-4.
An ompD gene deletion mutant strain derived from 01-4.
Construction of R200(Δstm1530) and determination of this mutant's susceptibility to antibiotics.
To examine the role of the stm1530 gene on antibiotic susceptibility, an stm1530 gene deletion mutant, R200(Δstm1530), was generated from the R200 strain, and the MICs of this strain to a number of antibiotics were then determined. As shown in Table 3, strain R200(Δstm1530) showed a >64-fold reduction in resistance to ceftriaxone compared to strain R200 (>256 μg ml−1 versus 4 μg ml−1). When cephalosporins other than ceftriaxone were examined, strain R200(Δstm1530) showed a >16-fold reduction in resistance to cephalothin and cefamandole compared to strain R200. In contrast, using the same strains, there was only a 2- to 4-fold reduction in resistance to streptomycin, gentamicin, and chloramphenicol and no effect on resistance to ampicillin, tetracycline, novobiocin, erythromycin, nalidixic acid, and rifampin. These results suggest that the putative outer membrane protein STM1530 plays an important role in the S. enterica serovar Typhimurium ceftriaxone resistance.
Table 3.
MICs for Salmonella enterica serovar Typhimurium strains R200, R200(ΔbaeSR), R200(ΔcpxAR), and R200(Δstm1530)
| Strain | MIC (μg ml−1) of the following druga: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CEF | FAM | CROb | AMP | TET | STR | GEM | NOV | ERY | NAL | RIF | CHL | |
| R200 | >256 | >256 | >256 | >512 | 8 | 256 | 1 | >512 | 4 | 1 | 16 | 64 |
| R200(ΔbaeSR)c | 8 | 4 | 0.25 | >512 | 2 | 128 | 0.5 | 256 | 2 | 0.5 | 8 | 16 |
| R200(ΔcpxAR)d | 8 | 4 | 0.125 | >512 | 2 | 16 | 0.25 | >512 | 2 | 1 | 8 | 16 |
| R200(Δstm1530)e | 16 | 16 | 4 | >512 | 8 | 64 | 0.5 | >512 | 4 | 1 | 16 | 32 |
CEF, cephalothin; FAM, cefamandole; CRO, ceftriaxone; AMP, ampicillin; TET, tetracycline; STR, streptomycin; GEM, gentamicin; NOV, novobiocin; ERY, erythromycin; NAL, nalidixic acid; RIF, rifampin; CHL, chloramphenicol.
The MIC was determined by Etest.
A baeSR gene deletion mutant strain derived from R200.
A cpxAR gene deletion mutant strain derived from R200.
An stm1530 gene deletion mutant strain derived from R200.
Outer membrane protein profile of R200(Δstm1530).
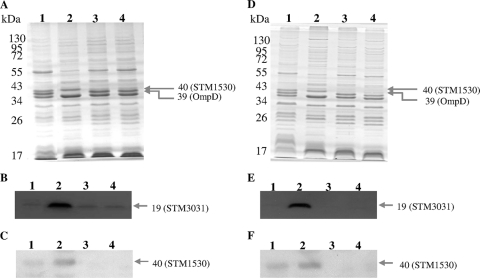
The outer membrane proteins from S. enterica serovar Typhimurium 01-4, R200, and R200(Δstm1530) were compared by SDS-PAGE analysis. Figure 1A shows that two protein bands, corresponding to molecular sizes of 40 and 39 kDa, were significantly altered. In strain R200(Δstm1530), the 40-kDa band, which had previously been identified to be the putative outer membrane protein STM1530, became undetectable compared to the level in strain R200 (lanes 2 and 4); and the 39-kDa band, which had previously been identified to be the putative outer membrane porin precursor OmpD, was returned to the level found in the 01-4 strain (lanes 1 and 4). The changes in these two proteins are exactly the same as those previously described when the R200(Δstm3031) strain was compared to R200 (lane 3). Furthermore, as shown in Fig. 1B, the STM3031 protein was significantly decreased in strain R200(Δstm1530) (lane 4) compared to strain R200 (lane 2) by Western blot analysis. It has previously been reported that the STM1530 protein was significantly decreased in strain R200(Δstm3031), and this was further confirmed by Western blot analysis in this study (Fig. 1C, lanes 2 and 3). Thus, these results suggest that the levels of expression of STM3031 and STM1530 are influenced by each other.
Fig. 1.
Comparison of the outer membrane protein profiles of different strains of S. enterica serovar Typhimurium. Each sample was resolved by 12% SDS-PAGE. The proteins were visualized by Coomassie blue staining (A and D) and Western blot analysis after probing with anti-STM3031 antibodies (B and E) and anti-STM1530 antibodies (C and F). (A to C) Lanes 1, strain 01-4; lanes 2, strain R200; lanes 3, strain R200(Δstm3031); lanes 4, strain R200(Δstm1530). (D to F) Lanes 1, strain 01-4; lanes 2, strain R200; lanes 3, R200(ΔbaeSR); lanes 4, R200(ΔcpxAR). The arrows indicate significantly changed proteins. Molecular size standards are shown on the left.
STM1530 is indeed an outer membrane protein.
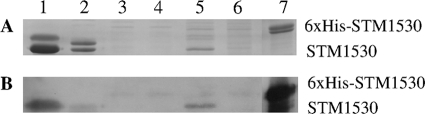
As shown in Fig. 2, by Western blot analysis, the STM1530 protein is highly abundant in the outer membrane fraction of strain R200 (lane 1), but only a very small, almost undetectable, amount is present in strain 01-4 (lane 2), and the protein is absent in strain R200(Δstm1530) (data not shown). Although the STM1530 protein was detectable in the inner membrane fraction (lane 5) of strain R200, this is likely to be due to incomplete separation of the membrane fractions, which has been reported previously (18). Thus, we conclude that STM1530 is indeed located in the outer membrane.
Fig. 2.
STM1530 is localized on the outer membranes of different strains of S. enterica serovar Typhimurium. The bacteria were fractionated into periplasmic, outer membrane, and inner membrane fractions by the procedure described in Materials and Methods. Each sample (6 μg of protein) was resolved by 12% SDS-PAGE. (A) Coomassie blue staining; (B) Western blot analysis after probing with anti-STM1530 antibodies. Lanes 1, 3, and 5 outer membrane, periplasmic, and inner membrane fractions of strain R200, respectively; lanes 2, 4, and 6, outer membrane, periplasmic, and inner membrane fractions of strain 01-4, respectively; lanes 7, recombinant protein 6×His-STM1530, which acts as a positive control.
Comparison of the susceptibilities of strains R200, R200(ΔcpxAR), and R200(ΔbaeSR) to various antibiotics.
To examine whether the CpxAR two-component system, which is involved in the bacterial envelope stress response, contributes to drug susceptibility, a mutant strain containing a double deletion of cpxA and cpxR, R200(ΔcpxAR), was generated from the R200 strain. The MICs of this strain for a number of antibiotics were then determined. As shown in Table 3, strain R200(ΔcpxAR) showed a >2,048-fold reduction in resistance to ceftriaxone compared to strain R200 (>256 μg ml−1 versus 0.125 μg ml−1). When other cephalosporin drugs were tested, strain R200(ΔcpxAR) was found to have >32-fold and >64-fold reductions in resistance to cephalothin and cefamandole, respectively, compared to strain R200. In contrast, for the same strains, there were only 2- to 16-fold reductions in resistance to tetracycline, streptomycin, chloramphenicol, gentamicin, erythromycin, and rifampin. Furthermore, there was no effect on resistance to ampicillin, novobiocin, and nalidixic acid. Our previous results have shown that expression of STM3031 is influenced by the regulator gene baeR, which is part of an S. enterica serovar Typhimurium two-component system gene pair (17). On the basis of this, a deletion mutant of the baeS sensor and baeR regulator genes, R200(ΔbaeSR), was generated. As shown in Table 3, strain R200(ΔbaeSR) shows a >1,024-fold reduction in resistance to ceftriaxone compared to strain R200 (>256 μg ml−1 versus 0.25 μg ml−1). When the other cephalosporin drugs are considered, strain R200(ΔbaeSR) showed >32-fold and >64-fold reductions in resistance to cephalothin and cefamandole, respectively, compared to strain R200. In contrast, for the same strains, there was only a 2- to 4-fold reduction in resistance to tetracycline, streptomycin, chloramphenicol, gentamicin, novobiocin, erythromycin, nalidixic acid, and rifampin. Furthermore, there was no effect on resistance to ampicillin. Taken together, these results suggest that both of the CpxAR and the BaeSR two-component signal transduction pathways seem to be directly or indirectly involved in the control of ceftriaxone sensitivity/resistance, as well as expression of several membrane proteins known or implicated in resistance to ceftriaxone. These two pathways may also be involved in cephalothin and cefamandole resistance.
Outer membrane profiles of R200(ΔbaeSR) and R200(ΔcpxAR).
The outer membrane proteins from S. enterica serovar Typhimurium strains 01-4, R200, R200(ΔbaeSR), and R200(ΔcpxAR) were compared by SDS-PAGE analysis. As shown in Fig. 1D, OmpD in strains R200(ΔbaeSR) and R200(ΔcpxAR) was returned to the level found in strain 01-4 (lanes 1, 3, and 4). In contrast, significant decreases in STM3031 (Fig. 1E) and STM1530 (Fig. 1F) were found in strains R200(ΔbaeSR) and R200(ΔcpxAR) (lanes 3 and 4) compared to R200 (lane 2). Taken together, these results suggest that the outer membrane proteins STM3031 and STM1530 both play important roles in ceftriaxone resistance and that the expression levels of both proteins are influenced by the BaeSR and CpxAR two-component systems, which are participants in the envelope stress response of S. enterica serovar Typhimurium.
Determination of mRNA levels of altered proteins in R200(ΔbaeSR) and R200(ΔcpxAR) by RT-PCR.
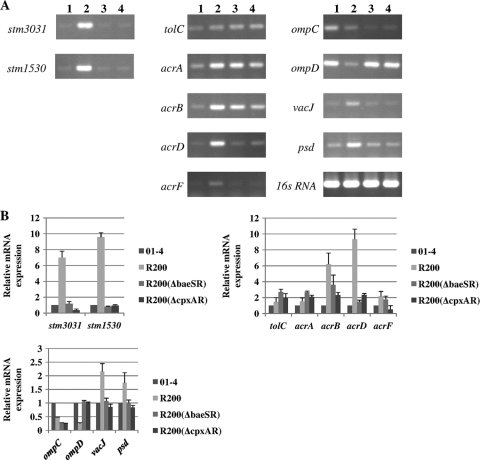
When the results for R200 shown in Fig. 3 are compared, significant decreases in the mRNA levels of STM3031 and STM1530 and significant increases in the mRNA levels of OmpD for strains R200(ΔbaeSR) and R200(ΔcpxAR) were detected; thus, the changes in translation levels of these proteins are mirrored by changes in the transcription levels of the same genes. In addition, the mRNA levels of ompC, vacJ, psd, and acrD in strains R200(ΔbaeSR) and R200(ΔcpxAR) were decreased relative to strain R200. The changes in the mRNA level pattern of the above seven genes in strains R200(ΔbaeSR) and R200(ΔcpxAR) are similar to those previously found in strain R200(Δstm3031). Among RND-type transporter genes other than acrD, the mRNA level of acrF was also significantly decreased in strain R200(ΔcpxAR), which resulted in a level lower than that in the 01-4 strain but not significantly changed relative to that in strain R200(ΔbaeSR). In contrast, the mRNA level of acrB was decreased in strains R200(ΔbaeSR) and R200(ΔcpxAR), but this decrease was significantly greater in R200(ΔcpxAR) than R200. The mRNA levels of the other components in the RND-type transporter gene group, acrA and tolC, were higher for strains R200(ΔbaeSR) and R200(ΔcpxAR) than strain R200. These changes in the mRNA patterns of strains R200(ΔbaeSR) and R200(ΔcpxAR) across these four genes are different from those previously found in strain R200(Δstm3031). Furthermore, the mRNA levels of the baeS and baeR genes were decreased in R200(ΔcpxAR); in contrast, the mRNA levels of the cpxA and cpxR genes were decreased in R200(ΔbaeSR) (data not shown). These results suggest that the BaeSR and the CpxAR two-component systems influence each other and that these two pathways are sensitive to and respond to ceftriaxone, an extracytoplasmic agent that is likely to induce cellular stress. They do this by upregulating the levels of STM3031, STM1530, and AcrD and downregulating the level of OmpD; the result of these changes is an increase in S. enterica serovar Typhimurium ceftriaxone resistance.
Fig. 3.
Quantification of the transcription levels of the genes encoding the various proteins with altered protein expression levels. (A) RT-PCR analysis of the mRNA expression levels of various genes. Lane 1, strain 01-4; lane 2, strain R200; lane 3, strain R200(ΔbaeSR); lane 4, strain R200(ΔcpxAR). (B) Relative mRNA expression levels (n-fold) determined from the results shown in panel A. The expression level of each mRNA in strain 01-4 represents 1-fold. The expression of the 16S RNA gene was used as an internal control. Each bar represents the average value of three independent experiments. Error bars are standard deviations.
DISCUSSION
In our previous study, we showed that a transposon inserted into the regulator gene baeR, which is part of a two-component system found in S. enterica serovar Typhimurium, resulted in a >4-fold reduction in resistance to ceftriaxone compared to ceftriaxone-resistant strain R200 (17). This is further supported in this study by the finding that deletion of both the sensor and regulator genes of the two-component system baeS and baeR results in a strain, R200(ΔbaeSR), that shows a >1,024-fold reduction in resistance to ceftriaxone (Table 3). In addition, a 2,048-fold reduction in resistance to ceftriaxone was found in strain R200(ΔcpxAR) (Table 3), which supports the hypothesis that the CpxAR two-component system also contributed to ceftriaxone resistance. Upon introduction of pBAD-baeSR into R200(ΔbaeSR) to give R200(ΔbaeSR)/pBAD-baeSR, there was a partial restoration of the ceftriaxone resistance (6-fold), as estimated by Etest MIC (data not shown). A similar finding whereby there was a partial restoration of ceftriaxone resistance (16-fold) was found for R200(ΔcpxAR) on complementation with pBAD-cpxAR to give R200(ΔcpxAR)/pBAD-cpxAR (data not shown). The fact that there was not complete restoration of ceftriaxone resistance might be due to the slight increase in the levels of STM3031 and STM1530 in both R200(ΔbaeSR)/pBAD-baeSR and R200(ΔcpxAR)/pBAD-cpxAR strains compared to R200 (data not shown). However, there are still other possibilities, too. First, it is well-known that autoregulation is a common feature of many regulatory pathways that control stress responses. For example, activation of the CpxAR pathway is tightly controlled by both autoamplification and feedback inhibition mechanisms, the function of which is to turn the CpxAR pathway on and off very rapidly (30). A second possibility is that there is cross talk or cross-regulation between the various bacterial two-component signal pathways involved in ceftriaxone resistance; for example, cross talk is known to occur between EnvZ-OmpR and CpxA-CpxR (34), and cross-regulation is also known to occur between ArcB-ArcA and EnvZ-OmpR (22).
It is well known that the expression of numerous genes is affected by more than one stress response pathway in E. coli, and this suggests that redundancy in these pathways probably also occurs in Salmonella. Nonetheless, a recent study using transcriptome analyses of E. coli showed that induction of specific genes via multiple pathways is not that common (5). Among the pathways where it does occur, an example is the two-component system regulators BaeR and CpxR; in E. coli these have been found to control several drug export systems that might be important to the export of toxic compounds during extracytoplasmic stress (16). The control is exerted via the binding of either one or the other, or both, of the regulators, namely, BaeR or CpxR, to the promoter region of the efflux pump acrD or to the promoter region of the operon mdtABC (16). A similar finding whereby BaeR increases multidrug resistance by directly binding to the promoter regions of acrD and mdtABC, thereby regulating AcrD and MdtABC, has also found in S. enterica serovar Typhimurium (26).
In this study, the finding that there was a significant reduction in the acrD mRNA level, together with a drastic decrease in ceftriaxone resistance in R200(ΔbaeSR) and R200(ΔcpxAR) (Fig. 3), provides additional evidence supporting the hypothesis that the BaeSR/CpxAR systems might upregulate AcrD and that this results in increased ceftriaxone resistance in Salmonella. Two possible mechanisms can be proposed for the above results. One is that BaeSR and CpxAR respond to ceftriaxone independently. Alternatively, one of these two pathways may be the main signaling system that responds to ceftriaxone, and after this signaling occurs, the other pathway functions as a modulator. Recent studies have suggested that CpxR may play an accessory or modulator role relative to other extracytoplasmic stress responders such as BaeR and PspF rather than acting as a stand-alone regulator (5). The results in this study showed that expression of one sensor and regulator pair, BaeSR or CpxAR, influences expression of the other one; this opens up the possibility that either pathway may function as a modulator of the other pathway. This final mechanism is the one that best explains the present findings.
Our previous studies have shown that both OmpD and OmpC are highly reduced at the protein level in the ceftriaxone-resistant strain R200 (18). OmpD is one of the most abundant outer membrane proteins of S. enterica serovar Typhimurium and is regulated by a multiplicity of environmental factors, including anaerobiosis, lower pH, and catabolite repression (33). As shown in Table 1, strain 01-4(ΔompD) showed a 4-fold elevation in resistance to ceftriaxone compared to strain 01-4; however, there was no difference in MIC value when ompC was deleted to give strain 01-4(ΔompC). In addition, a return in OmpD protein level to that of strain 01-4 paralleled a significant decrease (at least 64-fold) in ceftriaxone resistance in four mutants, strains R200(Δstm3031), R200(Δstm1530), R200(ΔbaeSR), and R200(ΔcpxAR) (Fig. 1A and 1D and Table 3) (18). This suggests that OmpD is highly likely to be functionally involved in allowing ceftriaxone to enter Salmonella because an increase in OmpD protein parallels a reduction in ceftriaxone resistance in these four mutants. Thus, OmpD would seem to also play an important role in the ceftriaxone resistance of S. enterica serovar Typhimurium.
Furthermore, we have confirmed in this study that the putative outer membrane protein STM1530 is indeed an outer membrane protein. Although the function of this protein remains unknown, a >64-fold reduction in resistance to ceftriaxone in strain R200(Δstm1530) suggests that this outer membrane protein plays a key role in ceftriaxone resistance. Protein structure prediction using the protein homology/analogy recognition engine (PHYRE) search tool suggests that STM1530 is a porin similar to Klebsiella pneumoniae OmpK36, E. coli PhoE, and E. coli OmpF. It has been reported that in S. enterica serovar Typhimurium, the porin OmpD together with a putative inner membrane efflux pump, YddG, is involved in resistance to methyl viologen and aromatic amino acids (8, 32). In addition, it has been suggested that OmpD in S. enterica serovar Typhimurium interacts with the periplasmic protein YdeI/OmdA to increase resistance to antimicrobial peptides (29). On the basis of the above models, we suggest that overexpression of the STM1530 protein in strain R200 may function in conjunction with a periplasmic protein(s) and/or an inner membrane efflux pump(s) to export ceftriaxone in a manner similar to that by OmpD. Interestingly, the level of STM3031 protein is significantly decreased in strain R200(Δstm1530) (Fig. 1B). In contrast, the level of STM1530 protein is significantly decreased in strain R200(Δstm3031) (Fig. 1C). Thus, it would seem likely that both of these outer membrane proteins, STM3031 and STM1530, play important roles in the ceftriaxone resistance of S. enterica serovar Typhimurium.
In conclusion, we identified the BaeSR and CpxAR two-component pathways to be important signal systems that are involved in conferring ceftriaxone resistance on S. enterica serovar Typhimurium and that they do this by controlling the levels of expression of STM3031, STM1530, OmpD, and AcrD. To our knowledge, this is the first evidence showing that STM1530 is an important protein involved in ceftriaxone resistance. Further studies are under way to explore how STM3031 and STM1530 influence each other, to explore whether STM3031and STM1530 are directly or indirectly regulated by BaeR and/or CpxR, and to discover how these regulatory mechanisms affect ceftriaxone resistance in S. enterica serovar Typhimurium.
ACKNOWLEDGMENTS
We thank Ying-Hsiu Lin for assistance with the construction of plasmid pKD46-apara.
This work was supported in part by grant NSC97-2320-B-010-009-MY3 from the National Science Council, Taiwan, and by a grant from the Ministry of Education of the Republic of China, Aim for the Top University Plan, Taiwan.
Footnotes
Published ahead of print on 6 June 2011.
REFERENCES
- 1. Arlet G., et al. 2006. Salmonella resistant to extend-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 8:1945–1954 [DOI] [PubMed] [Google Scholar]
- 2. Baranova N., Nikaido H. 2002. The BaeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 184:4168–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baucheron S., et al. 2004. AcrAB-TolC directs efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium DT104. Antimicrob. Agents Chemother. 48:3729–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beloin C., et al. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659–674 [DOI] [PubMed] [Google Scholar]
- 5. Bury-Mone S., et al. 2009. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 5:e1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Domenech-Sanchez A., et al. 2003. Role of Klebsiella pneumoniae OmpK35 porin in antimicrobial resistance. Antimicrob. Agents Chemother. 47:3332–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doroshenko V., et al. 2007. YddG from Escherichia coli promotes export of aromatic amino acids. FEMS Microbiol. Lett. 275:312–318 [DOI] [PubMed] [Google Scholar]
- 9. Dunne E. F., et al. 2000. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC β-lactamase. JAMA 284:3151–3156 [DOI] [PubMed] [Google Scholar]
- 10. Eaves D. J., Ricci V., Piddock L. J. V. 2004. Expression of acrB, acrF, acrD, marA, and soxS in Salmonella enterica serovar Typhimurium: role in multiple antibiotic resistance. Antimicrob. Agents Chemother. 48:1145–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fredericks C. E., Shibata S., Aiazwa S., Reimann S. A., Wolfe A. J. 2006. Acetyl phosphate-sensitive regulation of flagellar biosynthesis and capsule biosynthesis depends on the Rcs phosphorelay. Mol. Microbiol. 61:734–747 [DOI] [PubMed] [Google Scholar]
- 12. Giraud E., Cloeckaert A., Kerboeuf D., Chaslus-Dancla E. 2000. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 44:1223–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gonzalez-Sanz R., Herrera-Leon S., de la Fuente M., Arroyo M., Echeita M. A. 2009. Emergence of extended-spectrum β-lactamases and AmpC-type beta-lactamases in human Salmonella isolated in Spain from 2001-2005. J. Antimicrob. Chemother. 64:1181–1186 [DOI] [PubMed] [Google Scholar]
- 14. Hirakawa H., Nishino K., Hirata T., Yamaguchi A. 2003. Comprehensive studies of drug resistance mediated by overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Bacteriol. 185:1851–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hirakawa H., Nishino K., Yamada J., Hirata T., Yamaguchi A. 2003. β-Lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Antimicrob. Chemother. 52:576–582 [DOI] [PubMed] [Google Scholar]
- 16. Hirakawa H., Inazumi Y., Maasaki T., Hirata T., Yamaguchi A. 2005. Indole induced the expression of multidrug exporter genes in Escherichia coli. Mol. Microbiol. 55:1113–1126 [DOI] [PubMed] [Google Scholar]
- 17. Hu W. S., Li P. C., Cheng C. Y. 2005. Correlation between ceftriaxone resistance of Salmonella enterica Serovar Typhimurium and expression of outer membrane proteins OmpW and Ail/OmpX-like protein, which are regulated by BaeR of a two-component system. Antimicrob. Agents Chemother. 49:3955–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu W. S., Lin J. F., Lin Y. H., Chang H. Y. 2009. Outer membrane protein STM3031 (Ail/OmpX-like) plays a key role in the ceftriaxone resistance of Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 53:3248–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Laub M. T., Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41:121–145 [DOI] [PubMed] [Google Scholar]
- 20. Lee H. Y., et al. 2009. High rate of reduced susceptibility to ciprofloxacin and ceftriaxone among nontyphoid Salmonella clinical isolates in Asia. Antimicrob. Agents Chemother. 53:2696–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma D., et al. 1993. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175:6299–6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsubara M., Kitaoka S. I., Takeda S. I., Mizuno T. 2000. Turning of the porin expression under anaerobic growth conditions by His-to-Asp cross-phosphorelay through both the EnvZ-osmosensor and ArcB-anaerosensor in Escherichia coli. Genes Cells 5:555–569 [DOI] [PubMed] [Google Scholar]
- 23. Morita M., et al. 2010. Plasmid-mediated resistance to cephalosporins in Salmonella enterica serovar Typhi. Antimicrob. Agents Chemother. 54:3991–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nagakubo S., Nishino K., Hirata T., Yamaguchi A. 2002. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 184:4161–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. National Committee for Clinical Laboratory Standards 2003. Methods for dilution antimicrobial susceptibility tests for bacterial grow aerobically, 7th ed NCCLS document M7-A7. National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 26. Nishino K., Nikaido E., Yamaguchi A. 2007. Regulation of multidrug efflux systems involved in multidrug and metal resistance of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189:9066–9075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pages J. M., James C. E., Winterhalter M. 2008. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 6:893–903 [DOI] [PubMed] [Google Scholar]
- 28. Parry C. M., Threlfall E. J. 2008. Antimicrobial resistance in typhoidal and nontyphoidal Salmonellae. Curr. Opin. Infect. Dis. 21:531–538 [DOI] [PubMed] [Google Scholar]
- 29. Pilonieta M. C., Erickson K. D., Ernst R. K., Detweiler C. S. 2009. A protein important for antimicrobial peptide resistance, YdeI/OmdA, is in the periplasm and interacts with OmpD/NmpC. J. Bacteriol. 191:7243–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raivio T. L., Popkin D. L., Silhavy T. J. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J. Bacteriol. 181:5263–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rowley G., Spector M., Kormanec J., Roberts M. 2006. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat. Rev. Microbiol. 4:383–394 [DOI] [PubMed] [Google Scholar]
- 32. Santiviago C. A., et al. 2002. The Salmonella enterica sv. Typhimurium smvA, yddG, and ompD (porin) genes are required for the efficient efflux of methyl viologen. Mol. Microbiol. 46:687–698 [DOI] [PubMed] [Google Scholar]
- 33. Santiviago C. A., Toro C. S., Hildago A. A., Youderian P., Mora G. C. 2003. Global regulation of the Salmonella enterica serovar Typhimurium major porin, OmpD. J. Bacteriol. 185:5901–5905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Siryaporn A., Goulian M. 2008. Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol. Microbiol. 70:494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stock A. M., Robinson V. L., Goudreau P. N. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215 [DOI] [PubMed] [Google Scholar]
- 36. Whichard J. M., et al. 2007. Human Salmonella and concurrent decreased susceptibility to quinolones and extended-spectrum cephalosporins. Emerg. Infect. Dis. 13:1681–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]