Abstract
In many bacteria, including Staphylococcus aureus, progression from the logarithmic to the stationary phase is accompanied by conversion of most of bacterial membrane phosphatidylglycerol (PG) to cardiolipin (CL). Phagocytosis of S. aureus by human neutrophils also induces the conversion of most bacterial PG to CL. The genome of all sequenced strains of S. aureus contains two open reading frames (ORFs) predicting proteins encoded with ∼30% identity to the principal CL synthase (cls) of Escherichia coli. To test whether these ORFs (cls1 and cls2) encode cardiolipin synthases and contribute to CL accumulation in S. aureus, we expressed these proteins in a cls strain of E. coli and created isogenic single and double mutants in S. aureus. The expression of either Cls1 or Cls2 in CL-deficient E. coli resulted in CL accumulation in the stationary phase. S. aureus with deletion of both cls1 and cls2 showed no detectable CL accumulation in the stationary phase or after phagocytosis by neutrophils. CL accumulation in the stationary phase was due almost solely to Cls2, whereas both Cls1 and Cls2 contributed to CL accumulation following phagocytosis by neutrophils. Differences in the relative contributions of Cls1 and Cls2 to CL accumulation under different triggering conditions suggest differences in the role and regulation of these two enzymes.
INTRODUCTION
Staphylococcus aureus infections are associated with significant morbidity and mortality in community and health care settings. These infections are associated with a tremendous economic burden (14.5 billion dollars in the United States alone in 2003 [20]) and are further complicated by the increasing frequency of antibiotic resistance. The burden of disease from methicillin-resistant Staphylococcus aureus (MRSA) infections is growing and leading to worse patient outcomes. Approximately 100,000 invasive MRSA infections occurred in 2005 in the United States alone (20), leading to ca. 19,000 deaths, even more than that due to HIV infection.
Interactions of S. aureus with human hosts do not always lead to disease. In many individuals, S. aureus is part of the normal microbial flora (e.g., in the anterior nares), usually without significant pathological consequences (25). However, when the skin or a mucosal barrier is breached, colonizing S. aureus can penetrate across epithelial barriers to cause an array of local and more invasive infections, including skin and soft tissue infections, necrotizing pneumonia, osteomyelitis, sepsis, and infectious endocarditis (25). Once inside the host, invading S. aureus is challenged by professional phagocytes and by locally and systemically mobilized secreted antimicrobial compounds, including the group IIA phospholipase A2 (7, 11, 51, 52). S. aureus can adapt to the hostile host conditions by initiating expression of a variety of secreted and cell envelope-associated virulence factors (4, 30) and by adopting new physical and metabolic states (e.g., biofilms, endocardial vegetations, and small colony variants) that are associated with long-term bacterial persistence (9, 27, 28, 32, 36). What specific metabolic changes are needed for long-term survival are largely unknown.
Among the adaptive changes in the chemical composition of the cell envelope that have been well documented is a change in membrane phospholipid composition. In actively growing S. aureus, the predominant phospholipid species is phosphatidylglycerol (PG), whereas in stationary growth phase the predominant phospholipid is cardiolipin (CL) with a corresponding decline in PG content (23, 44). Similar bacterial phospholipid changes have been observed under conditions of osmotic stress, energy deprivation, or after phagocytosis by polymorphonuclear leukocytes (PMN) (7, 21, 24, 43, 44).
The synthesis of CL in bacteria is catalyzed by cardiolipin synthase, which catalyzes condensation of two PG molecules to yield CL and glycerol (43, 45). Cardiolipin synthase belongs to the phospholipase D class of enzymes and shares the same key residues forming the catalytic site of these enzymes (40). In E. coli and many other bacterial species, multiple genes are present that encode proteins with cardiolipin synthase activity and/or share homology with the principal E. coli cardiolipin synthase. In many cases, the relative role and meaning of the apparent multiplicity of cardiolipin synthases are not clear. For example, in E. coli, the principal cardiolipin synthase is encoded by the cls gene (Gen Bank accession number U15986) and is responsible for nearly all CL production in vivo. The activity of a second cardiolipin synthase, encoded by the f413 gene, has to date only been demonstrated in vitro (13, 31, 45). Similarly, three cls homologues have been identified in Bacillus subtilis, but to date only one of these genes has been shown to encode active cardiolipin synthase under the test conditions explored (12, 24, 37). Cardiolipin synthase activity has also been demonstrated in partially purified S. aureus membranes (43), but the genes encoding active CL synthases and the proteins responsible for CL accumulation in S. aureus were not known when the present study was initiated.
Accordingly, we have sought here to identify the gene(s) in S. aureus encoding active cardiolipin synthases and to establish the role of these enzymes in bacterial CL accumulation in the stationary phase and in bacteria ingested by neutrophils. We identified two genes encoding cardiolipin synthase that together accounted for essentially all net CL formation under these conditions and provide evidence suggesting that the regulation of these two enzymes is different.
MATERIALS AND METHODS
Materials.
Clinical grade dextran T500 (molecular mass, 500,000 Da) was purchased from Pharmacosmos A/S (Holbaek, Denmark). Sterile endotoxin-free H2O and 0.9% sterile endotoxin-free sodium chloride for patient use were purchased from Baxter Healthcare (Deerfield, IL). Ficoll-Paque Plus was purchased from GE Healthcare (formally Amersham Biosciences), Piscataway, NJ. HEPES, Hanks balanced salt solution (HBSS), and Dulbecco phosphate-buffered saline, with or without divalent cations, were obtained from Mediatech Cellgro (Manassas, VA). Chloroform, methanol, glacial acetic acid, and the HEMA-3 staining kit were purchased from Fisher Scientific. Endotoxin-free human serum albumin (25% [wt/vol]) was obtained from Talecris Biotherapeutics (Research Triangle Park, NC). Tryptic soy broth (TSB) and Bacto agar were purchased from BD Biosciences (Bedford, MA). [1-14C]palmitic acid (57 mCi/mmol) was purchased from Amersham Life Science, bovine serum albumin (BSA) was purchased from Boehringer Mannheim (Indianapolis, IN), and HP-TLC (high-performance thin-layer chromatography) plates were purchased from Merck (Germany). Purified lipid standards for PG and CL—1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] and tetraoleoyl cardiolipin, respectively—were purchased from Avanti-Polar Lipids (Alabaster, AL). DNA and RNA isolations were carried out using Qiagen kits (Valencia, CA). Lysostaphin was purchased from Sigma-Aldrich (St. Louis, MO). Mechanical disruption of S. aureus was carried out in FastPrep24 using blue matrix tubes (MP Biomedicals, Solon, OH). Taq polymerase, deoxynucleoside triphosphates, BP clonase, and RNasin were purchased from Invitrogen (Carlsbad, CA), and Phusion DNA polymerase was obtained from Finnzymes (Woburn, MA). Avian myeloblastosis virus reverse transcriptase (AMV-RT) and random hexamers were obtained from Roche (Palo Alto, CA). Sybr green PCR master mix was obtained from Applied Biosystems (Foster City, CA).
Construction of a cls deletion.
Markerless chromosomal deletions of cls1 and/or cls2 were constructed in both S. aureus SA113 (18) and LAC (USA300-0114 MRSA [50]) using the pKOR1 plasmid for allelic replacement as described by Bae and Schneewind (1). Briefly, flanking regions of cls1 were amplified by using the primer pairs TK161/TK162 and TK163/TK164 and of cls2 by using the primer pairs TK149/TK151 and TK150/TK152 (Table 1). The two PCR products were joined by overlap extension (49) and moved into pKOR1 using the Gateway cloning system (Invitrogen). The resulting plasmid was transformed into SA113 or LAC, and the gene deletions were constructed as described previously (26). Gene deletions were confirmed by PCR using primers annealing to flanking regions upstream and downstream of the deleted gene. To complement the deleted genes (cls1 or cls2) in trans, each structural gene, together with the upstream DNA region containing putative promoter sites, was amplified by PCR using the primer pairs TK167/TK168 for cls1 and TK169/TK170 for cls2. The resulting PCR product was digested by KpnI and SacI and ligated into the E. coli/S. aureus shuttle vector, pUC19/pC194. The resulting plasmids, pTKOcls1 and pTKOcls2 (Table 2), were introduced into S. aureus SA113 and LAC by electroporation (39).
Table 1.
Primers used in this study
| Primer | Sequence (5′-3′)a | Description | Source or reference |
|---|---|---|---|
| TK161 | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCCCAACTGGTTGCGAATCTAACA | Forward primer for amplification of DNA region upstream of cls1 with attB1 sites | This study |
| TK162 | CCTCTATAATCGAGACTCCTTACAAATAAAAGTCTTTTCTCCTATAAAGAAAGGCAC | Reverse primer with overlapping ends for amplification of DNA region upstream of cls1 | This study |
| TK163 | GTGCCTTTCTTTATAGGAGAAAAGACTTTTATTTGTAAGGAGTCTCGATTATAGAG | Forward primer with overlapping ends for amplification of DNA region downstream of cls1 | This study |
| TK164 | GGGGACCACTTTGTACAAGAAAGCTGGGTCGGAAACCACAATCCACCTAATACTGC | Reverse primer with attB2 for amplification of DNA region downstream of cls1 | This study |
| TK149 | GGGGACAAGTTTGTACAAAAAAGCAGGCTATGGTGGTAGTGCGCTATGGAGTA | Forward primer with attB1 sites for amplification of DNA region upstream of cls2 | This study |
| TK151 | AAAGTTACACTCCTCATATTTCTATTTGAAACCTCC CATCGAAAATC | Reverse primer with overlapping ends for amplification of DNA region upstream of cls2 | This study |
| TK150 | GATTTTCGATGGGAGGTTTCAAATAGAAATATGAGGAGTGTAACTTT | Forward primer with overlapping ends for amplification of DNA region downstream of cls2 | This study |
| TK152 | GGGGACCACTTTGTACAAGAAAGCTGGGTGCTCAGCATCAGCACTTGGTTTGT | Reverse primer with attB2 ends for amplification of DNA region downstream of cls2 | This study |
| TK167 | ACGTAGGTACCTTTAATAAGATAAACCAATTTCAAAACTAGTTCG | Forward primer for amplification of cls1 with the KpnI restriction site | This study |
| TK168 | AGTAGAGCTCGGCAAAATAAAAAGAGCCTCTATAATCGAGACTCCTTACAAATAAAT | Reverse primer for amplification of cls1 with the SacI restriction site | This study |
| TK169 | ACGTAGGTACCAGATGCTGGCAAAGTATTAATAATGTCTAA | Forward primer for amplification of cls2 with the KpnI restriction site | This study |
| TK170 | TATGTGAGCTCGCATTAAAGTTACACTCCTCATATTTCTAT | Reverse primer for amplification of cls2 with the SacI restriction site | This study |
| TK180 | CACAGAGCAGCAAAAGCGTTAG | Forward primer for cls1 real-time RT-PCR | This study |
| TK181 | CGCTTGCGAATTCCAGTCTAA | Reverse primer for cls1 real-time RT-PCR | This study |
| TK182 | GTGGAACAATTGGCGTTCAA | Forward primer for cls2 real-time RT-PCR | This study |
| TK183 | CCCATTCTTCGTCAGGACCA | Reverse primer for cls2 real-time RT-PCR | This study |
| TK23 | AGCCGACCTGAGAGGGTGA | Forward primer for 16S rRNA real-time RT-PCR | 22 |
| TK24 | TCTGGACCGTGTCTCAGTTCC | Reverse primer for 16S rRNA real-time RT-PCR | 22 |
| 1317 F | GGGGATTTAAATGGATCCAATTAAACTTAAAAC | Forward primer for cloning of Cls1 in pRB473 with the BamHI restriction site | This study |
| 1317 R | AATCGAGGATCCTTACAAATAAATTATAAAATTGGCG | Reverse primer for cloning of Cls1 in pRB473 with the BamHI restriction site | This study |
| 2088 F | CAGTCATAGCATGCACTCCTTCATTTACATTCC | Forward primer for cloning of Cls2 in pRB473 with the SphI restriction site | This study |
| 2088 R | TTGATGAATAGGATCCATATATTTTTTGGC | Reverse primer for cloning of Cls2 in pRB473 with the BamHI restriction site | This study |
Italics are used to indicate the attB1 and attB2 sites, boldfacing indicates overlapping ends, and underlining indicates restriction sites.
Table 2.
Plasmids used in this study
| Plasmid | Description | Restriction sites | Source or reference |
|---|---|---|---|
| pDB81 | pUC19/pC194 E. coli and S. aureus shuttle vector | D. J. Bartels | |
| pKORI | Original plasmid knockout vector | AttB/AttP | 1 |
| pKORcls1 | cls1 knockout vector | This study | |
| pKORcls2 | cls2 knockout vector | This study | |
| pTKOcls1 | pDB81 with cls1 (SAOUHSC_1310) including 460 bp upstream of translation start codon | KpnI/SacI | This study |
| pTKOcls2 | pDB81 with cls2 (SAOUHSC_2323) including 738 bp upstream of translation start codon | KpnI/SacI | This study |
| pRB474 | E. coli/S. aureus shuttle vector | 35 | |
| pRB474cls1 | pRB473 with cls1 (SAV1317) | BamHI/BamHI | This study |
| pRB474cls2 | pRB473 with cls2 (SAV2088) | BamHI/SphI | This study |
| pCM29 | sGFP expression under constitutive sarAP1 promoter | 33 |
Radiolabeling of S. aureus lipids during bacterial growth.
Bacterial lipids were radiolabeled during growth in subculture as previously described (8). Briefly, bacteria from overnight cultures in TSB were diluted ∼200-fold in fresh TSB supplemented with 1 μCi of [1-14C]palmitic acid/ml and 0.01% BSA and subcultured at 37°C to either mid-logarithmic phase (3 h) or to stationary phase (24 h). At the time of harvesting of the bacteria, BSA was added to a final concentration of 0.5% (wt/vol), and the bacteria were washed to remove any remaining bacterium-associated radiolabeled free fatty acid. The washed bacteria were resuspended to the desired concentration in the desired medium and used promptly, as described below.
Neutrophil isolation.
PMN from normal healthy volunteers were purified from peripheral blood as described earlier (7). Donor consent was obtained from each individual according to the protocol approved by the Institutional Review Board for Human Subjects at the University of Iowa. Isolated PMN were resuspended in sterile endotoxin-free HBSS without divalent cations, counted, and diluted to a final density of no greater than 2 × 107/ml. The PMN purity was ≥95%, as judged by microscopic examination of the cell suspension after staining with the HEMA-3 kit. PMN were kept at room temperature for no longer than 1 h before use in subsequent assays.
Phagocytosis.
S. aureus (5 × 107/ml) and PMN (1 × 107/ml) were separately incubated in HBSS containing divalent cations, 1% human serum albumin (HSA), and 10% pooled human serum at 37°C for 10 (PMN) or 20 (bacteria) min and then mixed 1:1 (vol/vol) to achieve a multiplicity of infection (MOI) of 5. Incubations were carried out in 5-ml polypropylene round-bottom tubes with shaking in a 37°C water bath. After a 30-min coincubation of bacteria and PMN, the samples were gently spun (500 × g) for 5 min in a Savant high-speed tabletop centrifuge to pellet the PMN and associated bacteria. The supernatant containing extracellular bacteria was discarded. The cell pellet was resuspended in the original volume of HBSS containing divalent cations supplemented with 1% HSA and 10% pooled human serum and further incubated at 37°C for specified intervals before assays of bacterial RNA and lipids, as described below. For all experiments with PMN, mid-logarithmic-phase S. aureus was used that was harvested after 3 h of metabolic labeling with [1-14C]palmitic acid. Bacterial uptake by PMN was quantified both by measurement of bacterial radioactivity recovered in PMN and in extracellular medium after sedimentation of PMN (see above) and by light microscopic evaluation of stained smears of resuspended PMN as previously described (6, 15) and confirmed in selected samples by fluorescence microscopy using green fluorescent protein (GFP)-expressing bacteria (6).
Assay of bleaching of bacterial cytosolic GFP in S. aureus ingested by neutrophils.
S. aureus (both wild type [wt] and cls1 cls2 mutant) carrying pCM29 encoding super bright sGFP (33) was grown and incubated with PMN as described above except that extracellular bacteria were removed 10 min after incubation with neutrophils by centrifugation at 500 × g for 5 min. At subsequent time points, the fluorescence of S. aureus-PMN suspensions was measured by flow cytometry as described previously (33). PMN without S. aureus were analyzed and gated as an GFP-S. aureus-negative control. PMN outside this gate were considered GFP positive. For each sample the mean fluorescent index (MFI) was calculated by multiplying the geometric mean of the fluorescence by the percentage of cells in the sample that were GFP positive (i.e., PMN containing ingested and still fluorescent S. aureus) At each time point, the MFI was calculated as a percentage of the MFI of the sample recovered and analyzed immediately after removal of the extracellular bacteria.
Characterization of 14C-labeled bacterial (S. aureus) phospholipids after incubation of [1-14C]palmitate-labeled S. aureus with neutrophils.
After incubation of [1-14C]palmitate-labeled S. aureus with neutrophils, cell suspensions containing 1-14C-labeled S. aureus and neutrophils were extracted via a modified Bligh-Dyer method (13, 32). Briefly, whole-cell suspensions (1 volume) were mixed with 6 volumes of chloroform-methanol (1:2 [vol/vol]) and stored at 4°C until analyzed. Samples were spun at 1,900 × g for 10 min, and the supernatant was transferred to a fresh glass tube. Then, 3 volumes of 50 mM potassium chloride and 2 volumes of chloroform were added to the recovered supernatant. The samples were vortexed and spun for 10 min at 1,900 × g to separate the upper water-methanol phase from the lower chloroform phase. The chloroform phase was removed and saved. The remaining aqueous phase was mixed with 4 volumes of chloroform, vortexed, and spun for 10 min at 1,900 × g. The recovered chloroform phase was combined with the previously recovered chloroform phase, dried under a stream of nitrogen, resuspended in chloroform-methanol (2:1 [vol/vol]), and applied to an HP-TLC plate. Lipids were resolved either in one dimension using either (i) chloroform-methanol-acetic acid (65:25:10 [vol/vol/vol]) or (ii) chloroform-methanol-petroleum ether-acetic acid (40:20:30:10 [vol/vol/vol/vol]) (38) or in two dimensions, using chloroform-methanol-acetic acid-water (first dimension, 65:25:4:1 [vol/vol/vol/vol]; second dimension, 80:18:12:5 [vol/vol/vol/vol] [21]). The resolved 14C-labeled lipids were visualized by exposure of the HP-TLC plates to tritium storage phosphor screens and analyzed by using an Amersham Typhoon 9410 variable mode imager (Amersham Biosciences). Quantification was done by using ImageQuant software from Molecular Dynamics. Bacterial [14C]PG and [14C]CL were identified by their comigration with purified PG and CL standards. The recovery of total 14C-labeled lipids (in counts per minute [cpm]) was similar from each of the bacterial strains studied, both before and after incubation with neutrophils.
Characterization of unlabeled and [14C]palmitate-labeled S. aureus phospholipids after growth in TSB.
Lipids of S. aureus harvested after growth in TSB were extracted and resolved by each of the various TLC systems described above. For visualization of total lipids from either ([1-14C]palmitate-labeled or unlabeled bacteria, TLC plates were sprayed with 3% cupric acetate-8% phosphoric acid and charred on a hot plate (120°C). Approximately 10 to 20 μg of total phospholipid was applied, corresponding to 1 × 109 to 2 × 109 bacteria. Images and quantification of charred samples were performed using the Kodak GelLogic system and accompanying software.
Real-time reverse transcription-PCR (RT-PCR).
RNA was isolated from S. aureus (5 × 107) after incubation of bacteria alone, with 10% serum, or incubation of the serum-pretreated (opsonized) bacteria plus PMN as described above. After sedimentation of the bacteria associated with PMN or bacteria incubated alone (with or without preopsonization by 10% serum), the cell pellets were resuspended in 700 μl of RLT (Qiagen) and transferred to 2-ml tubes filled with 0.1-mm silica matrix (MP Bio/Blue matrix). The bacteria were disrupted by using a FastPrep24 homogenizer for 45 s at the highest speed. The tubes were then cooled and spun at 14,000 rpm at 4°C. The recovered supernatant (ca. 350 μl) was transferred to a new tube and spun again to remove any remaining particulate matter before isolating RNA according to the RNeasy protocol (Qiagen). Contaminating DNA in the samples was removed by Turbo DNase according to the manufacturer's protocol (Ambion). Total RNA was reverse transcribed by using AMV-RT (Roche), 20 ng random hexamers (Roche)/μl, and 20 U of RNase inhibitor (Roche) at 40°C for 1 h. The PCR included Sybr green (Applied Biosystems), 200 nM concentrations of each forward and reverse primer (Table 1), and cDNA equivalent to 4 ng of input RNA. The reactions were performed with an ABI Prism 7000 detection system (Applied Biosystems). In each set of analyses, standard curves for each primer pair used were analyzed, together with the experimental samples, to facilitate quantification of the amplicons.
Isolation and analysis of E. coli lipids.
E. coli was grown in LB medium to stationary phase (18 to 20 h). After the various bacterial cultures were adjusted to the same optical density, bacterial polar lipids were extracted with chloroform-methanol by the Bligh-Dyer procedure (2), vacuum dried, and dissolved in chloroform-methanol (2:1 [vol/vol]). The concentrated polar lipid extracts were spotted onto HP-TLC plates (Merck, Darmstadt, Germany) by using a Linomat 5 sample application unit (Camag, Berlin, Germany) and developed with chloroform-methanol-acetic acid (65:25:10 [vol/vol/vol]). Lipid species containing phosphate groups were stained with molybdenum blue spray (Sigma).
RESULTS
Cloning and activity of S. aureus cls1 and cls2 in E. coli.
The gene encoding the principal cardiolipin synthase in E. coli was used to screen the genome of S. aureus NCTC 8325 and other strains available at NCBI for homologous open reading frames (ORFs). Two ORFs were identified in S. aureus NCTC 8325, annotated SAOUHC_01310 and SAOUHC_2323, that we named cls1 and cls2, respectively. Each of the homologs is present in each of the S. aureus strains in the genomic database (e.g., in S. aureus MU50 genes SAV1317 and SAV2088 encode for the cls1 and cls2 homologs, respectively). The putative S. aureus cardiolipin synthases encoded by cls1 and cls2 display ∼30% amino acid identity (∼50% similarity) to the E. coli cardiolipin synthase. S. aureus Cls1 and Cls2 share 53% amino acid identity and are predicted by HMMTOP and TopPred 2 software (47, 48; http://mobyle.pasteur.fr/cgi-bin/portal.py?form=toppred) (Fig. 1B) to have two similar transmembrane-spanning helices (TMH, residues 15 to 38 and residues 48 to 70; Fig. 1B). Cls1 and Cls2 also have two predicted phospholipase D domains, a characteristic of all prokaryotic cardiolipin synthases (45), residing within a region of the protein (residues 227 to 252 and residues 403 to 434) predicted to be outside the cell membrane (Fig. 1B). To investigate whether cls1 and cls2 actually encode cardiolipin synthases, we cloned individually S. aureus cls1 and cls2, together with their putative promoter regions, into an E. coli plasmid expression vector pRB474 and used these plasmids to transform the CL-deficient E. coli SD9 (42) that lacks both cls (cardiolipin synthase) and pss-1 (phosphatidylserine synthase) (42). After overnight growth in LB to stationary phase, bacterial lipids were extracted and analyzed as described in Materials and Methods. As expected, CL was present in the wt strain (E. coli DH5α) but not detectable in E. coli SD9pRB474 vector control (Fig. 2). In contrast, significant amounts of CL were present in transformants of E. coli SD9 containing plasmids encoding cls1 (pRB474cls1) or cls2 (pRB474cls2). These findings strongly suggest that both ORFs in S. aureus—cls1 and cls2—encode for functional cardiolipin synthases (Fig. 2).
Fig. 1.
(A) Evidence of chromosomal deletion of cls1 and/or cls2 in S. aureus cls1, cls2, and cls1 cls2 mutants by PCR. Primer pairs annealing upstream and downstream of the cls1 (TK161/164; upper line) and cls2 (TK149/152; lower line) structural genes were used to confirm cls1 and cls2 chromosomal deletions. The source of genomic DNA used as a template is depicted above each PCR product. wt, wild type; cls1, S. aureus cls1; cls2, S. aureus cls2; cls1/2, S. aureus cls1 cls2. (B) Amino acid sequence alignment of S. aureus Cls1 and Cls2 using CLUSTAL W. Asterisks denote sites of amino acid identity; colons, sites of amino acid similarity. Boxed regions correspond to predicted transmembrane helices (TMH-1 and TMH-2), using HMMTOP 2.0 and TopPredII secondary structure prediction software, and conserved domains of the active site [HxK(x)4D(x)6GSxN, shaded amino acids] of the phospholipase D (PLD) family, identified by NCBI Blastp.
Fig. 2.
S. aureus cls1 and cls2 induce accumulation of CL in CL-deficient E. coli (SD9). S. aureus cls1 and cls2 were cloned into the pRB474 vector and expressed in CL-deficient E. coli SD9. After overnight incubation, the bacteria were harvested, and lipids were extracted and analyzed as described in Materials and Methods. The migration of PE (phosphatidylethanolamine), PG, and CL in the strains tested, as determined by comigration of purified standards (only CL is shown), is indicated by arrows. The results shown are representative of three independent experiments.
CL accumulation in log and stationary phases of wt and mutant (cls) S. aureus.
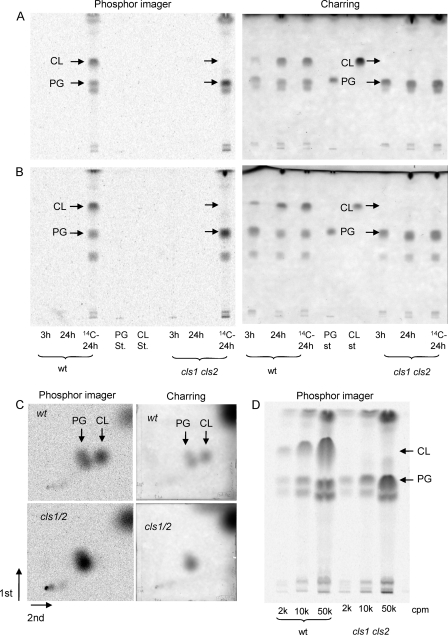
To test the role of cls1 and/or cls2 in accumulation of CL by S. aureus, we constructed isogenic mutants in both S. aureus SA113 and the USA300 LAC strain in which cls1 and/or cls2 were disrupted. Successful disruption of these genes was confirmed by PCR, using primers annealing outside the coding regions of these genes in wt and mutant SA113 (Fig. 1A). The same analyses and observations were made with wt and mutant strains of S. aureus LAC (data not shown). We initially compared the lipid content of wt and double mutant (cls1 cls2) S. aureus harvested after growth in TSB with or without [1-14C]palmitate at 37°C for 3 or 24 h. Growth in TSB of the wt and each of the mutant strains was essentially the same (not shown). Bacteria harvested at 3 h were in mid-log phase, whereas bacteria harvested at 24 h were in stationary phase. Bulk and metabolically ([1-14C]palmitate)-labeled phospholipids were resolved in three different TLC systems (Fig. 3) and visualized by charring or phosphorimage analysis (Fig. 3). The identities of bacterial PG and CL were confirmed by their comigration with purified PG and CL standards in each of these TLC systems (Fig. 3). Using the same TLC-based separation (solvent system chloroform-methanol-acetic acid, 65:25:10 [vol/vol/vol]), Tsai et al. (46) have confirmed the identity of CL by mass spectrometry. As shown in Fig. 3, the relative levels of CL to PG were substantially increased in wt S. aureus harvested from stationary (versus the log phases) (i.e., harvested after 24 h versus after 3 h of culture in TSB), a finding consistent with previous observations made in S. aureus and a number of other bacterial species (3, 10, 14, 15, 19, 24, 29, 34). In contrast, no accumulation of CL was observed in the cls1 cls2 double mutants of S. aureus LAC (Fig. 3A to C) and SA113 (Fig. 4) after 24 h of culture in TSB even when a 25-fold-higher load of lipids was applied (Fig. 3D).
Fig. 3.
Lipid profiles of S. aureus LAC (wt and cls1 cls2 double mutant) as determined by TLC. (A) S. aureus strains were grown with or without [1-14C]palmitic acid in TSB. Bacteria were harvested at 3 h (exponential phase; panels A and B) or at 24 h (stationary phase; panels A to D) of growth in TSB at 37°C, and lipids were extracted and resolved by one-dimensional (A, B, and D) or two-dimensional (C) TLC using chloroform-methanol-acetic acid (65:25:10 [vol/vol/vol]) (A), chloroform-methanol-petroleum ether-acetic acid (40:20:30:10 [vol/vol/vol/vol]) (B), or chloroform-methanol-acetic acid-water (65:25:4:1 [vol/vol/vol/vol] in the first dimension and 80:18:12:5 [vol/vol/vol/vol] in the second dimension) (C). (D) Increasing amounts (cpm) of lipids extracted from S. aureus LAC wt and the cls1 cls2 double-mutant strains after 24 h of growth in TSB were evaluated by TLC and resolved in one dimension using the chloroform-methanol-acetic acid (65:25:10 [vol/vol/vol]) solvent system. Purified lipid standards (PG and CL) were used to identify bacterial PG and CL in each of the solvent systems used. Total lipids were stained with 3% cupric acetate and 8% phosphoric acid and visualized after charring (A to C; right panels). 14C-labeled lipids were visualized after exposure to phosphorimager screen (A to C, left panels, and D). The arrows denote the migration of PG and CL. The chromatograms are representative of three or more experiments with strains derived from both S. aureus SA113 and LAC.
Fig. 4.
Ratio of [14C]CL to [14C]PG in wt and mutant S. aureus after subculture in TSB for 3 h (log) and 24 h (stat) or 2 h after phagocytosis of log-phase bacteria by PMN. [1-14C]palmitate-labeled PG and CL were resolved by TLC in two dimensions (chloroform-methanol-acetic acid-water (65:25:4:1 [vol/vol/vol/vol] in the first dimension and 80:18:12:5 [vol/vol/vol/vol] in the second dimension) and quantified by image analysis as described in Materials and Methods. The results shown represent the means ± the standard errors of the mean (SEM) of three to eight individual experiments with wt and mutant S. aureus SA113. Similar results were observed with wt and mutant S. aureus LAC. pcls1 and pcls2 refer to pTKOcls1 and pTKOcls2, respectively.
The above findings demonstrate a critical role of Cls1 and Cls2 in CL accumulation by stationary phase S. aureus. To define the relative roles of Cls1 and Cls2 in this lipid adaptive response, the lipid profiles of isogenic single-mutant (cls1 or cls2) S. aureus and double-mutant (cls1 cls2) bacteria complemented with either cls1 or cls2 were compared to wt and cls1 cls2 S. aureus (Fig. 4). The close similarity in the profiles of the bulk and metabolically [1-14C]palmitate-labeled lipids (Fig. 3) permitted the use of the metabolically labeled bacteria for more sensitive quantification of bacterial PG and CL. Both cls1 and cls2 S. aureus showed decreased CL levels in log and stationary phases, but the effects of disruption of cls2 were much more dramatic, with virtually no increased CL accumulation in stationary phase observed in cls2 bacteria (Fig. 4). Complementation studies in the double-mutant (cls1 cls2) strain by introduction of wt cls1 or cls2 in a high-copy-number plasmid (pTKcls1 and pTKcls2, respectively) confirmed that the absence of CL accumulation in cls1 cls2 S. aureus was due to disruption of these two genes and that, in both log and stationary phases, Cls2 contributed much more to bacterial CL accumulation than did Cls1 (Fig. 4).
Bacterial CL accumulation after phagocytosis of wt and cls mutant S. aureus by human neutrophils.
Accumulation of CL in S. aureus ingested by human neutrophils (PMN) has been previously observed (7, 16). To investigate the roles of Cls1 and Cls2 in accumulation of CL after phagocytosis of S. aureus by PMN, we analyzed the 14C-labeled lipids of [1-14C]palmitate-prelabeled bacteria before and after ingestion of S. aureus by PMN. The use of metabolically prelabeled bacteria permitted the unambiguous detection and quantification of bacterial PG and CL in the presence of the other unlabeled, nonbacterial (serum, PMN) lipids. Preopsonization of the wt and each of the mutant S. aureus strains with 10% pooled human serum had no appreciable effect on labeled bacterial lipids of the mid-log bacteria; PG remained the predominant labeled bacterial phospholipid and exceeded the levels of labeled CL by at least 4-fold (data not shown). Bacterial uptake by PMN of all of the strains tested was essentially the same: at an MOI of 5 cocci/PMN, >90% of each of the strains were ingested within 30 min, as judged both by assay of PMN-associated radiolabeled bacteria and by light microscopy (7, 16, 41; data not shown). As shown previously (7, 16), phagocytosis of wt S. aureus was followed by a roughly 10-fold increase in the relative levels of bacterial CL to PG (Fig. 4, open versus black bars). Changes in bacterial CL/PG were triggered within 30 min of phagocytosis but took 1 to 2 h for maximum changes to be manifest. This lipid change was modestly reduced in cls1 S. aureus and reduced to a greater but incomplete extent in cls2 S. aureus. Disruption of both cls1 and cls2 was needed to eliminate the PG to CL conversion during and after phagocytosis of S. aureus by PMN (Fig. 4). Consistent with that finding, complementation of cls1 cls2 with either cls1 or cls2 permitted substantial or virtually complete restoration of CL accumulation in S. aureus ingested by PMN, similar to that seen in cls2 and cls1 S. aureus, respectively (Fig. 4).
Fate of wt and cls double mutant S. aureus within PMN.
The absence of detectable conversion of bacterial PG to CL in cls1 cls2 S. aureus ingested by PMN made it possible to examine how this lipid adaptation influences the fate of S. aureus within human PMN. For this purpose, we measured the loss of fluorescence in bacteria expressing GFP after phagocytosis by PMN (33). The chlorination of GFP by the HOCl produced by the neutrophil results in ablation of the fluorescence of the chromophore (6). Whereas the overall extent of bleaching of cytoplasmic GFP is closely similar to the percentage of ingested S. aureus killed within PMN (33, 41), the kinetics of bleaching are somewhat slower than that of bacterial killing and therefore bleaching of bacterial cytosolic GFP might be more affected by the progressive accumulation of CL in wt S. aureus. However, as shown in Fig. 5, the rate and extent of GFP bleaching was essentially the same in ingested wt and cls1 cls2 S. aureus, as judged by flow cytometry. The uptake peaked at 10 min and was the same for wt and the double mutant (data not shown). The rate and extent of killing of wt and cls1 cls2 S. aureus by PMN were also the same (data not shown). These findings indicate that the conversion by S. aureus of PG to CL did not protect against neutrophil killing and myeloperoxidase-hydrogen peroxide-chloride-dependent bacterial alterations. However, induced CL accumulation could impact the subsequent fate of the remaining intracellular viable S. aureus and undigested bacterial phospholipids (see the Discussion).
Fig. 5.
Bleaching of sGFP-S. aureus after phagocytosis by PMN. wt and the double cls1 cls2 knockout S. aureus USA300 LAC (left) and S. aureus SA113 (right) expressing sGFP were opsonized and incubated with PMN for 10 min before extracellular bacteria were removed (time zero). The mean fluorescence index (MFI) of gated PMN population harboring sGFP expressing S. aureus was assessed at the indicated times by flow cytometry as described in Materials and Methods. The MFIs of samples at each time point are expressed as the percent MFI of the time zero sample for each strain. The data shown represent means ± the SEM of three (USA300 LAC) or five (SA113) independent experiments.
Differences in the regulation of cls1 and cls2 mRNA levels.
The findings described above strongly suggest a greater role of Cls2 versus Cls1 in bacterial CL accumulation both in the stationary phase and after phagocytosis by human PMN. This difference could reflect differences in cls1 versus cls2 gene expression, differences in Cls1 versus Cls2 activity, or both. To determine whether there were differences in levels of cls1 versus cls2 mRNA under these experimental conditions, we measured cls1 and cls2 mRNA in wt S. aureus (SA113) by quantitative RT-PCR after culture in TSB and before and after phagocytosis by PMN (Fig. 6). In early logarithmic growth (3 h), cls2 mRNA levels were ∼2.5-fold higher than that of cls1 (not shown). cls2 mRNA levels declined ca. 2- to 4-fold during the next several hours of culture in TSB but rebounded to the earlier (3 h) levels after 24 h culture in TSB. In contrast, the levels of cls1 mRNA remained virtually constant throughout the 24-h culture period in TSB (Fig. 6A). cls1 versus cls2 mRNA levels were also affected differently by phagocytosis by PMN: higher cls1 mRNA levels were temporarily induced shortly after phagocytosis, whereas cls2 mRNA levels declined ∼5-fold (Fig. 6B). These findings indicate different regulation of the synthesis and/or turnover of cls1 versus cls2 mRNA.
Fig. 6.
Levels of cls1 and cls2 mRNA in wt S. aureus SA113 during growth in TSB (A) and after phagocytosis by PMNs (B). Overnight cultures of S. aureus were diluted 100-fold in fresh TSB medium and, after the indicated times in subculture, the levels of cls1 and cls2 mRNA were measured by real-time RT-PCR as described in Materials and Methods, expressed relative to 16S rRNA, and normalized to the 3-h sample. S. aureus harvested after 3 h of subculture in TSB were opsonized by incubation in the presence of 10% human serum for 20 min at 37°C and then mixed with PMN at MOI of 5. Samples for the assay of cls1 and cls2 mRNA were taken before (0 min) and 30 and 120 min after phagocytosis by PMN. The data shown represent means ± the SEM of triplicate determinations from three independent experiments. *, Significant differences as determined by Student t test (P < 0.05).
DISCUSSION
Our findings demonstrate the existence of two ORFs in S. aureus, denoted cls1 and cls2, that each encode proteins that contribute to CL accumulation in S. aureus. We have demonstrated roles for both cls1 and cls2 in CL accumulation by: (i) creation of isogenic single and double mutants, which showed reduced CL accumulation in logarithmic and stationary phases of growth or after phagocytosis by PMN, and (ii) expression of cls1 or cls2 in either CL-deficient E. coli or a cls1 cls2 double mutant of S. aureus with restoration of bacterial CL accumulation under those conditions (Fig. 2 to 4). The presence of the same ORFs in each of the S. aureus strains for which complete genomic information is available indicates that these proteins are conserved elements of S. aureus biochemical machinery.
Both in the stationary phase and after ingestion by neutrophils, Cls2 displayed a much more prominent role than Cls1 in CL accumulation by S. aureus (Fig. 3 and 4). This was apparent both by the extent of induction of CL accumulation in single mutants (cls1 versus cls2) and the extent of restoration of CL accumulation by cls1 versus cls2 complementation of the cls1 cls2 double mutant (Fig. 4). These findings confirm and complement the recent findings of Tsai et al. (46) that were published while our study was in review. Of note, in each of the conditions in which Cls1, as well as Cls2, contributed to CL accumulation (i.e., during phagocytosis [Fig. 4] or during culture of the bacteria in high concentrations [15 to 25%] of NaCl, at pH 5, or under anaerobiosis [46]), there was a greater overall accumulation of CL and higher CL/PG ratio, suggesting that Cls1 may be needed and activated when more extensive conversion of PG to CL is required. In the case of long exposures of S. aureus to high salinity, activation of either Cls1 or Cls2 appeared to have adaptive value since either single mutant but not the cls1 cls2 double mutant showed near-normal long-term survival in 15 to 25% NaCl (46).
Together, our findings and those of Tsai et al. (46) clearly establish Cls1 and Cls2 as key elements of the PG→CL adaptive response of S. aureus. It should be noted, however, that neither study directly demonstrated the cardiolipin synthase activity of Cls1 or Cls2, i.e., the catalytic conversion of 2 PG to CL plus glycerol. Nevertheless, several considerations argue strongly that Cls1 and Cls2 are cardiolipin synthases: (i) the homology of the proteins encoded by cls1 and cls2 to bona fide prokaryotic cardiolipin synthases; (ii) the reduction in bacterial PG that paralleled accumulation of CL in S. aureus (Fig. 3A to D), a finding consistent with the conversion of PG to CL; (iii) the accumulation of CL in CL-deficient E. coli carrying plasmids encoding cls1 or cls2 (Fig. 2); (iv) the virtual absence of CL accumulation in the cls1 cls2 double mutant of S. aureus; and (v) the fact that in other bacterial species where the biochemical basis of the conversion of PG to CL has been characterized, it was mediated by cardiolipin synthases.
How the various conditions that induce PG→CL conversion in S. aureus (e.g., high salinity, hypertonicity, exposure to organic solvents, inhibitors of glycolysis and oxidative phosphorylation, nutrient deprivation, growth stasis, and phagocytosis) affect cardiolipin synthase activity is unknown. Our comparative analyses of cls1 versus cls2 mRNA under conditions favoring or not favoring CL accumulation (Fig. 6) suggest that neither the induction of Cls2-dependent CL accumulation in the stationary phase and during phagocytosis nor the greater contribution of Cls2 versus Cls1 under these conditions is regulated by modulating cardiolipin synthase transcript levels. One exception could be the elevation of cls1 mRNA levels observed during the phagocytosis of S. aureus that seems to parallel the greater contribution of Cls1 to CL accumulation in ingested S. aureus (Fig. 4). More definitive conclusions, however, must await the development of specific antibodies for Cls1 and Cls2 to permit the measurement of Cls1 and Cls2 protein levels and assays of Cls1 and Cls2 enzyme activity. The relatively acute effects of exposure to organic solvents, hypersalinity, energy poisons, and phagocytes seem most compatible with regulation of CL accumulation occurring being either by allosteric alterations of cardiolipin synthase, alterations of the physical presentation of PG, or both.
The different regulation of cls1 and cls2, at least as manifested by contrasting regulation of mRNA levels, suggests that Cls1 and Cls2 are designed to function in S. aureus CL metabolism under different circumstances. Why that would be is not yet clear. Studies in E. coli have revealed two different proteins, each with cardiolipin synthase activity, that differ in their ability to degrade CL and thus potentially differ in the extent to which they may assist the bacteria to reverse the effects of CL accumulation when extracellular conditions change (13). CL may also be needed in specialized membrane sites of growing bacteria for initiating DNA replication and protein secretion (5, 17, 53). Thus, diverse proteins may participate in bacterial CL metabolism as needed both for constitutive functions of growing bacteria and stress responses of nongrowing bacteria.
The significance of CL accumulation in nongrowing and ingested S. aureus is not known. The growth of S. aureus ingested by human neutrophils is rapidly arrested and the majority of the ingested bacteria is killed within 15 to 30 min, whereas most of conversion of bacterial PG to CL takes place 1 to 2 h after phagocytosis (7, 16, 41). Thus, it is not surprising that uptake, killing, and myeloperoxidase-H2O2-chloride-dependent bleaching of cytosolic GFP of wt and cls1 cls2 mutant S. aureus by neutrophils were essentially the same. However, neutrophils neither fully eradicate viable ingested S. aureus nor, in the absence of mobilized extracellular group IIA phospholipase A2, digest bacterial phospholipids (7, 16, 41). Therefore, the consequences of the conversion of S. aureus PG to CL after phagocytosis by neutrophils may not bear on the intracellular fate of S. aureus, as our findings indicate, but rather on subsequent interactions between remaining viable S. aureus and bacterial phospholipids with surrounding host tissue, including cellular and extracellular defenses mobilized later in the immune response (51). Studies to examine these possibilities are in progress.
ACKNOWLEDGMENTS
We thank Iris Fedke for constructing pRB474cls1 and pRB474cls2 and Jamie Schwartz for guidance in studies with neutrophils and for providing S. aureus LAC.
This study was supported in part by grants from the German Research Foundation (SFB766) and the German Ministry of Education and Research (NGFN2, SkinStaph) to A.P., U.S. Public Health Service grant AI 70958 to W.M.N., and American Heart Association Postdoctoral Fellowship 0725702Z and Marie Curie International Reintegration Grant 249285 to T.K. and with resources and use of facilities at the Iowa City Department of Veterans Affairs Medical Center, Iowa City, IA.
Footnotes
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Bae T., Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63 [DOI] [PubMed] [Google Scholar]
- 2. Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 3. Burritt M. F., Henderson T. O. 1975. Properties of a membrane-bound cardiolipin synthetase from Lactobacillus plantarum. J. Bacteriol. 123:972–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheung A. L., Bayer A. S., Zhang G., Gresham H., Xiong Y.-Q. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1–9 [DOI] [PubMed] [Google Scholar]
- 5. Dowhan W., Bogdanov M. 2009. Lipid-dependent membrane protein topogenesis. Annu. Rev. Biochem. 78:515–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Espey M. G., Xavier S., Thomas D. D., Miranda K. M., Wink D. A. 2002. Direct real-time evaluation of nitration with green fluorescent protein in solution and within human cells reveals the impact of nitrogen dioxide versus peroxynitrite mechanisms. Proc. Natl. Acad. Sci. U. S. A. 99:3481–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Femling J. K., Nauseef W. M., Weiss J. P. 2005. Synergy between extracellular group IIA phospholipase A2 and phagocyte NADPH oxidase in digestion of phospholipids of Staphylococcus aureus ingested by human neutrophils. J. Immunol. 175:4653–4661 [DOI] [PubMed] [Google Scholar]
- 8. Foreman-Wykert A. K., Weinrauch Y., Elsbach P., Weiss J. 1999. Cell wall determinants of the bactericidal action of group IIA phospholipase A2 against Gram-positive bacteria. J. Clin. Invest. 103:715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fowler V. G., Jr., et al. 2005. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA 293:3012–3021 [DOI] [PubMed] [Google Scholar]
- 10. Gould R. M., Lennarz W. J. 1970. Metabolism of phosphatidylglycerol and lysyl phosphatidylglycerol in Staphylococcus aureus. J. Bacteriol. 104:1135–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Graves S., Kobayashi S., DeLeo F. 2010. Community-associated methicillin-resistant Staphylococcus aureus immune evasion and virulence. J. Mol. Med. 88:109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo D., Tropp B. E. 1998. Cloning of the Bacillus firmus OF4 cls gene and characterization of its gene product. Biochem. Biophys. Acta 1389:34–42 [DOI] [PubMed] [Google Scholar]
- 13. Guo D., Tropp B. E. 2000. A second Escherichia coli protein with CL synthase activity. Biochem. Biophys. Acta 1483:263–274 [DOI] [PubMed] [Google Scholar]
- 14. Heber S., Tropp B. E. 1991. Genetic regulation of cardiolipin synthase in Escherichia coli. Biochem. Biophys. Acta 1129:1–12 [DOI] [PubMed] [Google Scholar]
- 15. Hiraoka S., Matsuzaki H., Shibuya I. 1993. Active increase in cardiolipin synthesis in the stationary growth phase and its physiological significance in Escherichia coli. FEBS Lett. 336:221–224 [DOI] [PubMed] [Google Scholar]
- 16. Hunt C. L., Nauseef W. M., Weiss J. P. 2006. Effect of d-alanylation of (lipo)teichoic acids of Staphylococcus aureus on host secretory phospholipase A2 action before and after phagocytosis by human neutrophils. J. Immunol. 176:4987–4994 [DOI] [PubMed] [Google Scholar]
- 17. Ichihashi N., Kurokawa K., Matsuo M., Kaito C., Sekimizu K. 2003. Inhibitory effects of basic or neutral phospholipid on acidic phospholipid-mediated dissociation of adenine nucleotide bound to DnaA protein, the initiator of chromosomal DNA replication. J. Biol. Chem. 278:28778–28786 [DOI] [PubMed] [Google Scholar]
- 18. Iordanescu S., Surdeanu M. 1976. Two restriction and modification systems in Staphylococcus aureus NCTC 8325. J. Gen. Microbiol. 96:277–281 [DOI] [PubMed] [Google Scholar]
- 19. Joyce G. H., Hammond R. K., White D. C. 1970. Changes in membrane lipid composition in exponentially growing Staphylococcus aureus during the shift from 37 to 25°C. J. Bacteriol. 104:323–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klevens R. M., et al. 2007. Invasive methicillin-resistant Staphylococcus aureus Infections in the United States. JAMA 298:1763–1771 [DOI] [PubMed] [Google Scholar]
- 21. Koch H. U., Haas R., Fischer W. 1984. The role of lipoteichoic acid biosynthesis in membrane lipid metabolism of growing Staphylococcus aureus. Eur. J. Biochem. 138:357–363 [DOI] [PubMed] [Google Scholar]
- 22. Koprivnjak T., et al. 2006. Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus. J. Bacteriol. 188:3622–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koprivnjak T., Peschel A., Gelb M. H., Liang N. S., Weiss J. P. 2002. Role of charge properties of bacterial envelope in bactericidal action of human group IIA phospholipase A2 against Staphylococcus aureus. J. Biol. Chem. 277:47636–47644 [DOI] [PubMed] [Google Scholar]
- 24. Lopez C. S., Alice A. F., Heras H., Rivas E. A., Sanchez-Rivas C. 2006. Role of anionic phospholipids in the adaptation of Bacillus subtilis to high salinity. Microbiology 152:605–616 [DOI] [PubMed] [Google Scholar]
- 25. Lowy F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 26. Meredith T. C., Swoboda J. G., Walker S. 2008. Late-stage polyribitol phosphate wall teichoic acid biosynthesis in Staphylococcus aureus. J. Bacteriol. 190:3046–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moreillon P., Que Y. A. 2004. Infective endocarditis. Lancet 363:139–149 [DOI] [PubMed] [Google Scholar]
- 28. Moreillon P., Que Y. A., Bayer A. S. 2002. Pathogenesis of streptococcal and staphylococcal endocarditis. Infect. Dis. Clin. N. Am. 16:297–318 [DOI] [PubMed] [Google Scholar]
- 29. Nishijima S., et al. 1988. Disruption of the Escherichia coli cls gene responsible for cardiolipin synthesis. J. Bacteriol. 170:775–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Novick R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429–1449 [DOI] [PubMed] [Google Scholar]
- 31. Ohta A., Obara T., Asami Y., Shibuya I. 1985. Molecular cloning of the cls gene responsible for cardiolipin synthesis in Escherichia coli and phenotypic consequences of its amplification. J. Bacteriol. 163:506–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Otto M. 2008. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 322:207–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pang Y. Y., et al. 2010. agr-dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J. Innate Immun. 2:546–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Patricia B., Jesús M.-R., Ana H., Juan L. R., Ana S. 2007. A Pseudomonas putida cardiolipin synthesis mutant exhibits increased sensitivity to drugs related to transport functionality. Environ. Microbiol. 9:1135–1145 [DOI] [PubMed] [Google Scholar]
- 35. Peschel A. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405–8410 [DOI] [PubMed] [Google Scholar]
- 36. Proctor R. A., et al. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4:295–305 [DOI] [PubMed] [Google Scholar]
- 37. Salzberg L. I., Helmann J. D. 2008. Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition. J. Bacteriol. 190:7797–7807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Samet J. M., Friedman M., Henke D. C. 1989. High-performance liquid chromatography separation of phospholipid classes and arachidonic acid on cyanopropyl columns. Anal. Biochem. 182:32–36 [DOI] [PubMed] [Google Scholar]
- 39. Schenk S., Laddaga R. A. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133–138 [DOI] [PubMed] [Google Scholar]
- 40. Schlame M. 2008. Thematic review series: glycerolipids—cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J. Lipid Res. 49:1607–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schwartz J., Leidal K. G., Femling J. K., Weiss J. P., Nauseef W. M. 2009. Neutrophil bleaching of GFP-expressing staphylococci: probing the intraphagosomal fate of individual bacteria. J. Immunol. 183:2632–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shibuya I., Miyazaki C., Ohta A. 1985. Alteration of phospholipid composition by combined defects in phosphatidylserine and cardiolipin synthases and physiological consequences in Escherichia coli. J. Bacteriol. 161:1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Short S. A., White D. C. 1972. Biosynthesis of cardiolipin from phosphatidylglycerol in Staphylococcus aureus. J. Bacteriol. 109:820–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Short S. A., White D. C. 1971. Metabolism of phosphatidylglycerol, lysylphosphatidylglycerol, and cardiolipin of Staphylococcus aureus. J. Bacteriol. 108:219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tropp B. E. 1997. Cardiolipin synthase from Escherichia coli. Biochem. Biophys. Acta 1348:192–200 [DOI] [PubMed] [Google Scholar]
- 46. Tsai M., et al. 2011. Staphylococcus aureus requires cardiolipin for survival under conditions of high salinity. BMC Microbiol. 11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tusnady G. E., Simon I. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489–506 [DOI] [PubMed] [Google Scholar]
- 48. Tusnady G. E., Simon I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850 [DOI] [PubMed] [Google Scholar]
- 49. Urban A., Neukirchen S., Jaeger K. E. 1997. A rapid and efficient method for site-directed mutagenesis using one-step overlap extension PCR. Nucleic Acids Res. 25:2227–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Voyich J. M., et al. 2005. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 175:3907–3919 [DOI] [PubMed] [Google Scholar]
- 51. Weiss J., Bayer A. S., Yeaman M. 2006. Cellular and extracellular defenses against staphylococcal infections, p. 544–549 In Fischetti V. A., et al. (ed.), Gram-positive pathogens, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 52. Weiss J., Inada M., Elsbach P., Crowl R. M. 1994. Structural determinants of the action against Escherichia coli of a human inflammatory fluid phospholipase A2 in concert with polymorphonuclear leukocytes. J. Biol. Chem. 269:26331–26337 [PubMed] [Google Scholar]
- 53. Zhang Y.-M., Rock C. O. 2008. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6:222–233 [DOI] [PubMed] [Google Scholar]