Abstract
To investigate the outermost structure of the Bacillus subtilis spore, we analyzed the accessibility of antibodies to proteins on spores of B. subtilis. Anti-green fluorescent protein (GFP) antibodies efficiently accessed GFP fused to CgeA or CotZ, which were previously assigned to the outermost layer termed the spore crust. However, anti-GFP antibodies did not bind to spores of strains expressing GFP fused to 14 outer coat, inner coat, or cortex proteins. Anti-CgeA antibodies bound to spores of wild-type and CgeA-GFP strains but not cgeA mutant spores. These results suggest that the spore crust covers the spore coat and is the externally exposed, outermost layer of the B. subtilis spore. We found that CotZ was essential for the spore crust to surround the spore but not for spore coat formation, indicating that CotZ plays a critical role in spore crust formation. In addition, we found that CotY-GFP was exposed on the surface of the spore, suggesting that CotY is an additional component of the spore crust. Moreover, the localization of CotY-GFP around the spore depended on CotZ, and CotY and CotZ depended on each other for spore assembly. Furthermore, a disruption of cotW affected the assembly of CotV-GFP, and a disruption of cotX affected the assembly of both CotV-GFP and CgeA-GFP. These results suggest that cgeA and genes in the cotVWXYZ cluster are involved in spore crust formation.
INTRODUCTION
The sporulation of Bacillus subtilis is initiated in response to nutrient limitations and involves a highly ordered program of gene expression and morphological changes (20, 24). The first morphological change in sporulation is the appearance of an asymmetrically positioned septum that divides the cell into a larger mother cell and a smaller forespore. Next, the mother cell membrane migrates around the forespore membrane during a phagocytic-like process called engulfment. The completion of engulfment involves the fusion of the mother cell membrane to pinch off the forespore within the mother cell. Compartment-specific gene expression brings about the maturation of the spore and its release upon the lysis of the mother cell (reviewed in reference 10). Mature spores remain viable during long periods of starvation and are resistant to heat, toxic chemicals, lytic enzymes, and other factors capable of damaging vegetative cells (17). Spores germinate and resume growth when nutrients become available (19).
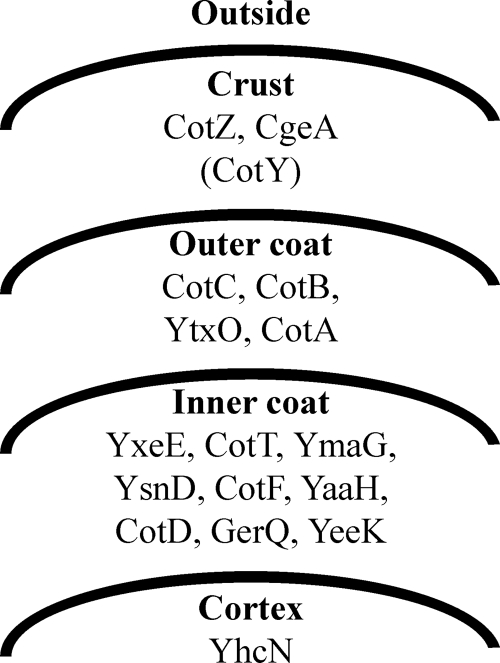
The outer portions of Bacillus spores consist of a cortex, a spore coat layer, and, in some cases, an exosporium. The cortex, a thick layer of peptidoglycan, is deposited between the inner and outer membranes of the forespore and is responsible for maintaining the highly dehydrated state of the core, thereby contributing to the extreme dormancy and heat resistance of spores. Spore coat assembly involves the deposition of at least 50 protein species (2, 13, 14) into two major layers: an electron-dense outer layer called the outer coat and a less electron-dense inner layer with a lamellar appearance, called the inner coat (Fig. 1) (26). These layers provide a protective barrier against bactericidal enzymes and chemicals such as lysozyme and organic solvents (17). A loose-fitting, balloon-like exosporium surrounds the spore coats of some species, including Bacillus anthracis, Bacillus cereus, and some clostridia. This structure is not observed in the B. subtilis spore, with the exception of a strain isolated from the human gastrointestinal tract (2, 3). Therefore, the spore coat has been thought to be the outermost structure of the B. subtilis spore. However, we recently found a layer located outside the outer coat composed of CgeA and CotZ (Fig. 1) (4). Using a similar method, McKenney and coworkers also identified an outermost spore layer composed of CotG, CotW, and CotZ and demonstrated that this layer, which they termed the spore crust, is absent in cotXYZ mutant spores (16). The identification of spore surface proteins is also becoming increasingly important for potential practical applications (9). In this study, we demonstrate that the spore crust is the externally exposed, outermost component of the B. subtilis spore, forming a layer that covers the spore coat. Furthermore, we identified genes involved in spore crust formation as well as additional components of the spore crust.
Fig. 1.
Schematic representation of the outer structure of the B. subtilis spore and proteins assigned to each layer. With the exception of CotY (in parentheses), assignments of proteins in each layer are based on our previous data (4). The localization of CotY in the spore crust is suggested by the results of this study.
MATERIALS AND METHODS
General methods and bacterial construction.
B. subtilis cells were cultured in LB medium and induced to sporulate by exhaustion in Schaeffer's medium (18) at 37°C for 24 h. Plasmid DNA for the transformation of B. subtilis was harvested from Escherichia coli strain JM109. Bacterial strains, plasmids, and primers used in this study are listed in Tables S1 and S2 in the supplemental material.
To construct a vector to introduce GFP (green fluorescent protein)-fused genes into the amyE locus, we amplified an internal fragment of amyE by PCR using genomic DNA of B. subtilis 168 as a template and primer pair AMYE980/AMYE1860R. The fragment was digested with HindIII and then cloned into HindIII-digested pGFP7C to yield plasmid pGFP7CA, in which the amyE fragment was downstream of GFP and in the same orientation.
To fuse GFP to the C terminus of CotV or CotY, we first amplified each gene and its 5′ promoter region (27) from B. subtilis 168 genomic DNA by PCR using primer pairs COTVM350/COTV383R and COTX40/COTY485R, respectively. Fragments were digested with BamHI and XhoI and then cloned into BamHI/XhoI-digested pGFP7CA to yield plasmids pCOTV8GA and pCOTY8GA. These plasmids were introduced into the amyE locus by a single-crossover event with selection for chloramphenicol resistance (5 μg/ml), yielding strains COTV8GA and COTY8GA (see Table S1 in the supplemental material). To introduce the cotZ-GFP fusion into the amyE locus without the cotY gene, we amplified the promoter region of the cotYZ and cotZ genes from B. subtilis 168 genomic DNA by PCR using primer pairs COTYM200/COTYM7R and COTZ1/COTZ443R, respectively. EcoRI/BamHI-digested PcotYZ and BamHI/XhoI-digested cotZ fragments were cloned into EcoRI/XhoI-digested pGFP7CA to yield the plasmid pCOTZ8GA. This plasmid was introduced into the amyE locus by a single-crossover event with selection for chloramphenicol resistance (5 μg/ml), yielding strain COTZ8GA (Table S1). All GFP fusions used in this study contain a hexahistidine tag at their C termini, but for simplicity, these constructs are referred to as GFP fusions.
To disrupt cotV, cotW, cotX, cotY, or cotZ, we amplified an internal fragment of each gene from B. subtilis 168 genomic DNA by PCR using primer pairs COTV19/COTV220R, COTW34/COTW230R, COTX20/COTX169R, COTY21/COTY184R, and COTZ97/COTZ311R, respectively. Fragments were digested with HindIII and BamHI and then cloned into HindIII/ BamHI-digested pMutin3 to yield the corresponding plasmids pCOTV5E, pCOTW5E, pCOTX5E, pCOTY5E, and pCOTZ5E. These plasmids were introduced into the cotV, cotW, cotX, cotY, or cotZ locus by a single-crossover event with selection for erythromycin resistance (0.5 μg/ml), yielding strains COTV5E, COTW5E, COTX5E, COTY5E, and COTZ5E, respectively.
Immunofluorescence staining of spores.
Following sporulation in Schaeffer's medium for 24 h, aliquots of cells were washed twice with phosphate-buffered saline (PBS) (pH 7.4). Pellets were resuspended in GTE-lysozyme buffer (50 mM glucose, 20 mM Tris-HCl [pH 7.5], 10 mM EDTA, 200 μg/ml lysozyme) and incubated for 20 min at room temperature. After washing twice with PBS, pellets were resuspended in 2% bovine serum albumin in PBS (BSA-PBS) and then incubated for 30 min. Rabbit polyclonal anti-GFP antibodies (12) or rabbit polyclonal anti-CgeA antibodies (11), diluted 1:500, were added, and the mixture was incubated for 30 min at room temperature. Pellets were washed twice with PBS and incubated with a 1:1,000 dilution of Alexa Fluor 594-conjugated goat anti-rabbit IgG (Invitrogen) in BSA-PBS for 30 min at room temperature. Thereafter, pellets were washed twice with PBS and transferred onto microscope slides.
Fluorescence microscopy.
The fluorescence of GFP and Alexa Fluor 594-conjugated goat anti-rabbit IgG (Invitrogen) was observed by using an Olympus BX51 fluorescence microscope fitted with mirror cube units for green (U- MGFPHQ) and red (U-MWG2) (Olympus, Japan), respectively. A UPlanApo 100× oil Iris 3Ph objective lens and a U-TV1X-2 camera adapter were used. Images were captured by using a cooled charge-coupled-device camera (CoolSNAP ES/OL; Roper Scientific). Images were generated by RS Image Express processing software, version 4.5. Images of GFP fusion proteins were captured by fluorescence microscopy using exposure times of 0.5 to 1 s, and red fluorescence was imaged by using an exposure time of 0.2 s. The autofluorescence of spores under the experimental conditions used to capture GFP fluorescence and red immunofluorescence was obtained from exposure times of 3 s and 1 s, respectively.
RESULTS
Surface exposure of spore crust proteins.
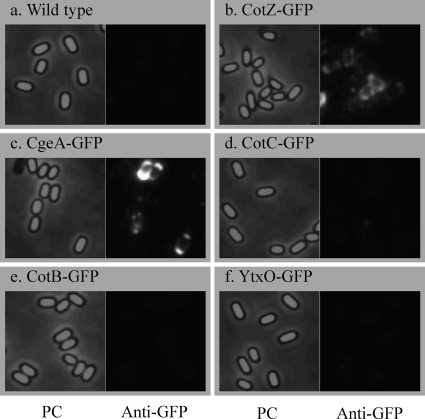
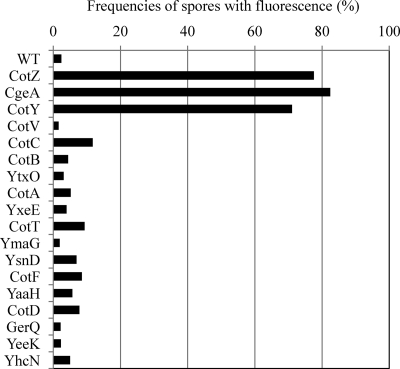
In order to obtain insights into the outermost structure of the B. subtilis spore, we first addressed whether the spore crust is indeed the outermost layer. The outermost layer is exposed on the surface of the spore and should thus be accessible to antibodies. We therefore carried out an immunofluorescence microscopy analysis using an anti-GFP antibody and B. subtilis strains expressing spore coat protein-GFP fusions described previously (Fig. 1) (4). In these strains, GFP was translationally fused to the 3′ end of the gene at the native locus so that the fusion gene was the only copy that was expressed. As expected, no fluorescent anti-GFP antibody signal was detected in wild-type spores, which do not express GFP (Fig. 2 a). In contrast, fluorescence surrounding spores was observed for CotZ-GFP and CgeA-GFP spores (Fig. 2b and c), indicating that anti-GFP antibodies bound specifically to GFP on the spores of these GFP fusion strains (see also Fig. S1 in the supplemental material for merged color images). However, strains expressing GFP fusions of other proteins, including four outer coat proteins, nine inner coat proteins, and one cortex protein (Fig. 1), showed no fluorescent anti-GFP antibody signal (Fig. 2 and Fig. S1). As shown in Fig. 3, a count of the number of spores with anti-GFP antibody immunofluorescence revealed that approximately 80% of spores of CotZ-GFP and CgeA-GFP strains were immunopositive (background in wild-type spores, <5%) (for CotY and CotV data, see below) (Fig. 3). In contrast, the majority of spores from strains containing GFP fusions of 14 proteins assigned to the outer coat, inner coat, and cortex exhibited no anti-GFP antibody fluorescence (Fig. 1 and 3 and Fig. S1). GFP fusion strains for all 16 proteins exhibited abundant GFP fluorescence (green) surrounding spores (4), indicating that all fusion proteins were expressed and located in spores; however, with the exception of CotZ-GFP and CgeA-GFP, none was accessible to antibodies. These results indicate that GFPs fused to CotZ and CgeA are exposed on the surface of spores, whereas GFPs fused to proteins assigned to the outer coat, inner coat, and cortex are not, consistent with the idea that the spore crust is the outermost layer of the B. subtilis spore.
Fig. 2.
Immunofluorescence microscopic detection of GFP-fused proteins in B. subtilis spores using anti-GFP antibodies. Strains 168 (a), COTZ8G (b), CGEA8G (c), COTC8G (d), COTB8G (e), and YTXO8G (f) were induced to sporulate by nutrient exhaustion and subjected to immunofluorescence microscopy with anti-GFP antibodies. Phase-contrast (PC) and immunofluorescence (Anti-GFP) images were captured by using exposure times of 1 s and 0.2 s, respectively.
Fig. 3.
Frequencies of spores stained with anti-GFP antibodies. The numbers of spores positive and negative for anti-GFP antibody staining in the experiments shown in Fig. 2 and Fig. S1 in the supplemental material were counted. The percentages of spores in the total population with immunofluorescence are shown. At least 100 spores were counted for each strain. Immunofluorescence images of all strains were captured by using an exposure time of 0.2 s. WT, wild type.
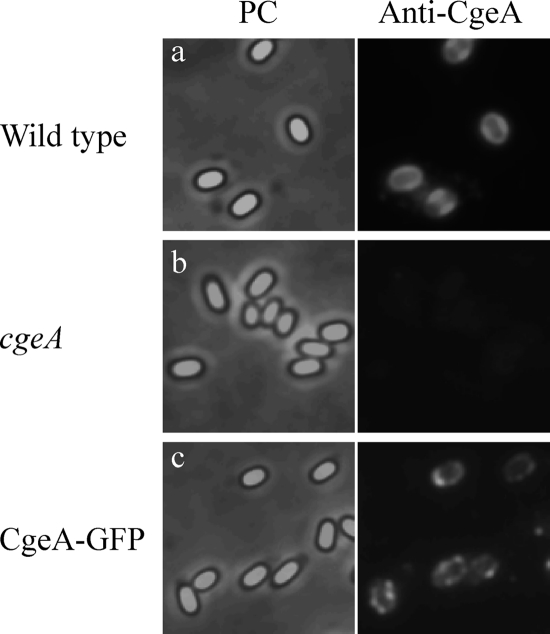
Although CotZ-GFP and CgeA-GFP were located on the surface of spores, it was conceivable that the surface exposure of these proteins was an artifact of the fusion to GFP. To rule out this possibility, we next performed immunofluorescence microscopy of wild-type spores using anti-CgeA antibodies. Anti-CgeA antibodies bound strongly to spores of both wild-type and CgeA-GFP strains but not to a cgeA mutant strain (Fig. 4; see also Fig. S2 in the supplemental material for merged color images). A count of the number of spores with anti-CgeA antibody immunofluorescence revealed that 3.17%, 72.2%, and 82.6% of spores of cgeA mutant, wild-type, and CgeA-GFP strains were immunopositive, respectively (at least 300 spores were counted). These results indicate that anti-CgeA antibodies bound specifically to the CgeA protein on the surface of the B. subtilis spore. Therefore, we conclude that the spore crust composed of CotZ and CgeA is the outermost structure of the B. subtilis spore and forms a layer that covers the spore coat.
Fig. 4.
Immunofluorescence microscopic analysis of B. subtilis spores using anti-CgeA antibodies. Strains 168 (a), MTB945 (b), and CGEA8G (c) were induced to sporulate by nutrient exhaustion and subjected to immunofluorescence microscopy using anti-CgeA antibodies. Phase-contrast (PC) and immunofluorescence (Anti-GFP) images were captured by using exposure times of 1 s and 0.2 s, respectively.
Proteins involved in spore crust formation.
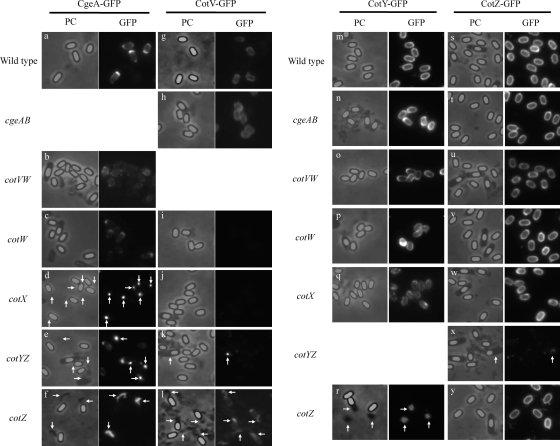
The localization of CotW-GFP to the spore crust and the assembly of CotW-GFP around the spore require cotXYZ genes, suggesting that CotX, CotY, and/or CotZ plays important roles in spore crust formation (16). cotVWXYZ is a cluster of genes transcribed at the late stage of sporulation in mother cells from the promoters PcotVWX, PcotX, and PcotYZ (27). Because CotW and CotZ are localized to the spore crust (4, 16), it is conceivable that the genes in the cluster are involved in spore crust formation. In order to identify genes important for spore crust formation, we monitored CgeA-GFP in the cotV, cotW, cotX, cotY, and cotZ mutant strains (Fig. 5). Because cotW and cotZ are cotranscribed with the upstream genes cotV and cotY, respectively, they are also disrupted in the cotV and cotY insertional mutant strains through a polar effect (Fig. 5). In the absence of the cotV and cotW genes, CgeA-GFP localized around the spore, indicating that CotV and CotW are dispensable for CgeA-GFP assembly on spores (Fig. 5a, b, and c). However, CgeA-GFP assembly around the spore was abolished by the cotX disruption; instead, CgeA-GFP was observed as spots adjacent to spores (Fig. 5d, arrows; see also Fig. S3 in the supplemental material for merged color images). This result indicates that CotX has an important role in spore crust formation. Because cotY and cotZ are paralogous genes and are cotranscribed from the promoter PcotYZ, it is conceivable that the disruption of cotY or cotZ individually might not affect the formation of the spore crust. Interestingly, the disruption of either the downstream gene cotZ or the upstream gene cotY abolished the assembly of CgeA-GFP around spores, indicating that CotZ plays a critical role in the formation of the spore crust and that CotY cannot fully compensate for the loss of CotZ function (Fig. 5e and f). Misassembled materials containing CgeA-GFP appeared as dark areas adjacent to spores by phase-contrast microscopy of the cotZ mutant strain but not cotY mutants (Fig. 5f, arrows; see also Fig. S3 in the supplemental material for merged color images), suggesting that spore crust-like material formed but failed to assemble around the spore of the cotZ mutant. The disruption of cgeA, cotV, cotW, cotX, cotY, or cotZ did not affect the localization of the other 14 proteins assigned to the outer coat, inner coat, or cortex (Fig. 1 and data not shown). These results indicate that among the tested genes, at least cotX and cotZ have important roles in spore crust formation but not in spore coat formation.
Fig. 5.
Localization of proteins in wild-type and mutant spores. Strains expressing cgeA-GFP (a to f), cotV-GFP (g to l), cotY-GFP (m to r), or cotZ-GFP (s to y) and those with insertional mutations in cgeA (h, n, and t), cotV (b, o, and u), cotW (c, i, p, and v), cotX (d, j, q, and w), cotY (e, k, and x), or cotZ (f, l, r, and y) were induced to sporulate by nutrient exhaustion. (Left) Phase-contrast images; (right) GFP fluorescence images. Arrows indicate misassembled materials.
Because CotW and CotZ are located in the spore crust (4, 16), we constructed strains expressing C-terminal GFP fusion proteins of CotV, CotX, or CotY. We introduced cotV-GFP, cotX-GFP, or cotY-GFP into the amyE locus (27). Therefore, the intact gene remains at the native locus in these strains. Although the fluorescence intensity of the CotX-GFP strain was below the level of detection (data not shown), CotV-GFP and CotY-GFP fluorescence located around the spores was clearly evident (Fig. 5g and m). CotV-GFP fluorescence was partially detached in cotZ mutant spores (Fig. 5l, arrows), indicating that normal CotV assembly requires CotZ. The localization of CotY-GFP around the spore was abolished in the cotZ mutant (Fig. 5r, arrows). Misassembled material that appeared dark by phase-contrast microscopy in cotZ mutant spores also contained CotV-GFP and CotY-GFP (Fig. 5l and r, arrows; see also Fig. S4 and S5 in the supplemental material for merged color images). We also carried out immunofluorescence microscopy of CotV-GFP and CotY-GFP strains using anti-GFP antibodies (Fig. 3 and Fig. S1). Although CotV-GFP was not exposed on the surface of spores, antibodies bound efficiently to spores of the CotY-GFP strain. Taken together, these data indicate that CotV-GFP partially depended on CotZ for its localization on the spore, whereas CotY-GFP, which is exposed on the surface of the spore, requires CotZ to surround the spore. These results further suggest that CotY and, possibly, CotV are additional components of the spore crust.
We next attempted to identify genes important for the assembly of CotV-GFP and CotY-GFP on spores. Because cgeA and cgeB are cotranscribed as an operon, an insertional mutation in cgeA affects cgeB expression through a polar effect. The disruption of cgeAB did not affect the localization of CotV-GFP or CotY-GFP (Fig. 5 h and n). CotV and CotW were dispensable for CotY-GFP assembly around the spore, but CotV-GFP failed to localize on spores of the cotW mutant, as indicated by the absence of obvious fluorescence (Fig. 5i, o, and p). Likewise, CotX was required for the localization of CotV-GFP, but not CotY-GFP, on spores (Fig. 5j and q). The mutation in cotY, which affects the expression of both cotY and cotZ, abolished CotV-GFP localization; fluorescence was observed as a spot in the mother cell (Fig. 5k, arrow). Misassembled material containing CgeA-GFP, CotV-GFP, and CotY-GFP, observed adjacent to free spores of the cotZ mutant (Fig. 5f, l, and r, arrows), was not found for the cotY mutant (Fig. 5e and k). Thus, CotY is presumably needed to form the misassembled spore crust-like material observed for cotZ mutant spores.
To investigate the importance of CotY, we introduced cotZ-GFP and the promoter region of cotZ upstream of cotY (without the cotY gene) into the amyE locus. CotZ-GFP expressed from the amyE locus surrounded spores in the absence of cgeAB, cotVW, cotW, and cotX, indicating that CotZ is able to localize on the spore independent of CgeA, CgeB, CotV, CotW, and CotX (Fig. 5s, t, u, v, and w; see also Fig. S6 in the supplemental material for merged color images). CotZ-GFP also localized appropriately on spores in the absence of the cotZ gene at the native allele (Fig. 5y). CgeA-RFP (red fluorescent protein) was detached from spores in the cotZ mutant strain, but this detachment was suppressed by the introduction of cotZ-GFP into the amyE locus (data not shown). These data indicate that CotZ-GFP is functional at least in terms of spore crust formation. Therefore, we could assess the importance of CotY using the strain possessing a cotYZ disruption and cotZ-GFP at the amyE locus (Fig. 5x). CotZ-GFP failed to localize around the spore, and only faint fluorescence in the mother cell was observed (Fig. 5x, arrow). These results indicate that CotY and CotZ depend on each other to localize on spores, and both proteins have critical roles in spore crust formation. Taken together, our results suggest that (i) CgeA, CotY, and CotZ are located in the spore crust; (ii) CotY and CotZ play critical roles in spore crust formation; (iii) CotW has an important role in the assembly of CotV-GFP; and (iv) CotX has an important role in the assembly of CgeA-GFP and CotV-GFP.
DISCUSSION
A loose-fitting, balloon-like exosporium surrounds the spore coats of some bacterial species, including B. anthracis, B. cereus, and some clostridia, whereas this feature is not observed for the B. subtilis spore, with the exception of a strain isolated from the human gastrointestinal tract (2, 3). The gene product of B. anthracis exsY, the B. subtilis cotY and cotZ ortholog, was identified in purified exosporium (21). Taken together with our finding that CotY and CotZ are components of the spore crust, this suggests the possibility that the spore crust of B. subtilis is a cognate of the B. anthracis exosporium layer. Some exosporium proteins of B. anthracis are glycosylated or associated with glycosylated material (23). Spore crust was stained by ruthenium red under an electron microscope, suggesting that the spore crust is composed of polysaccharide in addition to proteins (16). These observations suggest the possibility that the composition of the spore crust is similar to that of the exosporium. cgeA is part of the cgeABCDE cluster, whose transcription is controlled by σK and GerE, and spsA, the paralog of cgeD, encodes glycosyltransferase (22, 24). Since cgeABCDE and spsA, as well as cotVWXYZ, are expressed late during sporulation in the mother cell, these gene products might also be involved in spore crust formation.
The exosporium is dispensable for most laboratory tests of spore resistance; thus, its function is not well understood. It was suggested previously that the exosporium has a role in protecting spores of B. anthracis from macrophage-mediated killing (7). Also, in Bacillus thuringiensis, the exosporium may protect insecticidal crystal toxins (often enclosed within the structure) from the environment, thus enhancing pathogenicity (1). The function of the spore crust is also not yet clear, since cotZ mutant spores, which lack a spore crust, are resistant to heat and lysozyme treatment (data not shown). Zhang et al. reported previously that cotXYZ mutant spores have an unusual outer coat that is readily disrupted during fixation, embedding, or sectioning for electron microscopy (28), suggesting that the spore crust might contribute to the tight packing of the outer coat. cotXYZ mutant spores readily clump upon purification (28), and cotZ mutant spores also tended to clump during our experiments (data not shown). In addition, our attempts to identify proteins exposed on the surface of cotZ mutant spores by immunofluorescence microscopy failed, since antibodies bound nonspecifically to these spores (data not shown). These observations suggest that spores lacking a spore crust are abnormally adhesive; thus, in a natural environment, the spore crust might help to prevent clumping and allow spores to distribute.
Since there is no obvious phenotype for a crust-defective spore and the disruption of any one gene encoding a spore coat protein typically has little or no effect on spore resistance, morphology, or germination, it is difficult to know whether the GFP fusions used in this study are functional. Anti-CgeA antibody covered all around wild-type spores (Fig. 4a), whereas anti-GFP (Fig. 2c) and anti-CgeA (Fig. 4c) antibodies did not uniformly stain spores of the CgeA-GFP strain. These observations suggest the possibility that the GFP fusion affects CgeA localization or CgeA exposure on the surface of spores. Therefore, we cannot rule out the possibility that the proteins used in this study were affected by the fluorescent protein fusions. However, at least the localization and surface exposure of CgeA on spores were confirmed for the wild-type strain (Fig. 4a), and CotZ-GFP was functional in terms of forming the spore crust. Immunofluorescence microscopy of wild-type spores using anti-CotV, anti-CotY, and anti-CotZ antibodies or Western blot analysis of proteins extracted from purified spores of GFP fusion strains using specific antibodies for spore coat and spore crust proteins is desirable to check whether the GFP fusion of proteins used in this study is affecting the assembly of the coat or crust component.
Although cotY and cotZ are paralogous genes, their gene products could not compensate for each other in spore crust formation and instead depended on each other to assemble on spores (Fig. 5r and x). There are several possible reasons why these proteins required each other to assemble on spores. One possibility is that the proteins might have distinct, essential roles in spore crust formation and need the spore crust to localize on the spore. Alternatively, these proteins may form hetero-oligomers; therefore, both proteins are necessary to form a protein polymer in the spore crust. The latter possibility could be tested by using yeast two-hybrid experiments or pulldown assays, which would help reveal the interaction network of proteins in the spore crust.
The identification of spore surface proteins is becoming increasingly important for potential practical applications (9). It was proposed previously that CotA, CotB, CotC, and CotG are externally exposed on the surface of spores (6, 8, 15, 25), whereas CotA-GFP, CotB-GFP, and CotC-GFP were not efficiently exposed on spores in this study (Fig. 3). There are several possible reasons for the discrepancy. Purification procedures used in previous studies may have partially removed the crust and exposed the outer coat. GFP was fused to the C-terminal end of the coat and crust proteins in this study; however, GFP fused to the N-terminal end of these proteins might be exposed on spores more efficiently, as was the case in a previous study (5). The frequencies of spores with fluorescence signals in immunofluorescence microscopy using anti-GFP antibodies of CotA-GFP, CotB-GFP, and CotC-GFP strains were slightly higher than that of the wild type (Fig. 3). These results suggest that some outer coat proteins are present in the spore crust in small amounts that are at least detectable by methods used in previous studies (e.g., each antibody exhibits different sensitivities) (6, 8, 15, 25). However, our data demonstrated that the spore crust proteins CgeA, CotY, and CotZ were clearly more efficiently exposed externally on the spore surface than outer coat proteins (Fig. 3). Accordingly, CgeA, CotY, and CotZ would be preferred candidates for anchoring heterologous proteins to display on the surface of B. subtilis spores.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Science Research Promotion Fund and by a grant-in-aid for scientific research (C) (20580089) and a grant-in-aid for young scientists (B) (23780095) from the Japanese Society for the Promotion of Science.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Du C., Nickerson K. W. 1996. Bacillus thuringiensis HD-73 spores have surface-localized Cry1Ac toxin: physiological and pathogenic consequences. Appl. Environ. Microbiol. 62:3722–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Henriques A. O., Moran C. P., Jr 2007. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61:555–588 [DOI] [PubMed] [Google Scholar]
- 3. Hong H. A., et al. 2009. Bacillus subtilis isolated from the human gastrointestinal tract. Res. Microbiol. 160:134–143 [DOI] [PubMed] [Google Scholar]
- 4. Imamura D., Kuwana R., Takamatsu H., Watabe K. 2010. Localization of proteins to different layers and regions of Bacillus subtilis spore coats. J. Bacteriol. 192:518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Isticato R., Di Mase D. S., Mauriello E. M., De Felice M., Ricca E. 2007. Amino terminal fusion of heterologous proteins to CotC increases display efficiencies in the Bacillus subtilis spore system. Biotechniques 42:151–152,154, 156 [DOI] [PubMed] [Google Scholar]
- 6. Isticato R., et al. 2001. Surface display of recombinant proteins on Bacillus subtilis spores. J. Bacteriol. 183:6294–6301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kang T. J., et al. 2005. Murine macrophages kill the vegetative form of Bacillus anthracis. Infect. Immun. 73:7495–7501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim J. H., Lee C. S., Kim B. G. 2005. Spore-displayed streptavidin: a live diagnostic tool in biotechnology. Biochem. Biophys. Res. Commun. 331:210–214 [DOI] [PubMed] [Google Scholar]
- 9. Kim J., Schumann W. 2009. Display of proteins on Bacillus subtilis endospores. Cell. Mol. Life Sci. 66:3127–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kroos L. 2007. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. 41:13–39 [DOI] [PubMed] [Google Scholar]
- 11. Kuwana R., Ikejiri H., Yamamura S., Takamatsu H., Watabe K. 2004. Functional relationship between SpoVIF and GerE in gene regulation during sporulation of Bacillus subtilis. Microbiology 150:163–170 [DOI] [PubMed] [Google Scholar]
- 12. Kuwana R., Takamatsu H., Watabe K. 2007. Expression, localization and modification of YxeE spore coat protein in Bacillus subtilis. J. Biochem. 142:681–689 [DOI] [PubMed] [Google Scholar]
- 13. Kuwana R., et al. 2002. Proteomics characterization of novel spore proteins of Bacillus subtilis. Microbiology 148:3971–3982 [DOI] [PubMed] [Google Scholar]
- 14. Lai E.-M., et al. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J. Bacteriol. 185:1443–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mauriello E. M., et al. 2004. Display of heterologous antigens on the Bacillus subtilis spore coat using CotC as a fusion partner. Vaccine 22:1177–1187 [DOI] [PubMed] [Google Scholar]
- 16. McKenney P. T., et al. 2010. A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr. Biol. 20:934–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nicholson W. L., Munakata N., Horneck G., Melosh H. J., Setlow P. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nicholson W. L., Setlow P. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 19. Paidhungat M., Setlow P. 2001. Spore germination and outgrowth, p. 537–548 In Sonenshein A. L., Hoch J. A., Losick R. (ed.), Bacillus subtilis and its closest relatives. ASM Press, Washington, DC [Google Scholar]
- 20. Piggot P., Losick R. 2002. Sporulation genes and intercompartmental regulation, p. 483–517 In Sonenshein A. L., Hoch J. A., Losick R. (ed.), Bacillus subtilis and its closest relatives. ASM Press, Washington, DC [Google Scholar]
- 21. Redmond C., Baillie L. W., Hibbs S., Moir A. J., Moir A. 2004. Identification of proteins in the exosporium of Bacillus anthracis. Microbiology 150:355–363 [DOI] [PubMed] [Google Scholar]
- 22. Roels S., Losick R. 1995. Adjacent and divergently oriented operons under the control of the sporulation regulatory protein GerE in Bacillus subtilis. J. Bacteriol. 177:6263–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Steichen C., Chen P., Kearney J. F., Turnbough C. L., Jr 2003. Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J. Bacteriol. 185:1903–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stragier P., Losick R. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297–341 [DOI] [PubMed] [Google Scholar]
- 25. Tang J., et al. 2007. Atomic force microscopy imaging and single molecule recognition force spectroscopy of coat proteins on the surface of Bacillus subtilis spore. J. Mol. Recognit. 20:483–489 [DOI] [PubMed] [Google Scholar]
- 26. Warth A. D., Ohye D. F., Murrell W. G. 1963. The composition and structure of bacterial spores. J. Cell Biol. 16:579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang J., Ichikawa H., Halberg R., Kroos L., Aronson A. I. 1994. Regulation of the transcription of a cluster of Bacillus subtilis spore coat genes. J. Mol. Biol. 240:405–415 [DOI] [PubMed] [Google Scholar]
- 28. Zhang J., Fitz-James P. C., Aronson A. I. 1993. Cloning and characterization of a cluster of genes encoding polypeptides present in the insoluble fraction of the spore coat of Bacillus subtilis. J. Bacteriol. 175:3757–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.