Abstract
Campylobacter jejuni is the leading bacterial cause of human gastroenteritis worldwide. Despite stringent microaerobic growth requirements, C. jejuni is ubiquitous in the aerobic environment and so must possess regulatory systems to sense and adapt to external stimuli, such as oxidative and aerobic (O2) stress. Reannotation of the C. jejuni NCTC11168 genome sequence identified Cj1556 (originally annotated as a hypothetical protein) as a MarR family transcriptional regulator, and further analysis indicated a potential role in regulating the oxidative stress response. A C. jejuni 11168H Cj1556 mutant exhibited increased sensitivity to oxidative and aerobic stress, decreased ability for intracellular survival in Caco-2 human intestinal epithelial cells and J774A.1 mouse macrophages, and a reduction in virulence in the Galleria mellonella infection model. Microarray analysis of gene expression changes in the Cj1556 mutant indicated negative autoregulation of Cj1556 expression and downregulation of genes associated with oxidative and aerobic stress responses, such as katA, perR, and hspR. Electrophoretic mobility shift assays confirmed the binding of recombinant Cj1556 to the promoter region upstream of the Cj1556 gene. cprS, which encodes a sensor kinase involved in regulation of biofilm formation, was also upregulated in the Cj1556 mutant, and subsequent studies showed that the mutant had a reduced ability to form biofilms. This study identified a novel C. jejuni transcriptional regulator, Cj1556, that is involved in oxidative and aerobic stress responses and is important for the survival of C. jejuni in the natural environment and in vivo.
INTRODUCTION
Campylobacter jejuni infection is one of the most commonly identified bacterial causes of acute human gastroenteritis worldwide (1). The symptoms of campylobacteriosis are malaise, fever, severe abdominal pain, and diarrhea (12). C. jejuni infection has also been associated with postinfectious sequelae, including septicemia and neuropathies, such as Guillain-Barré syndrome (GBS) (52). C. jejuni is a commensal in avian species, and the consumption and handling of poultry products is a major source of human infection (4, 39). However, a diverse range of environmental sources, such as untreated water, raw or unpasteurized milk, vegetables, and transmission from pets, are also all recognized sources of infection (55). Despite specific microaerobic growth requirements, C. jejuni is ubiquitous in the aerobic environment and appears capable of withstanding different stresses, including suboptimal carbon source growth, temperature changes, and exposure to atmospheric oxygen (26). C. jejuni can also persist in the environment through survival in biofilms (36). During human infection, C. jejuni has to withstand a further range of stresses, including changes in pH and the host innate immune response (11). The last decade has seen major advances in our understanding of C. jejuni physiology, yet many unanswered questions remain regarding the pathogenesis and survival mechanisms of the bacterium. A more complete understanding of the regulation of C. jejuni response mechanisms to the diverse stresses encountered both during the infection cycle and within the natural environment is required to facilitate appropriate intervention strategies to reduce the burden of C. jejuni-associated disease (64).
Oxidative, nitrosative, and aerobic (O2) stresses are major factors that pathogens must counteract in order to survive within the host (4, 25, 57, 87). C. jejuni is a microaerophilic organism optimally suited to low levels of atmospheric oxygen; however, the bacterium is able to survive oxidative stresses in vivo (4). The incomplete reduction of oxygen to water creates reactive oxygen species (ROS) molecules, such as hydrogen peroxide (H2O2), that are used by the host against invading pathogens (20). ROS are also released by the immune system to combat invading microorganisms (4). An example of ROS release is the deposition of various oxygen species generated by the respiratory burst oxidase as the bacterium remains bound within an endosome (41). ROS can damage bacterial DNA (33). Reactive nitrogen species (RNS), such as nitric oxide, are a family of antimicrobial molecules produced by the enzymatic activity of inducible nitric oxide synthase 2 (iNOS) (34). Acidified nitrite kills C. jejuni, and expression of the NOS2 isoform is increased in macrophages upon exposure to the bacterium (34). RNS tend to interfere with respiration and DNA replication through inactivation of zinc metalloproteins (25). Both ROS and RNS are also derived from phagocytosis through the generation of superoxide and nitric oxide radicals via NADPH phagocyte oxidase and inducible nitric oxide synthase pathways, which are important pathways within polymorphonuclear phagocytes, including white blood cells and mononuclear phagocytes (25). Aerobic stress is caused by bacterial exposure to raised oxygen levels. Even though oxygen is considered a stress for C. jejuni, few studies have described specific phenotypic consequences of aerobiosis, and those that have vary in their conclusions (72). Exposure of C. jejuni to oxygen for 24 h accelerated the transition to the viable but nonculturable (VBNC) state or coccoid form (40). In contrast, another study identified the increased culturability of C. jejuni when exposed to oxygen for 15 h (48). Recently, it has been demonstrated that aerobic stress conditions promoted the production of C. jejuni biofilms (65).
C. jejuni possesses a variety of mechanisms for reacting to nitrosative, oxidative, and aerobic stresses. C. jejuni possesses a truncated hemoglobin (Ctb), along with a single-domain hemoglobin (Cgb). Both Ctb and Cgb have been characterized as part of the C. jejuni nitrosative stress response regulon (23, 78). This regulon is under the control of NssR (49). Previous studies have also implicated Ctb with a role in oxygen metabolism (77, 78). C. jejuni contains several genes encoding important oxidative stress response proteins. The superoxide dismutase SodB is involved in the breakdown of superoxide to H2O2 and O2 (61). The catalase KatA converts H2O2 to H2O and O2. In addition, the alkyl hydroperoxide reductase AhpC confers resistance to cumene hydroperoxide and aerobic stress (6). However, C. jejuni lacks an OxyR ortholog, which regulates ahpC and katA expression in response to oxidative stress in many enteric bacteria, such as Salmonella species and Escherichia coli (14). C. jejuni also lacks the classical SoxRS system, which mediates transcriptional activation of the oxidative stress regulon in response to superoxide-generating agents (2). In C. jejuni, the Fur homolog PerR was found to repress ahpC and katA transcription in an iron-dependent manner, thus making PerR a functional but not homologous substitute for OxyR (57, 75). In addition, C. jejuni proteins involved in responding to aerobic stress have also been identified. SodB and KatA have been shown to counteract the detrimental effects of aerobic stress (69). The heat shock protease HtrA and the regulator HspR have been shown to be important for short-term aerobic tolerance (3, 12). The fdxA gene upstream of ahpC encodes a ferrodoxin that has been identified as important for aerotolerance (74). Also, SpoT, which regulates the C. jejuni stringent response, was found to be important for low CO2 growth and aerobic survival (28). Cj1556 was identified as a member of the MarA family of transcriptional regulators through reannotation of the NCTC11168 genome sequence (29). In this study, further bioinformatics analysis indicated a role for Cj1556 in the C. jejuni stress responses, and a defined isogenic C. jejuni 11168H Cj1556 mutant was constructed in order to investigate this hypothesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The C. jejuni wild-type strain used in this study was 11168H (38), a hypermotile derivative of the original sequenced strain NCTC11168 that shows higher levels of cecal colonization in a chick colonization model (35). C. jejuni was grown at 37°C in a microaerobic chamber (Don Whitley Scientific, United Kingdom) containing 85% N2, 10% CO2, and 5% O2 either on blood agar plates containing Columbia agar base (Oxoid, United Kingdom) supplemented with 7% (vol/vol) horse blood (TCS Microbiology, United Kingdom) and Campylobacter Selective Supplement (Oxoid) or in brucella broth (Oxoid) with shaking at 75 rpm. C. jejuni strains were grown on blood agar plates for 24 h prior to use in coculture experiments. E. coli XL-2 Blue MRF′ competent cells (Stratagene) were used for cloning experiments and were grown at 37°C under aerobic conditions either on Luria-Bertani (LB) agar plates or in LB broth with shaking at 200 rpm. Appropriate antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; and chloramphenicol, 50 μg/ml for E. coli studies and 10 μg/ml for C. jejuni studies. All reagents were obtained from Invitrogen (United Kingdom) unless otherwise stated.
Construction of the C. jejuni 11168H Cj1556 mutant.
A defined isogenic 11168H Cj1556 mutant was constructed using previously published methods (35, 38, 43). Briefly, the primers Cj1556-F and Cj1556-R were designed for PCR detection of Cj1556 (Table 1). Using the pUC library from the C. jejuni NCTC11168 genome-sequencing project (58), plasmid cam25a2 (1489074.0.1490567), which contains a 1.494-kb insert including the coding sequences (CDSs) Cj1555c to Cj1560, was selected and designated pUC-Cj1556. The Cj1556 open reading frame (ORF) in pUC-Cj1556 was inactivated by insertion of an aph-3 (aminoglycoside 3′-phosphotransferase) kanamycin resistance (Kmr) cassette (73). The Kmr cassette was excised from pJMK30 (76) using BamHI. pUC-Cj1556 was digested with BclI and ligated with the Kmr cassette to form pUC-Cj1556-Kmr. pUC-Cj1556-Kmr was transformed into XL-2 Blue MRF′ competent cells, and transformants were selected on LB agar supplemented with ampicillin and kanamycin after 48 h of growth at 37°C. Transformants were screened by PCR using Cj1556-specific and Kmr-specific primers (Table 1). pUC-Cj1556-Kmr plasmids with the Kmr cassette in the same orientation as the Cj1556 gene were selected and electroporated into wild-type 11168H as described previously (35, 38). The electroporated bacteria were plated onto blood agar plates and incubated at 37°C under microaerobic conditions for 2 days. Cells were harvested and resuspended in 0.5 ml phosphate-buffered saline (PBS). Two hundred microliters of this suspension was spread onto blood agar plates containing kanamycin. Putative Cj1556 mutants were screened using PCR and sequencing.
Table 1.
Oligonucleotide primers used in this study
| Primer name | Sequence |
|---|---|
| Cj1556-F | ATCATTCTCTTTGTCCTAT |
| Cj1556-R | TAAGATGGATTCTAAACTATTG |
| Kmr forward-out | TGGGTTTCAAGCATTAGTCCATGCAAG |
| Kmr reverse-out | GTGGTATGACATTGCCTTCTGCG |
| Comp-Cj1556-F | CCCCCATGGATAAGGATTTATAATGAAAAAATATCATTCTCT |
| Comp-Cj1556-R | CCCGCTAGCTTAAACGATATTTTTATAGCTAT |
| Comp-Cj1556-R-HIS | CCCGCTAGCTTAATGATGATGATGATGATGAACGATATTTTTATAGCTAT |
| Upstream Cj1556-F | ATGCAATCTAGAAATTAT |
| Upstream Cj1556-R | GGACAAAGAGAATGATATT |
| Upstream flaA-F | ATCACAGCTTATATTAAAG |
| Upstream flaA-R | GTGTTAATACGAAATCCCAT |
| Upstream flgK-F | ATTTGTTCTTATTGTCAA |
| Upstream flgK-R | ATGTTCCAAAAATACCCAT |
Complementation of the C. jejuni Cj1556 mutant.
Complementation procedures were performed by inserting a copy of the Cj1556 gene into the Cj1556 mutant chromosome using a C. jejuni NCTC11168 complementation vector (31). The coding region for Cj1556 was amplified by PCR using primers Comp-Cj1556-F and Comp-Cj1556-R (Table 1), which introduced an NcoI site at the 5′ end and an NheI site at the 3′ end, as well as the native ribosome binding site of Cj1556 (72, 83). Following digestion with NheI and NcoI, this PCR product was ligated into the pDENNIS vector. The construct was checked by sequencing and electroporated into the Cj1556 mutant. Putative clones were selected on blood agar plates containing kanamycin and chloramphenicol. Confirmation of the presence of copies of both Cj1556 and Cj1556-Kmr was performed by PCR using the Comp-Cj1556-F and Comp-Cj1556-R primers, as well as the Cj1556-F and Cj1556-R primers, and also by sequencing. For isolation of recombinant Cj1556 protein, a 6×His tag sequence was cloned into a second construct using the primers Comp-Cj1556-F and Comp-Cj1556-R-HIS (Table 1).
Nitrosative, oxidative, and heat stress assays.
C. jejuni was grown on blood agar plates for 24 h at 37°C under microaerobic conditions. Bacterial cells were harvested into 1 ml PBS and adjusted to an optical density at 600 nm (OD600) of 1. For nitrosative stress assays, bacterial cells were exposed to acidified sodium nitrite (NaNO2) at a final concentration of 100 mM NaNO2 for 30 min and 10 mM NaNO2 for 75 min. For nitrosative stress assays, all media used were at pH 5 to allow formation of acidified NaNO2 to promote the production of nitric oxide radicals (22, 34). For oxidative-stress assays, bacterial cells were exposed to hydrogen peroxide (H2O2) at a final concentration of 10 mM for 15 min. Heat stress assays were performed at 42°C for 1 h, 55°C for 15 min, and 60°C for 5 min. Serial dilutions were prepared, 10 μl of the 10−1 to 10−6 dilutions were spotted onto blood agar plates and incubated for 48 h at 37°C under microaerobic conditions, and colonies were counted.
Cell culture procedures.
The Caco-2 human intestinal epithelial and J774A.1 mouse macrophage cell lines were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% (vol/vol) fetal calf serum (FCS) (Sigma-Aldrich, United Kingdom), 1% (vol/vol) nonessential amino acids, 100 μg/ml streptomycin, and 100 U/ml penicillin. The T84 human colonic epithelial cell line was maintained in a 1:1 mixture of DMEM and Ham's F-12 medium containing Glutamax, 2.5 mM l-glutamine, 15 mM HEPES, and 0.5 mM sodium pyruvate supplemented with 10% (vol/vol) FCS, 100 μg/ml streptomycin, and 100 U/ml penicillin. The cells were maintained at 37°C in 5% CO2 and 95% air. For Caco-2 cell coculture experiments, cells were seeded at 1 × 105/ml and grown in 24-well plates to >90% confluence (∼1 × 106 cells/ml). For T84 cell coculture experiments, cells were seeded at 5 × 105/ml and grown in 24-well plates to >90% confluence (∼5 × 106 cells/ml). For coculture experiments involving J774A.1 mouse macrophages, cells were seeded at 5 × 105/ml and grown in 24-well plates for 24 h. For enzyme-linked immunosorbent assay (ELISA) experiments, T84 cells were maintained in low-serum 1% (vol/vol) and antibiotic-free medium overnight prior to coculture. Infections were terminated by removing the supernatant from the cells, followed by two washes in PBS. Cell culture supernatants were stored at −80°C.
Interaction, invasion, and intracellular survival assays.
Interaction (adhesion and invasion) and invasion assays were performed using Caco-2 cells as described previously (12). Bacterial cells were harvested into 1 ml brucella broth and adjusted to an OD600 of 0.1. Serial dilutions were prepared, and 200-μl volumes were plated onto blood agar plates and incubated for 72 h at 37°C under microaerobic conditions. Colonies were counted to calculate the initial inoculum. C. jejuni (approximately 1 × 108 cells) in DMEM was added to a monolayer of approximately 1 × 106 Caco-2 cells (multiplicity of infection [MOI], 100:1) and incubated for 3, 6, or 24 h. The number of interacting bacteria was determined by washing the monolayers three times with PBS and then lysing the cells by addition of 0.2% (vol/vol) Triton X-100. The number of intracellular bacteria was determined by further incubating the monolayers after the initial interaction time point with DMEM containing gentamicin (150 μg/ml) for 2 h at 37°C to allow killing of extracellular bacteria. The monolayers were then washed three times in PBS, and the epithelial cells were lysed as described above. For intracellular survival assays, bacterial cells were cocultured with a monolayer of Caco-2 cells for 3 h, followed by washing the monolayers three times with PBS. The monolayers were then incubated in DMEM containing gentamicin (150 μg/ml) for 2 h and incubated in DMEM containing a reduced concentration of gentamicin (10 μg/ml) for 19 h. The monolayers were then washed three times in PBS, and the epithelial cells were lysed as described above. To ascertain whether the above results were due to a genuine Cj1556 mutant phenotype and not to increased sensitivity to Triton X-100, stress assays were performed on all three strains with 0.2% (vol/vol) Triton X-100. No difference was observed between the levels of sensitivity to Triton X-100 in the 11168H wild-type strain, the Cj1556 mutant, and the Cj1556 complement strains (data not shown). Experiments with survival in tissue culture medium from coculture were performed as described above, but after 24 h of coculture, the tissue culture medium alone was removed, followed by plating of serial dilutions to determine the CFU/ml. In all cases, serial dilutions, plating, and determination of bacterial numbers were performed as stated above.
Macrophage survival assay.
Macrophage survival assays were performed as described previously (80) using J774A.1 mouse macrophages (67). Briefly, C. jejuni was harvested into 1 ml brucella broth and adjusted to an OD600 of 0.1. C. jejuni cells (approximately 1 × 108 cells) in DMEM were added to a culture of approximately 5 × 105 J774A.1 mouse macrophages (MOI, 200:1) and incubated for 3 h. The cells were washed three times in PBS, followed by incubation in DMEM containing gentamicin (150 μg/ml) for 2 h to allow killing of extracellular bacteria. The macrophages were incubated in DMEM containing a reduced concentration of gentamicin (10 μg/ml), and bacterial survival was determined at 0, 4, and 16 h. At each time point, the macrophages were washed three times with PBS and lysed by adding 0.2% (vol/vol) Triton X-100 in PBS. Serial dilutions, plating, and enumeration of bacteria were performed as stated above.
IL-6 and IL-8 ELISAs.
Supernatants from uninfected T84 cells and T84 cells infected with C. jejuni at an MOI of 20:1 for 24 h were collected. The levels of interleukin-6 (IL-6) and IL-8 secretion were assessed using a commercially available sandwich ELISA kit according to the manufacturer's instructions (Peprotech, United Kingdom). Detection was performed using a Dynex MRX II 96-well plate reader at an absorbance of 405 nm (A405) and analyzed using Revelation software (Dynex).
Transcriptome studies: experimental design, template labeling, microarray hybridizations, data acquisition, and microarray data analysis.
Gene expression profiling of C. jejuni 11168H from the late log growth phase (16 h) was performed using an indirect-comparison method or type 2 experimental design (86). Replicate test sets of Cy5-labeled C. jejuni 11168H total RNA samples were combined with a common reference sample (Cy3-labeled C. jejuni 11168H genomic DNA) as described in previous studies (24, 44, 84). C. jejuni 11168H genomic DNA was isolated from bacteria grown on blood agar for 24 h using the Puregene DNA purification kit (Gentra, United Kingdom) and used as the common reference sample in all microarray experiments. C. jejuni RNA was isolated from 16-h cultures using the RNeasy Mini purification kit (Qiagen) and RNAprotect Bacteria Reagent (Qiagen) as described previously (37). Whole-genome C. jejuni NCTC11168 microarrays printed on UltraGAPS glass slides (Corning) constructed by the BμG@S Microarray Group (http://www.bugs.sgul.ac.uk/) were used in this study (37). The procedures used for Cy5 labeling of total RNA samples (37) and Cy3 labeling of 11168H genomic DNA (21) were as described previously. All hybridizations were performed as described previously (21, 37) with the following modifications. For probe hybridization, Cy5-labeled probes of C. jejuni 11168H total RNA (test) and Cy3-labeled common reference samples of C. jejuni 11168H DNA (control) were combined and purified using a MinElute PCR Purification kit (Qiagen). The final elution was made up to a volume of 50 μl with a final concentration of 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.3% (wt/vol) SDS. The hybridization mixture was denatured at 98°C for 2 min and cooled slowly to room temperature. A 22- by 25-mm LifterSlip coverslip (Erie Scientific) was placed over the reporter element area on the microarray, and the hybridization mixture was applied underneath the coverslip. The microarray slide was placed in a humidified hybridization cassette (Telechem International) and incubated in a water bath for 18 h at 65°C without shaking. The microarray slides were then washed as described previously (37). The microarray slides were scanned with an Affymetrix 418 array scanner (MWG Biotech, Germany) according to the manufacturer's guidelines. Signal and local background intensity readings for each spot were quantified using ImaGene software v8.0 (BioDiscovery). The quantified data were analyzed using GeneSpring GX software v7.3 (Agilent). Statistically significant up- and downregulated genes were selected when comparing gene expression against the 11168H wild-type strain using analysis of variance (ANOVA) with a Benjamini and Hochberg false-discovery rate as the multiple-testing correction (5, 18).
Electrophoretic mobility shift assays.
E. coli strains were grown overnight for 16 h at 37°C with shaking at 200 rpm. The cultures were spun at 4,000 rpm for 10 min at 4°C. The bacterial pellet was resuspended in 1 ml equilibration buffer (Sigma-Aldrich). The cells were sonicated according to the manufacturer's instructions (Diagenode, Belgium), followed by centrifugation for 5 min at 13,000 rpm. The supernatant containing lysed cell content was poured into a new 1.5-ml microcentrifuge tube. The lysed cells were incubated with Ni-nitrilotriacetic acid (NTA) (Qiagen) for 1 h at 4°C on a rotator. Elution was performed using a His-Select spin column (Sigma-Aldrich). To demonstrate the DNA binding properties of Cj1556, purified recombinant protein was hybridized to PCR-amplified fragments (140 to 180 bp) located upstream of the translation initiation sites of the Cj1556, flaA, and flgK genes (Tables 1 and 2). Recombinant native protein (2.5 μg) was hybridized with 20 ng of purified DNA, along with 2 μl hybridization solution (20% glycerol, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM dithiothreitol [DTT], 250 mM NaCl, 50 mM Tris-HCl, pH 7.5) and incubated at room temperature for 40 min. Samples were resuspended in Tris-glycine native sample buffer (Invitrogen) up to 10 μl and analyzed using a Tris-glycine gel under nondenaturing conditions (Invitrogen), followed by Western blot analysis.
Table 2.
DNA fragments used as promoter probes for electrophoretic mobility shift assays
| Fragment region | Purpose of selection | Location within genome (nucleotides) | Size of fragment (bp) |
|---|---|---|---|
| Upstream of Cj1556 | Proposed area of binding | 1489630–1489800 | 170 |
| Upstream of flaA | Negative control | 1271120–1270940 | 180 |
| Upstream of flgK | Negative control | 1400460–1400600 | 140 |
Biofilm assays.
C. jejuni was grown on blood agar plates for 24 h at 37°C under microaerobic conditions. Brucella broth (10 ml) was preincubated in a 50-ml flask at 37°C under microaerobic conditions 24 h prior to inoculation and then inoculated with the bacterial cells harvested into brucella broth to an OD600 of 0.1; 1 ml was then added to 24-well polystyrene plates (Corning) and incubated under microaerobic conditions with gentle agitation for 3 days. The wells were washed three times with PBS, followed by addition of 0.2% (wt/vol) crystal violet (Sigma-Aldrich) for 10 min. The wells were then washed three times with PBS, followed by dissolving the biofilm with 20% acetone-80% ethanol. Detection was performed using a Dynex MRX II 96-well plate reader at A595.
G. mellonella infection model.
Galleria mellonella larvae were obtained from LiveFoods Direct (United Kingdom) and kept on wood chips at 16°C. The larvae were injected with a 10-μl inoculum of a 24-h C. jejuni culture diluted to an OD600 of 0.1 by microinjection (Hamilton, Switzerland) in the right foremost leg, giving an infectious dose of approximately 106 CFU (17). Injections with PBS and no-injection controls were also performed. The larvae were incubated at 37°C, with survival and percentage survival recorded at 24-h intervals. For each experiment, 10 G. mellonella larvae were infected, and experiments were repeated three times.
Statistical analyses.
Data are presented as mean ± standard deviation (SD). All experiments represent at least three biological replicates performed in triplicate in each experiment. Statistical analyses were performed using Prism software (GraphPad Software). Variables were compared using Student's t test.
Microarray data accession numbers.
The array design is available in BμG@Sbase (accession no. A-BUGS-9) (http://bugs.sgul.ac.uk/A-BUGS-9) and also ArrayExpress (accession no. A-BUGS-9). Fully annotated microarray data have been deposited in BμG@Sbase (accession number E-BUGS-119) (http://bugs.sgul.ac.uk/E-BUGS-119) and also ArrayExpress (accession number E-BUGS-119).
RESULTS
Bioinformatics analysis indicates Cj1556 has a role in regulation of stress responses.
The 333-nucleotide predicted CDS of Cj1556 was originally annotated as a hypothetical protein in the genome sequence of C. jejuni NCTC11168 (29). Following reannotation, the updated product function indicated that Cj1556 is a transcriptional regulator based on the identification of a new Pfam motif (PF01638), defined as an HxlR-like helix-turn-helix motif (29). The HxlR-like helix-turn-helix motif is located 45 nucleotides into the CDS and encompasses the remainder of the CDS. The HxlR-like helix-turn-helix motif is part of the MarR family of transcriptional regulators, which includes proteins that control virulence factor production, bacterial responses to both antibiotics and oxidative stress, and also catabolism of environmental aromatic compounds (82, 85). The predicted function of Cj1556 was investigated further using the Campylobacter Protein Interaction Database (60), and putative interactions with Ctb (Cj0465c) were identified. Ctb is a group III truncated hemoglobin, and characterization studies in C. jejuni have already shown Ctb to be part of the nitrosative stress response regulon (49). Ctb has also been linked with moderating oxygen metabolism within C. jejuni (49). Collectively, these bioinformatics analyses suggest that Cj1556 has an important role as a stress response regulator.
Construction and characterization of a C. jejuni 11168H Cj1556 mutant.
To investigate the function of Cj1556, a defined isogenic 11168H Cj1556 mutant was constructed by insertion of a Kmr cassette, using standard mutagenesis techniques (35, 38), with Kmr in the same orientation as the Cj1556 CDS to obviate potential polar effects. To further confirm phenotypic changes, the Cj1556 mutant was complemented, verified by PCR/sequencing, and termed Cj1556 complement. Motility assays demonstrated that there were no significant differences in the motility of the Cj1556 mutant or Cj1556 complement compared to the wild-type strain 11168H at 24, 48, and 72 h (data not shown).
The Cj1556 mutant exhibits increased sensitivity to both oxidative and heat stresses.
Nitrosative stress assays were performed using acidified NaNO2. However, no differences between the survival of the wild-type strain 11168H and of the Cj1556 mutant were observed (data not shown). Oxidative stress assays were performed using H2O2. The Cj1556 mutant exhibited increased sensitivity to H2O2 compared to the 11168H wild-type strain (Fig. 1A). In addition, the Cj1556 complement restored the wild-type H2O2 sensitivity phenotype (Fig. 1A). Previous research has suggested a link between aerobic and heat stresses (12, 63). In order to investigate this further, a range of heat stress experiments were performed. No significant differences in survival were observed at 42°C/60 min or 55°C/15 min. However, the Cj1556 mutant displayed increased sensitivity compared to the wild-type strain at 60°C/5 min, and the Cj1556 complement restored the wild-type phenotype (Fig. 1B).
Fig. 1.
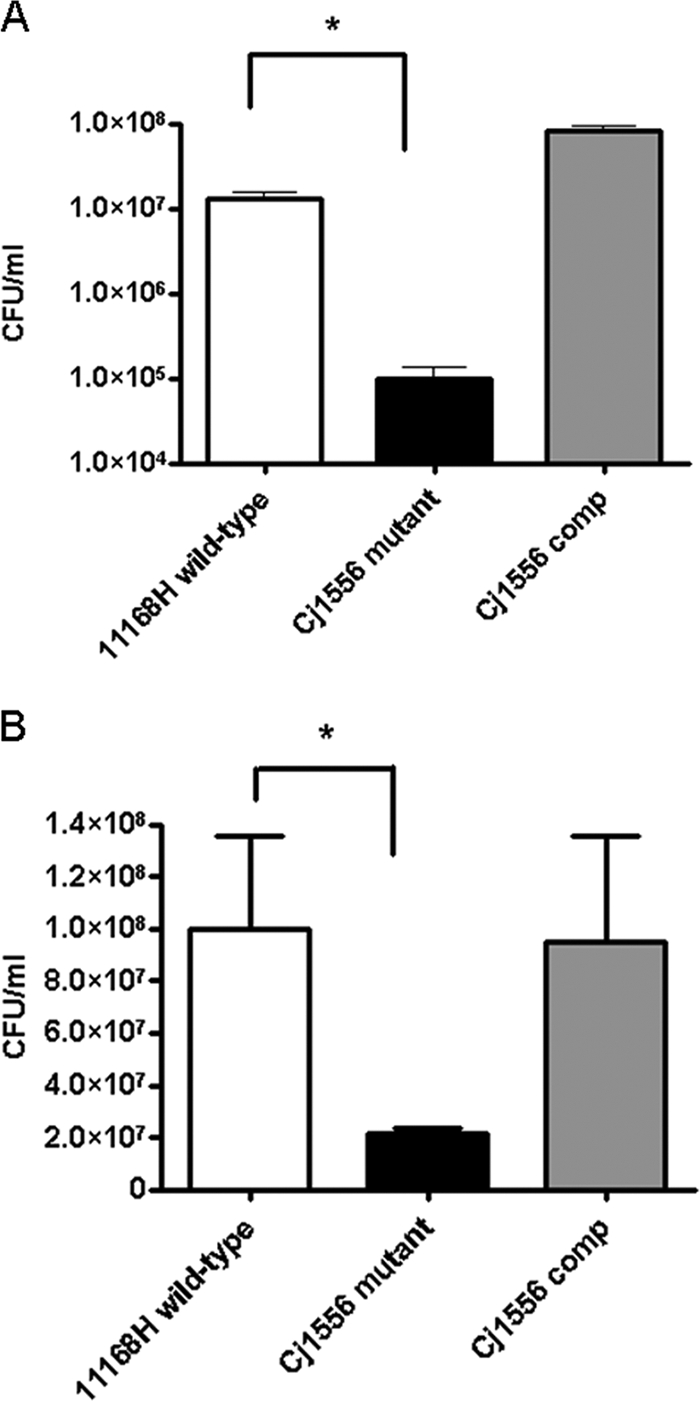
Effects of oxidative (A) and heat (B) stresses on the survival of the C. jejuni 11168H wild-type, Cj1556 mutant, and Cj1556 complement (Cj1556 comp) strains. The C. jejuni strains were incubated with 10 mM H2O2 for 15 min at 37°C (A) or at 60°C for 5 min (B), and bacterial survival was assessed. The asterisks denote a statistically significant difference (P < 0.05) for the Cj1556 mutant compared to the 11168H wild-type strain.
The Cj1556 mutant displays reduced ability to interact with and invade Caco-2 intestinal epithelial cells.
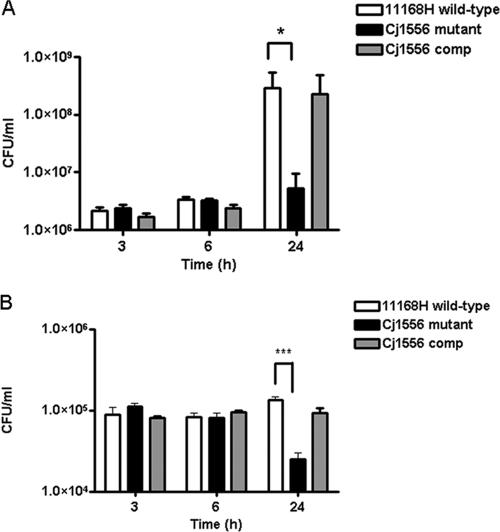
Interaction (adhesion and invasion) and invasion assays were performed using 11168H wild-type, Cj1556 mutant, and Cj1556 complement strains. No significant differences were observed when the levels of interaction at 3 h and 6 h were compared; however, the Cj1556 mutant displayed a reduced ability to interact with Caco-2 cells after 24 h of coculture compared with the 11168H wild-type and Cj1556 complement strains (Fig. 2A). The Cj1556 mutant also displayed a reduced ability to invade Caco-2 cells after 24 h of coculture compared with the 11168H wild-type and Cj1556 complement strains (Fig. 2B). No significant differences were observed when the levels of invasion at 3 h and 6 h were compared.
Fig. 2.
Interaction (adhesion and invasion) and invasion assays. The 11168H wild-type, Cj1556 mutant, and Cj1556 complement (Cj1556 comp) strains were cocultured with Caco-2 intestinal epithelial cells for 3, 6, or 24 h. The Caco-2 cells either were lysed and the numbers of interacting bacteria were assessed (A) or were incubated with gentamicin (150 μg/ml) for 2 h to kill extracellular bacteria and then lysed, and the numbers of intracellular bacteria were assessed. The asterisks denote a statistically significant difference (*, P < 0.05; ***, P < 0.001) for the Cj1556 mutant compared to the 11168H wild-type strain.
The Cj1556 mutant exhibits reduced intracellular survival in Caco-2 intestinal epithelial cells and in J774A.1 macrophage cells.
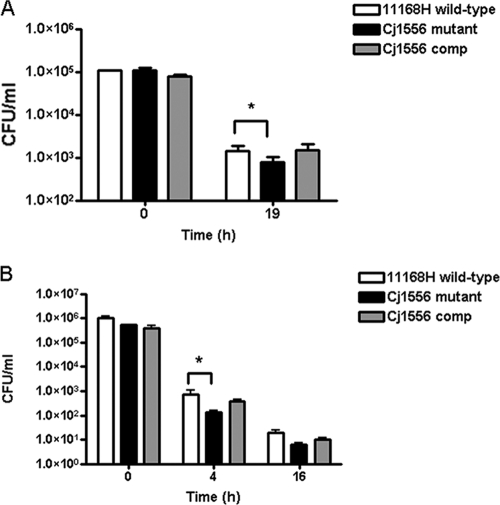
A modification of the interaction and invasion assays was used to analyze the level of intracellular survival in Caco-2 intestinal epithelial cells (53) in order to investigate the ability of C. jejuni to survive when exposed to intracellular stress conditions, such as ROS. There was a statistically significant reduction in the level of intracellular survival of the Cj1556 mutant compared to the 11168H wild-type and Cj1556 complement strains (Fig. 3A). Intracellular survival assays using macrophages were also performed to further investigate the survival rates of the 11168H wild-type, Cj1556 mutant, and Cj1556 complement strains. Macrophages internalize and destroy C. jejuni (80), and previous studies have shown that C. jejuni is killed within 24 h of internalization (80). There was a statistically significant reduction in the level of intracellular survival of the Cj1556 mutant compared to the 11168H wild-type strain (Fig. 3B).
Fig. 3.
Intracellular survival assays. The 11168H wild-type, Cj1556 mutant, and Cj1556 complement (Cj1556 comp) strains were cocultured with Caco-2 intestinal epithelial cells (A) or J774A.1 mouse macrophages (B) for 3 h and then incubated with gentamicin (150 μg/ml) for 2 h to kill extracellular bacteria, followed by further incubation with gentamicin (10 μg/ml). The cells were lysed, and the numbers of intracellular bacteria were assessed. The asterisks denote a statistically significant difference (P < 0.05) for the Cj1556 mutant compared to the 11168H wild-type strain.
The Cj1556 mutant exhibits reduced survival in both coculture media and an aerobic environment.
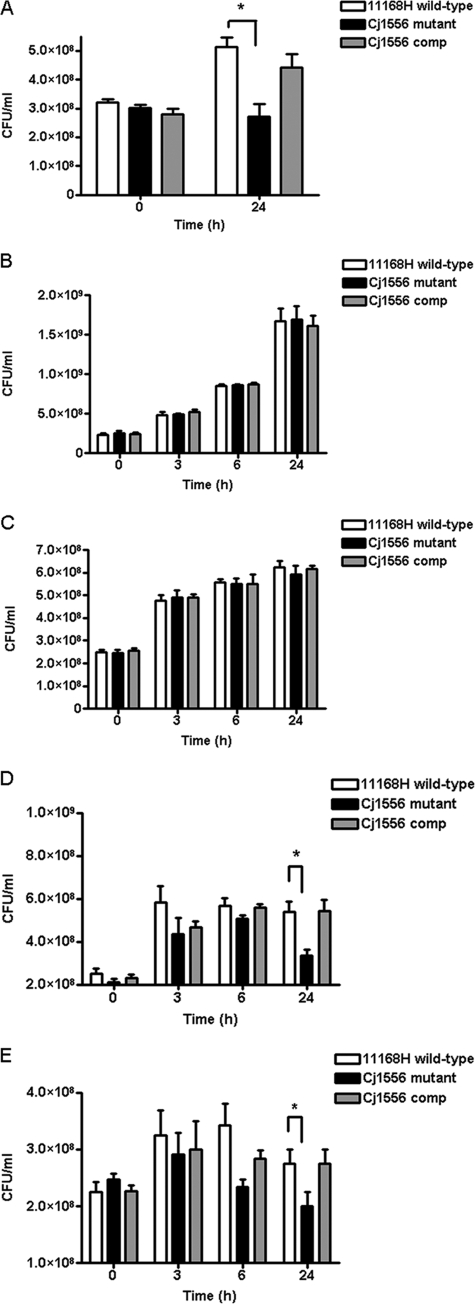
A further variation of the intracellular survival assay was used to assess the survival of C. jejuni in tissue culture medium. There was a statistically significant increase in the number of viable bacterial cells obtained from the supernatant after 24 h of coculture with Caco-2 cells when the 11168H wild-type and Cj1556 complement strains were compared to the Cj1556 mutant (Fig. 4A). Following the identification of significant differences between the 11168H wild-type strain and the Cj1556 mutant in response to oxidative stress and intracellular survival, further investigations of the abilities of these strains to survive aerobic stress were performed. The difference in the level of Cj1556 mutant survival between the interaction and intracellular assays suggested that additional stresses might affect C. jejuni during these assays. Survival assays with the 11168H wild-type, Cj1556 mutant, and Cj1556 complement strains were performed under either microaerobic or aerobic conditions in either brucella broth or tissue culture medium with no shaking to replicate the conditions for the coculture assays. A statistically significant reduction in the number of viable bacterial cells with the Cj1556 mutant compared to the 11168H wild-type strain in both types of media was observed after 24 h of incubation under aerobic conditions (Fig. 4D and E), but not under microaerobic conditions (Fig. 4B and C).
Fig. 4.
Survival assays. (A) The 11168H wild-type, Cj1556 mutant, and Cj1556 complement strains (Cj1556 comp) were cocultured with Caco-2 intestinal epithelial cells for 24 h, followed by assessment of the number of bacteria in the coculture medium. (B to E) Further survival assays were performed, in which the C. jejuni strains were grown under microaerobic (B and C) and aerobic (D and E) conditions in brucella broth (B and D) or tissue culture medium (C and E). Then, the numbers of viable bacteria were assessed. The asterisks denote a statistically significant difference (P < 0.05) for the Cj1556 mutant compared to the 11168H wild-type strain.
The Cj1556 mutant induces a reduced IL-6 response from T84 cells.
IL-6 and IL-8 are well-characterized markers denoting a host immune response against pathogens (56). Only minimal secretion of IL-6 and IL-8 was detected when the 11168H wild-type and Cj1556 mutant strains were cocultured with Caco-2 cells (data not shown). However, using the T84 cell line, significant levels of induction of both IL-6 and IL-8 by the 11168H wild-type strain and the Cj1556 mutant were observed (Fig. 5). There was no significant difference in the level of IL-8 induction by the Cj1556 mutant compared to that by the 11168H wild-type strain (Fig. 5A); however, a significant reduction in the level of IL-6 induction by the Cj1556 mutant was observed (Fig. 5B).
Fig. 5.
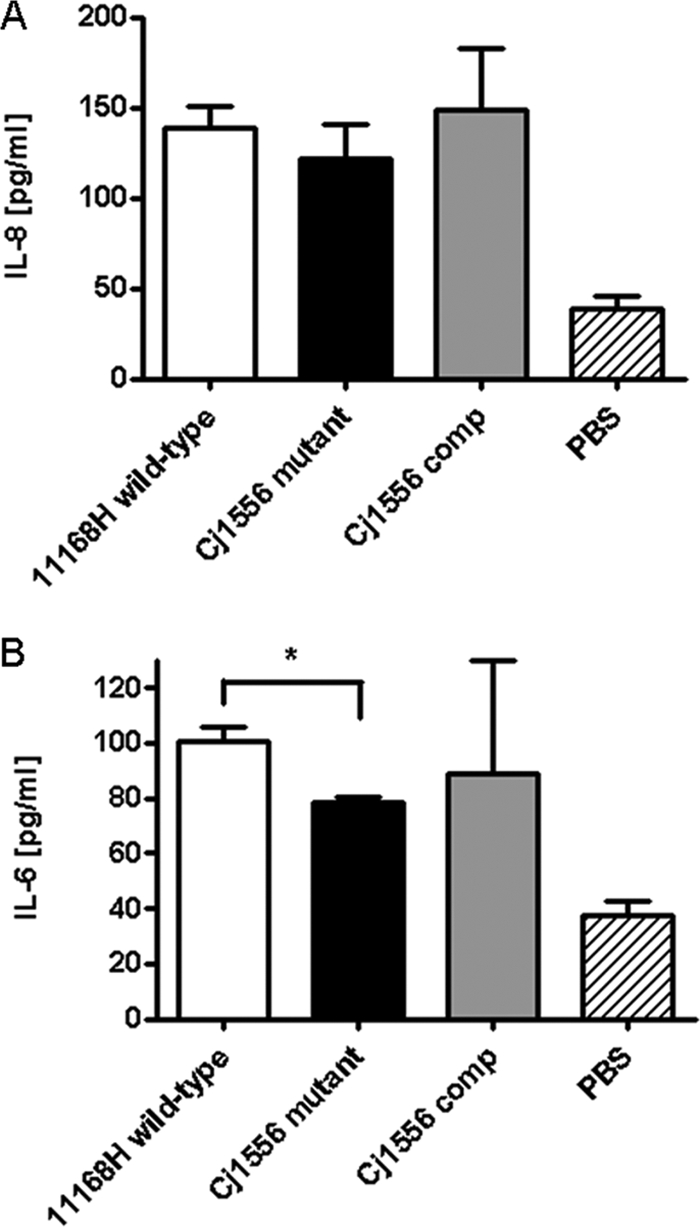
T84 intestinal epithelial cell responses to 24 h of coculture with the 11168H wild-type, Cj1556 mutant, and Cj1556 complement (Cj1556 comp) strains were assessed. The levels of IL-8 and IL-6 secreted during C. jejuni interaction with T84 cells were quantified using either a human IL-8 ELISA (A) or IL-6 ELISA (B). The asterisk denotes a statistically significant difference (P < 0.05) for the Cj1556 mutant compared to the 11168H wild-type strain.
Microarray analysis indicates negative autoregulation of Cj1556 expression.
To analyze the gene expression profile of the Cj1556 mutant compared to the 11168H wild-type strain, microarray experiments were performed using total-RNA samples isolated from C. jejuni grown to late log phase (16 h). A total of 91 genes were differentially expressed in the Cj1556 mutant compared to the 11168H wild type, with 73 genes upregulated and 18 genes downregulated based on an ANOVA selection methodology (5, 18). Interestingly, the gene with the most pronounced upregulation (10.4-fold) was Cj1556. Sequence analysis of the Cj1556 reporter element used on the arrays showed that this particular sequence was present upstream of the Kmr cassette in the Cj1556 mutant (data not shown). Usually, the mutated gene in a defined mutant would be expected to appear downregulated; however, the microarray data indicate that in the absence of the Cj1556 protein, Cj1556 gene expression is dramatically increased. This suggests that Cj1556 represses further expression of the Cj1556 gene, acting as a negative autoregulator. Further analysis of genes associated with oxidative and aerobic stress responses showed that many were downregulated in the Cj1556 mutant, including katA (5.13-fold), perR (5.05-fold), and hspR (2.07-fold) (Table 3), indicating potential reasons for the increased sensitivity of the Cj1556 mutant to these stresses.
Table 3.
Changes in expression of genes linked to the C. jejuni oxidative and aerobic stress responses in the Cj1556 mutant compared to the 11168H wild-type straina
| Gene name | Fold change | Product function |
|---|---|---|
| spoT | +1.26 | Putative guanosine-3′,5′-bis(diphosphate) 3′-pyrophosphohydrolase |
| sodB | +1.24 | Superoxide dismutase (Fe) |
| htrA | +1.21 | Serine protease (protease DO) |
| fdxA | +1.07 | Ferredoxin |
| dcuA | −1.17 | Anaerobic C4-dicarboxylate transporter |
| ahpC | −1.27 | Alkyl hydroperoxide reductase |
| dps | −1.36 | Putative bacterioferritin |
| hspR | −2.07 | Heat shock transcriptional regulator |
| perR | −5.05 | Peroxide stress regulator |
| katA | −5.13 | Catalase |
htrB showed no hybridization during microarray studies and was not included in this analysis.
Electrophoretic mobility shift assays indicate binding of Cj1556 to a DNA promoter probe upstream of the Cj1556 gene.
To investigate whether Cj1556 acts as a DNA binding protein and could potentially bind to the promoter region of the Cj1556 gene to repress further expression, as indicated by the microarray data, electrophoretic mobility shift assays were performed. The full-length Cj1556 protein was expressed and purified from E. coli. Binding of this recombinant Cj1556 protein to a 170-bp DNA fragment upstream of the Cj1556 gene was observed, indicating a protein-DNA complex (Fig. 6). Such binding of recombinant Cj1556 was not observed with DNA fragments representing the promoter regions of the negative-control genes flaA and flgK. These data indicate that Cj1556 acts as a DNA binding protein and also supports the microarray data that suggest a negative autoregulation system for the expression of Cj1556. Negative autoregulation is often a feature of the MarR family of transcriptional regulators (82).
Fig. 6.
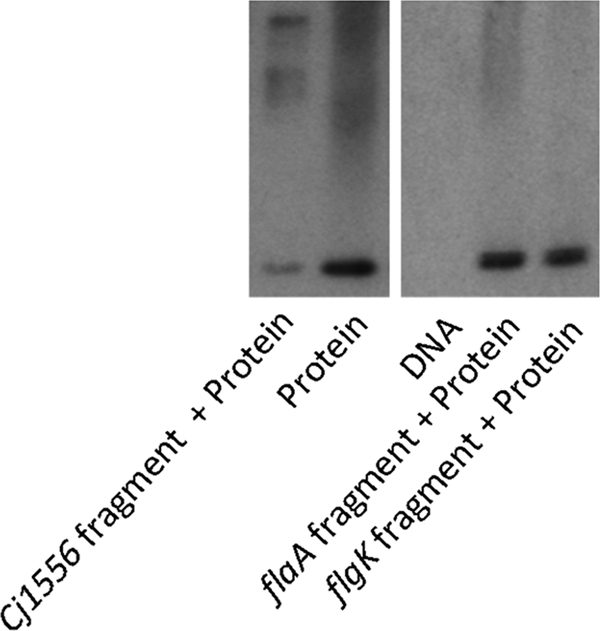
Electrophoretic mobility shift assays indicate that Cj1556 binds to a DNA promoter sequence upstream of the Cj1556 gene. Shown is a native Western blot for recombinant 6×His-tagged Cj1556 protein hybridized to DNA fragments representing the upstream promoter sequences of Cj1556 and flaA and flgK (both negative controls) following separation on a Tris-glycine gel under nondenaturing conditions.
The Cj1556 mutant exhibits reduced biofilm formation.
Biofilms are commonly defined as matrix-enclosed bacterial populations adherent to each other and/or to surfaces of interfaces (19). Studies have shown that C. jejuni can form biofilms (36) and that this may be an important factor in the survival of C. jejuni in the environment. Recent studies have also shown increased biofilm formation under aerobic stress conditions (65). The microarray data identified cprS as being 2.0-fold upregulated in the Cj1556 mutant compared to the 11168H wild-type strain. A cprS mutant has been shown to have enhanced and accelerated biofilm formation (71). Therefore, an increase in CprS production in the Cj1556 mutant was predicted to result in a decrease in biofilm formation. Analysis of the 11168H wild-type and Cj1556 mutant strains indicated a significant reduction in relative biofilm formation by the Cj1556 mutant (Fig. 7). Complementation of the Cj1556 mutation restored the wild-type phenotype (Fig. 7).
Fig. 7.
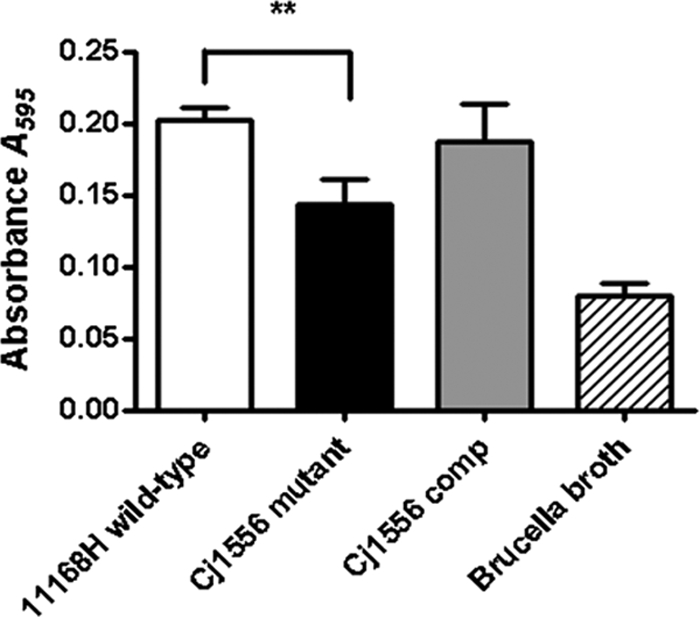
Biofilm assay of the 11168H wild-type, Cj1556 mutant, and Cj1556 complement (Cj1556 comp) strains. C. jejuni biofilms were grown for 3 days and rinsed three times with PBS, followed by crystal violet staining. The asterisks denote a statistically significant difference (P < 0.01) for the Cj1556 mutant compared to the 11168H wild-type strain.
G. mellonella larvae exhibit increased survival after infection with the Cj1556 mutant.
G. mellonella larvae have been used as a model to study infection by C. jejuni and other enteric pathogens (15, 17). Insect larvae are favorable to use as nonmammalian infection models, as they can be infected at 37°C and possess specialized phagocytic cells, termed hemocytes (8, 51). Hemocytes mimic the functions of phagocytic cells in mammals and are able to degrade bacterial pathogens and also generate bactericidal compounds, such as superoxide, via a respiratory burst (8, 42). Infection with the Cj1556 mutant resulted in a statistically significant increase in the survival of G. mellonella larvae compared to infection with the 11168H wild-type strain (Fig. 8). Complementation of the Cj1556 mutation restored the wild-type phenotype (Fig. 8). This suggests the Cj1556 mutant is more susceptible to the host immune mechanisms, resulting in reduced bacterial survival within G. mellonella.
Fig. 8.
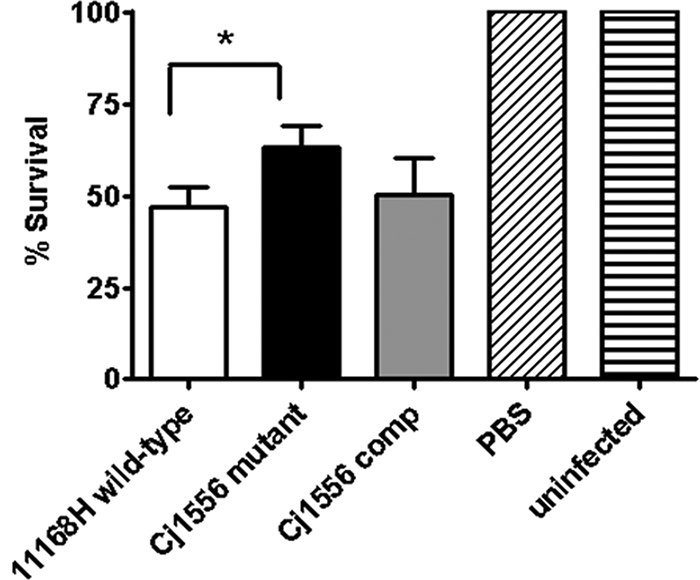
G. mellonella larvae were injected with a 10-μl inoculum of a 24-h C. jejuni culture diluted to an OD600 of 0.1 by microinjection in the right foremost leg, giving an infectious dose of approximately 106 CFU. The larvae were incubated at 37°C, with survival and appearance recorded at 24-h intervals. Brucella broth and no-injection controls were used. For each experiment, 10 G. mellonella larvae were infected, and the experiments were repeated in triplicate. The asterisk denotes a statistically significant difference (P < 0.05) for the Cj1556 mutant compared to the 11168H wild-type strain.
DISCUSSION
The human intestinal pathogen C. jejuni must survive diverse conditions in different hosts and also in the environment. The ability of C. jejuni to survive both oxidative and aerobic stress conditions is fundamental, considering the ubiquity of the bacterial pathogen. During reannotation of the C. jejuni NCTC11168 genome sequence (29), Cj1556 was identified as a putative transcriptional regulator. Based on motif and protein interaction data, we hypothesized that Cj1556 was an important C. jejuni stress response regulator and therefore investigated the ability of a Cj1556 mutant to survive different stresses and further explored the role of Cj1556 during host-pathogen interactions.
In addition to Cj1556, the C. jejuni NCTC11168 genome contains another CDS (Cj1546) with the MarR family motif. Cj1546 was also reannotated as a putative transcriptional regulator with 43.6% identity and 58.4% similarity to Cj1556. Analysis of a comparative genomics microarray data set containing 111 C. jejuni strains (16) identified Cj1546 in over 95% and Cj1556 in approximately 50% of these C. jejuni strains. One hypothesis as to the function of these MarR motif-containing proteins is that both perform similar roles in relation to aerobic and oxidative stresses; however, while all C. jejuni strains contain Cj1546, strains such as C. jejuni NCTC11168 and 81-176 that also contain Cj1556 may have a greater ability for survival within the human host due to greater resistance to oxidative stresses.
Oxidative stress assays showed that the Cj1556 mutant has an increased sensitivity to oxidative stress compared to the 11168H wild-type strain and that the wild-type level of sensitivity to oxidative stress was fully restored with complementation of the Cj1556 mutation. In fact, the Cj1556 complement demonstrated even greater resistance to H2O2 than the 11168H wild-type strain, possibly due to the strength of the promoter, as the complementation vector utilizes the constitutive chloramphenicol cassette promoter to express the Cj1556 gene and not the native Cj1556 promoter. C. jejuni proteins associated with heat stress responses, such as HspR, have also been linked to oxidative and aerobic stress (3). The Cj1556 mutant showed a greater level of sensitivity to 60°C stress than the wild-type strain. Heat stress above 55°C has been noted to accelerate the spiral-to-coccoid transition and results in cell death (54). Previous studies have identified numerous C. jejuni genes involved in heat shock response, and HtrA and HspR have also been shown to have roles in aerobic survival, host cell adherence, and invasion (12). Transcriptional analysis identified hspR as being approximately 2.0-fold downregulated in the Cj1556 mutant compared to the 11168H wild-type strain. It is interesting that the Cj1556 mutant has increased sensitivity to heat stress, and this may be due to Cj1556 interacting with HspR, suggesting a connection between the heat shock response and aerobic tolerance (3, 12).
The ability of the Cj1556 mutant to interact with (adhere and invade) and invade Caco-2 cells was investigated at 3-, 6-, and 24-h time points. Significant differences in both interaction and invasion were observed only at 24 h postinfection. This indicates that the Cj1556 mutant does not appear to have any defect in the ability to adhere to or invade Caco-2 cells but may have a reduced ability to survive contact with host cells over time. To further investigate longer-term survival, intracellular survival assays were performed. These assays indicated that the Cj1556 mutant has a reduced ability to survive within Caco-2 cells compared to the 11168H wild-type strain. The difference in the level of survival between the Cj1556 mutant and the 11168H wild-type strain in the intracellular survival assay at 24 h postinfection was approximately 0.5 log units (Fig. 3A), very similar to the difference in the number of invasive bacteria between the Cj1556 mutant and the 11168H wild-type strain at 24 h postinfection (Fig. 2B). However, the difference in the number of interacting bacteria between the Cj1556 mutant and the 11168H wild-type strain at 24 h postinfection was approximately 1.5 log units (Fig. 2A). This suggested that, in addition to a reduced ability for intracellular survival, the Cj1556 mutant was also more susceptible to extracellular stresses than the 11168H wild-type strain. The Cj1556 mutant exhibits increased sensitivity to H2O2 in vitro, so it is reasonable to suggest ROS released by Caco-2 cells during these experiments have an effect on C. jejuni survival. Standard coculture assays result in exposure of C. jejuni, not only to ROS released by Caco-2 cells, but also to aerobic stress, as the assays are performed in a CO2 incubator. The approximate atmospheric O2 and CO2 levels are around 21% and 0.04%, respectively. During coculture experiments, the level of CO2 is around 5%, so the O2 level will be around 16 to 18%. Based on the relative levels of survival between the interaction, invasion, and intracellular survival assays, we hypothesized that the greater level of sensitivity exhibited by the Cj1556 mutant during the interaction assay may be due in part to increased exposure of extracellular C. jejuni to aerobic stress. Aerobic survival assays were performed to replicate the conditions during the interaction, invasion, and intracellular survival assays by incubating C. jejuni in tissue culture medium, but in the absence of Caco-2 cells. A reduction in survival was observed for the Cj1556 mutant compared to the 11168H wild-type strain under these aerobic stress conditions, but not under microaerobic conditions. C. jejuni typically loses viability within intestinal epithelial cells over 24 h with no evidence of intracellular replication (41). Evidence to date suggests that C. jejuni resides in membrane-bound compartments termed C. jejuni-containing vacuoles (CCV), avoiding entry into lysosomes (81). C. jejuni engulfed by macrophages must resist a combination of unfavorable conditions, such as ROS. There are contradictory reports regarding the ability of C. jejuni to survive within macrophages, depending on the macrophage cell type and C. jejuni strain used (20, 79). In this study, the Cj1556 mutant exhibited reduced intracellular survival within the mouse macrophage J774A.1 cell line. Taken together, these data indicate that Cj1556 plays a multifactorial role in bacterial survival during adhesion to and invasion of human intestinal epithelial cells.
In this study, there was no significant difference in the levels of IL-8 induction by the Cj1556 mutant and the 11168H wild-type strain; however, a significant reduction in the level of IL-6 induction by the Cj1556 mutant compared to the 11168H wild-type strain was observed. IL-8 acts as a chemoattractant, allowing the recruitment of lymphocytes and neutrophils (32, 62), whereas IL-6 is believed to be important for epithelial cell integrity (27). It is possible that less IL-6 was induced when T84 cells were cocultured with the Cj1556 mutant than with the 11168H wild-type strain due to the decreased survival characteristic of the Cj1556 mutant strain. Based on data from this study, coculturing the Cj1556 mutant for 24 h in a 37°C CO2 incubator would result in decreased survival of the Cj1556 mutant based on the increased sensitivity of the strain compared to the 11168H wild-type strain. This may be a possible reason for the decreased IL-6 production. This result also suggests that IL-8 may be important for an extracellular response, as both the Cj1556 mutant and the 11168H wild-type strain elicited similar levels of IL-8 from T84 intestinal epithelial cells. However, IL-6 may be more important for an intracellular response, as the Cj1556 mutant was shown to invade less than the 11168H wild-type strain and so elicited less IL-6 from T84 intestinal epithelial cells.
The digestive secretion bile consists of around 50% bile salts, such as cholates and deoxycholates. Bile salts exhibit potent antibacterial properties, acting as detergents to disrupt cell membranes and as DNA-damaging agents (7). Although bacteria inhabiting the gastrointestinal tract are able to resist the antimicrobial effects of bile, a number of studies have also shown that bile increases the virulence potential of enteric pathogens (7). The bile salt sDOC has been shown to increase the virulence of C. jejuni, enhancing bacterial ability to invade epithelial cells (45). Growing C. jejuni in the presence of a physiologically relevant concentration of sDOC (0.1% [wt/vol]) changes the invasion kinetics so that maximal invasion of INT 407 cells occurs in under 30 min compared to 3 h for C. jejuni grown in the absence of sDOC (45). Microarray analysis has shown that a number of C. jejuni virulence factor genes are upregulated in the presence of 0.1% (wt/vol) sDOC, including ciaB, cmeABC, dccR, and tlyA (45). Interestingly, Cj1556 was also upregulated in the presence of sDOC, with transcription increased 2.8-fold (45). The transcriptional response of E. coli O157:H7 to bile treatment has also been investigated using microarrays and has identified bile-induced changes in transcription for genes encoding proteins affecting membrane structure and permeability, bile resistance, adhesion, and virulence potential (30). Most interestingly, these data indicate that bile induces expression of the marRAB operon by binding to the repressor protein MarR and thus preventing binding of MarR to the marRAB promoter site (30). Cj1556 is a member of the MarR family of transcriptional regulators, and further studies will be required to confirm whether bile can bind to the Cj1556 protein and thus prevent binding to the Cj1556 promoter site, resulting in the upregulation of Cj1556 in the presence of bile observed previously (45).
Microarray analysis of the Cj1556 mutant identified Cj1556 as the most upregulated gene. Analysis of the Cj1556 nucleotide sequence upstream of the Kmr cassette in the Cj1556 mutant confirmed that this was the sequence printed on the oligonucleotide array, suggesting that expression of Cj1556 is controlled by a negative autoregulation feedback mechanism. In the wild-type strain, basal levels of Cj1556 would block further expression of Cj1556 by inhibiting the binding of RNA polymerase to the Cj1556 promoter site. However, in the absence of Cj1556 in the Cj1556 mutant, expression of Cj1556 can continue. Such negative autoregulation is a feature of the MarR family of transcriptional regulators. In this study, experiments confirmed the binding of recombinant Cj1556 to a 170-bp DNA fragment upstream of the Cj1556 gene, confirming the DNA binding ability of Cj1556. To confirm that this was not a nonspecific artifact, two random negative-control promoter regions were selected (upstream of flaA and flgK). Both of the negative controls showed bands for the Cj1556 recombinant protein alone. The microarray data also indicated downregulation of katA, perR, and hspR in the Cj1556 mutant (Table 3). Reduced expression of KatA, PerR, and HspR would provide an explanation for the increased sensitivity of the Cj1556 mutant to oxidative, aerobic, and heat stresses observed in this study; however, further experiments are required to confirm this hypothesis.
The ability of C. jejuni to form biofilms goes some way to explain how a bacterium with such fastidious growth requirements remains ubiquitous in the environment (13, 36). C. jejuni can form three distinct forms of biofilm: cell-cell aggregates, pellicles at the air-liquid interface, and glass-attached flocs (36). Our understanding of the specific mechanisms underlying biofilm formation in C. jejuni is still limited (72). C. jejuni lacks the classical two-component regulatory systems involved in biofilm formation that are present in other bacteria, such as GacSA in Pseudomonas aeruginosa (59). Genes involved in biofilm formation have been linked to responses to oxidative and aerobic stress, and C. jejuni biofilm formation is increased under aerobic conditions (65). A C. jejuni spoT mutant has been found to overproduce a novel calcofluor white-reactive exopolysaccharide and to demonstrate enhanced biofilm formation (46). Interestingly, a C. jejuni cprS mutant has been shown to display growth defects and enhanced and accelerated biofilm formation and also to exhibit decreased oxidative stress tolerance (71). Transcriptional analysis of the Cj1556 mutant identified cprS as being upregulated, and the decrease in biofilm formation observed in this study indicates a potential link between CprS and Cj1556.
The G. mellonella insect model has been developed for potential identification of C. jejuni virulence determinants and was used to investigate the pathogenicity of the Cj1556 mutant (8). G. mellonella larvae possess specialized phagocytic cells, termed hemocytes. The insect immune system is subdivided into humoral and cellular defense responses. Humoral defenses include the production of antimicrobial peptides (47), reactive intermediates of oxygen or nitrogen (9), and the complex enzymatic cascades that regulate coagulation or melanization of hemolymph (50). Cellular defense refers to hemocyte-mediated immune responses, like phagocytosis, nodulation, and encapsulation (66, 70). Hemocytes perform many of the functions of phagocytic cells in mammals and are capable of ingesting bacterial pathogens and generating bactericidal compounds, such as superoxide, via a respiratory burst (8, 15). After infection of G. mellonella with Yersinia pseudotuberculosis, bacteria accumulate in hemocytes, thus suggesting that G. mellonella may be useful for the identification of other genes associated with intracellular survival (15). Infection with the Cj1556 mutant resulted in increased survival of G. mellonella larvae compared to survival after infection with the 11168H wild-type strain. This suggests the Cj1556 mutant is more susceptible to the host immune mechanisms, resulting in reduced bacterial survival within G. mellonella. At least six types of hemocytes have been identified in insects such as G. mellonella, with plasmatocytes and granulocytes the most abundant (10). Production of ROS has also been detected in hemocytes, with evidence of oxygen radicals and H2O2 both found in plasmatocytes of G. mellonella (68). These data link the increased sensitivity of the Cj1556 mutant to H2O2 stress in vitro with attenuation of virulence in vivo using the G. mellonella model of infection.
In summary, the basis of C. jejuni survival is dependent upon the ability to sense and respond to the different environments encountered within hosts and in the environment. This study has identified a novel C. jejuni transcriptional regulator, Cj1556, that is involved in oxidative and aerobic stress responses and is important for the survival of C. jejuni in the natural environment and in vivo.
ACKNOWLEDGMENTS
We thank Dennis Linton (University of Manchester, United Kingdom) for providing the C. jejuni NCTC11168 complementation vector. We thank Madeleine Moule, Sofia Lourenco, and Meredith Stewart (London School of Hygiene and Tropical Medicine) for technical support and guidance. We acknowledge BμG@S (the Bacterial Microarray Group at St George's, University of London) for supplying the microarray and advice.
Dominic Mills was supported by a Bloomsbury Colleges Ph.D. Studentship (2007 to 2010). We acknowledge the Wellcome Trust for funding the BμG@S multicollaborative microbial pathogen microarray facility under the Functional Genomics Resources Initiative.
Footnotes
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Allos B. M. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201–1206 [DOI] [PubMed] [Google Scholar]
- 2. Amabile-Cuevas C. F., Demple B. 1991. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 19:4479–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andersen M. T., et al. 2005. Diverse roles for HspR in Campylobacter jejuni revealed by the proteome, transcriptome and phenotypic characterization of an hspR mutant. Microbiology 151:905–915 [DOI] [PubMed] [Google Scholar]
- 4. Atack J. M., Kelly D. J. 2008. Contribution of the stereospecific methionine sulphoxide reductases MsrA and MsrB to oxidative and nitrosative stress resistance in the food-borne pathogen Campylobacter jejuni. Microbiology 154:2219–2230 [DOI] [PubMed] [Google Scholar]
- 5. Bacon J., et al. 2004. The influence of reduced oxygen availability on pathogenicity and gene expression in Mycobacterium tuberculosis. Tuberculosis 84:205–217 [DOI] [PubMed] [Google Scholar]
- 6. Baillon M. L., van Vliet A. H., Ketley J. M., Constantinidou C., Penn C. W. 1999. An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J. Bacteriol. 181:4798–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Begley M., Gahan C. G., Hill C. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29:625–651 [DOI] [PubMed] [Google Scholar]
- 8. Bergin D., Reeves E. P., Renwick J., Wientjes F. B., Kavanagh K. 2005. Superoxide production in Galleria mellonella hemocytes: identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect. Immun. 73:4161–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bogdan C., Rollinghoff M., Diefenbach A. 2000. Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Curr. Opin. Immunol. 12:64–76 [DOI] [PubMed] [Google Scholar]
- 10. Boman H. G., Hultmark D. 1987. Cell-free immunity in insects. Annu. Rev. Microbiol. 41:103–126 [DOI] [PubMed] [Google Scholar]
- 11. Boor K. J. 2006. Bacterial stress responses: what doesn't kill them can make then stronger. PLoS Biol. 4:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brondsted L., Andersen M. T., Parker M., Jorgensen K., Ingmer H. 2005. The HtrA protease of Campylobacter jejuni is required for heat and oxygen tolerance and for optimal interaction with human epithelial cells. Appl. Environ. Microbiol. 71:3205–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buswell C. M., et al. 1998. Extended survival and persistence of Campylobacter spp. in water and aquatic biofilms and their detection by immunofluorescent-antibody and -rRNA staining. Appl. Environ. Microbiol. 64:733–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cabiscol E., Tamarit J., Ros J. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3:3–8 [PubMed] [Google Scholar]
- 15. Champion O. L., et al. 2009. Galleria mellonella as an alternative infection model for Yersinia pseudotuberculosis. Microbiology 155:1516–1522 [DOI] [PubMed] [Google Scholar]
- 16. Champion O. L., et al. 2005. Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc. Natl. Acad. Sci. U. S. A. 102:16043–16048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Champion O. L., et al. 2010. Insect infection model for Campylobacter jejuni reveals that O-methyl phosphoramidate has insecticidal activity. J. Infect. Dis. 201:776–782 [DOI] [PubMed] [Google Scholar]
- 18. Corcionivoschi N., et al. 2009. Campylobacter jejuni alters surface capsular polysaccharide when co-cultured with epithelial cells. Infect. Immun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappin-Scott H. M. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 20. Day W. A., Jr., Sajecki J. L., Pitts T. M., Joens L. A. 2000. Role of catalase in Campylobacter jejuni intracellular survival. Infect. Immun. 68:6337–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dorrell N., et al. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duncan C., et al. 1995. Chemical generation of nitric oxide in the mouth from the enterosalivary circulation of dietary nitrate. Nat. Med. 1:546–551 [DOI] [PubMed] [Google Scholar]
- 23. Elvers K. T., et al. 2005. NssR, a member of the Crp-Fnr superfamily from Campylobacter jejuni, regulates a nitrosative stress-responsive regulon that includes both a single-domain and a truncated haemoglobin. Mol. Microbiol. 57:735–750 [DOI] [PubMed] [Google Scholar]
- 24. Eriksson S., Lucchini S., Thompson A., Rhen M., Hinton J. C. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118 [DOI] [PubMed] [Google Scholar]
- 25. Fang F. C. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2:820–832 [DOI] [PubMed] [Google Scholar]
- 26. Fields J. A., Thompson S. A. 2008. Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J. Bacteriol. 190:3411–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Friis L. M., Keelan M., Taylor D. E. 2009. Campylobacter jejuni drives MyD88-independent interleukin-6 secretion via Toll-like receptor 2. Infect. Immun. 77:1553–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gaynor E. C., Wells D. H., MacKichan J. K., Falkow S. 2005. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol. Microbiol. 56:8–27 [DOI] [PubMed] [Google Scholar]
- 29. Gundogdu O., et al. 2007. Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics 8:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hamner S., McInnerney K., Williamson K., Franklin M. J., Ford T. E. 2010. Bile salts influence the ordered expression of virulence genes in Escherichia coli O157:H7, abstr. ED12/12. SGM Spring 2010 Meet. Abstr. Society for General Microbiology, Reading, United Kingdom [Google Scholar]
- 31. Hitchen P., et al. 2010. Modification of the Campylobacter jejuni flagellin glycan by the product of the Cj1295 homopolymeric tract containing gene. Microbiology 158:1953–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hobbie S., Chen L. M., Davis R. J., Galan J. E. 1997. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J. Immunol. 159:5550–5559 [PubMed] [Google Scholar]
- 33. Imlay J. A., Linn S. 1986. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J. Bacteriol. 166:519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Iovine N. M., et al. 2008. Reactive nitrogen species contribute to innate host defense against Campylobacter jejuni. Infect. Immun. 76:986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jones M. A., et al. 2004. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect. Immun. 72:3769–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Joshua G. W., Guthrie-Irons C., Karlyshev A. V., Wren B. W. 2006. Biofilm formation in Campylobacter jejuni. Microbiology 152:387–396 [DOI] [PubMed] [Google Scholar]
- 37. Kamal N., et al. 2007. Deletion of a previously uncharacterized flagellar-hook-length control gene fliK modulates the sigma54-dependent regulon in Campylobacter jejuni. Microbiology 153:3099–3111 [DOI] [PubMed] [Google Scholar]
- 38. Karlyshev A. V., Linton D., Gregson N. A., Wren B. W. 2002. A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology 148:473–480 [DOI] [PubMed] [Google Scholar]
- 39. Ketley J. M. 1997. Pathogenesis of enteric infection by Campylobacter. Microbiology 143:5–21 [DOI] [PubMed] [Google Scholar]
- 40. Klancnik A., Botteldoorn N., Herman L., Mozina S. S. 2006. Survival and stress induced expression of groEL and rpoD of Campylobacter jejuni from different growth phases. Int. J. Food Microbiol. 112:200–207 [DOI] [PubMed] [Google Scholar]
- 41. Konkel M. E., Hayes S. F., Joens L. A., Cieplak W., Jr 1992. Characteristics of the internalization and intracellular survival of Campylobacter jejuni in human epithelial cell cultures. Microb. Pathog. 13:357–370 [DOI] [PubMed] [Google Scholar]
- 42. Lavine M. D., Strand M. R. 2002. Insect hemocytes and their role in immunity. Insect Biochem. Mol. Biol. 32:1295–1309 [DOI] [PubMed] [Google Scholar]
- 43. Linton D., et al. 2005. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 55:1695–1703 [DOI] [PubMed] [Google Scholar]
- 44. Lucchini S., Liu H., Jin Q., Hinton J. C., Yu J. 2005. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect. Immun. 73:88–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Malik-Kale P., Parker C. T., Konkel M. E. 2008. Culture of Campylobacter jejuni with sodium deoxycholate induces virulence gene expression. J. Bacteriol. 190:2286–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McLennan M. K., et al. 2008. Campylobacter jejuni biofilms up-regulated in the absence of the stringent response utilize a calcofluor white-reactive polysaccharide. J. Bacteriol. 190:1097–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meister M., Hetru C., Hoffmann J. A. 2000. The antimicrobial host defense of Drosophila. Curr. Top. Microbiol. Immunol. 248:17–36 [DOI] [PubMed] [Google Scholar]
- 48. Mihaljevic R. R., et al. 2007. Environmental stress factors affecting survival and virulence of Campylobacter jejuni. Microb. Pathog. 43:120–125 [DOI] [PubMed] [Google Scholar]
- 49. Monk C. E., Pearson B. M., Mulholland F., Smith H. K., Poole R. K. 2008. Oxygen- and NssR-dependent globin expression and enhanced iron acquisition in the response of campylobacter to nitrosative stress. J. Biol. Chem. 283:28413–28425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Muta T., Iwanaga S. 1996. The role of hemolymph coagulation in innate immunity. Curr. Opin. Immunol. 8:41–47 [DOI] [PubMed] [Google Scholar]
- 51. Mylonakis E., Casadevall A., Ausubel F. M. 2007. Exploiting amoeboid and non-vertebrate animal model systems to study the virulence of human pathogenic fungi. PLoS Pathog. 3:e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nachamkin I., Allos B. M., Ho T. 1998. Campylobacter species and Guillain-Barre syndrome. Clin. Microbiol. Rev. 11:555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Naito M., et al. 2010. Effects of sequential Campylobacter jejuni 81-176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J. Bacteriol. 192:2182–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nguyen H. T., Corry J. E., Miles C. A. 2006. Heat resistance and mechanism of heat inactivation in thermophilic campylobacters. Appl. Environ. Microbiol. 72:908–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Olson C. K., Ethelberg S., van Pelt W., Tauxe R. V. 2008. Epidemiology of Campylobacter jejuni infections in industrialized nations, p. 163–189 In Nachmkin I., Szymanski C. M., Blaser M. J. (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC [Google Scholar]
- 56. Oppenheim J. J., Zachariae C. O., Mukaida N., Matsushima K. 1991. Properties of the novel proinflammatory supergene “intercrine” cytokine family. Annu. Rev. Immunol. 9:617–648 [DOI] [PubMed] [Google Scholar]
- 57. Palyada K., et al. 2009. Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10:481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Parkhill J., et al. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668 [DOI] [PubMed] [Google Scholar]
- 59. Parkins M. D., Ceri H., Storey D. G. 2001. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 40:1215–1226 [DOI] [PubMed] [Google Scholar]
- 60. Parrish J., et al. 2007. A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 8:R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pesci E. C., Cottle D. L., Pickett C. L. 1994. Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect. Immun. 62:2687–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Philpott D. J., Yamaoka S., Israel A., Sansonetti P. J. 2000. Invasive Shigella flexneri activates NF-kappa B through a lipopolysaccharide-dependent innate intracellular response and leads to IL-8 expression in epithelial cells. J. Immunol. 165:903–914 [DOI] [PubMed] [Google Scholar]
- 63. Phongsisay V., Perera V. N., Fry B. N. 2007. Expression of the htrB gene is essential for responsiveness of Salmonella typhimurium and Campylobacter jejuni to harsh environments. Microbiology 153:254–262 [DOI] [PubMed] [Google Scholar]
- 64. Pittman M. S., et al. 2007. Growth of Campylobacter jejuni on nitrate and nitrite: electron transport to NapA and NrfA via NrfH and distinct roles for NrfA and the globin Cgb in protection against nitrosative stress. Mol. Microbiol. 63:575–590 [DOI] [PubMed] [Google Scholar]
- 65. Reuter M., Mallett A., Pearson B. M., van Vliet A. H. 2010. Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl. Environ. Microbiol. 76:2122–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schmidt O., Theopold U., Strand M. 2001. Innate immunity and its evasion and suppression by hymenopteran endoparasitoids. Bioessays 23:344–351 [DOI] [PubMed] [Google Scholar]
- 67. Sikic Pogacar M., et al. 2009. Survival of stress exposed Campylobacter jejuni in the murine macrophage J774 cell line. Int. J. Food Microbiol. 129:68–73 [DOI] [PubMed] [Google Scholar]
- 68. Slepneva I. A., Glupov V. V., Sergeeva S. V., Khramtsov V. V. 1999. EPR detection of reactive oxygen species in hemolymph of Galleria mellonella and Dendrolimus superans sibiricus (Lepidoptera) larvae. Biochem. Biophys. Res. Commun. 264:212–215 [DOI] [PubMed] [Google Scholar]
- 69. Stead D., Park S. F. 2000. Roles of Fe superoxide dismutase and catalase in resistance of Campylobacter coli to freeze-thaw stress. Appl. Environ. Microbiol. 66:3110–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Strand M. R., Pech L. L. 1995. Immunological basis for compatibility in parasitoid-host relationships. Annu. Rev. Entomol. 40:31–56 [DOI] [PubMed] [Google Scholar]
- 71. Svensson S. L., et al. 2009. The CprS sensor kinase of the zoonotic pathogen Campylobacter jejuni influences biofilm formation and is required for optimal chick colonization. Mol. Microbiol. 71:253–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Svensson S. L., Frirdich E., Gaynor E. C. 2008. Survival strategies of Campylobacter jejuni: stress responses, the viable but nonculturable state, and biofilms, p. 571–590 In Nachmkin I., Szymanski C. M., Blaser M. J. (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC [Google Scholar]
- 73. Trieu-Cuot P., Gerbaud G., Lambert T., Courvalin P. 1985. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 4:3583–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. van Vliet A. H., Baillon M. A., Penn C. W., Ketley J. M. 2001. The iron-induced ferredoxin FdxA of Campylobacter jejuni is involved in aerotolerance. FEMS Microbiol. Lett. 196:189–193 [DOI] [PubMed] [Google Scholar]
- 75. van Vliet A. H., Baillon M. L., Penn C. W., Ketley J. M. 1999. Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181:6371–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. van Vliet A. H., Wooldridge K. G., Ketley J. M. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 180:5291–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wainwright L. M., Elvers K. T., Park S. F., Poole R. K. 2005. A truncated haemoglobin implicated in oxygen metabolism by the microaerophilic food-borne pathogen Campylobacter jejuni. Microbiology 151:4079–4091 [DOI] [PubMed] [Google Scholar]
- 78. Wainwright L. M., Wang Y., Park S. F., Yeh S. R., Poole R. K. 2006. Purification and spectroscopic characterization of Ctb, a group III truncated hemoglobin implicated in oxygen metabolism in the food-borne pathogen Campylobacter jejuni. Biochemistry 45:6003–6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wassenaar T. M., Engelskirchen M., Park S., Lastovica A. 1997. Differential uptake and killing potential of Campylobacter jejuni by human peripheral monocytes/macrophages. Med. Microbiol. Immunol. 186:139–144 [DOI] [PubMed] [Google Scholar]
- 80. Watson R. O., Galan J. E. 2008. Campylobacter jejuni survives within epithelial cells by avoiding delivery to lysosomes. PLoS Pathog. 4:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Watson R. O., Galán J. E. 2008. Interaction of Campylobacter jejuni with host cells, p. 289–296 In Nachmkin I., Szymanski C. M., Blaser M. J. (ed.). Campylobacter, 3rd ed. ASM Press, Washington, DC [Google Scholar]
- 82. Wilkinson S. P., Grove A. 2004. HucR, a novel uric acid-responsive member of the MarR family of transcriptional regulators from Deinococcus radiodurans. J. Biol. Chem. 279:51442–51450 [DOI] [PubMed] [Google Scholar]
- 83. Wosten M. M., Boeve M., Gaastra W., van der Zeijst B. A. 1998. Cloning and characterization of the gene encoding the primary sigma-factor of Campylobacter jejuni. FEMS Microbiol. Lett. 162:97–103 [DOI] [PubMed] [Google Scholar]
- 84. Wosten M. M., et al. 2006. The Campylobacter jejuni PhosS/PhosR operon represents a non-classical phosphate-sensitive two-component system. Mol. Microbiol. 62:278–291 [DOI] [PubMed] [Google Scholar]
- 85. Wösten M. M. S. N., Mourik A. V., Putten J. P. M. V. 2008. Regulation of genes in Campylobacter jejuni, p. 611–624 In Nachmkin I., Szymanski C. M., Blaser M. J. (ed.). Campylobacter, 3rd ed. ASM Press, Washington, DC [Google Scholar]
- 86. Yang I. V., et al. 2002. Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol. 3:research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zaki M. H., Akuta T., Akaike T. 2005. Nitric oxide-induced nitrative stress involved in microbial pathogenesis. J. Pharmacol. Sci. 98:117–129 [DOI] [PubMed] [Google Scholar]