Abstract
Highly conserved amino acid residues in the C subunits of the germinant receptors (GRs) of spores of Bacillus and Clostridium species have been identified by amino acid sequence comparisons, as well as structural predictions based on the high-resolution structure recently determined for the C subunit of the Bacillus subtilis GerB GR (GerBC). Single and multiple alanine substitutions were made in these conserved residues in three regions of GerBC, and the effects of these changes on B. subtilis spore germination via the GerB GR alone or in concert with the GerK GR, as well as on germination via the GerA GR, were determined. In addition, levels of the GerBC variants in the spore inner membrane were measured, and a number of the GerBC proteins were expressed and purified and their solubility and aggregation status were assessed. This work has done the following: (i) identified a number of conserved amino acids that are crucial for GerBC function in spore germination via the GerB GR and that do not alter spores' levels of these GerBC variants; (ii) identified other conserved GerBC amino acid essential for the proper folding of the protein and/or for assembly of GerBC in the spore inner membrane; (iii) shown that some alanine substitutions in GerBC significantly decrease the GerA GR's responsiveness to its germinant l-valine, consistent with there being some type of interaction between GerA and GerB GR subunits in spores; and (iv) found no alanine substitutions that specifically affect interaction between the GerB and GerK GRs.
INTRODUCTION
The ability to form spores is a remarkable property shared by many Bacillus species (23). Spores of these bacteria are formed in sporulation, a process triggered by starvation for one or more nutrients. These spores are metabolically dormant and extremely resistant to all manner of environmental stress factors (22, 23) and can remain in this dormant, resistant state for years. However, when conditions are favorable for growth, spores can rapidly return to life in the process of germination followed by outgrowth (11, 19, 21). A major signal that promotes germination is the presence of specific nutrients called germinants in the spore's environment. These nutrients are sensed by germinant receptors (GRs) located in the spore's inner membrane, and it is very likely that the binding of a nutrient germinant with its cognate GR triggers subsequent germination events.
Bacillus subtilis spores contain three major GRs, termed GerA, GerB, and GerK (22, 23). The GerA GR responds to l-alanine or l-valine, while the GerB and GerK GRs cooperate to respond to a mixture of l-asparagine, d-glucose, d-fructose, and K+ ions (termed AGFK). Each of these GRs is encoded by a tricistronic operon, and the three polypeptides encoded by each operon (termed A, B, and C) are essential for specific GR activities and most likely form a multisubunit complex. The A and B subunits of each GR are likely to be polytopic integral membrane proteins, while the C subunit is a peripheral membrane protein held in the membrane by a diacylglycerol anchor.
While the GRs are essential for spore germination in response to specific nutrients, little is known about their mechanism(s) of action. The A, B, and C subunits of different GRs both within and across species exhibit a high degree of amino acid sequence identity (20), but primary sequence-based BLAST and protein family searches indicate that GR subunits as a group share at most minimal sequence homology to other proteins and thus do not provide any notable conclusions about the function of these proteins. However, the sequences of GR B subunits show weak homology with a subfamily of the bacterial single-component membrane transporter APC (9, 12). Limited site-directed and random mutagenesis studies have suggested that the A and B subunits of GRs are somehow involved in GR-GR cooperativity and in recognizing specific germinants (1, 3–6, 13, 16). However, such analyses, as well as possible conclusions about the effects of such mutations, are greatly hampered by the lack of significant information on the structure of any GR A or B protein. To date, specific amino acid changes in C proteins that alter GR function other than to eliminate it have not yet been identified. Recently, a high-resolution structure of the C subunit of the B. subtilis GerB receptor, termed GerBC, was determined by X-ray crystallography (10). Importantly, both sequence alignment and secondary structure prediction analyses suggest that the overall structure of GerBC is conserved among C protein homologs in species ranging from the Bacillaceae to the Clostridiales. In this study, we use a structure-guided site-directed mutagenesis approach to identify and characterize a set of functionally important and evolutionally conserved amino acid residues in GerBC. Our structural and functional analyses have defined three regions of GerBC important for its function and have shown that substitution for residues in these regions has profound effects on germination via the GerB GR and maintenance of normal GerBC levels in spores and also have effects on l-valine germination via the GerA GR.
MATERIALS AND METHODS
BLAST search and sequence conservation analysis.
The B. subtilis GerBC and Clostridium perfringens GerKC amino acid sequences were used as query sequences for the initial BLAST search of spore-forming members of the Bacillales and Clostridiales orders on the NCBI genomic BLAST server (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). All the top hits (E value < e-45 and query coverage > 90%) were confirmed to have the recognition sequence for addition of a diacylglycerol anchor near the N termini of these proteins. The best hit for each species was then used as the query sequence to further search its own genome in order to find all possible homologs in the various species. A ClustalW alignment of GerBC homologs was performed using DNASTAR Lasergene, suite 8 (DNASTAR Inc., Madison, WI).
B. subtilis strains used and spore preparation.
Four isogenic B. subtilis strains were used in this work: (i) PS832 (wild type), a laboratory derivative of strain 168; (ii) FB8, PS832 with a single missense mutation that results in a G297S substitution in the gerBA gene that allows the spores of this strain to germinate with l-asparagine alone (16); (iii) FB10, PS832 with a single missense mutation that results in an F269I substitution in the gerBB gene that allows the spores of this strain to germinate with l-asparagine alone (16); and (iv) FB60, PS832 with a deletion of the great majority of the gerB coding region and its replacement with a chloramphenicol resistance (Cmr) (3 μg/ml) cassette (18).
Luria broth (LB) medium was used for vegetative growth of B. subtilis strains at 37°C (15). B. subtilis spores were prepared at 37°C on 2× Schaeffer's sporulation medium plates without antibiotics, and spores were harvested and purified as described previously (14, 15). All spores were stored at 4°C, protected from light, and were at least >98% free from growing or sporulating cells, germinated spores, and cell debris as determined by phase-contrast microscopy.
Site-directed mutagenesis.
Mutations were introduced into gerBC (1,125 bp) using an overlap PCR method (8). The PCR products were then cloned into a modified pBluescript II KS(−) vector carrying a large insert from the gerBC region, extending from 180 bp inside the upstream gerBB gene to 500 bp into the sequence downstream of gerBC (see Fig. S1 in the supplemental material) in order to provide homology for a double crossover with the chromosomal DNA. In addition, a chloramphenicol resistance cassette cloned from plasmid pDG364 (7) was introduced between the gerBC gene and its immediate downstream sequence. The mutation sites in the plasmids were confirmed by DNA sequencing. The mutagenized plasmids were used to transform PS832, FB8, or FB10 competent cells, with selection for Cmr (2). Transformants in which the mutagenized gerBC gene had integrated into the chromosome with replacement of the wild-type gerBC gene were identified by PCR, and the PCR-amplified regions were sequenced to confirm the presence of the mutation(s).
Spore germination assays.
Spores at an optical density at 600 nm (OD600) of 2.0 were heat activated at 75°C for 30 min and cooled on ice before germination was initiated. Spore germination was monitored by real-time measurement of the release of the spores' large depot (∼10% of spore dry weight) of dipicolinic acid (DPA) based on fluorescence emission of the Tb3+-DPA complex at 545 nm (excited at 270 nm) using a Gemini EM microplate fluorescence reader (Molecular Devices, Sunnyvale, CA) as described previously (24–26). Briefly, the assays were carried out in 96-well plates in a total volume of 200 μl/well consisting of 25 mM Na-HEPES (pH 7.4), 50 μM terbium chloride, 10 mM germinants (l-valine, l-asparagine, or the AGFK mixture with equal concentrations of all four components) or as indicated, and heat-activated spores (OD600 of 0.5) at 37°C for 4 h. Each reaction mixture was tested in quadruplicate, and the background detected at zero time was used as a blank. For all spores examined, the maximum number of relative fluorescence units (RFU) recorded for their l-valine-mediated germination was considered to be 100%, and the percentage of DPA released at each time point was calculated against this maximum RFU. The germination reactions of spores of all strains in the absence of the germinant were used as negative controls. The percentages of spores that had germinated by the end of germination incubations were also routinely checked by phase-contrast microscopy. These measurements invariably agreed with those determined from RFU values. Germination rates (percentage of DPA release min−1) were calculated as the slope of the linear segment of DPA release that followed the lag phase immediately after addition of germinants.
To determine kinetic parameters of spores' germination with l-valine, germination was carried out in the presence of different l-valine concentrations. For AGFK-mediated germination, assays were carried out at various concentrations of l-asparagine supplemented with 10 mM (each) d-glucose, d-fructose, and KCl (GFK). For germination of spores with the FB8 or FB10 genetic background, assays were carried out at various l-asparagine concentrations but without GFK. All curves generated by plotting the germinant concentration versus the germination rate were fitted using nonlinear regression to the Hill equation (n = 1) of the OriginPro 7.5 software program (OriginLab Corporation, Northampton, MA) to determine the maximum initial rate of germination (Vmax) and the germinant concentration needed to achieve 50% of the maximal rate (C50).
Expression and purification of GerBC proteins.
The various B. subtilis gerBC genes were cloned into a modified pET15b plasmid containing a tobacco etch virus (TEV) protease cleavage site between the N-terminal His6 tag and the target gene (10). The GerBC variants were expressed in Escherichia coli, and the resulting proteins contained an additional four N-terminal residues (Gly-Gly-Gly-Arg) prior to the native GerBC sequence. These proteins (residues 25 to 374 and lacking the signal sequence and signal for diacylglycerol addition) were purified by Ni2+-nitriloacetic acid affinity chromatography, followed by TEV protease cleavage to remove the His6 tag, and then by cation exchange and gel filtration (SD200; GE Healthcare) chromatography as described previously (10).
Preparation of spore inner membrane fractions.
Inner membranes of spores were prepared as described previously (17). Briefly, purified spores of various gerBC strains with the wild-type background (∼15 mg dry weight) were decoated in 5 ml of decoating buffer (0.1 M NaCl, 0.1 M NaOH, 1% sodium dodecyl sulfate [SDS], and 0.1 M dithiothreitol) at 70°C for 2 h and then washed 10 times with distilled water by centrifugation. Decoated spores (6 to 9 mg, dry weight) were resuspended in 0.5 ml of TEP buffer (50 mM Tris-HCl [pH 7.4], 5 mM EDTA, 1 mM phenylmethanesulfonylfluoride) containing 1 mg of lysozyme, 1 μg (each) of RNase A and DNase I, and 20 μg of MgCl2 and incubated at 37°C for 5 min. Treated spore suspensions were then incubated on ice for 20 min and sonicated with 100 mg of ∼ 0.1-mm glass beads 5 times with 15-s pulses to shear the spore inner membrane from the peptidoglycan cortex and the germ cell wall. The whole sample was then centrifuged at 16,000 × g at 4°C for 5 min to remove the integument fraction and intact spores. The supernatant fluid was saved, and the pellet fraction was washed with 0.5 ml of TEP buffer and centrifuged at 16,000 × g at 4°C for 5 min. The two supernatant fluids were pooled and then centrifuged for 1 h at 100,000 × g in a Beckman TLS 100.2 rotor, generating a soluble fraction and a pellet fraction; the latter contains the inner membrane fragments and was resuspended in 40 to 80 μl of TEP buffer containing 1% Triton X-100.
Production of GerBC antibody and Western blot analysis.
The wild-type B. subtilis GerBC protein (residues 25 to 374) was expressed and purified as described above. Purified protein (∼2 mg) was used to raise polyclonal antibodies in two New Zealand White female rabbits (Pocono Rabbit Farm and Laboratory, Canadensis, PA). Anti-GerBC antibodies were detected by Western blotting in a bleed 2 months after the initial injection, at which time the animals were exsanguinated. The specificity of the antiserum was examined by Western blot analysis using purified GerAC, GerBC, and GerKC proteins, and the GerBC antiserum did not detect GerAC or GerKC or either of these proteins in spores lacking the gerB operon (see Results; also data not shown). Western blot analysis of purified wild-type and GerBC protein variants (2 ng), as well as GerBC in spores' inner membrane, was as described previously (17). Inner membrane fractions from ∼1 mg (dry weight) of decoated spores were run on SDS-PAGE, and proteins were transferred to membranes for Western blot analysis. Two nanograms of purified wild-type GerBC and the inner membrane fraction from 1 mg (dry weight) of decoated wild-type spores were used as a positive control, and the inner membrane fraction from 1 mg (dry weight) of decoated FB62 spores (ΔgerB) was used as a negative control.
RESULTS
Structure-based mutational analysis of GerBC.
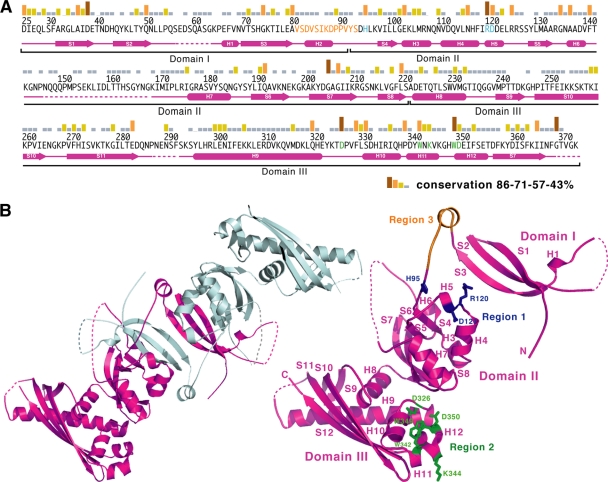
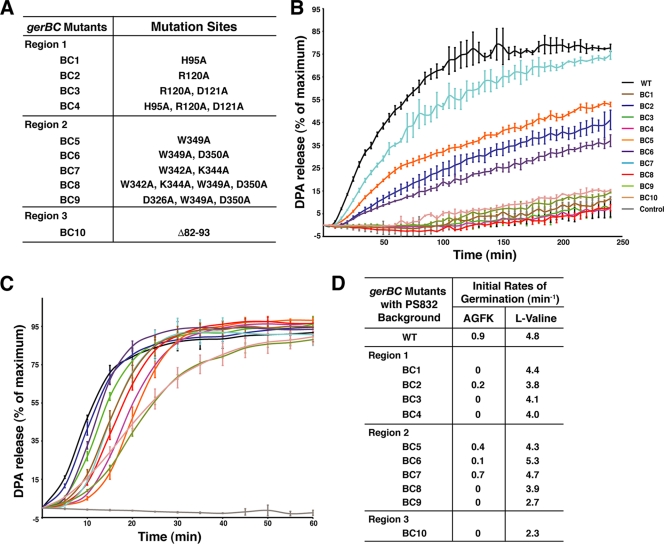
We previously identified two highly conserved surface areas (regions 1 and 2) in seven closely related GerBC homologs based on the crystal structure of B. subtilis GerBC (10) (Fig. 1). Region 1 (surface residues His95, Arg120, As121, Arg125, and Pro173) is located on the helical corner of domain II of GerBC, while region 2 (Asp326, Trp342, Lys344, Trp349, and Asp350) is located on the helical base of domain III; the two regions are about 50 Å from each other (Fig. 1B). To determine to what extent these two regions are conserved in all GerBC homologs, we performed a broad BLAST search using the B. subtilis GerBC protein as the query sequence against 21 completed spore-forming Bacillaceae genomes. A total of 109 gerBC homologs were identified, with 1 to 24 genes per species (see Table S1 in the supplemental material). In addition, a search using a characterized GerBC homolog from Clostridium perfringens (termed GerKC) identified 53 gerBC homologs in 23 annotated Clostridiales genomes, with 1 to 4 genes per species (see Table S1 in the supplemental material). All the GerBC homologs contain a likely site for diacylglycerol addition near their N termini and share ∼14 to 94% pairwise sequence identities with B. subtilis GerBC. The ClustalW alignment of 109 GerBC homologs reveals that most of the residues clustered in region 1 and 2 are also highly conserved among all C proteins (Fig. 1A), reinforcing the notion that they play important roles in C protein function. There are other conserved residues that are located throughout the GerBC sequences. However, in this study, we decided to first replace the conserved residues that are clustered together in an attempt to identify functionally important regions of the C proteins. To this end, we chose three residues (His95, Arg120, and Asp121) in region 1 and five residues (Asp326, Trp342, Lys344, Trp349, and Asp350) in region 2 for our initial structural and functional analyses, and nine gerBC mutant strains carrying single or multiple mutations giving rise to alanine substitutions have been constructed (Fig. 2 A). Mutation to alanine was selected because of its small size and because its methyl side chain cannot participate in hydrogen bonding, thus minimizing potential aggregation and/or misfolding problems in the resultant GerBC variants.
Fig. 1.
Sequence conservation between B. subtilis GerBC and its homologs in Bacillales species. (A) Amino acid sequence of the B. subtilis GerBC protein. Sequence conservation among the 109 GerBC homologs in the Bacillales is shown as a bar graph. Secondary-structure assignments of GerBC (10) are shown as magenta cylinders (α helices) and arrows (β strands), while disordered regions are shown as broken lines. The amino acid residues that are mutated in this work are shown in blue (region 1), green (region 2), and orange (region 3). (B) Overview of the interlocked dimeric B. subtilis GerBC structure (10) (left) and the location of the mutated regions in the GerBC monomer structure (right). The two monomers of GerBC are colored in magenta and grey. The residues that are mutated in regions 1 and 2 are shown as ball-and-stick models and are colored as in panel A. The residues deleted in region 3 are also colored in orange.
Fig. 2.
Effects of gerBC mutations on germination of spores with the wild-type background. (A) List of gerBC mutant strains grouped based on the location of the mutations (see Fig. 1). (B) Germination of wild-type and mutant spores with the wild-type (PS832) background in the presence of AGFK (10 mM [each] l-asparagine, d-glucose, d-fructose, and KCl). The percentage of DPA release of spores of each strain was normalized against the maximum RFU obtained for their l-valine-mediated germination as described in Materials and Methods. The germination curve of the spores with wild-type GerBC in the absence of germinants is shown and is used as the negative control. Data represent means ± SD for at least three independent measurements. (C) Germination of wild-type and mutant spores with the PS832 background in the presence of 10 mM l-valine. The percentages of DPA release were normalized as described in Materials and Methods, and the maximum number of RFU that each germination assay reached was set at 100%. Data are the means ± SD from at least three independent measurements. Curves are colored as in panel B. (D) Initial rates of germination of all spores. Rates were calculated as described in Materials and Methods. All values reported have been corrected for the rate of DPA release in the absence of germinants; this correction was always <0.1.
One of the more notable features of the GerBC structure is that domain I is positioned well away from domain II by an extended loop, and two GerBC molecules assemble into an interlocked dimer in the crystal via the interchange of domain I between two monomers (10) (Fig. 1B). Because GerBC is anchored in the spores' inner membrane via a diacylglycerol moiety in thioether linkage to the N-terminal cysteine residue immediately upstream of domain I, it is possible that the flexible linker between domains I and II might allow domains II and III to interact with other GR subunits and/or downstream effectors. To directly test this notion, we also generated a linker mutant, BC10, by deleting the entire 13-residue linker (Fig. 1B and 2A). As expected, all mutant strains with different genetic backgrounds (wild-type, FB8, and FB10) sporulated normally (data not shown), and their spores were prepared and purified as described in Materials and Methods.
Effects of gerBC mutations on AGFK-mediated spore germination.
Given the essential role of the GerB GR in spore germination with AGFK, we first examined the germination of spores of the various gerBC mutant strains with this germinant mixture. Perhaps not surprisingly, spores of most gerBC mutant strains exhibited severe defects in AGFK germination (Fig. 2B and D), and the various mutants can be divided roughly into three groups. Group 1 includes only BC7, which contains two amino acid substitutions (W342A and K344A) in region 2. BC7 spores had essentially the same phenotype as wild-type spores, since with 10 mM AGFK BC7 spores released only slightly less DPA and had only a slightly lower initial germination rate than wild-type spores (Fig. 2B and D). Group 2 mutants (BC2, BC5, and BC6) exhibited an extended lag phase following AGFK addition, released 37 to 53% of the DPA of wild-type spores, and had initial germination rates 2.5- to 10-fold lower than that of wild-type spores (Fig. 2D). In addition, the group 1 and 2 mutants (BC7, BC2, BC5, and BC6) did not germinate with GFK alone (data not shown), suggesting these mutations do not disrupt the sensing of l-asparagine. Group 3 mutants (BC1, BC3, BC4, BC8, BC9, and BC10) showed essentially complete loss of germination with AGFK (Fig. 2D). BC1, which contains an H95A substitution in region 1, is the only mutant in this category carrying a single mutation, indicating that His95 plays a crucial role in GerBC function. Multiple alanine substitutions in GerBC also generally exerted a significant influence on GerB GR function. For example, BC3 and BC4 have double and triple mutations in region 1, respectively, and BC8 and BC9 carry quadruple and triple mutations in region 2, respectively. Invariably, the multiple mutations examined had greater effects than individual mutations tested alone, suggesting that the multiple mutations had cumulative effects. Deletion of the flexible linker in GerBC (BC10) also eliminated AGFK-mediated germination. These data confirm that all three highly conserved regions in GerBC are extremely important for GerBC function and thus for GerB GR function in spore germination.
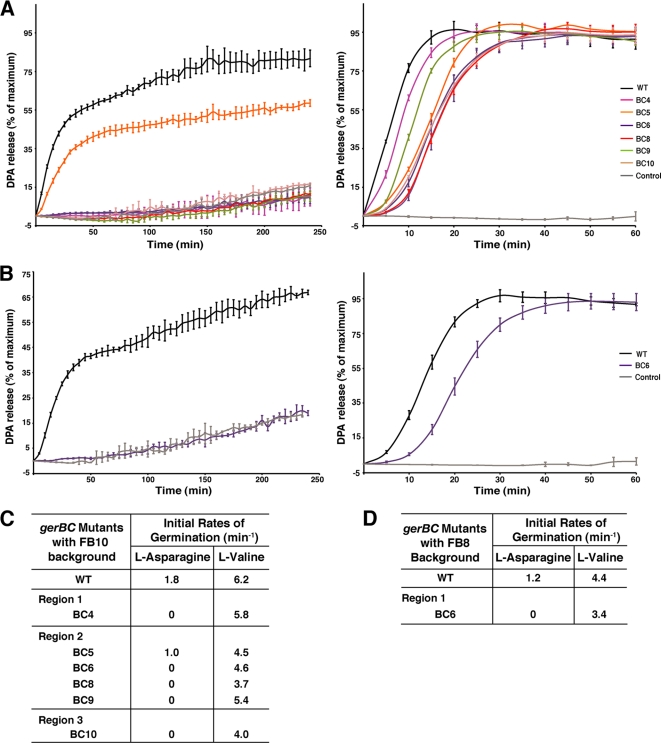
Effects of gerBC mutations in combination with gerBA or gerBB mutations that allow the GerB GR to function alone.
Since AGFK germination of spores with the wild-type background requires both the GerB and GerK GRs, it was possible that some gerBC mutations might somehow specifically interfere with the GerB GR's interaction with the GerK GR. We previously showed that any of several mutations in the gerB operon, termed gerB* mutations, allow the GerB GR to trigger germination with l-asparagine alone and independently of the GerK GR (16). Consequently, to examine the function of the GerB GR alone, a number of the gerBC mutants were introduced into B. subtilis strains containing single gerB* mutations in either gerBA (FB8) or gerBB (FB10), and the germination of the spores of these mutant strains with l-asparagine alone was examined (Fig. 3). The importance of the mutated GerBC residues in all three regions for GerB GR function alone was indicated by the complete loss of l-asparagine germination for spores of all gerBC mutants tested in the FB8 or FB10 background except for BC5, which showed 59% of maximum DPA release compared to wild-type spores (81%), and with a slightly reduced initial germination rate (Fig. 3).
Fig. 3.
Effects of gerBC mutations on germination of spores with the FB10 or FB8 background. (A) Germination of spores with the FB10 background in the presence of l-asparagine (left) or l-valine (right). The percentages of DPA release were normalized as described in Materials and Methods, and the maximum number of RFU obtained for each l-valine-mediated germination reaction was considered to be 100%. The germination curve of wild-type spores in the absence of germinant is shown and used as the negative control. Data represent means ± SD for at least three independent measurements. (B) Germination of spores with the FB8 background in the presence of l-asparagine (left) or l-valine (right). The percentages of DPA release were normalized as described above. The germination curve of spores with wild-type GerBC in the absence of germinants is shown. Data represent means ± SD for at least three independent measurements. (C) Initial rates of germination of spores with the FB10 background. Rates were calculated as described in the legend for Fig. 1D. (D) Initial rates of germination of spores with the FB8 background.
Kinetic analysis of gerBC mutant spore germination with l-asparagine.
As a further assessment of the effects of the gerBC mutations on GerBC and GerB GR function, we also examined the germination of a number of gerBC mutant spores with the wild-type or FB10 background with various l-asparagine concentrations and with or without a constant high level of GFK, respectively. The relative maximum initial germination rates (Vmax) and the l-asparagine concentrations needed to achieve 50% of Vmax (C50) for the wild-type and mutant spores were obtained from hyperbolic curves of germination rates with l-asparagine with or without GFK (Table 1). Based on these analyses, the mutant strains carrying the GerBC substitutions can be classified into two groups (Table 1). Spores of four gerBC mutants with the wild-type background (BC2, BC5, BC6, and BC7) and one with the FB10 background (BC5) had essentially the same C50 and Vmax values for l-asparagine germination as the wild-type or parental gerB* spores, respectively, consistent with their relatively good germination with AGFK (see Fig. 2 and 3). However, there was no l-asparagine germination of all other mutant spores, even at the highest concentration tested (15 mM). These results support the notion that multiple substitutions in highly conserved regions of GerBC, especially the triple and quadruple mutations, have a marked influence on GerBC and thus GerB GR functionality.
Table 1.
Kinetic analysis of germination of wild-type and mutant gerBC spores with l-valine, AGFK, or l-asparagine alonea
| gerBC strain | Kinetics of germination with: |
|||
|---|---|---|---|---|
|
l-Asparagineb |
l-Valine |
|||
| C50 (mM)c | Vmax (min−1) | C50 (mM) | Vmax (min−1) | |
| PS832 | ||||
| WT | 1.3 ± 0.4 | 117 ± 9 | 1.6 ± 0.2 | 113 ± 5 |
| BC1 | NAd | 0 | 2.8 ± 0.5 | 120 ± 7 |
| BC2 | 1.7 ± 0.6 | 98 ± 11 | 2.1 ± 0.2 | 115 ± 3 |
| BC3 | NA | 0 | 3.3 ± 0.4 | 121 ± 6 |
| BC4 | NA | 0 | 3.1 ± 0.5 | 125 ± 6 |
| BC5 | 0.7 ± 0.1 | 105 ± 3 | 4.2 ± 0.4 | 126 ± 4 |
| BC6 | 2.0 ± 0.5 | 104 ± 7 | 2.3 ± 0.2 | 114 ± 4 |
| BC7 | 1.1 ± 0.1 | 104 ± 2 | 5.6 ± 0.7 | 135 ± 7 |
| BC8 | NA | ∼0 | 1.5 ± 0.4 | 113 ± 8 |
| BC9 | NA | ∼0 | 2.1 ± 0.3 | 114 ± 4 |
| BC10 | NA | ∼0 | 2.7 ± 0.5 | 122 ± 8 |
| FB10 | ||||
| WT | 0.5 ± 0.04 | 107 ± 2 | 1.7 ± 0.1 | 111 ± 2 |
| BC4 | NA | 0 | 2.4 ± 0.3 | 118 ± 4 |
| BC5 | 0.3 ± 0.1 | 109 ± 3 | 1.7 ± 0.2 | 111 ± 4 |
| BC6 | NA | 0 | 4.0 ± 0.4 | 124 ± 5 |
| BC8 | NA | 0 | 4.8 ± 0.6 | 131 ± 7 |
| BC9 | NA | 0 | 2.5 ± 0.2 | 118 ± 3 |
| BC10 | NA | 0 | 2.6 ± 0.3 | 116 ± 4 |
Kinetic parameters of spore germination were determined as described in Materials and Methods and the legends to Fig. 2 and 3.
GFK at 10 mM each were present in the l-asparagine germination of spores with the PS832 (wild-type) background but not in the germination of spores with the FB10 background.
C50, germinant concentration that stimulates the half-maximal initial rate of germination.
NA, not applicable, since the germination rate was zero for these spores.
Effects of gerBC mutations on spore germination with l-valine.
While the results noted above indicated that the various gerBC mutations have major impacts on GerB GR functionality, it is possible that alterations in GerB GR function could also affect the function of other GRs, as has been suggested by previous work (1, 2). Consequently, we also determined the effects of the gerBC mutations on l-valine-triggered spore germination via the GerA GR. With 10 mM l-valine, spores of all gerBC mutant strains with the wild-type background released 100% of their DPA within 60 min (Fig. 2C), but some with an extended lag time following germinant addition, and >95% spores of all strains had germinated after 4 h (data not shown). To compare the l-valine germination of these various spores more quantitatively, we examined the initial rate of these spores' L-valine germination. As shown in Fig. 2D, of the spores of strains with point mutations in GerBC regions 1 and 2, only BC9 spores with three mutations in region 2 had a markedly reduced initial rate of germination with l-valine (∼50% of that of the wild-type spores) (Table 2). The l-valine germination rate of BC10 spores, in which GerBC lacks the linker between domains I and II, was also ∼50% lower (Fig. 2D; Table 2). These observations were unexpected since GerBC is assumed to play a role only in AGFK-mediated germination. Like spores of the gerBC mutants in the wild-type background, spores of the mutant strains with either the FB8 or FB10 background also germinated with an extended lag time and slightly slower than the parental spores with l-valine, although only at ≥60% of the rate of the parental spores, and released essentially all of their DPA within 60 min (Fig. 3; Table 2).
Table 2.
Germination properties of gerBC mutant spores and GerBC protein behaviora
| gerBC mutant | Relative rate of spore germinationb |
Level of GerBC in spore inner membranec | Protein solubility in E. coli (ΔΝ24)d | |||
|---|---|---|---|---|---|---|
| PS832 background |
FB10 (FB8) background |
|||||
| AGFK | l-Valine | l-Asparagine | l-Valine | |||
| WT | 100 | 100 | 100 (100) | 100 (100) | +++++ | Yes |
| Region 1 | ||||||
| BC1 | 0 | 92 | —e | — | +++++ | − |
| BC2 | 22 | 79 | — | — | +++++ | − |
| BC3 | 0 | 85 | — | — | +++++ | Yes |
| BC4 | 0 | 83 | 0 | 94 | + | Yes |
| Region 2 | ||||||
| BC5 | 44 | 90 | 56 | 73 | ++ | − |
| BC6 | 11 | 110 | 0 (0) | 74 (77) | ++ | No |
| BC7 | 78 | 98 | — | — | +++++ | − |
| BC8 | 0 | 81 | 0 | 60 | ND | − |
| BC9 | 0 | 56 | 0 | 87 | ND | No |
| Region 3 | ||||||
| BC10 | 0 | 48 | 0 | 65 | ND | Yes |
Rates of spore germination, levels of GerBC variants in spores' inner membrane, and folding of GerBC variants expressed in E. coli were determined as described in Materials and Methods.
The initial rates of the germination of the wild-type spores were set at 100.
Levels of GerBC in spores with the wild-type background were estimated from the Western blotting results (Fig. 4) as described in Methods. +++++, ≥80% of wild-type level; ++, 20 to 30% of wild-type level; +, 10 to 20% of wild-type level; ND, not detected by Western blotting.
Solubility of purified GerBC proteins (residues 25 to 374; ΔΝ24) was assessed by proteins' presence in the supernatant or pellet fractions from disrupted E. coli cells expressing the GerBC variants.
—, not determined.
Kinetic analysis of gerBC mutant spores in l-valine germination.
To further characterize the effects of gerBC mutations on spore germination via the GerA GR, the Vmax and C50 values for the wild-type and mutant spores germinating with l-valine were obtained as described above for l-asparagine-mediated germination (Table 1). Since the GerB GR is thought to be responsible only for AGFK germination, the responses of the various mutant spores to l-valine are striking. Although all gerBC mutant spores germinated well with 10 mM l-valine (>90% germination after 2 h), nine out of 10 mutant spores displayed increased C50 values for l-valine (1.4- to 3.6-fold) (Table 1). In contrast, the Vmax values deviated only slightly from that for wild-type spores, with the maximal difference being a 1.2-fold increase for BC7 spores with the wild-type background and BC8 spores with the FB10 background (Table 1). Of the region 1 mutants, the double (BC3) and triple (BC4) mutations, as well as the single mutation in H95 (BC1), decreased the sensitivity of the spores to l-valine by 1.8- to 2.5-fold, whereas the single mutation of R120 (BC2) induced less change in the C50 (Table 1).
The responses of the region 2 gerBC mutant spores to l-valine were more complicated, the most notable being the 3.6-and 2.7-fold reductions in responsiveness to l-valine with BC7 and BC5 spores, respectively. This observation was unexpected since both BC7 and BC5 spores had essentially the same germination rates with l-valine as wild-type spores and ≥44% of the wild-type rate of AGFK germination (Fig. 1D; Table 2). On the other hand, the C50 values for spores with the triple (BC9) and quadruple (BC8) mutations were very similar to that for wild-type spores. The region 3 mutant, BC10, exhibited a modest 1.8-fold increase in the C50 value compared to that of wild-type spores.
For the spores with the FB10 background, the influence of the gerBC mutations on l-valine germination was relatively mild, but a different trend from that of those with the wild-type background was observed. The BC8 and BC6 mutations, which had no effect and a modest effect on C50 with the wild-type background, respectively, exerted the largest effects on spores' sensitivity to l-valine (2.9- and 2.4-fold increases in C50, respectively), whereas the spores of the other three mutants tested displayed no or only slight increases in C50. Overall, while the effects of the GerBC mutations on l-valine germination were generally not huge, they were certainly significant and were also reproducible. Combined with effects of these mutants on the initial rate of germination in l-valine (Table 2), these kinetic data suggest the following: (i) that GerBC or the GerB GR somehow influences the GerA GR's response to l-valine, and (ii) that both regions 2 and 3 in GerBC are the most important in the effects of GerBC on the responsiveness of the GerA GR to l-valine.
Effects of gerBC mutations on levels of GerBC in spores' inner membrane.
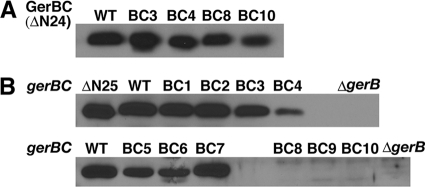
Since some severe germination defects were observed for the gerBC mutant spores, an obvious question is whether the alanine substitutions affect GerBC protein solubility and/or protein level in the spore's inner membrane. To this end, we expressed and purified several GerBC proteins that lack the N-terminal 24 amino acid residues but contain substitutions in region 1 (BC3 and BC4) or 2 (BC6 and BC8) or the deletion of the linker region (BC10). Analysis of the behavior of these various proteins showed that GerBCs with the BC3, BC4, and BC10 mutations were largely soluble after cell lysis (data not shown) and eluted at essentially the same retention volume as wild-type GerBC on gel filtration (see Fig. S2 in the supplemental material). In contrast, GerBCs with the region 2 BC6 and BC8 mutations were completely insoluble, suggesting that these proteins were likely misfolded and thus aggregated.
In order to compare GerBC levels in the spores' inner membrane, we raised rabbit polyclonal antiserum against purified GerBC. This antiserum was specific for GerBC, since it did not react with purified GerAC and GerKC in Western blots (data not shown), and it detected no protein at molecular masses of the latter proteins in the inner membrane fraction from spores lacking the GerB GR (see below). However, this antiserum readily detected purified GerBC variants that were either soluble (BC3, BC4, and BC10) or insoluble (BC8; purified with 6 M urea present), and wild-type as well as variant GerBC proteins all gave similar reactivities in Western blot analysis (Fig. 4A). As shown in Fig. 4B, Western blot analysis with the GerBC antiserum also detected a band of the predicted size for GerBC in the inner membrane fraction from wild-type spores, and this band was missing from the inner membrane fraction from ΔgerB spores, as expected (18). Note also that bands at the expected molecular masses of GerAC (42.4 kDa; GerBC is 42.5 kDa) and GerKC (45.8 kDa) were not detected by this antiserum in ΔgerB spores' inner membrane. The inner membrane fractions of spores with three of the GerBC variants with alanine substitutions in region 1 (BC1, BC2, and BC3) had essentially the same levels of GerBC as wild-type spores. In contrast, BC4 spores had only ∼10% of the GerBC of wild-type spores (Fig. 4B), indicating that even though the BC4 protein is soluble in purified form, this protein failed to assemble normally in the spores' inner membrane. For the region 2 mutants, BC7 was at wild-type levels in the spores' inner membrane, BC5 and BC6 were present at approximately one-third of the wild-type amount, and no GerBC was detected in the inner membrane of the BC8 and BC9 spores (Fig. 4B). This was also the case for the linker mutant BC10 (Fig. 4B). Interestingly, although BC6 was insoluble when expressed alone in our protein expression analysis, it appears that this protein assembled normally in the spores' inner membrane.
Fig. 4.
Reactivities of GerBC protein variants with anti-GerBC antiserum and levels of wild-type and variant GerBC proteins in the inner membrane of spores with the wild-type background. (A) Detection of purified wild-type and variant GerBC proteins (residues 25 to 374; 2 ng each) by Western blotting with the polyclonal anti-GerBC antiserum. (B) Detection of wild-type and variant GerBC proteins in spores' inner membrane by Western blotting with the polyclonal anti-GerBC antiserum. Inner membranes extracted from ∼1 mg dry decoated spores were run on SDS-PAGE, and purified GerBC protein (residues 25 to 374; ΔΝ24) was used as a positive control. The inner membrane fraction from spores of the gerB-null mutant (ΔgerB) (18) was used as a negative control. Note that GerBC in the inner membranes migrated slightly slower than purified GerBC, since the latter lacks the N-terminal 24 residues and cannot be diacylglycerylated.
DISCUSSION
GRs function at the heart of the spore germination process and are responsible for much of the specificity and diversity of nutrient-mediated spore germination. In this study, we have utilized available structural and bioinformatic information on the B. subtilis GerBC protein, and we have reported for the first time on the identification and characterization of key amino acid residues that play significant roles in the function of a GR C subunit in spore germination.
We initially identified three regions in GerBC where the amino acid sequence and/or most likely the structure is very well conserved in all GerBC homologs as initial targets for mutational and functional analyses. The sequence alignment of known GerBC homologs alone was not very instructive, because most of the highly conserved residues are located throughout the GerBC homologs' sequences. Since GerBC adopts an unusual configuration in which each of the three distinct domains possesses a unique fold (10), the structure by itself was also not very informative. However, the combination of the sequence conservation and structural analyses revealed that the majority of conserved residues are clustered into two distinct regions, 1 and 2, in GerBC. Hence, we posit that these residues might work together in mediating C protein function. In addition, region 3, which bridges domains I and II of GerBC, was also selected for analysis based on the notion that this linker region may be important for maintaining the assembly of and interactions between these domains from either the same C protein subunit or different ones. We must also emphasize that this unique feature of the GerBC structure appears to be conserved among all C proteins (10).
As was perhaps not unexpected, all mutants with individual or multiple alanine substitutions in these three regions had various effects on the spore germination with specific germinants. For region 1 mutants, the single mutation in BC1 (H95A), the double mutation in BC3 (R120A and D121A), and the combined triple mutation in BC4 markedly affected germination properties, including abolishing germination with AGFK and elevating C50 values with l-valine. Importantly, both BC1 and BC3 spores retained the same levels of the GerBC protein as wild-type spores, although BC4 spores had only low GerBC levels. Interestingly, recent work has shown that some alanine substitutions in the B. subtilis GerAA and GerAB proteins result in the loss of the GerAC protein from spores (6, 13). An obvious question, then, is whether reduction in GerBC levels in spores also has comparable effects on levels of GerBA and GerBB. Unfortunately, at present we do not know to what extent defects in GerBC influence the assembly of the entire GerB receptor because we do not have specific antibodies for GerBA or GerBB. However, since GerBC proteins bearing either the BC1 mutation or the double BC3 mutation likely maintain the stability and folding of the protein based on the solubility and the lack of notable aggregation of these two GerBC variants, these results suggest that residues H95 and R120/D121 are directly involved in mediating C protein function. Interestingly, the phenotypes and properties of the BC10 deletion mutant in region 3 mirror those of the region 1 mutants except that the BC10 mutation has a more severe effect on germination with l-valine and the BC10 GerBC protein was absent from the spore inner membrane. In the crystal structure of GerBC, region 3 forms a stem-like structure to position domain I well away from domain II, whereas the three residues in region 1 pack into a platform that is anchored tightly on the base of this stem (Fig. 1B). These observations lead us to postulate that both regions 1 and 3 are crucial for preserving GerBC domain structure, thereby anchoring GerBC to the membrane and promoting intra- or intersubunit interactions in GerB and perhaps other GRs (see below).
Our biochemical studies further suggest that of the five conserved residues in region 2, three core residues, Asp326, Trp349, and Asp350, seem to play an important role in GerB GR function. In the GerBC structure, the side chains of these three residues form a partially buried crevice stabilized by a hydrogen bond shared by the carboxylate group of Asp326 and the indole nitrogen of Trp349, while two other conserved residues, Trp342 and Lys344, lie on the rim of the crevice and are exposed to the solvent (Fig. 1B). We found that replacing the three core residues with alanine (BC9) abolished spore germination with AGFK, while the double mutant (W342A and K344A; BC7) spores exhibited near-normal germination. In addition, BC9 spores contained no GerBC protein in their inner membrane, while the GerBC level in BC7 spores was essentially the same as that in wild-type spores. Since the GerBC proteins carrying either double (BC6; Trp349 and Asp350) or triple (BC9) alanine mutations in the core residues in region 2 were insoluble and thus likely misfolded when overexpressed in E. coli, it is possible that these three residues may play an important role in defining the overall fold of GerBC. However, it also appears likely that interactions with the GerBA and/or GerBB proteins can stabilize GerBC protein variants, because the double mutant BC6 spores retained about one-third of the GerBC protein in their membrane, even though this GerBC protein was insoluble when expressed in E. coli.
In addition to recognizing a specific nutrient germinant or germinants, the GerB GR also must interact in some fashion with the GerK GR in order to trigger spore germination with AGFK (1). By characterizing the effects of gerBC mutations in combination with the gerB* mutations in gerBA or gerBB that allow the GerB* GR to trigger spore germination with l-asparagine alone, we hoped to identify specific mutations that eliminated AGFK germination but not germination with l-asparagine alone. However, none of the mutations of residues in region 1, 2, or 3 had this effect. While BC5 mutant spores with the wild-type background had an ∼2-fold-lower rate of spore germination with AGFK, spores with the BC5 mutation in the FB10 (gerBB*) background also had a similarly reduced rate of germination with l-asparagine alone. With one exception, the spores of the other mutants tested completely lost germination with l-asparagine in both the wild-type and gerB* backgrounds. The exception was spores of mutant BC6, which retained a low rate of germination with AGFK in the wild-type background but exhibited no germination with l-asparagine alone in the gerB* background; the reason for this effect is not clear. However, together all these data indicate that the gerBC mutations made in this work do not affect GerK-mediated spore germination.
One of the most surprising results in this work was that some alterations in the GerB receptor also had effects on l-valine mediated germination, even though l-valine germination is thought to be mediated by the GerA GR alone. The kinetic analyses of l-valine germination of gerBC mutant spores clearly showed that almost all the gerBC mutants, particularly those in region 2, had significantly increased C50 values for l-valine but relatively constant Vmax values, indicating that the mutated side chains influence the responsiveness of the GerA GR to l-valine rather than downstream signal transduction events. In the absence of structural and biochemical information about the overall organization and arrangement of different GRs in spores' inner membrane, it is difficult to explain why the integrity of the GerB receptor affects the activity of the GerA receptor. However, the recent observation that all GRs are colocalized in a small cluster in the spore inner membrane suggests that GRs may function in concert with others of different specificity (K. K. Griffiths, J. Zhang, A. E. Cowan, J. Yu and P. Setlow, unpublished data). Indeed, strong cooperativity between GRs has been observed when two or three GRs are activated with a combination of nutrients at suboptimal concentrations, such that the resulting germination rate is much higher than the rates triggered by individual nutrients acting on these same GRs (X. Yi and P. Setlow, unpublished data). However, pulldown analyses have so far failed to demonstrate any direct interactions between purified His6-tagged GerBC and either purified GerAC or a purified large soluble N-terminal domain of GerAA (Y. Li and B. Hao, unpublished data). Clearly, immediate future objectives are the following: (i) to produce anti-GerBA and anti-GerBB antisera, as well as specific antisera against the subunits of the GerA and GerK GRs, in order to measure and quantify the abundance of those proteins in spores; and (ii) to determine the interactions between the subunits of different GRs in the spore inner membrane.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Multi-University Research (MURI) award from the U.S. Department of Defense (to P.S. and B.H.).
We are grateful to Keren Griffiths, Xuan Yi, Barbara Setlow, Sonali Ghosh, and Jay Vyas for advice and help in construct design, spore germination, and bioinformatics analyses.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 17 June 2011.
REFERENCES
- 1. Atluri S., Ragkousi K., Cortezzo D. E., Setlow P. 2006. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J. Bacteriol. 188:28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cabrera-Martinez R. M., Tovar-Rojo F., Vepachedu V. R., Setlow P. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J. Bacteriol. 185:2457–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Christie G., Gotzke H., Lowe C. R. 2010. Identification of a receptor subunit and putative ligand-binding residues involved in the Bacillus megaterium QM B1551 spore germination response to glucose. J. Bacteriol. 192:4317–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Christie G., Lazarevska M., Lowe C. R. 2008. Functional consequences of amino acid substitutions to GerVB, a component of the Bacillus megaterium spore germinant receptor. J. Bacteriol. 190:2014–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Christie G., Lowe C. R. 2008. Amino acid substitutions in transmembrane domains 9 and 10 of GerVB that affect the germination properties of Bacillus megaterium spores. J. Bacteriol. 190:8009–8017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cooper G. R., Moir A. 2011. Amino scid residues in the GerAB protein important in the function and assembly of the alanine spore germination receptor of Bacillus subtilis 168. J. Bacteriol. 193:2261–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cutting S. M., Vander Horn P. B. 1990. Genetic analysis, p. 22–74In Harwood C. R., Cutting S. M. (ed.), Molecular biological methods for Bucillus. John Wiley and Sons, Chichester, United Kingdom [Google Scholar]
- 8. Heckman K. L., Pease L. R. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2:924–932 [DOI] [PubMed] [Google Scholar]
- 9. Jack D. L., Paulsen I. T., Saier M. H. 2000. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146(Pt. 8):1797–1814 [DOI] [PubMed] [Google Scholar]
- 10. Li Y., Setlow B., Setlow P., Hao B. 2010. Crystal structure of the GerBC component of a Bacillus subtilis spore germinant receptor. J. Mol. Biol. 402:8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moir A. 2006. How do spores germinate? J. Appl. Microbiol. 101:526–530 [DOI] [PubMed] [Google Scholar]
- 12. Moir A., Kemp E. H., Robinson C., Corfe B. M. 1994. The genetic analysis of bacterial spore germination. J. Appl. Bacteriol. 77:9S–16S [PubMed] [Google Scholar]
- 13. Mongkolthanaruk W., Cooper G. R., Mawer J. S., Allan R. N., Moir A. 2011. Effect of amino acid substitutions in the GerAA protein on the function of the alanine-responsive germinant receptor of Bacillus subtilis spores. J. Bacteriol. 193:2268–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nicholson W. L., Setlow P. 1990. Sporulation, germination and outgrowth, p. 391–450In Harwood C. R., Cutting S. M. (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, United Kingdom [Google Scholar]
- 15. Paidhungat M., Setlow B., Driks A., Setlow P. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J. Bacteriol. 182:5505–5512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paidhungat M., Setlow P. 1999. Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant D-alanine. J. Bacteriol. 181:3341–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paidhungat M., Setlow P. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J. Bacteriol. 183:3982–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paidhungat M., Setlow P. 2000. Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182:2513–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paidhungat M., Setlow P. 2002. Spore germination and outgrowth, p. 537–548In Sonenshein A. L., Hoch J. A., Losick R. (ed.), Bacillus subtilis and its relative: from genes to cells. American Society for Microbiology, Washington, DC [Google Scholar]
- 20. Paredes-Sabja D., Setlow P., Sarker M. R. 2011. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 19:85–94 [DOI] [PubMed] [Google Scholar]
- 21. Setlow P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550–556 [DOI] [PubMed] [Google Scholar]
- 22. Setlow P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101:514–525 [DOI] [PubMed] [Google Scholar]
- 23. Setlow P., Johnson E. A. 2007. Spores and their significance, p. 35–67In Doyle M. P., Beuchat L. R. (ed.), Food microbiology: fundamentals and frontiers, 3rd ed. ASM Press, Washington, DC [Google Scholar]
- 24. Yang W. W., Ponce A. 2009. Rapid endospore viability assay of Clostridium sporogenes spores. Int. J. Food. Microbiol. 133:213–216 [DOI] [PubMed] [Google Scholar]
- 25. Yi X., Setlow P. 2010. Studies of the commitment step in the germination of spores of Bacillus species. J. Bacteriol. 192:3424–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yung P. T., Ponce A. 2008. Fast sterility assessment by germinable-endospore biodosimetry. Appl. Environ. Microbiol. 74:7669–7674 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.