Abstract
We recently reported that HIV-1 resistant to 3′-azido-3′-deoxythymidine (AZT) is not cross-resistant to 3′-azido-2′,3′-dideoxypurines. This finding suggested that the nucleoside base is a major determinant of HIV-1 resistance to nucleoside analogs. To further explore this hypothesis, we conducted in vitro selection experiments by serial passage of HIV-1LAI in MT-2 cells in increasing concentrations of 3′-azido-2′,3′-dideoxyguanosine (3′-azido-ddG), 3′-azido-2′,3′-dideoxycytidine (3′-azido-ddC), or 3′-azido-2′,3′-dideoxyadenosine (3′-azido-ddA). 3′-Azido-ddG selected for virus that was 5.3-fold resistant to 3′-azido-ddG compared to wild-type HIV-1LAI passaged in the absence of drug. Population sequencing of the entire reverse transcriptase (RT) gene identified L74V, F77L, and L214F mutations in the polymerase domain and K476N and V518I mutations in the RNase H domain. However, when introduced into HIV-1 by site-directed mutagenesis, these 5 mutations only conferred ∼2.0-fold resistance. Single-genome sequencing analyses of the selected virus revealed a complex population of mutants that all contained L74V and L214F linked to other mutations, including ones not identified during population sequencing. Recombinant HIV-1 clones containing RT derived from single sequences exhibited 3.2- to 4.0-fold 3′-azido-ddG resistance. In contrast to 3′-azido-ddG, 3′-azido-ddC selected for the V75I mutation in HIV-1 RT that conferred 5.9-fold resistance, compared to the wild-type virus. Interestingly, we were unable to select HIV-1 that was resistant to 3′-azido-ddA, even at concentrations of 3′-azido-ddA that yielded high intracellular levels of 3′-azido-ddA-5′-triphosphate. Taken together, these findings show that the nucleoside base is a major determinant of HIV-1 resistance mechanisms that can be exploited in the design of novel nucleoside RT inhibitors.
INTRODUCTION
There is no cure available yet for HIV-1 infection. As a result, HIV-infected patients require lifelong antiretroviral therapy (ART) (29). First-line ART regimens typically include two or more nucleoside reverse transcriptase (RT) inhibitors (NRTIs) combined with a nonnucleoside RT inhibitor or a boosted protease inhibitor (15, 18, 22). Unfortunately, ART is often limited by the development of HIV-1 drug resistance and by acute or chronic drug toxicity (6). Therefore, it is essential that there be continued discovery and development of new antiretrovirals that are potent and nontoxic and that require several mutations to confer HIV-1 resistance (i.e., a high genetic barrier to resistance).
Due to its essential role in virus replication, HIV-1 RT is a key drug target (31). In this regard, the NRTIs are an important therapeutic group of drugs that target the DNA polymerase activity of the enzyme. To date, 7 NRTIs have been approved by the FDA for the treatment of HIV-1 infection. These include zidovudine (3′-azido-3′-deoxythymidine [AZT]), lamivudine (3TC), emtricitabine (FTC), didanosine (ddI), stavudine (d4T), abacavir sulfate (ABC), and tenofovir (TFV) disoproxil fumarate (TDF). Once metabolized in cells to their triphosphate forms by cellular kinases (19, 27), NRTI-triphosphates (NRTI-TP) inhibit HIV-1 reverse transcription by termination of the nascent viral DNA chains (24).
Two phenotypic mechanisms of HIV-1 resistance to NRTIs have been described, specifically, NRTI-TP discrimination and NRTI-monophosphate (NRTI-MP) excision. With the discrimination phenotype, NRTI resistance mutations (e.g., K65R, K70E, L74V, Q151M [with A62V, V75I, F77L, F116Y], and M184V) allow HIV-1 RT to preferentially bind and incorporate the natural deoxynucleoside triphosphate (dNTP) substrates over the NRTI-TP (7, 26). With the excision phenotype, NRTI resistance mutations, also known as thymidine analog mutations (e.g., M41L, D67N, K70R, L210W, T215F/Y, and K219Q/E), enhance HIV-1 RT's ability to excise the chain-terminating NRTI-MP from the 3′-terminus of the DNA primer by using ATP as the pyrophosphate donor (1, 12, 13, 30).
Previously, our group reported that the nucleoside base was a major determinant for HIV-1 resistance mechanisms (17, 23, 25). Specifically, we demonstrated that AZT-resistant HIV-1 did not exhibit cross-resistance to the 3′-azido-2′,3′-dideoxypurines: 3′-azido-2′,3′-dideoxyguanosine (3′-azido-ddG) and 3′-azido-2′,3′-dideoxyadenosine (3′-azido-ddA) (17, 23). In this study, we further explored the relationship between NRTI base and resistance mutation patterns by conducting in vitro selection for viruses resistant to 3′-azido-ddG, 3′-azido-2′,3′-dideoxycytidine (3′-azido-ddC), and 3′-azido-ddA.
MATERIALS AND METHODS
Nucleosides.
3′-azido-ddA and 3′-azido-ddG were obtained from Berry Associates, Inc. (Ann Arbor, MI). AZT and ddI were obtained from Sigma Chemical Corporation (St. Louis, MO). ABC was provided by GlaxoSmithKline (Research Triangle Park, NC). TFV was obtained from the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH). 3′-azido-ddC, 3TC, and d4T were kindly provided by Raymond Schinazi (Emory University). 3′-azido-ddA-5′-triphosphate was synthesized as previously described (8). All NRTIs were prepared as 40 mM stock solutions in dimethyl sulfoxide or sterile water and stored at −20°C.
Cells and viruses.
MT-2 cells (AIDS Research and Reference Reagent Program) were cultured in RPMI 1640 with 2 mM l-glutamine (Lonza, Walkersville, MD) supplemented with 10% fetal bovine serum (HyClone Laboratories, Inc., Logan, UT), 10 mM HEPES buffer (Gibco, Grand Island, NY), and 50 IU/ml of penicillin and 50 μg/ml of streptomycin (Gibco). The P4/R5 reporter cell line (provided by Nathaniel Landau, Salk Institute, La Jolla, CA), which expresses the β-galactosidase gene under the control of the HIV-1 long terminal repeat promoter that is transactivated by HIV-1 tat, was maintained in phenol red-free Dulbecco's modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum, 50 IU/ml of penicillin, 50 μg/ml of streptomycin, and 0.5 μg/ml of puromycin (Clontech, Palo Alto, CA). Stock viruses were prepared in MT-2 cells as described previously (17). Briefly, 5 μg of plasmid DNA was electroporated into 1.3 × 107 MT-2 cells. Cell-free supernatants were collected 5 to 7 days posttransfection at peak cytopathic effect (CPE) and stored at −80°C. The infectivities of the virus stocks were determined by 3-fold end point dilution in P4/R5 cells, and the 50% tissue culture infectivity dose was calculated using the Reed and Muench equation (20).
Selection of drug-resistant HIV-1.
Resistant virus was selected by serial passage of wild-type (WT) xxHIVLAI in MT-2 cells in increasing concentrations of 3′-azido-ddG, 3′-azido-ddC, or 3′-azido-ddA. To initiate each selection experiment, MT-2 cells (1 × 106) were pretreated for 2 h with twice the concentration of drug required to inhibit viral replication by 50% (EC50) of the nucleoside analog before inoculation with virus. Viral replication was monitored by CPE. At three or four syncytia per field at 100× magnification, the cell-free supernatant was harvested and 0.1 ml of supernatant was added to fresh MT-2 cells to initiate a new passage. Remaining supernatant was stored frozen at −80°C. The concentration of drug was doubled every 5 to 15 passages, as viral growth permitted. Mean EC50 values (n = 3) were determined at every five passages to identify changes in drug susceptibility. Fold resistance was calculated by dividing the mean EC50 of the passaged virus by the mean EC50 of wild-type HIV-1LAI. The population genotype of the virus was determined every 10 passages by standard automated sequencing.
Drug susceptibility assays.
NRTI susceptibility was determined in P4/R5 cells as described previously (17). Briefly, 3-fold dilutions of inhibitor were added to P4/R5 cells in triplicate, and the cells were infected with an amount of virus that produced 100 relative light units (RLU) in no-drug virus control wells. After 48 h, the cells were lysed (Gal-Screen; Tropix/Applied Biosystems, Foster City, CA) and the RLU were measured using a ThermoLab Systems luminometer (Waltham, MA). The EC50 and fold resistance were calculated as described above. 3′-azido-ddC is not well phosphorylated in P4/R5 cells, and the 3′-azido-ddC-selected virus did not grow well in P4/R5 cells. Susceptibility to 3′-azido-ddC was therefore determined in MT-2 cells, and viral replication was quantified by measuring p24 antigen production (Perkin-Elmer, Inc., Waltham, MA).
HIV-1 population sequencing.
To confirm the genotypes of virus populations, viral RNA was extracted from culture supernatants by using oligo(dT)25 Dynabeads (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions and treated with 1 IU/μl of DNase I for 2 h. The RNA was converted into cDNA and amplified using the SuperScript III one-step RT-PCR system with Platinum Taq DNA polymerase (Invitrogen). The entire coding region of RT was amplified using the forward primer 5′-GCTCTATTAGATACAGGAGCAGATGAT-3′ and the reverse primer 5′-CCTTCTAAATGTGTACAATCTAGTTGCCAT-3′. PCR products were purified (Wizard SV Gel and PCR Clean-Up system; Promega, Madison, WI) and sequenced using a Big Dye terminator kit (v.3.1) on an ABI 3130 automated DNA sequencer (Applied Biosystems, Foster City, CA). The following forward primers were used for sequencing RT: 5′-GGACCTACACCTGTCAAC-3′, 5′-GTTCCCTTAGATGAAGAC-3′, 5′-GAGGAACCAAAGCACTAA-3′, and 5′-CACCCTAACTGACACACC-3′.
Construction of mutant recombinant HIV-1.
Mutant RT clones were generated by site-directed mutagenesis (QuikChange Lightning site-directed mutagenesis kit; Stratagene, La Jolla, CA) using the p6HRT-MO plasmid. This plasmid contains the entire RT and protease coding sequence and four silent restriction sites (XmaI, MluI, XbaI, and NgoMIV from the 5′ to 3′ end of RT at codons 14, 358, 490, and 554, respectively) (3). After site-directed mutagenesis, the mutated RT was ligated into pxxHIV-1LAI-MO, which contains the entire genome of HIV-1LAI and the same silent restriction sites in RT as p6HRT-MO. Infectious virus was generated by electroporation of the mutated xxHIV-1LAI-MO plasmid into MT-2 cells as described above. All mutations in recombinant viruses were confirmed by full-length sequencing of the entire RT coding region. Plasmids were kindly provided by Jessica Brehm (University of Pittsburgh).
SGS and generation of recombinant infectious viruses.
Full-length RT sequences from single viral genomes in passaged virus were obtained by isolating viral RNA (as described above) and synthesizing cDNA with SuperScript III reverse transcriptase (Invitrogen) and primer 4232 (5′-TTCCCTACAATCCCCAAAGTCAAGG-3′). The cDNA was then serially diluted to yield 30% positive PCRs as described elsewhere for single-genome sequencing (SGS) (16). First-round PCR was performed using AmpliTaq Gold (Applied Biosystems), primer 4232, and primer Bcl (5′-AGGAAGAATGGAAACCAAAAATGATAG-3′). Second-round PCR was performed using primers Bcl and 3908 (5′-CAAAAGAAATAGTAGCCAGCTGTG-3′). Positive reactions were determined by gel electrophoresis, purified by treatment with ExoSAP-IT (USB, Cleveland, OH), and sequenced as described above (16). To generate infectious recombinant clones derived from single viral genomes, full-length RT was amplified from purified first-round PCR product using the following primers containing restriction sites: 1989Bcl, 5′-GTTTTATCAAAGTAAGACAGTATGATCAGATAC-3′; Sgr, 5′-TAACCTGCCACCGGTGGTAG-3′. The purified products were concatemerized, digested with BclI and SgrAI, and cloned into pxxLAI-3D {pxxLAI-MO with 4 additional silent restriction sites: BclI (nucleotide [nt] 2011), BstBI (nt 3096), HpaI (nt 3383), and SgrAI (nt 3897)}. The recombinant plasmid was used to produce infectious virus as described above. Primers and plasmid were kindly provided by Jessica Brehm (University of Pittsburgh).
Cellular metabolism of 3′-azido-ddA.
MT-2 cells (5 × 106) were incubated for 4 h with 3′-azido-ddA at 5, 12.5, 25, and 50 μM 3′-azido-ddA. The cells were then centrifuged for 10 min at 350 × g at 4°C, and the pellet was resuspended and washed three times with cold phosphate-buffered saline. Nucleoside triphosphates were extracted by incubation overnight at −20°C with 1 ml 60% methanol–water. The supernatants were then collected and centrifuged at 16,000 × g for 5 min. The pellets were then reextracted for 1 h on ice by using an additional 200 μl of 60% methanol in water, followed by centrifugation at 16,000 × g for 5 min. The extracts were combined, dried under a gentle filtered airflow, and stored at −20°C. The residues were resuspended in 100 μl of water prior to liquid chromatography (LC)-tandem mass spectrometry analysis.
The LC system was an UltiMate 3000 modular system (Dionex, Sunnyvale, CA) consisting of a quaternary pump, vacuum degasser, thermostated autosampler (4°C), and thermostated column compartment (28°C). An API5000 mass spectrometer (AB/SCIEX, Foster City, CA) was used as a detector. LC Analyst software version 1.4.2 was used to control both the LC and the mass spectrometer and for the data analysis and quantification. Phosphorylated 3′-azido-ddA was quantified by ion exchange, with the separation performed on a 5-μm-particle-size Biobasic C18 (50- by 1.0-mm) column (Thermo Electron, Bellefonte, PA) using a gradient. The mobile phase consisted of A (10 mM ammonium acetate), B (ammonia buffer [pH 9.6]), and C, acetonitrile. Sample volumes of 5 μl were injected onto the column.
The flow was diverted to waste for the initial 1 min of the analysis. Initial composition of the mobile phase was 70% A and 30% C with a gradient at 3.5 min to 70% B and 30% C for 5 min. The mass spectrometer was operated in positive ionization mode with a spray voltage of 5 kV, gas 1 at 15 (arbitrary units), gas 2 at 20 (arbitrary units), and source temperature of 230°C. A standard curve was prepared by spiking 8 standards of synthesized 3′-azido-ddA-5′-TP in the range of 1 nM to 500 nM in cell lysate. The precursor-product ion transitions, declustering potential (DP, in V), collision energies (CE, in V), and exit potential (CXP, in V) for 3′-azido-ddA-DP and -TP were m/z 437.2 → m/z 136; DP 121, CE 29, CXP 14; m/z 517.2 → m/z 136; DP 121, CE 31, CXP 14, respectively.
RESULTS
Selection of HIV-1 resistance to 3′-azido-ddG.
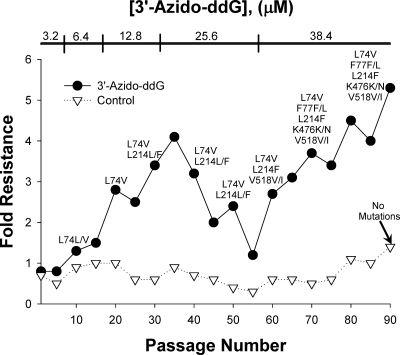
Wild-type HIV-1LAI was serially passaged in MT-2 cells in increasing concentrations of 3′-azido-ddG. After every fifth passage, 3′-azido-ddG susceptibility of the passaged virus was determined in a single-cycle replication inhibition assay in P4/R5 cells (Fig. 1). Full-length RT genotype analysis of the virus population (i.e., population genotype) was determined every 10th passage (Fig. 1). L74V was the first mutation to emerge at passage 10, followed by L214F at passage 30, V518I at passage 60, and K476N and F77L at passage 90 (Fig. 1). The final virus population contained all 5 mutations and was 5.3-fold more resistant than a control xxHIV-1LAI that was passaged in parallel in the absence of drug. No RT mutations were detected in the control virus. Next, we tested the susceptibility of 3′-azido-ddG-resistant virus from passage 88 to a panel of NRTIs in P4/R5 cells (Table 1). The 3′-azido-ddG-resistant HIV-1 showed cross-resistance to ddI (6.2-fold), 3TC (4.8-fold), and ABC (4.2-fold). Low-level cross-resistance was also noted for TFV (2.5-fold), D4T (1.5-fold), and 3′-azido-ddA (1.5-fold). 3′-azido-ddG-resistant HIV-1 remained sensitive to AZT.
Fig. 1.
3′-azido-ddG-resistant xxHIV-1LAI was selected by serial passage of virus in increasing concentrations of 3′-azido-ddG. Passaged virus was phenotyped for 3′-azido-ddG susceptibility (EC50, 10.1 ± 2.5 μM at passage 88) and compared to the control wild-type xxHIV-1LAI passaged in parallel without 3′-azido-ddG (EC50, 1.91 ± 1.16 μM at passage 88) to determine resistance at every fifth passage. Standard population sequencing was performed at every 10th passage and identified the indicated RT mutations that differed from the starting virus population.
Table 1.
Resistance and cross-resistance of wild-type (WT) and 3′-azido-ddG-selected virus
| Drug | EC50 (μM)a |
Fold resistanced | P valuee | |
|---|---|---|---|---|
| WTb | Selected virusc | |||
| ddI | 4.3 ± 0.2 | 26.6 ± 1.2 | 6.2 | <0.0001 |
| 3′-azido-ddG | 1.9 ± 1.2 | 10.1 ± 2.5 | 5.3 | <0.001 |
| 3TC | 2.2 ± 1.8 | 10.5 ± 4.1 | 4.8 | <0.05 |
| ABC | 8.9 ± 0.5 | 37.2 ± 6.3 | 4.2 | <0.05 |
| TFV | 2.1 ± 0.7 | 5.4 ± 0.7 | 2.5 | <0.05 |
| D4T | 14.1 ± 3.0 | 23.2 ± 1.1 | 1.5 | <0.01 |
| 3′-azido-ddA | 3.7 ± 0.6 | 5.6 ± 0.2 | 1.5 | <0.05 |
| AZT | 0.3 ± 0.2 | 0.4 ± 0.3 | 1.3 | NS |
Means ± standard deviations of three experiments.
WT virus passaged in parallel with the 3′-azido-ddG selection virus in the absence of drug; genotype was confirmed by sequencing.
Virus isolated from passage 88 of the 3′-azido-ddG selection that contained L74V, F77L, L214F, K476N, and V518I by standard population sequencing.
Fold resistance compared with wild-type xxHIV-1LAI.
Statistical significance compared to wild-type xxHIV-1LAI, determined by a two-sample Student's t test. NS, not significant.
3′-azido-ddG susceptibilities of viruses containing different combinations of L74V, F77L, L214F, K476N, and V518I.
Recombinant viruses were generated by site-directed mutagenesis to contain different combinations of the L74V, F77L, L214F, K476N, and V518I mutations. Surprisingly, many of the recombinant viruses (e.g., K476N, L74V/L214F/K476N/V518I, and L74V/F77L/L214F/K476N/V518I) were growth defective, precluding further assessments of NRTI susceptibility (Table 2). Drug susceptibility assays in P4/R5 cells revealed that the F77L, L214F, and V518I mutations alone did not confer 3′-azido-ddG resistance (Table 2). Interestingly, the remaining recombinant viruses only displayed low levels (1.5- to 2.1-fold over WT) of 3′-azido-ddG resistance. This finding suggested that population sequencing did not provide an adequate representation of the mutant variants within the selected virus population. Specifically, the results suggested that mutations identified by population sequencing showed variable linkage on viral genomes and that other important mutations may not have been detected.
Table 2.
3′-Azido-ddG and 3′-azido-ddC susceptibilities of HIV-1LAI mutants
| Susceptibility category and virusa | EC50 (μM)b | Fold resistancec | P valued |
|---|---|---|---|
| 3′-azido-ddG susceptibility | |||
| Wild type | 1.5 ± 0.4 | ||
| L74V | 2.2 ± 0.5 | 1.5 | NS |
| L74V/L214F | 2.8 ± 0.4 | 1.9 | <0.05 |
| L74V/L214F/K476N | 3.1 ± 0.3 | 2.1 | <0.005 |
| L74V/L214F/V518I | 2.2 ± 0.9 | 1.5 | NS |
| L74V/F77L/L214F/K476N | 2.8 ± 0.2 | 1.9 | <0.05 |
| K476Ne | |||
| L74V/L214F/K476N/V518Ie | |||
| L74V/F77L/L214F/K476N/V518Ie | |||
| 3′-azido-ddC susceptibility | |||
| Wild type | 0.6 ± 0.2 | ||
| V75I | 1.6 ± 0.8 | 2.6 | <0.05 |
Mutants were created in pxxLAI-MO by site-specific mutagenesis, and infectious virus was produced by transfection of MT-2 cells.
Means ± standard deviations of three experiments.
Fold resistance was determined versus the wild-type xxHIV-1LAI-MO clone.
Statistical significance compared to wild-type xxHIV-1LAI-MO, determined by two-sample Student's t test. NS, not significant.
The mutant virus was growth defective.
Single-genome sequencing and cloning of RT from the 3′-azido-ddG-resistant HIV-1.
We performed single-genome sequencing analyses of full-length RT from virus at passage 90 to genetically characterize individual mutant variants within the selected virus population. This approach permitted more detailed analysis of selected viruses, including mutation linkage. Table 3 shows the mutational patterns observed in 22 single genome sequences. Interestingly, all of the sequences contained the L74V and L214F mutations. The F77L (91% of sequences), K476N (68%), and V106I (50%) mutations were also frequently observed in the single genome sequences. By contrast, the frequencies of other mutations varied considerably (Table 3), and no obvious pattern of linkage emerged from this study.
Table 3.
Predicted amino acid changes from wild-type RT in single genome sequences derived from 3′-azido-ddG-selected virus (passage 90)
| RT residue | Sequence no. |
% of clones | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | ||
| L74 | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V | 100 |
| F77 | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | 91 | ||
| V106 | I | I | I | I | I | I | I | I | I | I | I | 50 | |||||||||||
| E122 | K | K | K | K | K | K | K | K | 36 | ||||||||||||||
| E169 | K | 5 | |||||||||||||||||||||
| L214 | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F | 100 |
| R277 | K | K | K | K | K | 23 | |||||||||||||||||
| R284 | K | 5 | |||||||||||||||||||||
| G333 | E | 5 | |||||||||||||||||||||
| V435 | I | 5 | |||||||||||||||||||||
| S447 | N | N | N | 14 | |||||||||||||||||||
| R461 | K | 5 | |||||||||||||||||||||
| K476 | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | 68 | |||||||
| P510 | T | T | T | 14 | |||||||||||||||||||
| K512 | R | 5 | |||||||||||||||||||||
| E514 | D | 5 | |||||||||||||||||||||
| V518 | I | I | I | I | I | I | 27 | ||||||||||||||||
| V531 | I | G | 5 I/G | ||||||||||||||||||||
| L533 | S | M | 5 S/M | ||||||||||||||||||||
To assess the phenotype of the different RT sequences, recombinant infectious viruses were created by ligating single-genome-derived full-length RT amplicons into a wild-type vector (pxxLAI-3D). Infectious virus was produced and assayed for 3′-azido-ddG susceptibility in P4/R5 cells (Table 4). Some clones (36%) were growth defective and could not be phenotyped. All of the replication-competent viruses showed similar levels of 3′-azido-ddG resistance, with an average fold resistance of 3.6 ± 0.3 (range, 3.2 to 4.0). Of note, bulk cloning of full-length RT amplified from the virus population into pxxLAI-3D produced virus exhibiting only 2.2-fold resistance to 3′-azido-ddG.
Table 4.
3′-Azido-ddG susceptibility of recombinant HIV-1 with RT derived from single-genome amplifications
| Clonea | Genotypeb | EC50 (μM)c | Fold resistanced | P valuee |
|---|---|---|---|---|
| 1 | L74V/F77L/V106I/E122K/L214F/R277K/S447N/V518I/V531I | 4.5 ± 1.2 | 3.2 | <0.05 |
| 2 | L74V/F77L/V106I/E122K/L214F/K476N | 4.8 ± 1.0 | 3.4 | <0.01 |
| 3 | L74V/F77L/V106I/L214F/R277K/K476N | 5.3 ± 1.8 | 3.8 | <0.05 |
| 6 | L74V/F77L/V106I/E122K/L214F/S447N/P510T/L533M | 5.0 ± 1.6 | 3.6 | <0.05 |
| 7 | L74V/F77L/E122K/L214F/K476N | 4.7 ± 0.7 | 3.4 | <0.01 |
| 10f | L74V/F77L/L214F/K476N | 5.6 ± 1.5 | 4.0 | <0.01 |
| 13 | L74V/F77L/L214F/R277K/V435I/P510T/V518I | 5.4 ± 0.9 | 3.8 | <0.01 |
| Bulk | Mixture | 2.31 ± 0.01 | 2.2 | <0.05 |
Recombinant clones were produced by amplifying full-length RT from single genomes or as a population (bulk) and cloning into the xxHIVLAI-3D vector for infectious virus production by electroporation into MT-2 cells.
Genotypes identified by single-genome sequencing (Table 3) that are not listed were cloned but determined to be growth defective.
Means ± standard deviations of three experiments.
Fold resistance was determined compared to the xxHIV-1LAI clone (EC50, 1.40 ± 0.05 μM).
Statistical significance compared to wild-type xxHIV-1LAI-MO, determined by using a two-sample Student's t test.
Clone 10 contains a silent G-to-A transition at nucleotide 423 that encodes G141 in the HIV-1xxLAI background.
Selection of HIV-1 resistance to 3′-azido-ddC.
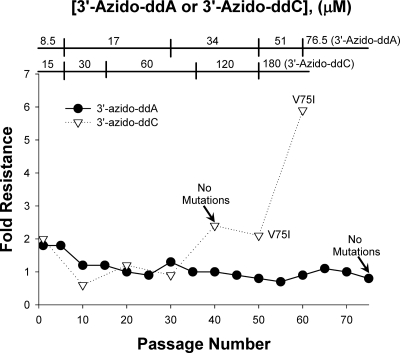
Following 55 passages of HIV-1LAI in increasing concentrations of 3′-azido-ddC, we selected virus that was 5.9-fold resistant to 3′-azido-ddC compared to wild-type virus (Fig. 2). Population sequencing of the entire HIV-1 RT identified the V75I mutation in the DNA polymerase domain of the enzyme. When introduced into the backbone of the wild-type HIV-1LAI virus by site-directed mutagenesis, the V75I mutation conferred 2.6-fold resistance to 3′-azido-ddC in MT-2 cells (Table 2).
Fig. 2.
Drug-resistant xxHIV-1LAI was selected by serial passage of virus in increasing concentrations of 3′-azido-ddC (EC50, 86.7 ± 16.9 μM at passage 60) or 3′-azido-ddA (EC50, 3.32 ± 1.06 μM at passage 75). Fold resistance and RT genotype were determined as described for Fig. 1.
Selection of HIV-1 resistance to 3′-azido-ddA.
After 75 passages of HIV-1LAI in increasing concentrations of 3′-azido-ddA (8.5 μM to 76.5 μM), we were unable to select for HIV-1 resistance to 3′-azido-ddA (Fig. 2). This inability to select for drug-resistant HIV-1 was not due to inefficient metabolism of 3′-azido-ddA to 3′-azido-ddATP. In Table 5, we show from the quantitative mass spectrometry analyses that 3′-azido-ddA was metabolized to the active TP form in MT-2 cells in a dose-dependent manner.
Table 5.
Levels of 5′-phosphorylated 3′-azido-ddA in MT-2 cells after 4 h of incubation with 3′-azido-ddA at 5, 12.5, 25, or 50 μM
| 3′-azido-ddA concn (μM) | Level of metabolized 3′-azido-ddA (pmol/106 cells)a |
Level of metabolized 3′-azido-ddA (μM)d |
||
|---|---|---|---|---|
| TPb | DPc | TP | DP | |
| 5 | 0.13 ± 0.02 | 0.030 ± 0.002 | 0.240 ± 0.004 | 0.054 ± 0.003 |
| 12.5 | 0.22 ± 0.01 | 0.08 ± 0.01 | 0.40 ± 0.02 | 0.15 ± 0.02 |
| 25 | 0.36 ± 0.05 | 0.17 ± 0.04 | 0.66 ± 0.09 | 0.30 ± 0.07 |
| 50 | 0.77 ± 0.07 | 0.25 ± 0.07 | 1.41 ± 0.14 | 0.45 ± 0.13 |
Means ± standard deviations of three experiments.
TP is 3′-azido-ddA-5′-triphosphate.
DP is 3′-azido-ddA-5′-diphosphate.
Means ± standard deviations of three experiments, determined using an average diameter for MT-2 cells of 12.3 μm.
DISCUSSION
This study clearly shows that the base component of 3′-azido nucleosides strongly influences the pattern of resistance mutations in HIV-1 RT that are selected in vitro. Previously, we reported that AZT selected for the classical thymidine analog mutations D67N, K70R, and T215F in the DNA polymerase domain of the enzyme as well as A371V and Q509L in the connection and RNase H domains, respectively (3). The experimental approach used previously to select AZT-resistant HIV-1 was essentially the same as that described here. In contrast to AZT, 3′-azido-ddG selected for different combinations of L74V, F77L, and L214F in the DNA polymerase and K476N and V518I in the RNase H domains of HIV-1 RT, respectively, and 3′-azido-ddC selected only the V75I mutation in the DNA polymerase domain. Of note, we have been unable to select for 3′-azido-ddA resistance in MT-2 cells (Fig. 2) or in primary human lymphocytes (data not shown).
The level of resistance selected in vitro to 3′-azido-ddG is lower than that observed for AZT. AZT-resistant HIV-1, selected after 65 passages in MT-2 cells, contained up to 5 mutations and exhibited >16,200-fold resistance to AZT (3). By contrast, 3′-azido-ddG-resistant HIV-1, selected after 90 passages in MT-2 cells, also contained up to 5 mutations, but it exhibited only 5.3-fold resistance to 3′-azido-ddG. Interestingly, the virus population selected by 3′-azido-ddG was exceptionally diverse (Table 3), and the resistant phenotype could not be recapitulated through the construction of site-directed mutants that were based on the population genotype (Table 2). This finding suggests that population genotype analyses may not be definitive when complex mixtures of viruses are present because mutation linkage cannot be assessed. Indeed, our phenotypic analyses of single genome sequences provided a more accurate characterization of the resistant variants that emerged under 3′-azido-ddG selective pressure (Table 4). Importantly, these analyses also demonstrated that no single mutation or set of mutations is able to confer sufficient 3′-azido-ddG resistance such that it can become the dominant species in the population.
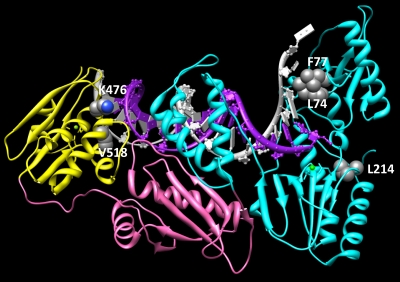
To date, the phenotypic mechanisms responsible for 3′-azido-ddG and 3′-azido-ddC resistance have not been elucidated, although ongoing biochemical studies are addressing this issue. The selection of the L74V and F77L mutations (part of the Q151M complex) by 3′-azido-ddG likely suggests a discrimination mechanism. However, the mechanisms of resistance for K476N and V518I are uncertain. Both residues reside in the RNase H domain and may interact with the T/P positioning or affect RNase H cleavage activity. Figure 3 shows the locations of these mutations in the crystal structure of HIV-1 RT. Of note, other resistance mutations in the connection (e.g., N348I and A360V) and RNase H (e.g., Q509L) domains of HIV-1 RT influence drug susceptibility via an indirect RNase H-mediated effect on the NRTI-MP excision phenotype (2, 4, 5, 21, 32). However, we have also shown that a mutation in the connection domain of HIV-1 RT (G333E) directly impacts the enzyme's ability to incorporate 3TC-TP (33).
Fig. 3.
Locations of 3′-azido-ddG resistance mutations in the p66 subunit of HIV-1 RT (pdb 1n6q). The DNA polymerase, connection, and RNase H domains are colored cyan, pink, and yellow, respectively. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco.
The V75I mutation identified in the 3′-azido-ddC-resistant HIV-1 has also been observed in selection experiments with acyclovir (ACV) and a monophosphorylated prodrug of ACV (9, 28). Biochemical studies showed that mutant V75I RT did not alter binding efficiency of ACV-triphosphate but increased incorporation of dGTP versus ACV-5′-triphosphate through a discrimination mechanism (28). This suggests that a discrimination mechanism may also be responsible for the observed HIV-1 resistance to 3′-azido-ddC. Studies of cross-resistance of the V75I mutant in primary activated CD4+ lymphoblasts have shown that V75I increases the EC50 for FTC, 3TC, ddI, and ABC, does not change the EC50 for TDF or d4T, and causes hypersusceptibility to AZT (10).
We were unable to select 3′-azido-ddA-resistant HIV-1 in vitro. Virus from passage 75 in media containing 3′-azido-ddA concentrations up to 75.6 μM did not show mutations in RT and did not exhibit any change in 3′-azido-ddA susceptibility. It is not clear why resistance did not emerge and how passaged virus was able to replicate despite high 3′-azido-ddA concentrations. We showed that 3′-azido-ddA was efficiently metabolized to the active 5′-triphosphate form in MT-2 cells that were used for selection (Table 5). Nevertheless, there may be subsets of cells that do not efficiently metabolize 3′-azido-ddA to the active 5′-triphosphate and allow virus to replicate without developing resistance. Such a mechanism has been shown in vitro through isolation of cell clones that are not protected from HIV-1 infection by AZT (11). The cell cycle can transiently alter the amount of intracellular NRTI-TP or the levels of competing natural dNTPs. Cellular resistance, or permanent alterations in NRTI metabolism or uptake, is more likely to develop during prolonged exposure of cells to an NRTI (14). We reduced this latter possibility by using new cells at the start of each passage, although the 5- to 7-day duration of each passage could have allowed outgrowth of cells with altered NRTI metabolism. An alternative explanation for HIV-1 replication in the presence of 3′-azido-ddA is the emergence mutations outside of RT that could increase viral infectivity such that a subset of cells with subinhibitory 3′-azido-ddATP concentrations could be infected efficiently.
In summary, NRTIs with different base structures, but the same 3′-azido-2′,3′-dideoxyribose sugar, selected for divergent resistance mutations in RT in vitro. These findings indicate that the base component of NRTIs can be modified to alter the mechanisms and genetic barrier to HIV-1 drug resistance. These insights should prove useful in the synthesis of novel NRTIs that pose a high genetic barrier to HIV-1 resistance.
ACKNOWLEDGMENTS
This work was supported by award numbers 3R01AI071846-04S1 and T32 AI065380 from the National Institute of Allergy and Infectious Diseases, 2P30-AI-050409 from the National Institutes of Health for the Emory Center for AIDS Research, and the Department of Veterans Affairs. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR001081).
R.F.S. is a founder and major shareholder of RFS Pharma, LLC. J.W.M. is a consultant for Merck, Gilead Sciences, and RFS Pharma, LLC, and holds share options in RFS Pharma, LLC. All other authors have no competing interests.
Footnotes
Published ahead of print on 6 June 2011.
REFERENCES
- 1. Boyer P. L., Sarafianos S. G., Arnold E., Hughes S. H. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brehm J. H., Mellors J. W., Sluis-Cremer N. 2008. Mechanism by which a glutamine to leucine substitution at residue 509 in the RNase H domain of HIV-1 reverse transcriptase confers zidovudine resistance. Biochemistry 47:14020–14027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brehm J. H., et al. 2007. Selection of mutations in the connection and RNase H domains of human immunodeficiency virus type 1 reverse transcriptase that increase resistance to 3′-azido-3′-dideoxythymidine. J. Virol. 81:7852–7859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ehteshami M., et al. 2008. Connection domain mutations N348I and A360V in HIV-1 reverse transcriptase enhance resistance to 3′-azido-3′-deoxythymidine through both RNase H-dependent and -independent mechanisms. J. Biol. Chem. 283:22222–22232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hachiya A., et al. 2008. Amino acid mutation N348I in the connection subdomain of human immunodeficiency virus type I reverse transcriptase confers multiclass resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J. Virol. 82:3261–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hammer S. M., et al. 2006. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society USA panel. JAMA 296:827–843 [DOI] [PubMed] [Google Scholar]
- 7. Krebs R., Immendorfer U., Thrall S. H., Wohrl B. M., Goody R. S. 1997. Single-step kinetics of HIV-1 reverse transcriptase mutants responsible for virus resistance to nucleoside inhibitors zidovudine and 3TC. Biochemistry 36:10292–10300 [DOI] [PubMed] [Google Scholar]
- 8. Ludwig J., Eckstein F. 1989. Rapid and efficient synthesis of nucleoside 5′-O-(1-thiotriphosphates), 5′-triphosphates and 2′,3′-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J. Org. Chem. 54:631–635 [Google Scholar]
- 9. Matamoros T., Kim B., Menéndez-Arias L. 2008. Mechanistic insights into the role of Val75 of HIV-1 reverse transcriptase in misinsertion and mispair extension fidelity of DNA synthesis. J. Mol. Biol. 375:1234–1248 [DOI] [PubMed] [Google Scholar]
- 10. McMahon M. A., Siliciano J. D., Kohli R. M., Siliciano R. F. 2010. Sensitivity of V75I HIV-1 reverse transcriptase mutant selected in vitro by acyclovir to anti-HIV drugs. AIDS 24:319–323 [DOI] [PubMed] [Google Scholar]
- 11. Medina D. J., et al. 1995. Sanctuary growth of human immunodeficiency virus in the presence of 3′-azido-3′-deoxythymidine. J. Virol. 69:1606–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meyer P. R., Matsuura S. E., Mian A. M., So A. G., Scott W. A. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35–43 [DOI] [PubMed] [Google Scholar]
- 13. Meyer P. R., Matsuura S. E., So A. G., Scott W. A. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. U. S. A. 95:13471–13476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nyce J., Leonard S., Canupp D., Schulz S., Wong S. 1993. Epigenetic mechanisms of drug resistance: drug-induced DNA hypermethylation and drug resistance. Proc. Natl. Acad. Sci. U. S. A. 90:2960–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Painter G., Almond M., Mao S., Liotta D. 2004. Biochemical and mechanistic basis for the activity of nucleoside analogue inhibitors of HIV reverse transcriptase. Curr. Top. Med. Chem. 4:1035–1044 [DOI] [PubMed] [Google Scholar]
- 16. Palmer S., et al. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43:406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parikh U. M., Koontz D. L., Chu C. K., Schinazi R. F., Mellors J. W. 2005. In vitro activity of structurally diverse nucleoside analogs against human immunodeficiency virus type 1 with the K65R mutation in reverse transcriptase. Antimicrob. Agents Chemother. 49:1139–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parniak M. A., Sluis-Cremer N. 2000. Inhibitors of HIV- I reverse transcriptase. Adv. Pharmacol. 49:67–109 [DOI] [PubMed] [Google Scholar]
- 19. Piliero P. J. 2004. Pharmacokinetic properties of nucleoside/nucleotide reverse transcriptase inhibitors. J. Acquir. Immune Defic. Syndr. 37:S2–S12 [DOI] [PubMed] [Google Scholar]
- 20. Reed L. J., Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. (Lond.) 27:439–497 [Google Scholar]
- 21. Schuckmann M. M., et al. 2010. The N348I mutation at the connection subdomain of HIV-1 reverse transcriptase decreases binding to nevirapine. J. Biol. Chem. 285:38700–38709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sharma P. L., Nurpeisov V., Hernandez-Santiago B., Beltran T., Schinazi R. F. 2004. Nucleoside inhibitors of human immunodeficiency virus type 1 reverse transcriptase. Curr. Top. Med. Chem. 4:895–919 [DOI] [PubMed] [Google Scholar]
- 23. Sluis-Cremer N., et al. 2005. The 3′-azido group is not the primary determinant of 3′-azido-3′-deoxythymidine (AZT) responsible for the excision phenotype of AZT-resistant HIV-1. J. Biol. Chem. 280:29047–29052 [DOI] [PubMed] [Google Scholar]
- 24. Sluis-Cremer N., Arion D., Parniak M. A. 2000. Molecular mechanisms of HIV-1 resistance to nucleoside reverse transcriptase inhibitors (NRTIs). Cell. Mol. Life Sci. 57:1408–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sluis-Cremer N., et al. 2009. Anti-human immunodeficiency virus activity, cross-resistance, cytotoxicity, and intracellular pharmacology of the 3′-azido-2′,3′-dideoxypurine nucleosides. Antimicrob. Agents Chemother. 53:3715–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sluis-Cremer N., et al. 2007. Molecular mechanism by which the K70E mutation in human immunodeficiency virus type 1 reverse transcriptase confers resistance to nucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 51:48–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stein D., Moore K. 2001. Phosphorylation of nucleoside analog antiretrovirals: a review for clinicians. Pharmacotherapy 21:11–34 [DOI] [PubMed] [Google Scholar]
- 28. Tchesnokov E. P., et al. 2009. Mechanisms associated with HIV-1 resistance to acyclovir by the V75I mutation in reverse transcriptase. J. Biol. Chem. 284:21496–21504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thompson M. A., et al. 2010. Antiretroviral treatment of adult HIV infection. JAMA 304:321–333 [DOI] [PubMed] [Google Scholar]
- 30. Tu X., et al. 2010. Structural basis of HIV-1 resistance to AZT by excision. Nat. Struct. Mol. Biol. 17:1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Whitcomb J. M., Hughes S. H. 1992. Retroviral reverse transcription and integration: progress and problems. Annu. Rev. Cell Biol. 8:275–306 [DOI] [PubMed] [Google Scholar]
- 32. Yap S. H., et al. 2007. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 4:1887–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zelina S., Sheen C.-W., Radzio J., Mellors J. W., Sluis-Cremer N. 2008. Mechanisms by which the G333D mutation in the human immunodeficiency virus type 1 reverse transcriptase facilitates dual resistance to zidovudine and lamivudine. Antimicrob. Agents Chemother. 52:157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]