Abstract
Integrative and conjugative elements (ICEs), which are chromosomal mobile elements, can conjugatively transfer between bacteria. Recently, we identified a genomic island of Proteus mirabilis, a common agent of catheter-associated urinary tract infection (UTI), that possesses all the properties consistent with an ICE. This element, designated ICEPm1, is highly conserved in other causative agents of UTI, suggesting its mobility. We demonstrate that ICEPm1 can actively excise from the chromosome in a clonal population of bacteria and that this excision is integrase dependent. Although in P. mirabilis HI4320, ICEPm1 is annotated as integrated into the phenylalanine tRNA gene pheV, we show that ICEPm1 can integrate into either pheV or pheU. We determined that ICEPm1 transfers at a frequency of 1.35 × 10−5 transconjugants/donor to ICEPm1-deficient P. mirabilis using plate mating assays with clinical isolates. Insertional inactivation of a putative integrase gene on ICEPm1 decreased transfer frequencies of ICEPm1 to below the limit of detection. Mutation of the relaxase of ICEPm1 also eliminates transfer and demonstrates that this element is indeed self-transmissible and not transferred in trans, as are some mobilizable genomic islands. Together, these findings clearly demonstrate that ICEPm1 can actively excise from the chromosome in an integrase-dependent manner, dynamically integrate into both phenylalanine tRNA genes, and transfer into clinical strains using its own conjugation machinery.
INTRODUCTION
Comparative genomics and advances in sequencing technology have revealed the diversity in bacterial genomes within a single species. Most organisms require a core set of genes for survival, the core genome, which can be further supplemented by the accessory genome, genes conferring fitness properties for an organism in a specific niche (11, 33). Strain-to-strain variability in genetic content is in large part due to the presence or absence of genomic islands (GIs) (25, 28). GIs often contain genes that contribute to virulence, antibiotic resistance, metabolism, or other fitness properties (17, 33). Specific properties of GIs that distinguish them from the surrounding bacterial chromosome include a distinct GC content, association with tRNA genes or other genes conserved between species, and a tendency to carry mobility genes (15, 18).
Genomic islands are the result of horizontal gene transfer (HGT) events, which play a critical role in the evolution of bacterial species (32). Gain and loss of genomic islands result in the greatest and most rapid changes in the pathogenic potential of an organism and contribute to the emergence of pathogens from commensal organisms (15). Recently, it has become appreciated that many GIs encode a conjugative type IV secretion system (T4SS) and thus may have the ability to self-transfer (2, 23, 24). The conjugative ability that T4SSs confer to plasmids is well described, yet the discovery of T4SSs within GIs resulted in the establishment of a new class of mobile elements known as integrative and conjugative elements (ICEs) (41, 46).
ICEs, a subset of GIs, have a conserved modular structure composed of three functional units carrying genes important for ICE function: the integration, regulation, and conjugation modules (7, 45). ICEs are flanked by identical direct repeats attL and attR that can recombine with the aid of an integrase and excisionase to form a circularized, extrachromosomal form of the ICE (6). This circularized form carries the identical direct repeat, attI, which can then recombine with the reconstituted integration site on the chromosome, attB, and result in ICE reintegration. When the ICE-encoded integrase is activated, the ICE can excise from the chromosome, form a circular intermediate, and subsequently transfer to a recipient cell via a mating pore formed by the ICE-encoded T4SS (40, 46). Thus, ICEs site-specifically excise and integrate into the chromosome like many temperate bacteriophages, yet they are self-transmissible, similar to the case for conjugative plasmids. Because HGT contributes to the greatest and most rapid changes in the bacterial chromosome, the discovery of ICEs has given new insights into bacterial evolution.
Interspersed among the conserved modules needed for ICE function are cargo genes that generally encode specific functions that allow for adaptation to the surrounding environment (7, 46). For instance, the cargo genes of the clc element of Pseudomonas aeruginosa encode important metabolic properties necessary for growth on 3-chlorobenzoate, while the SXT element of Vibrio cholerae, the most extensively described ICE, carries genes conferring resistance to sulfamethoxazole, trimethoprim, and chloramphenicol (14, 21, 43, 46). While ICEs encode niche-specific functions in their cargo genes, the integration, regulation, and conjugation modules of ICEs are fairly conserved within ICE families.
We previously identified a genomic island, ICEPm1, that is conserved in Proteus mirabilis, Providencia stuartii, and Morganella morganii, using a comparative genomics hybridization array (13). These organisms are all urease-producing etiologic agents of catheter-associated urinary tract infection, a disease that is typically polymicrobial (22, 42, 44). ICEPm1 contains core modules and a syntenic structure consistent with prototypical ICEs (7, 46). It is flanked by identical 52-bp direct repeats and integrated into the 3′ end of a phenylalanine tRNA gene. Genes for a putative tyrosine-like recombinase, a putative helicase that could act as a relaxase, and a putative T4SS were all identified in this element by in silico analysis (13). Known cargo genes that are interspersed between the core modules of ICEPm1 encode a yersiniabactin-related iron acquisition system and an adhesin/protease that both contribute to virulence in the mouse model of ascending urinary tract infection (1, 19, 31). ICEPm1 was present in all P. mirabilis urinary isolates yet was heterogeneously distributed in commensal P. mirabilis strains that colonize other body sites (13). Additionally, several genes within ICEPm1 demonstrated 100% sequence identity between P. mirabilis, P. stuartii, and M. morganii. Therefore, because of the modular structure of ICEPm1 and its widespread prevalence among multiple bacterial species, we hypothesize that ICEPm1 is an active ICE.
The goal of this study was to demonstrate the ability of ICEPm1 to excise from the chromosome, form a circularized intermediate, and conjugatively transfer to ICEPm1-deficient clinical strains. We demonstrate that ICEPm1 can actively excise from the chromosome in an integrase-dependent and site-specific manner and subsequently transfer to clinical P. mirabilis isolates. Disruption of the putative relaxase gene (PMI2550), the integrase gene (intP), and a gene encoding a putative chromosome-partitioning protein (PMI2642) prevented detectable transfer. Mutations in the putative ICEPm1-encoded T4SS highlighted the self-transmissibility of ICEPm1. We demonstrate the dynamic nature of ICEPm1, as it can integrate into both pheU and pheV tRNA genes. Finally, we demonstrate that ICEPm1 can be transferred to Escherichia coli; however, possession of the identical attB sequence is not the only component necessary for this transfer.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. mirabilis HI4320 was cultured from the urine of a patient catheterized for ≥30 days, presenting with bacteriuria (>105 CFU/ml) (44). P. mirabilis 378L was cultured from the groin of a resident from a skilled-nursing facility (30). Additional strains with defined mutations are listed in Table 1. All strains were cultured in modified Luria-Bertani (LB) broth (containing, per liter, 0.5 g NaCl, 10 g tryptone, and 5 g yeast extract) or LB agar (with 15 g agar). Overnight broth cultures were inoculated from a single colony and incubated for 18 h at 37°C with aeration. All strains were maintained at −80°C in 25% glycerol. Antibiotics were used at the following concentrations: kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml; trimethoprim, 32 μg/ml; and nalidixic acid, 50 μg/ml.
Table 1.
Bacterial strains and plasmids used in the study of the function of ICEPm1
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Proteus mirabilis strains | ||
| HI4320 | Urine isolate from catheterized patient | 44 |
| ICEPm1::kan mutant | HI4320, ICEPm1 marked Kanr | This study |
| int::kan mutant | HI4320, PMI2549::kan insertion mutant | This study |
| rlxS::kan mutant | HI4320, PMI2550::kan insertion mutant | This study |
| traP::kan mutant | HI4320, PMI2594::kan insertion mutant | This study |
| parA::kan mutant | HI4320, PMI2642::kan insertion mutant | This study |
| 2476Ω ICEPm1::kan mutant | HI4320, PMI2476 (traI of SXT) insertion, ICEPm1 marked Kanr | This study |
| 378L | P. mirabilis groin isolate; Trir | 30 |
| 378L ICEPm1::kan mutant | ICEPm1-containing transconjugant | This study |
| Escherichia coli strains | ||
| C | Nalr | 3a |
| C ICEPm1::kan mutant | ICEPm1-containing transconjugant; Kanr Nalr | This study |
| CFT073 | Uropathogenic pyelonephritis isolate; Nalr | 29a |
| K-12 | MG1655; Nalr | |
| Plasmids | ||
| pACD4K-C | Targetron vector containing group II intron and kanamycin resistance cassette; Camr | Sigma |
| pACD4K-C-loxP | Targetron vector with loxP sites flanking the kanamycin resistance cassette; Camr | Sigma |
| pAR1219 | IPTG-inducible T7 polymerase; Ampr | Sigma |
| pQL123 | IPTG-inducible cre recombinase; Ampr | Sigma |
Molecular techniques.
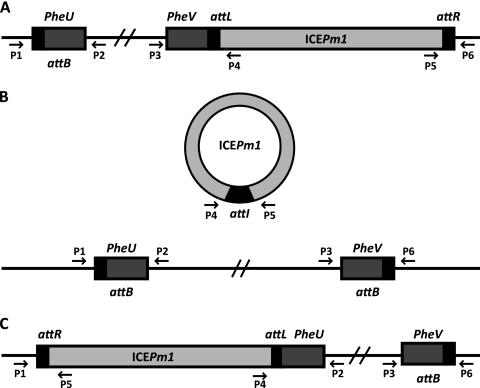
PCR was used to detect the integrated and excised forms of ICEPm1. Oligonucleotide primer sequences are listed in Table 2, and their targeted genomic locations are illustrated in Fig. 1. Oligonucleotides were specific for the flanking regions of pheU and pheV and thus could specifically identify ICEPm1 attachment sites at these locations. Reactions were performed in a final volume of 50 μl containing 1× PCR buffer, 0.5 μl Taq polymerase, and 200 μM deoxynucleoside triphosphates (dNTPs). PCR conditions were as follows: (i) 3 min (10 min for colonies) at 94°C; (ii) 30 cycles of 30 s at 94°C, 30 s at 56°C, and 1 min at 72°C; and (iii) 7 min at 72°C. For detection of amplification products from clonal populations, colony PCR was performed by inoculating a single colony with a pipette tip. When genomic DNA was used as template, 100 ng total DNA was used. To isolate genomic DNA, bacteria were collected by centrifugation (13,000 rpm, 10 min) from 500 μl of overnight culture. Genomic DNA was purified over a DNeasy column (Qiagen), treated with RNase A, and eluted in 200 μl elution buffer. For confirmation of transconjugants, multiplex PCR analysis was performed using a Qiagen multiplex PCR kit. Primers designed for amplification of PMI2641 (located within ICEPm1) and the attTn7 site were used together in each reaction. Multiplex colony PCR was performed in a final volume of 50 μl containing 2× Qiagen multiplex PCR master mix and each primer at 2 μM. Primers designed for amplification of PMI2551 and PMI2602 were also used for confirmation of transconjugants. PCR amplification products were purified using a QIAquick column and sequenced on an ABI model 3730 instrument. Sequences were aligned using the Wilbur-Lipman method in MegAlign software (Lasergene).
Table 2.
DNA sequences of oligonucleotides used in this study
| Function and oligonucleotide name | Sequence |
|---|---|
| ICE identification primers | |
| P1 | CAACTCTTGTTTGCCCTCTCAG |
| P2 | TGAGATCGGGTTTAATACGC |
| P3 | CGTTGACGCATCACGCTGAATA |
| P4 | CCGAGCGTTGGAGATCTCACTTCC |
| P5 | GATATAATTGGAACAATTCTGGTC |
| P6 | CTCAGTGACTTAACTCACTGACG |
| SXT identification primers | |
| SXT-P1 | GTTCCTGCACGTTGGATAGCTT |
| SXT-P2 | CGACAAGCTATCATCGAT |
| SXT-P3 | GCCACAGCTTGTTTCGTGTA |
| SXT-P4 | CGCAATGCTCGTTCATTATCT |
| Transconjugant confirmation primers | |
| Tn7F | AGATGCTGGCTTTGAAGAAAGTG |
| Tn7R | CACAGCATAACTGGACTGATTTC |
| 2551F | CAGAAGATTACATGAATAATG |
| 2551R | GAGAGTGTGATGAGATGTGAAT |
| 2602F | GCGAATGAACTTCACCA |
| 2602R | GCCACTAATCAGAGGGAGT |
| 2641F | GCACGCTCTGCTCCGCC |
| 2641R | CGGGAGGTGCGTCCATG |
| rplCF | CGTTGATGCTCTGATGCGTCT |
| rplCR | CGACTACTTGATGCACAAGCGC |
Fig. 1.
Locations of primers used in this study to detect ICEPm1 integration and excision. Oligonucleotide sequences are listed in Table 2. pheU and pheV are located on opposite DNA strands, share an identical nucleotide sequence, and are 73 bp in length. Boxes represent the 52-bp direct repeat (black), phenylalanine tRNA genes (dark gray), and ICEPm1 (light gray). The 3 possible conformations of ICEPm1 are as follows: integrated into pheV (A), excised from the chromosome and not integrated into either phe tRNA (B), and integrated into pheU (C). The figure is not to scale.
Construction of mutants.
A kanamycin-resistant marked strain, P. mirabilis HI4320 (ICEPm1::kan), was constructed to serve as the donor strain in mating assays using the TargeTron gene knockout system (Sigma) following a modified protocol (34). Briefly, mutagenic oligonucleotides were synthesized using the TargeTron design site for the intergenic region between PMI2624 and PMI2625 (Table 2) and used to retarget the group II intron using PCR. This PCR product was ligated into pACD4K-C (Camr) harboring a T7 promoter, subcloned into E. coli DH5α, and confirmed by sequencing. Plasmids of the correct sequence were then introduced by electroporation into P. mirabilis already containing the helper plasmid pAR1219 (Ampr), which expresses T7 polymerase under the control of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter. Chloramphenicol- and ampicillin-resistant colonies were cultured, and expression of the intron was induced by addition of 500 μM IPTG. Kanamycin-resistant colonies were picked, and specific insertional mutations were confirmed using PCR primers flanking the expected insertion site (Table 2). PCR products were visualized on a 1.0% agarose gel stained with ethidium bromide. Products appearing to be ∼2 kb greater in size than those of the wild type were confirmed as mutants. To determine the roles of various ICE-carried genes, insertional mutations were similarly constructed in PMI2549, PMI2550, PMI2594, and PMI2642 (Table 1). Additionally, an insertional mutation was constructed in PMI2476, the putative relaxase gene of the SXT-like ICE. The kanamycin cassette was removed from the intron by IPTG induction of the Cre recombinase encoded on plasmid pQL123, and a second mutation was made by inserting the kanamycin cassette into the intergenic region between PMI2624 and PMI2625, creating the ΩtraISXT ICEPm1::kan strain.
Conjugation assay.
Mating assays were performed to determine the frequency of ICEPm1 transfer to ICEPm1-deficient cells. Overnight cultures of donor and recipient cells were mixed 1:1 and spotted onto the center of an LB agar plate. Plates were dried for 15 min and incubated for 6 h at 37°C. Bacteria were harvested by flooding the plate with 1 ml LB, and cells were suspended using a cell spreader. Serial dilutions were plated onto LB agar supplemented with kanamycin, trimethoprim, or both drugs to identify donors, recipients, and transconjugants, respectively (Autoplate 4000; Spiral Biotech). Viable colony counts were enumerated and expressed as CFU/ml, with a limit of detection of 200 CFU/ml (20 colonies on a plate) (Qcount; Spiral Biotech). The frequency of transfer was calculated by dividing the number of transconjugants formed by the number of donor bacterial cells present in each mating. The rate of spontaneous resistance of P. mirabilis HI4320 to trimethoprim (32 μg/μl) was found to be <1 × 10−9 and therefore did not interfere with calculating ICEPm1 transfer frequencies. For E. coli mating assays, nalidixic acid (50 μg/ml) was used to screen for recipients and nalidixic acid and kanamycin to screen for transconjugants.
RESULTS
ICEPm1 actively excises from the chromosome in a clonal population of P. mirabilis HI4320.
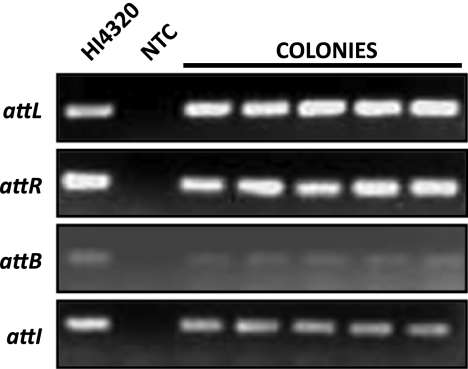
In the genome sequence annotation of P. mirabilis HI4320, ICEPm1 is integrated into the 3′ end of a phenylalanine tRNA gene, pheV, between loci PMI2548 and -2549 (genomic location, 2793767 to 2886300). To demonstrate that ICEPm1 can actively excise from the chromosome, we developed a PCR-based assay to identify ICEPm1 in both integrated and episomal states (Fig. 1). We used primers anchored in the chromosome and within the ICE to amplify attL (P3 and P4) and attR (P5 and P6), 52-bp direct repeats that are attachment sites of the ICE. Because primers P3 and P6 bind outside the ICE, in the chromosome, amplification of attL and attR is possible only when ICEPm1 is integrated into pheV (Fig. 1A). ICEPm1 can excise from the chromosome and form a closed circularized, episomal form that can be detected using primers that bind, and face outward, from the outermost ends of ICEPm1 (P4 and P5). These primers will amplify a product only when the 93-kb ICE has circularized to form site attI (Fig. 1B). When ICEPm1 is in this episomal form, a PCR amplicon for pheV can also be detected using primer pairs that bind in the chromosome at regions flanking the ICE (P3 and P6). An amplification product from these primers is detectable only if the ICE has excised, thus allowing for the shorter PCR amplification of the attB site located within pheV. All amplification products can be detected from the same chromosomal genomic DNA preparation as well as from colonies obtained from a plating of the parental strain (Fig. 2). This demonstrates that ICEPm1 is dynamically excising, forming a circular intermediate, and reintegrating into the chromosome in a clonal population.
Fig. 2.
ICEPm1 actively excises from the chromosome. The following primer pairs were used for amplification of attachment sites: attL, P3 and P4; attR, P5 and P6; attB, P3 and P6; and attI, P4 and P5. Lane 1, HI4320 genomic DNA; lane 2, no-template control (NTC). Colony PCR is shown for 5 isolated colonies obtained from a plating of the parental strain, P. mirabilis HI4320.
Direct sequencing of PCR amplicons resulted in the expected sequence of products for attL and attR as annotated (Fig. 3), as well as the expected sequence for attI and attB, given that ICEPm1 has precisely excised from the chromosome (Fig. 3). Sequencing of the last two products demonstrates that precise excision from the chromosome of ICEPm1 is achieved and that excision results in restoration of the full pheV sequence (which contains attB). Additionally, the identical 52-bp repeat observed at the flanking ends of ICEPm1 is created in the episomal form (attI).
Fig. 3.
ICEPm1 precisely excises from the chromosome at pheV. Sequences obtained by direct sequencing of PCR products for attL, attB, attI, and attR are aligned. All four products share the same 52-bp direct repeat (DR, shaded in black). Alignments also show the intact pheV-tRNA sequence maintained when ICEPm1 is integrated into the chromosome (attL) and when it is excised (attB).
P. mirabilis has another tRNA gene that transports phenylalanine, located approximately halfway around the genome from pheV, namely, pheU (genomic location, 375914 to 375986). The phenylalanine tRNAs PheU and PheV share the same anticodon (GAA), and although ICEPm1 is not annotated as being integrated at pheU, we attempted to determine whether this integration was possible. An amplification product was obtained using the primer pair P1 and P2, specific to chromosomal DNA flanking pheU, demonstrating an empty attB site within pheU. Upon direct sequencing, the attB-pheU site was identical to the attB-pheV site. Amplification products obtained with primer pairs P2 and P4 for attL-pheU and P1 and P5 for attR-pheU display the integration of ICEPm1 into pheU (Fig. 1C and data not shown). Integration into both pheU and pheV in a clonal population of ICEPm1 demonstrates not only that ICEPm1 is actively excising from the chromosome but that it can excise and reintegrate into either Phe-tRNA gene. Additionally, we were unable to obtain an amplification product using primer pair P6 and P4 or P1 and P4 (Fig. 1 and data not shown), revealing that integration of ICEPm1 is directional, a property characteristic of tyrosine-like recombinases (37).
ICEPm1 is transmissible to a clinical ICEPm1-deficient P. mirabilis strain and capable of excision in the recipient.
Genes encoding the kanamycin resistance cassette were introduced into an intergenic region of ICEPm1, between loci PMI2624 and PMI2625, which are fimbrial and ornithine decarboxylase pseudogenes, respectively. This serves as a marker for ICEPm1 acquisition without disrupting gene function in ICEPm1. P. mirabilis 378L, a clinical isolate already shown to be devoid of the ICE, was used as a recipient in mating assays to demonstrate ICEPm1 transfer. Mating assays were performed on LB agar plates or in LB broth with an incubation period of 6 h at 37°C. ICEPm1 transferred at a frequency of 1.35 × 10−5 transconjugants/donor on LB agar plates, while transconjugants were undetectable in the broth mating experiments.
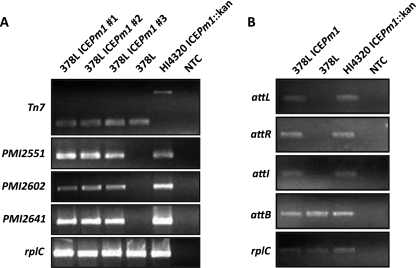
To confirm transfer, genomic DNA was extracted and purified from three colonies formed on dual-antibiotic-containing plates. Amplification of the attTn7 site was used to differentiate between P. mirabilis strains HI4320 and 378L; the size of the amplification product differs in these strains (Fig. 4A). Amplification of ICE genes for the beginning, middle, and end of the ICE (PMI2551, PMI2602, and PMI2641) by PCR demonstrated the presence of the ICE in the P. mirabilis 378L recipient cells, indicating that transfer had occurred (Fig. 4A).
Fig. 4.
Confirmation of ICEPm1 transfer to a clinical P. mirabilis commensal isolate and its retained function. (A) Mating assays were performed, and three transconjugants were selected for isolation and extraction of genomic DNA to confirm transfer of ICEPm1 by PCR. Amplification products were obtained for the beginning (PMI2551), middle (PMI2602), and end (PMI2641) of ICEPm1 to demonstrate complete transfer of the ICE. The attTn7 site, which produces products of different sizes in the donor (P. mirabilis HI4320 ICE::kan) and the recipient (P. mirabilis 378L) strains, was used to differentiate between the donor and recipient genetic backgrounds. (B) Amplification products for attI and attB in the transconjugant (378L ICEPm1) demonstrate that ICEPm1 is capable of excising in the recipient host background. NTC, no-template control.
To determine whether ICE function was maintained upon formation of the transconjugant, PM378L-ICEPm1::kan, PCR assays for attL-pheV, attR-pheV, attB-pheV, and attI were conducted; these demonstrated that ICEPm1 was able to actively excise from and reintegrate into the recipient chromosome, in a manner similar to that observed in the P. mirabilis HI4320 host genetic background (Fig. 4B). Additionally, amplification products were obtained for attL-pheU, attR-pheU, and attB-pheU, showing that ICEPm1 was also capable of integrating into pheU in the recipient.
intP is required for ICEPm1 excision and subsequent transfer.
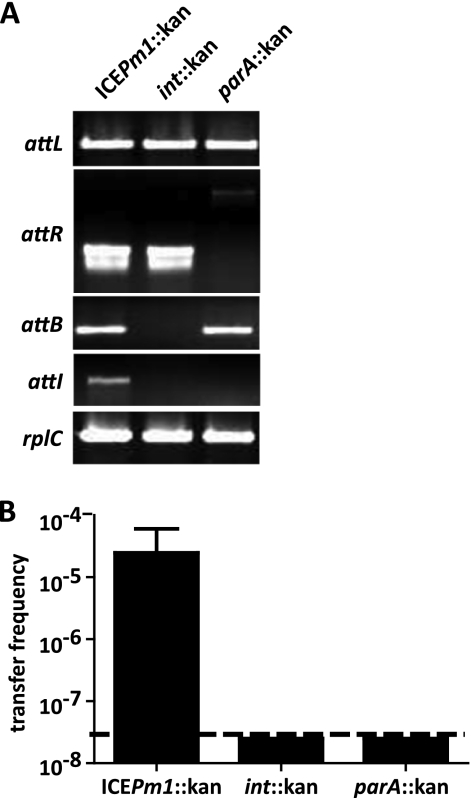
An insertional inactivation mutation was constructed in the ICEPm1 gene adjacent to attL, PMI2549, encoding a putative site-specific recombinase of the tyrosine recombinase family. Integrases that are necessary for ICE function are generally encoded near the attL or attR sites of these elements (5). Disruption of PMI2549 resulted in an ICEPm1 that was no longer capable of excising from the chromosome and forming a circular intermediate. Amplifications of attL and attR were positive, but no product was observed for attI or attB, indicating that ICEPm1 had lost its ability to excise from the chromosome (Fig. 5A). Based on the loss of this activity, PMI2549 was designated intP (integrase ICEPm1).
Fig. 5.
intP is necessary for ICEPm1 excision and transfer. (A) Amplification products for attL, attR, attB, and attI from ICEPm1::kan, intP::kan (PMI2549::kan), and parA::kan (PMI2642::kan) mutants. No amplification products were observed for attB or attI in the intP::kan mutant because ICEPm1 is unable to excise from the chromosome. attR produces a different-size amplification product in the parA::kan mutant because the primers bind outside where the kanamycin cassette is inserted. No product was observed for attI in the parA::kan mutant. (B) Frequencies (transconjugants/donor) of ICEPm1 transfer obtained with P. mirabilis 378L as the recipient and ICEPm1::kan, int::kan, and parA::kan mutants as donors. Limit of detection, 2.5 × 10−8 transconjugants/donor.
In addition to “locking” ICEPm1 into the chromosome by disrupting the function of its integrase, mutation of the integrase allowed us to demonstrate that ICEPm1 is present only in either pheU or pheV in a single bacterium. Because the introduction of the kanamycin cassette occurred in one bacterial cell and thus “locked” ICEPm1 in its current genomic state, we observed amplification products only for ICE locked into pheV. While amplification of attB-pheU was positive in the intP::kan mutant, no amplification products were observed for attL-pheU or attR-pheU, indicating that ICEPm1 is integrated only into pheV (data not shown). Furthermore, we created 29 independent intP::kan mutations, and in each we were able to amplify only attL-pheV products; no attL-pheU products were obtained. This verifies that ICEPm1 can be integrated into only one phe tRNA gene in a single cell and that ICEPm1 preferentially integrates into pheV, as originally annotated (35).
Mating assays were performed with P. mirabilis HI4320 intP::kan as the donor and P. mirabilis 378L as the recipient (Fig. 5B). Transconjugant formation was not observed, indicating that disruption of the ICE-encoded integrase is required for excision from the chromosome and that without excision, subsequent transfer is abolished.
parA is required for efficient transfer.
BlastP identified PMI2642 as a putative chromosome-partitioning protein. PMI2642 is located near attR in ICEPm1, which is commonly where proteins important for ICE stability are located; its homolog, the Soj protein of P. aeruginosa PAPI-1, has been shown to be important for stability of this ICE in its extrachromosomal form (36). Insertional inactivation of PMI2642 did not have an effect on ICEPm1 excision but resulted in no detectable amplification product for the attI site, suggesting loss of stability of the episomal form (Fig. 5A). Furthermore, disruption of PMI2642 resulted in a decreased frequency of transfer of ICEPm1 into P. mirabilis 378L to below the limit of detection (Fig. 5B).
ICEPm1 carries a T4SS that is necessary for conjugative transfer.
The PMI2569-PMI2592 gene cluster encodes a series of proteins with predicted signal peptide sequences and transmembrane domains that share homology with the T4SS encoded in ICEHin1056 of Haemophilus influenzae (13, 23). To verify that ICEPm1 carries a functional T4SS, which is necessary for its self-transmissibility, we disrupted the gene loci of two proteins predicted as being important in DNA mobility, PMI2550 (traI) and PMI2594 (traP). PMI2550 is a predicted helicase and, based on its genomic location adjacent to intP, is hypothesized to be the relaxase of ICEPm1, traI. Relaxases recognize the origin of transfer (oriT) of plasmids and ICEs and are thus necessary for mobility (27, 41). PMI2594 is part of the conjugative pilus superfamily of proteins (TIGR03748) and is 52% similar to tfc2 (loci, p1056.31), a protein encoded on ICEHin1056. Disruption of this gene in H. influenzae resulted in a 100,000-fold reduction in transfer frequency of the ICE and abolished pilus formation (23). When we disrupted PMI2594 (transfer Pilus), expected amplification products for all attachments sites were observed (Fig. 6A). Additionally, the frequency of ICEPm1 transfer was reduced 1,000-fold to 2.8 × 10−8 transconjugants/donor (Fig. 6B). Similarly, disruption of PMI2550 (traI) had no effect on ICE excision (Fig. 6A) yet decreased ICEPm1 transfer significantly to 4.2 × 10−8 transconjugants/donor. This confirms the roles of traI (PMI2550) and traP (PMI2594) in conjugative transfer of ICEPm1.
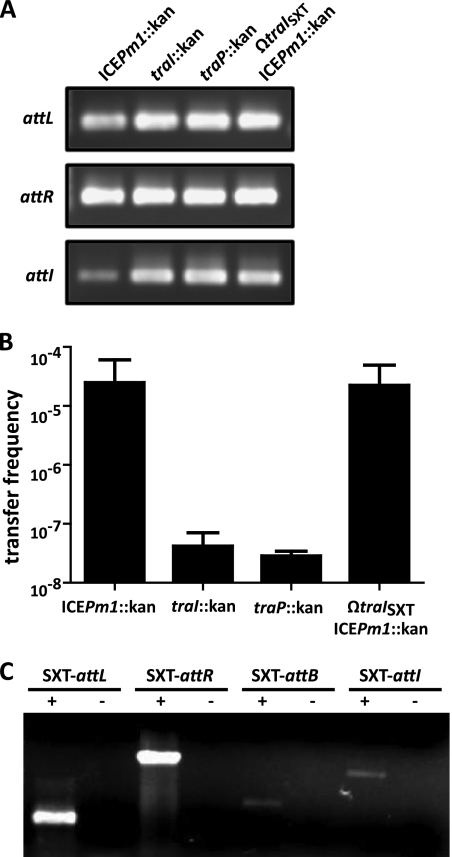
Fig. 6.
ICEPm1-carried T4SS genes are important for conjugative transfer. (A) Disruption of the ICEPm1 relaxase gene (traI) and conjugative pore-forming gene (traP) had no effect on the ability of ICEPm1 to excise from the chromosome, as all amplification products that represent excised and integrated forms are obtained. Additionally, insertional inactivation of the relaxase of the SXT-like ICE (traISXT) also did not affect excision. (B) Mating assays performed with the traI::kan and traP mutants decreased the transfer frequency of ICEPm1 by approximately 1,000-fold. The transfer frequency of ICEPm1 in the SXT-like relaxase mutant (ΩtraISXT ICEPm1::kan) was similar to wild-type levels, suggesting that the SXT-like ICE cannot transfer ICEPm1 in trans. Limit of detection, 2.5 × 10−8 transconjugants/donor. (C) Active excision of the SXT-like ICE (ICEPmiUsa1) from the chromosome of P. mirabilis HI4320. Primers were designed in a similar way as for the PCR assay for detection of ICEPm1 excision. Primer pair SXT-P1 and SXT-P2 was used to amplify SXT-attL. Primer pair SXT-P3 and SXT-P4 was used to amplify SXT-attR. Primer pairs SXT-P1 with SXT-P4 and SXT-P2 with SXT-P3 were used to amplify the attB and attI sites, respectively.
Another ICE showing significant homology to the SXT ICE of V. cholerae and thus a member of the R391/SXT family of ICEs has been reported in P. mirabilis HI4320 (genomic location, 2651089 to 2730732) (45). The SXT-like ICE, named ICEPmiUsa1, is located approximately 100 kb downstream from ICEPm1 and was identified by in silico analysis. Therefore, we were interested in determining whether this element, like ICEPm1, actively excises from the chromosome. We demonstrated excision of the SXT-like ICE from the chromosome using a PCR-based assay similar to that used to identify ICEPm1 active excision (Fig. 6C). Similar to the case for conjugative plasmids, ICE T4SSs have been shown to be able to transfer other mobile genomic islands (MGIs) in trans (9, 41). To confirm that the T4SS encoded on ICEPm1 was responsible for its self-transmissibility, we first demonstrated that the T4SS of the SXT-like ICE is not involved in the conjugative transfer of ICEPm1.
We insertionally inactivated the relaxase gene of the SXT-like ICE (traISXT), excised the kanamycin cassette, and remarked ICEPm1 with the kanamycin resistance cassette in the same intergenic region as before. The traISXT gene was recently demonstrated to be necessary for transfer of a nearby MGI in trans in V. cholerae (9). This mutant in P. mirabilis HI4320 maintained excision properties of ICEPm1 (Fig. 6A) as well as wild-type levels of conjugative transfer of the ICEPm1 mobile element at 2.2 × 10−5 transconjugants/donor (Fig. 6B). This suggests that ICEPm1 is self-transmissible and is not being transferred by the T4SS of the SXT-like ICE.
ICEPm1 transfer to Escherichia coli is strain dependent.
The DNA sequences of pheU and pheV of P. mirabilis HI4320 are conserved in E. coli strains CFT073 and MG1655 (K-12). In silico sequence analysis of E. coli CFT073 shows two known genomic islands integrated into the pheU and pheV sites (Fig. 7A). In comparison, the genes surrounding the pheU and pheV attB sites in a K-12 strain are genes that are adjacent to the terminal ends of the GIs carried by E. coli CFT073. This suggests that the CFT073 attB integration sites already carry genomic islands but that the K-12 attB sites are empty and could thus potentially receive ICEPm1.
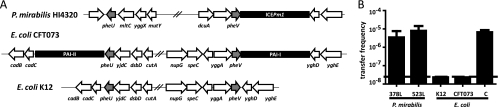
Fig. 7.
Transfer of ICEPm1 to E. coli. (A) The annotated genomic sequences of P. mirabilis HI4320, E. coli CFT073, and E. coli MG1655 (K-12) show conserved DNA sequences for pheU and pheV tRNAs yet variability in whether a pathogenicity island (PAI) is integrated into these tRNAs. The pheU site in P. mirabilis is empty, as previously described. Both pheU and pheV carry genetically distinct PAIs in E. coli CFT073 that carry important virulence factors for adhesion and red blood cell lysis (10, 16). No PAIs in the pheU and pheV sites of K-12 have been reported. Each tRNA is surrounded by those genes flanking the respective PAIs in CFT073, thus suggesting that these attB sites are “empty.” (B) Transfer frequency (transconjugants/donor) in mating experiments with ICEPm1::kan as the donor and P. mirabilis (378L or 523L) or E. coli (K-12, CFT073, or C) as the recipient. All E. coli strains contain an attB sequence identical to that in P. mirabilis. Plate matings were conducted with a 1:1 ratio of donor to recipient at 37°C for 6 h. The limit of detection is 2 × 10−8 transconjugants/donor.
We performed mating assays between P. mirabilis HI4320-ICEPm1::kan and the E. coli strains CFT073, K-12, and C to determine the specificity of ICEPm1 transfer. No transconjugants were observed in the mating assays with E. coli K-12 or CFT073 as the recipient, yet transfer with a restriction-modification-negative strain, E. coli C, as the recipient yielded transfer frequencies of 6.6 × 10−6, similar to those for Proteus-to-Proteus intraspecies matings (Fig. 7B). Furthermore, E. coli C ICEPm1::kan can act as a donor for conjugative transfer of ICEPm1::kan to the P. mirabilis ICE-deficient strain PM378L. Mating assays with another P. mirabilis commensal isolate, P. mirabilis 523L, as the recipient also showed transfer frequencies similar to those for matings with P. mirabilis 378L as the recipient, further confirming transfer into clinical strains (Fig. 7B).
DISCUSSION
ICEPm1 was previously predicted to belong to a class of genomic islands known as integrative and conjugative elements based on in silico and comparative genomics analyses (13). In this study we show that ICEPm1 is a functional integrative and conjugative element that can excise from the chromosome, form a detectable circular intermediate, and subsequently transfer to clinical ICEPm1-deficient strains. Mutation of the ICE-carried integrase gene, intP, renders ICEPm1 unable to excise from the chromosome and decreases transfer to undetectable frequencies. ICEPm1 directionally integrates into either pheU or pheV, although it is integrated predominately at pheV. Insertional inactivation of the relaxase gene and a mating pore formation gene resulted in a 1,000-fold decrease in transfer efficiency, verifying that ICEPm1 is indeed self-transmissible.
Many tRNA genes can serve as integration sites for genomic islands, yet phenylalanine tRNA genes are especially recognized as hot spots for foreign DNA integration (8, 39). Several bacteriophages and pathogenicity islands (PAIs) are known to be integrated at these sites, yet we believe that ICEPm1 and ICEMISymR7A are the first functionally characterized ICEs integrating at these loci (26, 38). One reason that integration into phenylalanine tRNAs occurs frequently may be related to their conserved sequence across species. However, mating assays failed with two E. coli strains, CFT073 and MG1655, that contain both pheU and pheV genes identical to those in P. mirabilis. This suggests that additional factors besides an identical attB site are required for ICE transfer and integration into a recipient strain.
After screening multiple separate mutants with mutations in the integrase gene, which eliminates ICEPm1 excision from the chromosome, we never observed ICEPm1 locked into pheU, despite identifying integration at pheU in the parental strain. This observation, together with the observation that ICEPm1 is annotated as integrated at pheV in HI4320, suggests that the conformation in which ICEPm1 is integrated at pheV likely dominates over integration at pheU or existence as an episome (35). Further studies are needed to determine why preferential integration occurs at pheV, as pheU and pheV are identical.
We did not observe any transconjugants when mating assays were performed in broth, while we saw frequent transfer on agar plates, suggesting that a solid surface may be necessary for mating. Conjugative type IV pili that lack the ability to mate in broth are generally more rigid and thick than pili that allow for broth mating (4). The requirement of a solid surface for transfer is also interesting in the context of the pathogens that harbor ICEPm1. We previously showed that P. mirabilis, P. stuartii, and M. morganii all harbor ICEPm1 and that the ICE is predominant in P. mirabilis urinary isolates. Biofilms facilitate horizontal gene transfer between bacteria (12). All of these organisms commonly colonize the catheterized urinary tract and are also known for forming biofilms on Foley catheters (42). Additionally, these isolates were cultured from catheterized patients with urinary tract infections (13). Thus, the catheter could serve multiple functions in the wide dissemination of ICEPm1 among these organisms: as a solid surface to bring bacteria in close proximity, promoting cell-to-cell contact (necessary for conjugal transfer), and facilitating biofilm formation, which protects the bacteria from the surrounding environment (42). In support of this notion, we show transfer of ICEPm1 between an organism that was isolated from the catheterized urinary tract and an organism that was colonizing the groin of a patient. This suggests that if these organisms can come into close contact within a patient, HGT could occur, thus disseminating important virulence factors, such as iron acquisition, to the commensal organism. Further studies are required to understand what induces ICEPm1 transfer under these conditions.
Similarly to plasmids, ICEs have the ability to transfer other mobile elements in trans (9, 20). The relaxase produced by an ICE can recognize similar oriT sequences located within mobilizable genomic islands (MGI) or a plasmid and therefore recruit these other mobile genetic elements to the mating pore (9, 41). In silico analysis of P. mirabilis HI4320 identified an SXT-like ICE located approximately 100 kb 5′ to pheV where ICEPm1 is annotated (45). We show that the SXT-like ICE cannot transfer ICEPm1 because the ICEPm1-carried relaxase gene (traI) and a mating pore formation gene (traP) are necessary for transfer. To confirm that the SXT-like ICE is incapable of transferring ICEPm1 in trans, we constructed insertional mutations in the relaxase gene of the SXT-like ICE, traISXT. Mating assays performed with this strain as a donor demonstrate wild-type frequencies of ICEPm1 transfer, thus confirming that ICEPm1 is self-transmissible. Further studies are needed to investigate transfer of the SXT-like ICE to determine whether transfer occurs at the same frequency and under the same conditions as for ICEPm1 as well as to understand the role that these elements play in lateral transfer of other genomic components in P. mirabilis.
Exclusion mechanisms that can limit ICE acquisition when an ICE is already present have been reported for both SXT in V. cholerae and ICEBs1 in Bacillus subtilis (3, 29, 47). Therefore, it is interesting that P. mirabilis HI4320 harbors two ICEs, both of which can actively integrate and excise from the chromosome. The T4SS genes of ICEPm1 are distinct from the SXT/R391 family and share homology with genes from the P. aeruginosa PAPI island, the H. influenzae ICEHin1056 island, and the Salmonella enterica serovar Typhi SPI-7 islands (13). In a phylogenetic analysis of the T4SS genes, these islands were evolutionarily distinct from P-like, F-like and I-like T4SSs, and they were denoted GI-like T4SSs, for genomic island-like type IV secretion systems (23). The T4SSs from the SXT/R391 family of ICEs cluster with the F-like systems. Furthermore, intP of ICEPm1 encodes an integrase of the XerC/D family of tyrosine-like recombinases, while the integrases encoded on ICEs of the SXT/R391 family are more similar to the P4-type lineage of tyrosine-like recombinases. This suggests that these two ICEs segregate into two different ICE families, which therefore could explain why the two exist in the same cell. Additionally, although ICEPm1 shows similarity to ICEHin1056 and PAPI-1, the amino acid similarity is rarely greater than 70%. This limited homology differs dramatically from homology among SXT/R391 ICEs, where most of the integrases are 99% identical. Therefore, ICEPm1 may fall into a third, yet-to-be-described family of ICEs.
To our knowledge, this is the first report of a single bacterium harboring two self-transmissible ICEs. The active integration and excision as well as transfer of these elements imply that the chromosome of P. mirabilis HI4320 is quite dynamic. Additionally, understanding how genome location and ICE conformation affect expression of the genes carried on the ICE as well as the genes surrounding pheU or pheV will provide insight into the effect that the dynamic nature of ICEs has on gene expression. P. mirabilis HI4320 also carries a 36-kb plasmid, thus suggesting that mobile genetic elements are common in its genome, which is relatively small at 4.1 Mb compared to those of other enterobacterial pathogens. It would not be surprising to find other MGIs that are transferred in trans by either of these ICEs or the plasmid, suggesting that the mobilome of P. mirabilis is substantial. This study demonstrates the activity of ICEs and thus the potential that these mobile elements have for disseminating virulence determinants and antibiotic resistance genes among clinical isolates.
ACKNOWLEDGMENTS
This study was supported in part by Public Health Service grant AI059722 from the National Institutes of Health. Additionally, E.F. was supported by a Rackham Dissertation award.
Footnotes
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Alamuri P., et al. 2010. Adhesion, invasion, and agglutination mediated by two trimeric autotransporters in the human uropathogen Proteus mirabilis. Infect. Immun. 78:4882–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ambur O. H., et al. 2009. Genome dynamics in major bacterial pathogens. FEMS Microbiol. Rev. 33:453–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Auchtung J. M., Lee C. A., Garrison K. L., Grossman A. D. 2007. Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Mol. Microbiol. 64:1515–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a. Bertani G., Weigle J. J. 1953. Host controlled variation in bacterial viruses. J. Bacteriol. 65:113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bradley D. E., Taylor D. E., Cohen D. R. 1980. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J. Bacteriol. 143:1466–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burrus V., Pavlovic G., Decaris B., Guedon G. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601–610 [DOI] [PubMed] [Google Scholar]
- 6. Burrus V., Waldor M. K. 2003. Control of SXT integration and excision. J. Bacteriol. 185:5045–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burrus V., Waldor M. K. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376–386 [DOI] [PubMed] [Google Scholar]
- 8. Chen N., et al. 2010. The pheV phenylalanine tRNA gene Klebsiella pneumoniae clinical isolates is an integration hotspot for possible niche-adaptation genomic islands. Curr. Microbiol. 60:210–216 [DOI] [PubMed] [Google Scholar]
- 9. Daccord A., Ceccarelli D., Burrus V. 2010. Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol. Microbiol. 78:576–588 [DOI] [PubMed] [Google Scholar]
- 10. Dobrindt U., et al. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365–6372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dobrindt U., Hochhut B., Hentschel U., Hacker J. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414–424 [DOI] [PubMed] [Google Scholar]
- 12. Ehrlich G. D., et al. 2010. The distributed genome hypothesis as a rubric for understanding evolution in situ during chronic bacterial biofilm infectious processes. FEMS Immunol. Med. Microbiol. 59:269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flannery E. L., Mody L., Mobley H. L. 2009. Identification of a modular pathogenicity island that is widespread among urease-producing uropathogens and shares features with a diverse group of mobile elements. Infect. Immun. 77:4887–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaillard M., Pernet N., Vogne C., Hagenbuchle O., van der Meer J. R. 2008. Host and invader impact of transfer of the clc genomic island into Pseudomonas aeruginosa PAO1. Proc. Natl. Acad. Sci. U. S. A. 105:7058–7063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gal-Mor O., Finlay B. B. 2006. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell. Microbiol. 8:1707–1719 [DOI] [PubMed] [Google Scholar]
- 16. Hacker J., et al. 1990. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb. Pathog. 8:213–225 [DOI] [PubMed] [Google Scholar]
- 17. Hacker J., Carniel E. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2:376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hacker J., Kaper J. B. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641–679 [DOI] [PubMed] [Google Scholar]
- 19. Himpsl S. D., et al. 2010. Proteobactin and a yersiniabactin-related siderophore mediate iron acquisition in Proteus mirabilis. Mol. Microbiol. 78:138–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hochhut B., Marrero J., Waldor M. K. 2000. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J. Bacteriol. 182:2043–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hochhut B., Waldor M. K. 1999. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32:99–110 [DOI] [PubMed] [Google Scholar]
- 22. Jones B. D., Mobley H. L. 1987. Genetic and biochemical diversity of ureases of Proteus, Providencia, and Morganella species isolated from urinary tract infection. Infect. Immun. 55:2198–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Juhas M., et al. 2007. Novel type IV secretion system involved in propagation of genomic islands. J. Bacteriol. 189:761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Juhas M., Crook D. W., Hood D. W. 2008. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 10:2377–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Juhas M., et al. 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33:376–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lawrence J. G., Ochman H. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. U. S. A. 95:9413–9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee C. A., Grossman A. D. 2007. Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J. Bacteriol. 189:7254–7261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lloyd A. L., Rasko D. A., Mobley H. L. 2007. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J. Bacteriol. 189:3532–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marrero J., Waldor M. K. 2007. Determinants of entry exclusion within Eex and TraG are cytoplasmic. J. Bacteriol. 189:6469–6473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a. Mobley H. L. T., Green D. M., Trifillis A. L., Johnson D. E., Chippendale G. R., Lockatell C. V., Jones B. D., Warren J. W. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mody L., Maheshwari S., Galecki A., Kauffman C. A., Bradley S. F. 2007. Indwelling device use and antibiotic resistance in nursing homes: identifying a high-risk group. J. Am. Geriatr. Soc. 55:1921–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nielubowicz G. R., Smith S. N., Mobley H. L. 2008. Outer membrane antigens of the uropathogen Proteus mirabilis recognized by the humoral response during experimental murine urinary tract infection. Infect. Immun. 76:4222–4231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ochman H., Lawrence J. G., Groisman E. A. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304 [DOI] [PubMed] [Google Scholar]
- 33. Pallen M. J., Wren B. W. 2007. Bacterial pathogenomics. Nature 449:835–842 [DOI] [PubMed] [Google Scholar]
- 34. Pearson M. M., Mobley H. L. 2007. The type III secretion system of Proteus mirabilis HI4320 does not contribute to virulence in the mouse model of ascending urinary tract infection. J. Med. Microbiol. 56:1277–1283 [DOI] [PubMed] [Google Scholar]
- 35. Pearson M. M., et al. 2008. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 190:4027–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qiu X., Gurkar A. U., Lory S. 2006. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 103:19830–19835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rajeev L., Malanowska K., Gardner J. F. 2009. Challenging a paradigm: the role of DNA homology in tyrosine recombinase reactions. Microbiol. Mol. Biol. Rev. 73:300–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramsay J. P., Sullivan J. T., Stuart G. S., Lamont I. L., Ronson C. W. 2006. Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol. Microbiol. 62:723–734 [DOI] [PubMed] [Google Scholar]
- 39. Rumer L., et al. 2003. Dissemination of pheU- and pheV-located genomic islands among enteropathogenic (EPEC) and enterohemorrhagic (EHEC) E. coli and their possible role in the horizontal transfer of the locus of enterocyte effacement (LEE). Int. J. Med. Microbiol. 292:463–475 [DOI] [PubMed] [Google Scholar]
- 40. Seth-Smith H., Croucher N. J. 2009. Genome watch: breaking the ICE. Nat. Rev. Microbiol. 7:328–329 [DOI] [PubMed] [Google Scholar]
- 41. Smillie C., Garcillan-Barcia M. P., Francia M. V., Rocha E. P., de la Cruz F. 2010. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74:434–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stickler D. J. 2008. Bacterial biofilms in patients with indwelling urinary catheters. Nat. Clin. Pract. Urol. 5:598–608 [DOI] [PubMed] [Google Scholar]
- 43. Waldor M. K., Tschape H., Mekalanos J. J. 1996. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J. Bacteriol. 178:4157–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Warren J. W., Tenney J. H., Hoopes J. M., Muncie H. L., Anthony W. C. 1982. A prospective microbiologic study of bacteriuria in patients with chronic indwelling urethral catheters. J. Infect. Dis. 146:719–723 [DOI] [PubMed] [Google Scholar]
- 45. Wozniak R. A., et al. 2009. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5:e1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wozniak R. A., Waldor M. K. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8:552–563 [DOI] [PubMed] [Google Scholar]
- 47. Wozniak R. A., Waldor M. K. 2009. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 5:e1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]