Abstract
Echinocandins represent a new antifungal group with potent activity against Candida species. These lipopeptides inhibit the synthesis of β-1,3-glucan, the major cell wall polysaccharide. Acquired resistance or reduced echinocandin susceptibility (RES) is rare and associated with mutations in two “hot spot” regions of Fks1 or Fks2, the probable β-1,3-glucan synthases. In contrast, many fungi demonstrate intrinsic RES for reasons that remain unclear. We are using Saccharomyces cerevisiae to understand the basis for RES by modeling echinocandin-Fks interaction. Previously characterized mutations confer cross-RES; we screened for mutations conferring differential RES, implying direct interaction of that Fks residue with a variable echinocandin side chain. One mutant (in an fks1Δ background) exhibited ≥16-fold micafungin and anidulafungin versus caspofungin RES. Sequencing identified a novel Fks2 mutation, W714L/Y715N. Equivalent W695L/Y696N and related W695L/F/C mutations in Fks1 generated by site-directed mutagenesis and the isolation of a W695L-equivalent mutation in Candida glabrata confirmed the role of the new “hot spot 3” in RES. Further mutagenesis expanded hot spot 3 to Fks1 residues 690 to 700, yielding phenotypes ranging from cross-RES to differential hypersusceptibility. Fks1 sequences from intrinsically RES Scedosporium species revealed W695F-equivalent substitutions; Fks1 hybrids expressing Scedosporium prolificans hot spot 3 confirmed that this substitution imparts RES.
INTRODUCTION
The incidence of invasive fungal infections has increased dramatically in recent decades due to a substantial increase in the number of immunocompromised individuals. The most common agents of these infections are the yeast Candida albicans and the mold Aspergillus fumigatus (15, 28, 31). However, in recent years there has been a relative increase in fungal infections caused by non-albicans yeasts, such as Candida glabrata, and previously uncommon molds, including Fusarium and Scedosporium species (4, 24, 31). This epidemiological shift can be attributed, at least in part, to the increasing use of azole antifungals (12, 29, 34). Azoles effectively treat most C. albicans and A. fumigatus infections; in contrast, C. glabrata, Fusarium solani, and the Scedosporium species S. prolificans and S. apiospermum exhibit intrinsically low susceptibility or resistance to azoles (4, 17, 20).
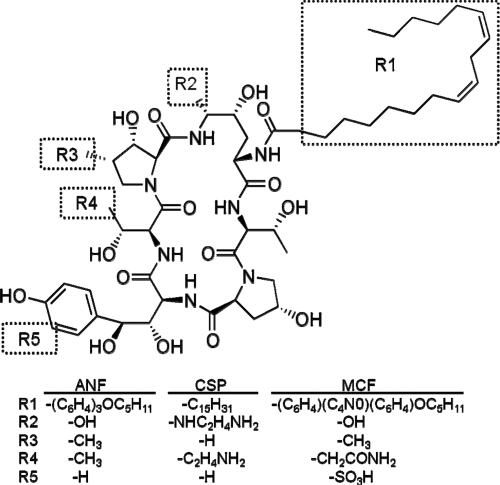
The echinocandins, including caspofungin (CSP), micafungin (MCF), and anidulafungin (ANF), represent the newest additions to the antifungal arsenal. These semisynthetic agents are lipopeptides which share a cyclic hexapeptide core linked to a hydrophobic side chain (Fig. 1) but differ in terms of the composition of this lipid chain (alkyl or aryl-alkyl) and modifications to the hexapeptide (e.g., an additional amino group on CSP and sulfate on MCF). Echinocandins exhibit potent fungicidal activity against most Candida species, with the exception of Candida parapsilosis, and fungistatic activity against A. fumigatus and related molds (6, 16, 19). Based on these activities and on their negligible toxicity, echinocandins have recently been elevated to first-line agents for treating invasive fungal infections in neutropenic patients and candidiasis or candidemia where azole resistance is likely (25).
Fig. 1.
Structures of echinocandins ANF, CSP, and MCF relative to that of natural product echinocandin B.
In susceptible fungi, echinocandins inhibit the synthesis of β-1,3-glucan, the major cell wall polysaccharide in ascomycetous yeast. The responsible enzyme is a membrane-associated complex consisting of at least two proteins: Fks1, a large integral membrane protein believed to represent the catalytic subunit, and the small GTPase Rho1, believed to represent its regulatory subunit (7, 27). This complex catalyzes a glucosyltransferase reaction using cytoplasmic UDP-glucose as the substrate to produce extracellular β-1,3-glucan. Echinocandins are noncompetitive inhibitors, implying that they do not interact with the active site. Few additional details of the β-1,3-glucan synthase reaction are understood, and it has been reproduced in vitro only with partially purified membrane preparations. While most molds encode a single Fks1, Candida species and Saccharomyces cerevisiae encode three. The FKS1 gene is essential in C. albicans (32), but in S. cerevisiae, fks1Δ disruptants remain viable due to calcineurin-dependent expression of the paralog FKS2 (22). S. cerevisiae fks1Δ fks2Δ double disruptants are nonviable, as are fks1Δ single disruptants treated with the calcineurin inhibitor FK506 (the gene name FKS1 derives from FK506 sensitivity) (22, 26).
Acquired resistance or reduced echinocandin susceptibility (RES) has been observed in clinical isolates of multiple Candida species and, to a limited extent, has been modeled in laboratory mutants of S. cerevisiae. These resistant strains harbor mutations in two “hot spot” regions of Fks1 (or its paralog Fks2), encompassing residues F639 through P647 for hot spot 1 and residues W1354 and R1357 for hot spot 2 (7, 27) (for simplicity, S. cerevisiae Fks1 numbering is used here and below, except where noted). Even with increasing echinocandin use, acquired RES has remained rare in normally susceptible fungi (2). On the other hand, the intrinsic low- or high-level RES of clinically common pathogens, such as C. parapsilosis and Cryptococcus neoformans, as well as emerging pathogens, such as Rhizopus, Fusarium, and Scedosporium species, represents a significant limitation to echinocandin use (3). While the basis for intrinsic RES may be multifactorial, recent studies using S. cerevisiae as a model suggest that the naturally occurring hot spot 1 substitutions P647A in C. parapsilosis (10) and F639Y in F. solani (14) contribute substantially to this phenotype.
It remains unclear whether or not Fks mutational hot spots represent echinocandin binding sites and, if they do, how this binding inhibits the β-1,3-glucan synthase reaction. One attempt to directly map the echinocandin binding site by cross-linking an azido derivative to membrane preparations failed to identify Fks1 (30). The Fks1 mutations described to date confer relatively uniform echinocandin cross-RES (2, 11, 27) and, thus, may confer this RES indirectly rather than through a direct effect on echinocandin binding. We reasoned that mutations conferring differential RES would, in contrast, imply a direct interaction between the mutated residue and a CSP-, MCF-, or ANF-specific side chain. Consequently, such mutations could be used to genetically map the echinocandin binding site(s). Using S. cerevisiae as a model, we identified two mutations conferring differential RES, one adjacent to hot spot 2 and the second (also identified in C. glabrata) within a previously uncharacterized region of Fks1. Site-directed mutagenesis of this “hot spot 3” confirmed its role in acquired RES; furthermore, sequence analysis and hybrid Fks1 construction implicated hot spot 3 in the intrinsic RES of Scedosporium species.
MATERIALS AND METHODS
Strains, media, drugs, and reagents.
S. cerevisiae strain BY4742 was obtained from Research Genetics (Invitrogen, Carlsberg, CA); S. prolificans 07-1208 and S. apiospermum 07-1632 were obtained from A. Fothergill (University of Texas Health Science Center at San Antonio). The partial deletion strain BY4742 fks1Δ453-649 was described previously (10). Plasmid pRS416 (URA3) was obtained from J. Nickels (Drexel University College of Medicine). The medium was YPD (1% yeast extract, 2% peptone, 2% dextrose) or, where indicated, SD-ura (DOB plus CSM-ura; Sunrise Science Products, San Diego, CA) or RPMI-S (RPMI 1640 supplemented with glutamine, 2% dextrose, 0.165 M MOPS [morpholinepropanesulfonic acid], pH 7, and 1× CSM). MCF (Mycamine) was obtained from Astellas (Deerfield, IL), CSP (Cancidas) from Merck (Rahway, NJ), ANF (Eraxis) from Pfizer (New York, NY), and FK506 from Tecoland (Edison, NJ). All drug stocks were prepared in 100% dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO) and stored at −20°C. DNA primers (Table 1) were obtained from IDT (Coralville, IA).
Table 1.
DNA primers used in this study
| Function and primera | Sequence |
|---|---|
| FKS hot spot amplification and sequencing | |
| FKS1c425F | 5′-GATTAGGCGATGTCGTCTGG |
| FKS1c583F | 5′-GGGGGTTGTTTACGTCATAT |
| FKS1c759R | 5′-GAAATAATGATGGCATTCCATACT |
| FKS2c484F | 5′-GTATTGTGCTTACAATGCTCC |
| FKS2c976R | 5′-AACCCCGAGATTGTGCGATA |
| FKS2c1166F | 5′-TGTTGCAATTGTCGGCGCTA |
| FKS2d49R | 5′-TGCGTGATAAACTTGCTTAGAA |
| Generation of fks1Δ453-697::URA3 and fks1Δ692-712::URA3 disruptantsb | |
| FKS1c453-URAF | 5′-AAAGAGACCCGTACTTGGTTACATTTGGTCACCAACTTCAGAGTGCACCATACCACAGCT |
| FKS1c697-URAR | 5′-ATAGAAAGATTTCCCAACGAGGAAAATGGTATTCACAATAGGTATTTCACACCGCATAGG |
| FKS1c692-URAF | 5′-TTTGGTTATCGCTACCGACTTCATTCTTTTCTTCTTGGATGAGTGCACCATACCACAGCT |
| FKS1c712-URAR | 5′-ATCTTGTGAAGATATTTCTCCATGGTGTTAAGATAGAAATGGTATTTCACACCGCATAGG |
| FKS1 mutagenesis | |
| FKS1m687XF | 5′-ACCCAAGATTGTCTTAGGTTTGGTTATCGCTACCGACTTCATTNTTTTCTTCTTGGATAC |
| FKS1m688XF | 5′-CAAGATTGTCTTAGGTTTGGTTATCGCTACCGACTTCATTCTTTNCTTCTTGGATACCTA |
| FKS1m689XF | 5′-TGTCTTAGGTTTGGTTATCGCTACCGACTTCATTCTTTTCYNCTTGGATACCTACTTATG |
| FKS1m690XF | 5′-CTTAGGTTTGGTTATCGCTACCGACTTCATTCTTTTCTTCNNGGATACCTACTTATGGTA |
| FKS1m691XF | 5′-CTTAGGTTTGGTTATCGCTACCGACTTCATTCTTTTCTTCTTGNATACCTACTTATGGTA |
| FKS1m692XF | 5′-TAGGTTTGGTTATCGCTACCGACTTCATTCTTTTCTTCTTGGATNCCTACTTATGGTACA |
| FKS1m693XF | 5′-TTGGTTATCGCTACCGACTTCATTCTTTTCTTCTTGGATACCTNCTTATGGTACATTATT |
| FKS1m694XR | 5′-AAAGATTTCCCAACAGAGAAAATGGTATTCACAATAATGTACCATNNGTAGGTATCCAAG |
| FKS1m695XR | 5′-ATAGAAAGATTTCCCAACAGAGAAAATGGTATTCACAATAATGTAMMATAAGTAGGTATC |
| FKS1m695L696NR | 5′-ATAGAAAGATTTCCCAACAGAGAAAATGGTATTCACAATAATGTTCAATAAGTAGGTATC |
| FKS1m696XR | 5′-ATAGAAAGATTTCCCAACAGAGAAAATGGTATTCACAATAATGSSCCATAAGTAGGTATC |
| FKS1m696XR2 | 5′-ATAGAAAGATTTCCCAACAGAGAAAATGGTATTCACAATAATGWWCCATAAGTAGGTATC |
| FKS1m697XR | 5′-TAAATAGAAAGATTTCCCAACAGAGAAAATGGTATTCACAATANNGTACCATAAGTAGGT |
| FKS1m697VR | 5′-AATAGAAAGATTTCCCAACAGAGAAAATGGTATTCACAATAACGTACCATAAGTAGGTAT |
| FKS1m698XR | 5′-CCTAAATAGAAAGATTTCCCAACAGAGAAAATGGTATTCACANNAATGTACCATAAGTAG |
| FKS1m698XR2 | 5′-AATAGAAAGATTTCCCAACAGAGAAAATGGTATTCACAKKAATGTACCATAAGTAGGTAT |
| FKS1m699XR | 5′-ATACCTAAATAGAAAGATTTCCCAACAGAGAAAATGGTATTCNNAATAATGTACCATAAG |
| FKS1m699XR2 | 5′-AAATAGAAAGATTTCCCAACAGAGAAAATGGTATTSMAAATAATGTACCATAAGTAG |
| FKS1m700XR | 5′-ACCTAAATAGAAAGATTTCCCAACAGAGAAAATGGTSYTCACAATAATGTACCATAAG |
| Generation of fks1::Sp692-712 and fks1::Sp692-712-F695W hybridsc | |
| FKS1-Sp692F | 5′-TTGTAAAGTGCAACCCAAGATTGTCTTAGGTTTGGTTATCGCTACCGACTTCATTCTTTTCTTCCTGGATACTTATCTC |
| FKS1-Sp712R | 5′-GGAGTATATTCTTTTTGGCAATCTTGTGAAGATATTTCTCCATGGTGTTAAGATAGAAATTCCCAGGTAAAACGATCGAG |
| FKS1-F695WSp692F | 5′-AGATTGTCTTAGGTTTGGTTATCGCTACCGACTTCATTCTTTTCTTCCTGGATACTTATCTCTGGTACGTGCTGGCAAAT |
| Amplification and sequencing of S. apiospermum FKS1 | |
| conFKS1c311F | 5′-TGCTGGGGTGAGGCTAACCAGGT |
| SpFKS1c884R | 5′-CCACTCATGAGGGTGAAGCTGCTT |
| SaFKS1c824F | 5′-AGTCGCTTTCCACGCCCATT |
| SpFKS1c1127R | 5′-ACCTCATCTCTTCAAACTCGGC |
| SaFKS1c1093F | 5′-ACCGTGGTGAGTATATTCAAC |
| conFKS1c1468R | 5′-CCAGACKGTRAGKGTRGCGAARAGRAGCATCAT |
F, forward primer; R, reverse primer; c, codon; d, downstream; m, mutation.
Underlining indicates portion of primer complementary to URA3 flanking sequence.
Underlining indicates portion of primer complementary to S. prolificans FKS1; homology extends into the S. cerevisiae FKS1 portion due to sequence conservation.
UV mutagenesis and colony screening.
BY4742 wild type (WT) and fks1Δ453-649 (1 × 107 cells) were plated on YPD agar containing 1 μg/ml MCF or CSP, exposed to a UV germicidal lamp (15 W for 5 s at a distance of 15 cm), and incubated at 30°C for 72 h. Control plates indicated a survival rate of approximately 50%. Colonies were streaked for isolation on drug-free YPD prior to screening by broth microdilution.
Drug susceptibility testing.
Broth microdilution assays in YPD medium (or, where indicated, RPMI-S) were used to determine MICs as previously described (13, 14), with the following exception: for mutants of the slower-growing fks1Δ453-649 strain, the starting cell density was increased from 1 × 104 to 1 × 105 cells/ml. Following incubation at 30°C for 24 h, the absorbance at 630 nm was read in a microplate reader. The MIC was defined as the lowest concentration inhibiting growth by ≥80% relative to the growth of drug-free controls.
DNA sequencing and PCR screens.
For FKS sequencing, colony PCR was employed with Taq polymerase (New England BioLabs) as recommended by the manufacturer with primer pair FKS2c484F-FKS2c976R, FKS2c1166F-FKS2d49R, or FKS1c425F-FKS1c759R, annealing temperature of 54°C, and 30 cycles of amplification. Following treatment of PCR products with ExoSAP-IT (USB, Cleveland, OH) or exonuclease I plus thermosensitive alkaline phosphatase (New England BioLabs), sequencing was performed by Genewiz (South Plainfield, NJ) using primer FKS2c484F, FKS2c1166F, or FKS1c583F. For PCR screening, the same conditions were employed with primer pair FKS1c425F-FKS1c759R; a portion was analyzed by gel electrophoresis and the remainder sent for sequencing as described above.
Confirmation of FKS2 mutation by transformation.
Genomic DNA was prepared from a late-log culture by phenol extraction and ethanol precipitation as described previously (14, 35). PCR employed Phusion polymerase (New England BioLabs, Ipswich, MA) as recommended by the manufacturer, with primer pair FKS2c484F-FKS2c976R, annealing at 54°C, and 28 cycles. Products were purified (E.Z.N.A. cycle pure kit; Omega Bio-Tek, Norcross, VA) and transformed into BY4742 fks1Δ453-649 using a Frozen-EZ Yeast Transformation II kit (Zymo Research, Orange, CA) as recommended by the manufacturer. Cells were plated on YPD medium containing 1 μg/ml MCF and incubated at 30°C for 72 h. Colonies were screened by broth microdilution, followed by PCR and DNA sequencing to confirm incorporation of the desired mutation.
Site-directed mutagenesis of FKS1.
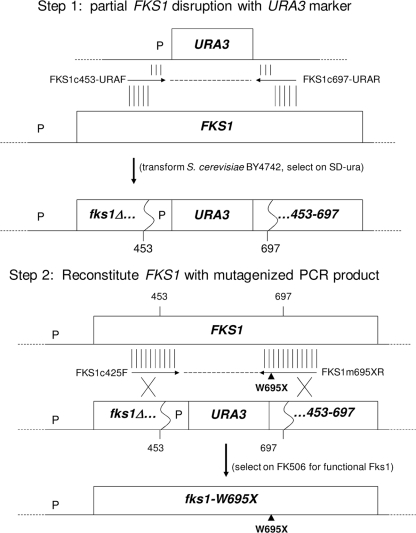
Mutations were incorporated into an otherwise unaltered chromosomal copy of FKS1 by a modification of the previously described (10, 14) PCR/reconstitution-based site-directed mutagenesis (PRSM) (Fig. 2). In step 1, a PCR product was generated using URA3 plasmid pRS416 as template and primers FKS1c453-URAF and FKS1c697-URAR, which include 20 bases at their 3′ ends complementary to URA3 flanking sequences and 40 bases at their 5′ ends complementary to FKS1 codons 440 to 453 and 697 to 710, respectively. Following purification and transformation into BY4742 as described above, cells were plated on SD-ura and colonies screened by replica plating on YPD with and without 1 μg/ml FK506, followed by sequencing to confirm fks1Δ453-697::URA3 disruption. In step 2, FKS1 was reconstituted by transformation of this disruptant with a purified PCR product generated using forward primer FKS1c425F and one of the FKS1 mutagenic reverse primers (e.g., FKS1m695XR) listed in Table 1. Transformants were selected on YPD containing 1 μg/ml FK506, screened by replica plating on YPD and SD-ura media for loss of URA3, and sequenced to confirm reconstitution and identify the specific mutation. Additional mutants were constructed with step 1 disruptant fks1Δ692-712::URA3 (generated as described above except using primers FKS1c692-URAF and FKS1c712-URAR) and step 2 transformations with products amplified with mutagenic forward primers (e.g., FKS1m693XF) and reverse primer FKS1c759R.
Fig. 2.
PRSM method for site-directed mutagenesis of S. cerevisiae Fks1 hot spot 3 (see Materials and Methods for details). P, promoter region. Vertical lines above and below primer arrows represent regions of homology.
Amplification and sequencing of S. apiospermum FKS1.
Primers representing both conserved and S. prolificans-specific FKS1 sequences were used to amplify S. apiospermum FKS1 fragments essentially as described for S. prolificans FKS1 (14). The template was S. apiospermum genomic DNA prepared by phenol extraction and ethanol precipitation, and products were purified and sequenced as described above. The initial sequences were used to design additional S. apiospermum-specific primers to complete a sequence equivalent to S. cerevisiae FKS1 codons 311 to 1468.
Construction of hybrid Fks1.
S. cerevisiae Fks1 hybrids incorporating specific regions of S. prolificans Fks1 were generated by a modification of the PRSM method essentially as described for F. solani hybrids (14). Disruptant fks1Δ692-712::URA3 (see above) was transformed with a PCR product generated with S. prolificans genomic DNA template and primers FKS1-Sp692F and FKS1-Sp712R. Selection on FK506, screening on SD-ura and by PCR, and sequence confirmation were as previously described.
Nucleotide sequence accession number.
The S. apiospermum FKS1 sequence was deposited in GenBank with accession number HQ412797.
RESULTS
Identification of mutants exhibiting differential RES.
Screens to identify mutations conferring differential RES were performed following UV mutagenesis of haploid S. cerevisiae BY4742 WT and fks1Δ strains. Mutants were selected on YPD plates containing 1 μg/ml echinocandin, which is 32-fold above the MIC of 0.03 μg/ml for WT cultures. Mutants were streaked for isolation on drug-free plates, and MICs in YPD determined. For representative mutants (all with MICs of ≥0.5 μg/ml for one or more echinocandins), the initial sequencing focused on hot spot 1 regions of FKS1 or FKS2 in, respectively, WT or fks1Δ strains. Of the 11 characterized WT mutants, all exhibited cross-RES (≤4-fold differential RES) and mutations in Fks1 hot spot 1 (S. Katiyar, M. Johnson, and T. Edlind, unpublished data). Of the 14 fks1Δ mutants characterized, 12 exhibited cross-RES, and all of these were similarly mutated in hot spot 1 of Fks2.
The remaining two mutants showed >8-fold differential RES relative to the susceptibility of the WT. Specifically, for mutant CR, the CSP MIC increased to 16 μg/ml while the MCF and ANF MICs remained near or below the WT levels. Conversely, for mutant MR, the MCF and ANF MICs increased to >16 and 4 μg/ml, respectively, while the CSP MIC was comparable to the WT CSP MIC. Intriguingly, both lacked hot spot 1 mutations. Rather, mutant CR exhibited the Fks2 mutation S1380P, which is only 4 residues downstream of the previously described hot spot 2 (equivalent to Fks2 residues W1373 to R1376).
Mutant MR exhibited the Fks2 double mutation W714L/Y715N in a region not previously associated with RES. In light of this novel location and since UV mutagenesis could introduce additional mutations that alter susceptibility, it was necessary to directly test the role of the W714L/Y715N mutation. This was accomplished by amplification of the mutated FKS2 region, transformation into the fks1Δ parent strain, and selection of transformants on YPD containing 1 μg/ml MCF. Sequencing identified transformants which had specifically incorporated the double W714L/Y715N mutation, as well as the single mutation W714L, the latter presumably a consequence of partial DNA repair. Susceptibility assays revealed that both exhibited the same pattern of differential RES as the original UV-mutagenized isolate (Table 2). Henceforth, we refer to the region defined by the Fks2 W714L mutation as hot spot 3.
Table 2.
Echinocandin susceptibilities of hot spot 3 mutants and hybrid constructs
| Genotypea | MIC (μg/ml) on: |
|||||
|---|---|---|---|---|---|---|
| YPD |
RPMI-S |
|||||
| MCF | CSP | ANF | MCF | CSP | ANF | |
| FKS1 FKS2 | 0.03 | 0.03 | 0.03 | 0.06 | 0.5 | 0.12 |
| fks1Δ FKS2 | 0.06 | 0.002 | 0.004 | |||
| fks1Δ fks2-W714L/Y715N | >16 | 0.06 | 4 | |||
| fks1Δ fks2-W714L | >16 | 0.06 | 4 | |||
| fks1-F688C | 0.008 | 0.008 | 0.008 | |||
| fks1-L690A | 0.004 | 0.008 | 0.008 | |||
| fks1-L690E | 0.12 | 0.06 | 0.06 | |||
| fks1-D691T | 0.12 | 0.12 | 0.03 | |||
| fks1-T692A | 0.25 | 0.25 | 0.5 | |||
| fks1-Y693C | 0.12 | 0.03 | 0.12 | |||
| fks1-L694A | 0.25 | 0.12 | 0.12 | |||
| fks1-W695L/Y696N | 0.25 | 4 | 4 | |||
| fks1-W695C | 0.06 | 1 | 0.03 | 0.06 | 8 | 0.25 |
| fks1-W695L | 0.25 | 8 | 4 | 2 | 16 | 4 |
| fks1-W695F | 0.25 | 0.03 | 4 | 0.5 | 0.5 | 4 |
| fks1-Y696I | 0.004 | 0.004 | 0.016 | 0.03 | 0.5 | 0.12 |
| fks1-Y696N | 0.25 | 0.06 | 0.25 | 0.25 | 1 | 1 |
| fks1-I697T | 0.25 | 0.25 | 0.25 | |||
| fks1-I697C | 0.03 | 0.06 | 0.25 | |||
| fks1-I697F | >16 | 0.06 | 2 | >16 | 1 | 8 |
| fks1-I697R | 0.25 | 0.25 | 0.25 | |||
| fks1-I698H | 0.001 | 0.008 | 0.002 | 0.03 | 0.5 | 0.03 |
| fks1-I698P | 0.004 | 0.004 | 0.002 | |||
| fks1-I698N | 0.004 | 0.016 | 0.008 | |||
| fks1-V699W | 0.03 | 0.12 | 0.25 | |||
| fks1-V699C | 0.03 | 0.016 | 0.12 | |||
| fks1-V699Q | 0.03 | 0.06 | 0.12 | |||
| fks1-N700S | 0.12 | 0.06 | 0.25 | |||
| fks1::Sp692-712 | 0.25 | 0.016 | 0.5 | 0.5 | 1 | 2 |
| fks1::Sp692-712-F695W | 0.03 | 0.016 | 0.03 | 0.12 | 0.5 | 0.25 |
Additional mutants with the following Fks1 mutations were isolated but exhibited minimally altered MICs (≤2-fold): F689H, F689S, L690G, L690W, D691A, T692S, Y693F, Y693S, L694T, Y696F, Y696W, Y696A, I697D, I697V, I698S, I698V, V699A, V699S, and V699T.
PRSM of Fks1 hot spot 3.
Fks2 is largely but not fully redundant with Fks1, as evidenced by the modest growth defect exhibited by fks1Δ cells (22). It was therefore important to construct and test Fks1 mutants equivalent to Fks2 W714L and W714L/Y715N; i.e., Fks1 W695L and W695L/Y696N. To do this, a method was employed that we previously used to test the role in RES of a hot spot 1 residue (10) and have termed PCR/reconstitution-based site-directed mutagenesis (PRSM) (Fig. 2). Here, PCR products spanning FKS1 codons 425 to 712 and incorporating the desired mutations into the reverse primer were transformed into partial disruptant fks1Δ453-697::URA3. Transformants were selected on FK506-containing medium for reconstitution of functional Fks1 (fks1Δ strains are FK506 susceptible due to their requirement for FKS2 expression, whereas reconstituted FKS1 transformants are FK506 resistant). FK506-resistant transformants were screened on SD-ura plates for loss of URA3 and by PCR for reconstitution of chromosomal FKS1, followed by sequencing to confirm the incorporation of the desired mutations.
Consistent with the original Fks2 mutant, the equivalent Fks1 W695L and W695L/Y696N mutants generated by PRSM exhibited both RES and differential susceptibility, though in a different pattern. Specifically, these mutants displayed 8-fold increased MCF MICs but ≥128- and 128-fold increased CSP and ANF MICs, respectively (Table 2). These data confirm a role for hot spot 3 in both Fks1 and Fks2 and further support its direct interaction with echinocandin. The basis for the change in differential RES of these initial hot spot 3 mutations—specifically, from CSP sensitive in the Fks2 mutants to CSP resistant in the Fks1 mutants—is unknown but potentially reflects underlying differences in cell wall stress responses between WT and fks1Δ strains or differences in the expression and localization of Fks1 and Fks2. Furthermore, in parallel studies with a Candida glabrata fks2Δ strain (unpublished data), the Fks1 mutation W681L (equivalent to S. cerevisiae Fks1 W695L or Fks2 W714L) was identified following UV mutagenesis and selection on 1 μg/ml ANF-containing medium. This mutant exhibited moderate CSP and ANF RES (MICs increased 16-fold compared to the MICs of the parent), similar to the S. cerevisiae Fks1 mutant.
To further explore hot spot 3, PRSM was used to generate additional Fks1 mutations at residues W695 and Y696 (typically, 2 or 4 different bases were randomly incorporated into the first two positions of the codon to be mutated, theoretically yielding up to 16 different mutations). In addition to the W695L mutation, two other mutants were generated, both of which elicited unique echinocandin susceptibility phenotypes: W695F, which conferred MCF and, especially, ANF-specific RES, and W695C, which conferred CSP-specific RES (Table 2). These differential effects again support a direct interaction between echinocandin and hot spot 3.
A distinctly different result was obtained with additional mutations of Fks1 Y696. The Y696N mutation resulted in 8-fold-increased MICs for MCF and ANF, while three other mutations had no effect on echinocandin susceptibility (Table 2). However, a fifth mutation, Y696I, conferred 8-fold hypersusceptibility to MCF and CSP, while ANF susceptibility was minimally affected. This hypersusceptibility was not due to decreased function, as shown by retention of FK506 resistance (MIC > 2 μg/ml). These differential effects of Y696 mutations further support a direct interaction between echinocandin and hot spot 3.
Extension of hot spot 3.
Having confirmed a role for Fks1 residues 695 and 696 in echinocandin activity, we next examined the adjacent residues to gauge the limits of hot spot 3. Residues 687 to 700 were subjected to PRSM, producing a total of 43 mutants. Over half (25 of 43) of these displayed echinocandin sensitivities similar to those of the WT; i.e., the MICs increased or decreased by ≤4 fold (Table 2). Of the remaining 18 mutants, a variety of phenotypes were observed, including reduced susceptibility to one (5 of 43), two (3 of 43), or all three (5 of 43) echinocandins. Similarly, some mutants exhibited hypersusceptibility to one (2 of 43), two (2 of 43), or all three (1 of 43) echinocandins. This hypersusceptibility was not a consequence of reduced Fks1 function, as these mutants exhibited normal growth in the absence of drug and WT levels of resistance to FK506 (MIC > 2 μg/ml).
Definitive limits to this hot spot have not yet been established. Modest 4-fold cross-hypersusceptibility was observed with the F688 mutation, while mutations conferring various degrees of RES extended from L690 to N700. Repeated attempts to mutate residue L687 proved unsuccessful; indeed, all 3 mutants generated exhibited the WT residue L687 (from a theoretical total of 16 different substitutions), suggesting an essential role for this residue in Fks function.
Sequence analysis implicates hot spot 3 in intrinsic RES.
BLAST and clustal analyses of Fks sequences from diverse fungal pathogens revealed that hot spot 3 is relatively well conserved among both yeasts and molds of the ascomycete, basidiomycete, and zygomycete phyla (Fig. 3). In particular, the hot spot 3-defining residue that is equivalent to S. cerevisiae Fks1 W695 is found in all of these fungi, with the notable exceptions of Scedosporium prolificans Fks1, which has the equivalent of F695, and Blastomyces dermatitidis, which has the equivalent of C695. Scedosporium species are opportunistic molds which exhibit intrinsic echinocandin RES (5, 31). The dimorphic fungus B. dermatitidis also exhibits intrinsic RES in the clinically relevant yeast phase (8), while its mold phase is MCF susceptible (23); unfortunately, mold-phase data for CSP and ANF are not available. As described above, a PRSM-generated W695C mutation conferred CSP-specific RES, while a W695F mutation conferred MCF- and, particularly, ANF-specific RES (Table 2). We hypothesize that these substitutions contribute to the intrinsic RES of these fungi.
Fig. 3.
Alignment of hot spot 3 regions of Fks1 homologs. The underlined segment indicates the currently defined limits of hot spot 3. The hot spot 3-defining residue W695 of S. cerevisiae Fks1 and the equivalent F695 residues of Scedosporium species are indicated in bold. Residues 692 to 712 of S. cerevisiae Fks1 and the equivalent region of S. prolificans used to generate the fks1::Sp692-712 hybrid construct are highlighted in gray. MIC values for S. cerevisiae are from this study; all others are published MIC90 values (5, 8). B. dermatitidis MICs are for the yeast phase; for the mold phase only MCF MICs (range, 0.008 to 0.031 μg/ml) are available (23). The S. prolificans and S. apiospermum sequences are unnumbered since their amino termini have not been determined. R. oryzae, Rhizopus oryzae.
A second Scedosporium species increasingly implicated in opportunistic infections and exhibiting RES is S. apiospermum (teleomorph Pseudallescheria boydii) (31). Using primers based on conserved Fks1 sequences, we amplified PCR products from S. apiospermum genomic DNA. Based on their sequences, additional PCR primers were designed that ultimately generated an S. apiospermum FKS1 sequence extending from the equivalent of codon 311 to 1468 (GenBank accession number HQ412797). BLASTP analysis indicates that the predicted Fks1 sequence is, as expected, most closely related to S. prolificans Fks1 (93% identity). Most relevant to our studies here, S. apiospermum Fks1, like S. prolificans Fks1, includes the equivalent of F695 (Fig. 3).
Hybrid Fks1 supports the role of hot spot 3 in S. prolificans intrinsic RES.
Ideally, the role of the F695-equivalent residue in intrinsic RES would be tested directly in Scedosporium species by generating an F695W-equivalent mutation, predicted to confer echinocandin susceptibility. However, genetic manipulation of these species is not currently feasible, and so, we employed the S. cerevisiae hybrid Fks1 approach previously used to test the role of F. solani hot spot 1 in intrinsic RES (14). Here, we replaced two different segments of S. cerevisiae Fks1 (residues 675 to 697 or 692 to 712) with the equivalent segments from S. prolificans Fks1. The hybrid construction employed a modification of the PRSM method.
The first hybrid construct, fks1::Sp675-697, displayed a severe growth defect (possibly due to replacement of the critical L687 residue; see above) and, hence, was not a suitable model for studying echinocandin susceptibility. In contrast, the alternative construct fks1::Sp692-712 grew comparably to the WT parent and was fully FK506 resistant, consistent with a functional Fks1. Importantly, this hybrid exhibited an echinocandin susceptibility profile similar to that of the W695F mutant described above; specifically, the ANF and MCF MICs increased 8- and 16-fold, respectively, relative to those of the WT parent (Table 2). To confirm the role of the F695 residue in this phenotype, an otherwise identical fks1::Sp692-712 hybrid was constructed which incorporated an F695W mutation. This mutation reduced the MICs to WT levels for all three echinocandins (Table 2), further implicating this hot spot 3 residue in Scedosporium's intrinsic RES.
Comparison of MICs obtained in YPD and supplemented RPMI.
Our mutant selections and susceptibility assays employed YPD, the standard medium for auxotrophic laboratory strains of S. cerevisiae. For comparison, representative fks1 mutants and the WT parent were also assayed in RPMI 1640 (supplemented to compensate for the auxotrophies), as routinely used with fungal clinical isolates. The CSP activity was reduced 16-fold in this medium relative to its activity in YPD, while the MCF and ANF activities were reduced only 2- to 4-fold (Table 2). When these differences in the parent susceptibilities are taken into account, the results obtained in YPD and RPMI for diverse mutants were qualitatively similar (Table 2).
DISCUSSION
Previous studies of laboratory mutants of S. cerevisiae and clinical isolates of diverse Candida species have localized RES-conferring mutations to hot spot 1 or, less commonly, hot spot 2 of Fks1 or its paralog Fks2 (27). With few exceptions, these mutations confer relatively uniform cross-RES to CSP, MCF, and ANF. This cross-RES may reflect interaction of the mutated Fks residue with a structural element common to all three echinocandins; e.g., the cyclic hexapeptide or the alkyl portion of the lipid side chain (Fig. 1). Alternatively, it could reflect an indirect effect of the mutation on echinocandin susceptibility. Indeed, to date there has been no evidence that echinocandins directly interact with Fks. By analogy, the majority of mutations conferring azole resistance in C. glabrata confer fluconazole-itraconazole cross-resistance and localize to transcription factor Pdr1, clearly distinct from the azole target, which is 14α-sterol demethylase (9, 35). Mutations that confer differential fluconazole versus itraconazole resistance have, however, been characterized in other fungi and, as expected, localize to the target enzyme (33, 36). This rationale led us to screen S. cerevisiae mutants selected on CSP- or MCF-containing plates for differential RES. Both of the mutants identified (in an fks1Δ strain) exhibited an Fks2 mutation, confirming Fks as an echinocandin target. However, both involved residues not previously implicated in RES, specifically, S1380P, 4 residues downstream of the previously characterized hot spot 2, and W714L, about 50 residues downstream of hot spot 1. The latter mutation defines the new hot spot 3, the focus of this study.
Site-directed mutagenesis of Fks1 confirmed a role in RES for residue W695, the equivalent of Fks2 W714. This mutation did not exhibit the same MCF/ANF RES and CSP susceptibility but, rather, a distinct pattern of differential RES. This suggests that the hot spot 3 environment differs somewhat for Fks1 (in the WT background) and Fks2 (in a Δfks1 background) in terms of its interaction with other Fks regions, other proteins, or perhaps, the lipid membrane. Further site-directed mutagenesis implicated several adjacent residues in RES or, intriguingly, echinocandin hypersusceptibility. The latter result suggests that echinocandin-Fks interaction even in susceptible organisms, such as S. cerevisiae, is not fully optimized; i.e., echinocandin modifications that further enhance activity are feasible.
Despite the expanded clinical use of echinocandins, acquired RES remains rare. A more pressing concern surrounding echinocandin use is the intrinsic RES of many emerging fungal pathogens. Our central hypothesis is that specific substitutions in otherwise conserved Fks1 hot spot residues contribute to intrinsic RES. In support of this, evidence has been presented that hot spot 1 substitutions P647A and F639Y contribute to the low- and high-level RES of C. parapsilosis and F. solani, respectively (10, 14). Here, we present evidence for a similar role for the W695F substitutions within hot spot 3 of both S. prolificans and S. apiospermum, as this mutation in S. cerevisiae decreased MCF and ANF susceptibilities by 8- and 128-fold, respectively. On the other hand, this single mutation does not fully account for the RES profile of Scedosporium species; substitutions elsewhere within Fks1 (e.g., F639Y) (14) and other factors, such as relatively low reliance on β-1,3-glucan versus α-1,3-glucan, most likely play a role. Our S. cerevisiae data also predict that the W696C substitution in B. dermatitidis would confer CSP-specific RES to the mold phase of this dimorphic fungus. The MCF susceptibility of this phase (23), in contrast to the intrinsic RES of the yeast phase, further supports a role for other factors in echinocandin susceptibility.
Mutations conferring differential RES are potentially powerful reagents for modeling Fks-echinocandin interaction. However, since Fks is an integral membrane protein and since echinocandins have lipid side chains that are likely to be membrane embedded (analogous to the antibacterial agent daptomycin) (1), these models will need to incorporate Fks topology. For example, the hot spot 3-defining residue W695 is robustly predicted by the widely used topology algorithm TMHMM (18) to fall within a transmembrane helix (data not shown), where it could interact with the echinocandin lipid chains. If this model is correct, the differential RES conferred by W695 replacement with L, F, or C would reflect their differential binding to the alkyl (CSP) versus aryl (MCF and ANF) components of these lipid chains. Topological analysis of Fks to more rigorously model Fks-echinocandin-membrane interaction is in progress.
Following completion of this work, Martins et al. (21), in their studies of the Fks1 homolog Bgs4 from fission yeast Schizosaccharomyces pombe, described a mutation in residue W760, which is equivalent to the S. cerevisiae hot spot 3-defining residues Fks1 W695 and Fks2 W714. The mutation, W760S, conferred resistance to CSP but not aculeacin A, an echinocandin structurally most similar to ANF; this is analogous to the differential RES effects of the Fks1 W695C mutation (Table 2). These data provide further, independent support for the role of hot spot 3 in echinocandin activity. Since this S. pombe mutant was selected on papulacandin B, which shares the lipid but not the peptide moieties of the echinocandins, it also supports the model above in which hot spot 3 is membrane embedded.
ACKNOWLEDGMENTS
We thank A. Fothergill for generously providing strains.
This work was supported by National Institute of Allergy and Infectious Disease grant AI075272 and by Pfizer Investigator Initiated Research grant WS428829.
Footnotes
Published ahead of print on 16 May 2011.
REFERENCES
- 1. Baltz R. H. 2009. Daptomycin: mechanisms of action and resistance, and biosynthetic engineering. Curr. Opin. Chem. Biol. 13:144–151 [DOI] [PubMed] [Google Scholar]
- 2. Castanheira M., et al. 2010. Low prevalence of fks1 hot spot 1 mutations in a worldwide collection of Candida strains. Antimicrob. Agents Chemother. 54:2655–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cornely O. 2008. Aspergillus to zygomycetes: causes, risk factors, prevention, and treatment of invasive fungal infections. Infection 36:296–313 [DOI] [PubMed] [Google Scholar]
- 4. Cortez K. J., et al. 2008. Infections caused by Scedosporium spp. Clin. Microbiol. Rev. 21:157–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cuenca-Estrella M., et al. 2009. Activity profile in vitro of micafungin against Spanish clinical isolates of common and emerging species of yeasts and molds. Antimicrob. Agents Chemother. 53:2192–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Denning D. W. 2003. Echinocandin antifungal drugs. Lancet 362:1142–1151 [DOI] [PubMed] [Google Scholar]
- 7. Douglas C. M. 2001. Fungal β-(1,3)-d-glucan synthesis. Med. Mycol. 39(Suppl. 1):55–66 [DOI] [PubMed] [Google Scholar]
- 8. Espinel-Ingroff A. 2003. In vitro antifungal activities of anidulafungin and micafungin, licensed agents and the investigational triazole posaconazole as determined by NCCLS methods for 12,052 fungal isolates: review of the literature. Rev. Iberoam. Micol. 20:121–136 [PubMed] [Google Scholar]
- 9. Ferrari S., et al. 2009. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog. 5:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garcia-Effron G., Katiyar S. K., Park S., Edlind T. D., Perlin D. S. 2008. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 52:2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garcia-Effron G., Lee S., Park S., Cleary J. D., Perlin D. S. 2009. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-β-d-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob. Agents Chemother. 53:3690–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Groll A. H., Walsh T. J. 2001. Uncommon opportunistic fungi: new nosocomial threats. Clin. Microbiol. Infect. 7:8–24 [DOI] [PubMed] [Google Scholar]
- 13. Jain P., Akula I., Edlind T. 2003. Cyclic AMP signaling pathway modulates susceptibility of Candida species and Saccharomyces cerevisiae to antifungal azoles and other sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 47:3195–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Katiyar S. K., Edlind T. D. 2009. Role for Fks1 in the intrinsic echinocandin resistance of Fusarium solani as evidenced by hybrid expression in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 53:1772–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kauffman C. A. 2006. Fungal infections. Proc. Am. Thorac. Soc. 3:35–40 [DOI] [PubMed] [Google Scholar]
- 16. Kauffman C. A., Carver P. L. 2008. Update on echinocandin antifungals. Semin. Respir. Crit. Care Med. 29:211–219 [DOI] [PubMed] [Google Scholar]
- 17. Kontoyiannis D. P., Lewis R. E. 2002. Antifungal drug resistance of pathogenic fungi. Lancet 359:1135–1144 [DOI] [PubMed] [Google Scholar]
- 18. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 19. Kurtz M. B., Rex J. H. 2001. Glucan synthase inhibitors as antifungal agents. Adv. Protein Chem. 56:423–475 [DOI] [PubMed] [Google Scholar]
- 20. Malani A. N., Kauffman C. A. 2007. Changing epidemiology of rare mould infections: implications for therapy. Drugs 67:1803–1812 [DOI] [PubMed] [Google Scholar]
- 21. Martins I. M., et al. 2011. Differential activities of three families of specific β(1,3)glucan synthase inhibitors in wild-type and resistant strains of fission yeast. J. Biol. Chem. 286:3484–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mazur P., et al. 1995. Differential expression and function of two homologous subunits of yeast 1,3-β-d-glucan synthase. Mol. Cell. Biol. 15:5671–5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakai T., et al. 2003. In vitro antifungal activity of micafungin (FK463) against dimorphic fungi: comparison of yeast-like and mycelial forms. Antimicrob. Agents Chemother. 47:1376–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nucci M., Anaissie E. 2007. Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 20:695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pappas P. G., et al. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 48:503–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parent S. A., et al. 1993. Calcineurin-dependent growth of an FK506- and CsA-hypersensitive mutant of Saccharomyces cerevisiae. J. Gen. Microbiol. 139:2973–2984 [DOI] [PubMed] [Google Scholar]
- 27. Perlin D. S. 2007. Resistance to echinocandin-class antifungal drugs. Drug Res. Updat. 10:121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pfaller M. A., Diekema D. J. 2010. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36:1–53 [DOI] [PubMed] [Google Scholar]
- 29. Pfaller M. A., Diekema D. J. 2004. Rare and emerging opportunistic fungal pathogens: concern for resistance beyond Candida albicans and Aspergillus fumigatus. J. Clin. Microbiol. 42:4419–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Radding J. A., Heidler S. A., Turner W. W. 1998. Photoaffinity analog of the semisynthetic echinocandin LY303366: identification of echinocandin targets in Candida albicans. Antimicrob. Agents Chemother. 42:1187–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Richardson M., Lass-Flörl C. 2008. Changing epidemiology of systemic fungal infections. Clin. Microbiol. Infect. 14:5–24 [DOI] [PubMed] [Google Scholar]
- 32. Roemer T., et al. 2003. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 50:167–181 [DOI] [PubMed] [Google Scholar]
- 33. Sanglard D., Ischer F., Koymans L., Bille J. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanglard D., Odds F. C. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73–85 [DOI] [PubMed] [Google Scholar]
- 35. Vermitsky J.-P., Edlind T. D. 2004. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-Like transcription factor. Antimicrob. Agents Chemother. 48:3773–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xiao L., et al. 2004. Three-dimensional models of wild-type and mutated forms of cytochrome P450 14α-sterol demethylases from Aspergillus fumigatus and Candida albicans provide insights into posaconazole binding. Antimicrob. Agents Chemother. 48:568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]