Abstract
Bacillus megaterium is deep-rooted in the Bacillus phylogeny, making it an evolutionarily key species and of particular importance in understanding genome evolution, dynamics, and plasticity in the bacilli. B. megaterium is a commercially available, nonpathogenic host for the biotechnological production of several substances, including vitamin B12, penicillin acylase, and amylases. Here, we report the analysis of the first complete genome sequences of two important B. megaterium strains, the plasmidless strain DSM319 and QM B1551, which harbors seven indigenous plasmids. The 5.1-Mbp chromosome carries approximately 5,300 genes, while QM B1551 plasmids represent a combined 417 kb and 523 genes, one of the largest plasmid arrays sequenced in a single bacterial strain. We have documented extensive gene transfer between the plasmids and the chromosome. Each strain carries roughly 300 strain-specific chromosomal genes that account for differences in their experimentally confirmed phenotypes. B. megaterium is able to synthesize vitamin B12 through an oxygen-independent adenosylcobalamin pathway, which together with other key energetic and metabolic pathways has now been fully reconstructed. Other novel genes include a second ftsZ gene, which may be responsible for the large cell size of members of this species, as well as genes for gas vesicles, a second β-galactosidase gene, and most but not all of the genes needed for genetic competence. Comprehensive analyses of the global Bacillus gene pool showed that only an asymmetric region around the origin of replication was syntenic across the genus. This appears to be a characteristic feature of the Bacillus spp. genome architecture and may be key to their sporulating lifestyle.
INTRODUCTION
Bacillus megaterium was first described by Anton De Bary more than 1 century ago in 1884 (14). Named for its large size, a “megat(h)erium” (Greek for big animal) of 1.5 by 4 μm, this microorganism is the largest of all bacilli. Long before Bacillus subtilis was introduced as a Gram-positive model organism, B. megaterium was used for studies on biochemistry as well as bacteriophages (13). The French microbiologist Maurice Lemoigne in 1925 discovered the polyester polyhydroxybutyrate in B. megaterium as an important energy storage molecule in bacteria (32), and Andre Lwoff discovered UV induction of bacteriophage in a lysogenic B. megaterium strain (35). Due to its large cell size, B. megaterium is well-suited for research on cell morphology, such as cell wall and cytoplasmic membrane biosynthesis, sporulation, spore structure and cellular organization, DNA partitioning, and protein localization (10, 61). In the 1960s, B. megaterium was used to study sporulation, since it sporulates and germinates efficiently (18). Because of its biotechnological use in the production of several substances, the nonpathogenic B. megaterium is of general interest to industry (7). In contrast to Gram-negative organisms like Escherichia coli, B. megaterium does not produce endotoxins associated with the outer membrane, which, combined with its growth on a variety of carbon sources and simple media, has made it a workhorse in food and pharmaceutical production processes for decades (i.e., α- and β-amylases used for starch modification in the baking industry and penicillin acylases essential for the synthesis of novel β-lactam antibiotics, among others [43]). Further, it is one of the most efficient producers of vitamin B12 (63, 64). B. megaterium has been extensively studied genetically and is amenable to genetic manipulation (63, 66). Hundreds of auxotrophs, division mutants, antibiotic-resistant, and UV-sensitive mutants have been characterized and have been previously mapped in QM B1551 (17, 20, 31, 55, 57, 62, 65). Strain QM B1551 is second only to B. subtilis in the number of multiply marked, characterized strains that are available from the Bacillus Genetic Stock Center (Vary collection, BGSC, Ohio State University; http://www.bgsc.org/). Several biotechnological mutants have been constructed in strain DSM319 and are commercially available (38, 40, 72). Recent analysis using phylogenetic analysis based on 16S rRNA genes led to the division of the genus Bacillus into 4 different families and 37 genera (34). Even after the taxonomic reorganization, Bacillus is a diverse genus with G+C content ranging from 34 to 35% (Bacillus cereus and related pathogens) to 44% (B. subtilis) and species that differ radically in lifestyles and metabolic properties. Most of the sequenced Bacillus genomes are closely related to B. cereus or B. subtilis. Recently, Porwal et al. showed that B. megaterium (with a G+C content of 38 to 39%) is only distantly related to the B. cereus and B. subtilis groups and that it is more deeply rooted in the phylogenetic tree than previously thought (44, 51). To gain insights into the genome evolution and the metabolic versatility that facilitate biotechnological applications, we have sequenced the complete genomes of B. megaterium strains QM B1551 and DSM319. We have used this information to examine the genetic diversity, genome dynamics, and phylogenetic relationships within B. megaterium and among members of the genus in great detail. Our genomic analysis reveals new genetic traits not previously seen in Bacillus and has resulted in a refined model for genome evolution and adaptation of B. megaterium.
MATERIALS AND METHODS
Bacterial strains.
Freeze-dried spores of B. megaterium QM B1551 prepared in 1967 by James C. Vary were provided by Patricia S. Vary (Northern Illinois University). These spores are closest in time to the original QM B1551 (Quarter Master Bacterium 1551; ATCC 12872) isolation by Hillel Levinson at the Pioneering Research Division, Quartermaster and Engineering Center, Natick, MA. B. megaterium DSM319 is a naturally plasmidless strain isolated by Stahl and Esser (53) and obtained from DSMZ (http://www.dsmz.de/). The plasmidless QM B1551 derivative, B. megaterium strain PV361 (56), was used for high-throughput phenotype arrays. PV361 was grown on rich and minimal media and showed no differences in sporulation compared to QM B1551. Germination of PV361 was performed on rich medium because it lacks pBM700, which carries key germination genes (55). Strain PV586, a Lac− derivative of PV361 (29), was used as transformation recipient in the replicon analysis of pBM600.
Detailed materials and methods describing genome sequencing, assembly, annotation, comparative genome analysis, pangenome computation, and the different experimental assays can be found in the supplemental material.
Nucleotide sequence accession numbers.
The B. megaterium complete genome sequences (QMB1551 project ID 30165; DSM319 project ID 33377) have been deposited in the NCBI GenBank under accession numbers CP001983 (chromosome, QM B1551), CP001984 to CP001990 (pBM100 to pBM700, respectively), and CP001982 (chromosome, DSM319).
RESULTS AND DISCUSSION
Genome architecture.
The chromosomes of B. megaterium strains QM B1551 and DSM319 are circular molecules of 5,097,129 bp and 5,097,447 bp, respectively, with an average G+C content of 38.2%. A well-defined G+C bias was observed, with the right-hand replichore significantly enriched in G+C relative to the left-hand replichore (Fig. 1 and Table 1). In addition, strain QM B1551 contains seven indigenous plasmids, pBM100 to pBM700, with sizes from 5.4 kb to over 164 kb, while strain DSM319 is a naturally plasmidless isolate (53) (Fig. 2 and Table 2; see also Fig. S1 in the supplemental material). The plasmids have significantly lower G+C contents than the chromosomes (33.0 to 36.5 versus 38.2%). The chromosome of strain QM B1551 contains 5,284 genes, and that of strain DSM319 contains 5,272 genes. The two chromosomes display a high level of genome conservation, having a nucleotide sequence identity of more than 95% over 83.3% of the length of the two chromosomes. The genomes are mostly colinear (Fig. 3), with only a single rearrangement larger than three genes: a 17-gene block in QM B1551 (Fig. 3, red arrows) is inverted and displaced by 1.7 Mbp relative to DSM319 (genes BMQ_1765 to BMQ_1781 are homologous with genes BMD_3632 to BMD_3616). The rearranged region does not appear to be a single functional unit: genes are on both strands and are involved in several different biological processes. Most of the genetic differences between the two genomes are due to insertions or deletions (indels) of single genes or small groups of genes at scattered and independent genomic locations throughout both chromosomes. We cataloged a total of 300 and 254 isolate-specific genes that are organized in 96 and 106 independent clusters for strains QM B1551 and DSM319, respectively (see Table S1 in the supplemental material). The distribution of these indels is not random (Fig. 1, circles 8 and 9): we found that the gene insertions in both chromosomes are rare in a 2-Mb region around the origin of replication (ori) (Fig. 1 and Fig. 3; see also Table S1). Compared to the function of genes common to both strains, strain-specific genes are increased in functions affecting interactions with the environment: cell envelope, transport, signal transduction, and gene regulation (Fig. 4; see also Fig. 9, below). In contrast, relatively fewer strain-specific genes are associated with basic cellular processes, such as amino acid, nucleotide, or cofactor biosynthesis, central intermediary metabolism, fatty acid metabolism, protein synthesis or degradation, or transcription. The isolate-specific genes are also more frequently annotated as conserved hypothetical genes or genes coding for enzymes of unknown specificity.
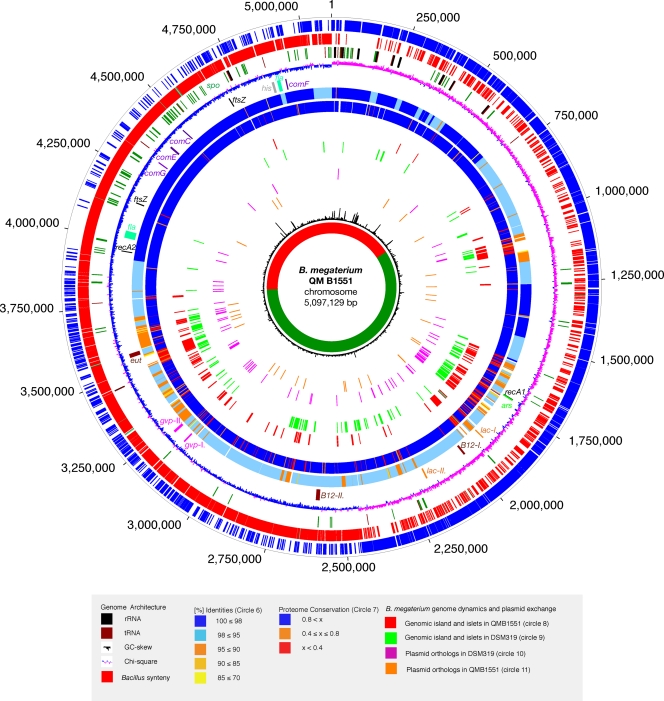
Fig. 1.
Circular representation of the B. megaterium chromosome. Circles (numbered 1 to 13, from outer to inner circle): circles 1 and 2, predicted open reading frames encoded on the QM B1551 plus (circle 1, blue) and minus (circle 2, red) strands; circle 3, predominant genes in conserved Bacillus region, showing tRNAs (brown), rRNA clusters (black), and sporulation and germination (dark green); circle 4, GC skew; circle 5, chromosomal regions of interest, including gas vesicle operons I and II (gvp; magenta), vitamin B12 operons I and II (B12; brown), flagellar operons (fla; aqua), RecA genes (rec; black), arsenate resistance operons (ars; green), β-galactosidase genes (lac; orange), ethanolamine utiliazation (eut; brown), histidine biosynthesis (his; gray), and competence operons CEFG (com; purple); circles 6 and 7, comparative analysis of the QM B1551 chromosome identities (circle 6) and proteome (circle 7) to DSM319; circles 8 and 9, nonrandom distribution of genomic islands and islets in QM B1551 (circle 8) and DSM319 (circle 9); circles 10 and 11, chromosomal genes with QM B1551 plasmid-borne orthologs in QM B1551 (circle 10) and DSM319 (circle 11); circle 12, chi-square values to show GC deviations; circle 13, region of conserved synteny (red) neighboring the ori identified by the comparative study of genome architectures in related Bacilli (see Fig. S4 in the supplemental material), with the remainder of the genome colored green.
Table 1.
Genomic properties of B. megaterium compared to related species
| Species | Strain | Genome size (Mbp) | % GC | No. of protein-coding genes | No. of tRNAs | No. of rRNA operons | Median BSR w/QM B1551 |
|---|---|---|---|---|---|---|---|
| B. megaterium | QM B1551 | 5.1 | 38.2 | 5,130, plus 499 on plasmids | 120, plus 19 on plasmids | 11, plus 1 on pBM400 | 1.00 |
| B. megaterium | DSM319 | 5.1 | 38.2 | 5,124 | 115 | 11 | 0.987 |
| B. subtilis | 168 | 4.2 | 43.5 | 4,176 | 85 | 10 | 0.335 |
| B. amyloliquifaciens | FZB42 | 3.9 | 46.5 | 3,813 | 86 | 10 | 0.299 |
| B. licheniformis | ATCC 14580 | 4.2 | 46.2 | 4,192 | 72 | 7 | 0.325 |
| B. pumilus | SAFR-032 | 3.7 | 41.3 | 3,678 | 69 | 7 | 0.295 |
| B. cereus | ATCC 10987 | 5.2 | 35.6 | 5,602 | 97 | 12 | 0.341 |
| B. anthracis | AO465 | 5.2 | 35.4 | 5,040 | 95 | 11 | 0.332 |
| B. thuringiensis | Al Hakam | 5.3 | 35.4 | 4,736 | 104 | 14 | 0.349 |
| B. weihenstephanensis | KBAB4 | 5.3 | 35.6 | 5,155 | 109 | 14 | 0.337 |
| B. halodurans | C-125 | 4.2 | 43.7 | 4,065 | 78 | 8 | 0.267 |
| B. clausii | KSM-K16 | 4.3 | 44.8 | 4,096 | 74 | 7 | 0.258 |
| Oceanobacillus iheyensis | HTE 831 | 3.6 | 35.7 | 3,500 | 69 | 7 | 0.265 |
| Listeria monocytogenes | EGD-e | 2.9 | 38.0 | 2,846 | 67 | 6 | 0.195 |
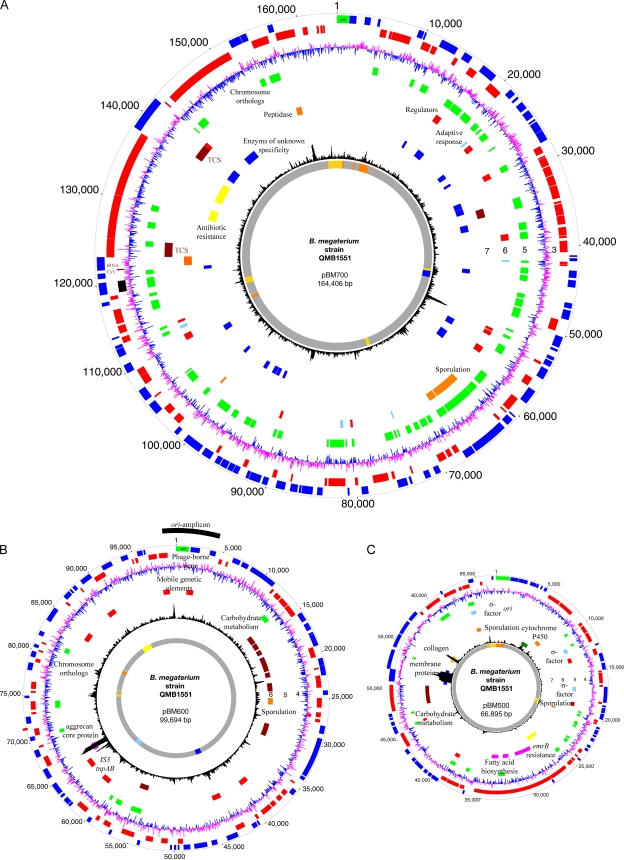
Fig. 2.
Circular representation of the B. megaterium plasmids pBM500, pBM600, and pBM700. Circles 1 and 2, predicted open reading frames encoded on the QM B1551 plus (circle 1, blue) and minus (circle 2, red) strands with the replication gene dnaA (green); circle 3 (circle 4 in pBM700 representation), GC skew; circle 4 (circle 5 in pBM700 representation), plasmid genes with chromosomal orthologs (green) in strains QM B1551 and DSM319. The two innermost circles are the chi square deviations from the average trinucleotide distribution and the results of a comparative analysis of each plasmid with the other seven indigenous plasmids of QM B1551, showing nucleotide identities (Fig. 1). (A) Regions and genes of interest in pBM700. Circle 3, tRNA-Cys and mobile genetic elements (black); circle 6, regulatory functions, including transcription regulators (red), two-component sensor histidine kinase systems (TCS; brown), adaptive response (light blue), and sporulation (orange); circle 7, metabolic features, including carbohydrate metabolism (brown) and two peptidases (gold); circle 8, several enzymes of unknown specificity (blue) and for antibiotic resistance (yellow). (B) Regions and genes of interest in pBM600. The origin amplicon is shown in black, with the identified replication gene in green. Circle 5, mobile genetic elements (red). The IS3 transposase is adjacent to an aggrecan core (proteoglycan) protein that is further distinguished by a deviating GC content, which suggests horizontal acquisition; circle 6, carbohydrate metabolism (brown) and sporulation genes (orange). (C) Regions and genes of interest in pBM500. Circle 5, regulatory functions, including transcription regulators (red) and three sigma factors (light blue); circle 6, metabolism features, including carbohydrate metabolism (brown) and fatty acid biosynthesis (magenta); circle 7, sporulation (orange) and erm(B) macrolide resistance (yellow); circle 8, regions of deviating GC content, membrane protein (blue), collagen-like protein (gold), and cytochrome P450 (green).
Table 2.
Plasmid content of B. megaterium strain QM B1551
| Plasmid | Size (bp) | Read count | Read coverage | Copy no. | CDS | rRNA | tRNA | % GC | Replicon |
|---|---|---|---|---|---|---|---|---|---|
| pBM100 | 5,428 | 253 | 40× | 3 | 8 | 0 | 0 | 34.8 | Rolling circle |
| pBM200 | 9,098 | 1,826 | 169× | 14 | 13 | 0 | 0 | 34.5 | Rolling circle |
| pBM300 | 26,587 | 7,052 | 224× | 18 | 38 | 0 | 0 | 35.5 | Theta |
| pBM400 | 53,865 | 7,849 | 123× | 10 | 60 | 3 | 18 | 36.5 | Theta |
| pBM500 | 66,985 | 5,163 | 65× | 5 | 94 | 0 | 0 | 33.9 | Theta |
| pBM600 | 99,694 | 2,334 | 20× | 2 | 111 | 0 | 0 | 33.0 | Theta (this study) |
| pBM700 | 164,406 | 2,365 | 12× | 1 | 175 | 0 | 1 | 33.5 | Theta |
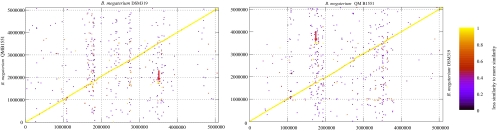
Fig. 3.
Genome synteny of B. megaterium. The genomes of strains QM B1551 and DSM319 are mostly colinear, but the distribution of isolate-specific genes is not random: gene insertions are rare in a 2-Mbp region around the origin of replication (ori). Each protein of the reference genome plotted on the x axis was used as the query sequence with BLASTP against the y axis subject genome. The N-terminal coordinates of the best hit proteins were plotted. The colors represent the level of similarity of the match, expressed by the BLAST score ratio (48).
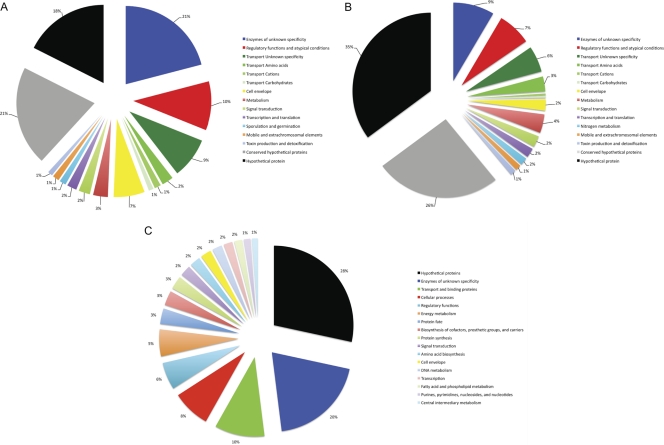
Fig. 4.
Functional classification of the isolate-specific gene inventory of B. megaterium strains QM B1551 and DSM319 compared to the DSM319 chromosome. Unique genes in strains QM B1551 (A) and DSM319 (B) are indicated. (C) Functional classes of total chromosomal genes of DSM319.
Fig. 9.
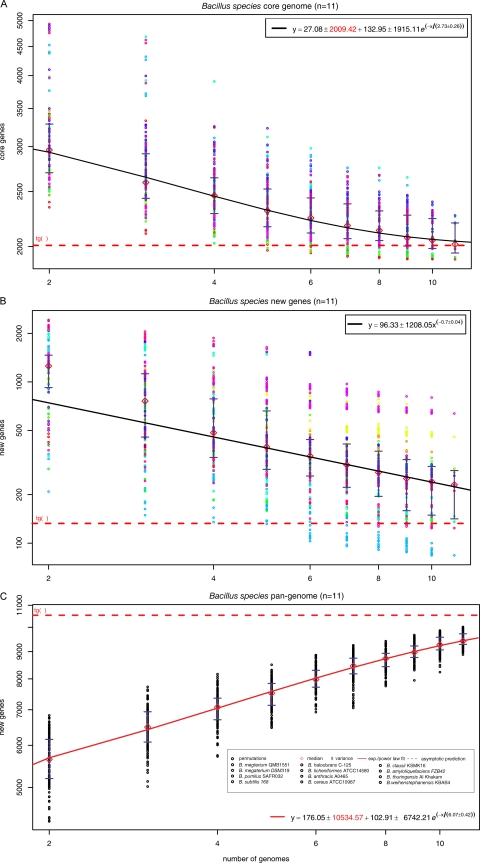
Pan-genome analysis and genomic plasticity in the Bacillus group of organisms. (A) Core genes. For each reported number of genomes (n), the circles represent the number of genes in common in different randomly chosen combinations of Bacillus species, with a sampling size of 1,000. Diamonds show the median values for each distribution. The curve represents the exponential regression of the least squares fit of the function Fcore(n) = κc exp[−n/τc] + tgc(θ), based on the medians of the distribution. The extrapolated core genome size is shown as a horizontal dashed red line. (B) Gene discovery. By using the same sampling method as for panel A, the number of new genes found was plotted for increasing values of n. A power law regression for new genes discovered was fitted to the means of new gene counts (diamonds) for each value of n. The curve is the least squares fit of the exponential decay equation Fnew(n) = κn exp[−n/τn] + tgn(θ), based on the means of the distribution. The value of tgn(θ) shown in this figure represents the number of new genes asymptotically predicted for further genome sequencing. (C) The Bacillus pan-genome. The total numbers of genes found according to the pan-genome analyses are shown for increasing values of the number (n) of Bacillus genomes sequenced, using medians and an exponential fit. Red diamonds indicate the means of the distributions. The dashed line represents the asymptotic prediction of the total number of genes expected to be found in the Bacillus pan-genome.
Plasmid analyses.
Plasmids make up 11% of the QM B1551 genome (24). This strain harbors seven indigenous plasmids with plasmid copy numbers ranging between 1 and 18 copies (Table 2). Since many other strains of B. megaterium carry multiple plasmids (63), this is a critical part of the genome analysis for this species. The three largest plasmids are shown in Fig. 2. Plasmids pBM100 and pBM200 of QM B1551 were previously sequenced and deposited in GenBank. Plasmids pBM300 and pBM400 have been analyzed (30, 52, 63) and have been resequenced to higher coverage and reannotated for this study (see Fig. S1 in the supplemental material). The plasmids carry a variety of genes, including genes for sporulation, germination, regulation, transport, and lantibiotic synthesis, as well as erythromycin and rifampin resistance. There are also genes for fatty acid metabolism, cell wall hydrolysis, sigma factors, and cell division, as well as integrons, insertion sequence (IS) elements, and transposons. Because B. megaterium is frequently found with Pseudomonas species in contaminated environments, it has long been suspected of having the capability of metabolizing unusual substrates of possible bioremedial use (30, 52, 61). To that extent, B. megaterium plasmids carry genes for heavy metal resistance, including Cu and Cd export, and genes such as styrene monooxygenase are present. Several metabolic genes on the larger plasmids are organized in what appear to be functional operon structures and may enable this strain to survive in unusual habitats (Fig. 2). The possible substrates for associated transporters cannot be deduced from genomic annotation, but the identification of these regions provides the basis for further experimental characterization.
Of special note is that all seven indigenous plasmids appear to be unique to B. megaterium compared to a broad panel of diverse Bacillus spp. plasmids. Besides similarities in insertion sequence genes among the plasmids, we found three genes similar to the pXO1 virulence plasmid of B. anthracis, including a reverse transcriptase (52) (see Fig. S1 in the supplemental material). This finding may indicate either a common phylogenetic origin or genetic exchange among these plasmid-borne genes. Plasmid pBM500 (Fig. 2) is intriguing because of the presence of three possible sigma factors (BMQ_pBM50015, BMQ_pBM50023, and BMQ_pBM50090), a collagen-like gene (BMQ_pBM50081), and an erm(B) resistance gene (BMQ_pBM50045). In addition, a cytochrome P450 gene (BMQ_pBM50008) is plasmid-borne. The P450 enzymes of B. megaterium have long been used as a model system (15, 20, 21). There is a gene cluster that appears to be responsible for the biosynthesis of a fatty acid compound (BMQ_pBM50048 to BMQ_pBM50053). Plasmid pBM600 seems to be a depository for mobile genetic elements, since it carries four transposases, three integrase genes, and a group II intron. It also has eight glycosyltransferases and other glucose carrier proteins within a 25-kb region (kb 13.9 to 39.6). Plasmid pBM700 encodes several transferases and transporters, transcriptional regulators, and bacteriocin and antibiotic synthesis operons. In addition, two two-component histidine kinase regulatory systems are present (BMQ_pBM70140/141 and BMQ_pBM70155/156), strengthening the role of this plasmid in metabolic activities and strain-specific niche adaptations. Previous studies on the pBM700 germination (ger) operons have helped identify specific amino acid residues that define receptor specificity to germinant compounds in Bacillus (11, 12). Strains of QM B1551 cured of pBM700 can no longer germinate on single germinant compounds (55), and the complete germination operon, with gerUA, gerUC, and gerUB and including the downstream monocistronic gene gerVB (BMQ_pBM70070 to BMQ_pBM70073), has been identified (10, 12).
Plasmid replicon analyses.
With the exception of pBM600 (Fig. 2), all replicons have been previously identified and functionally analyzed (30, 52, 64). A region containing a putative replicon gene, BMQ_pBM60001, was identified and cloned in pJM103, a plasmid that cannot replicate in B. megaterium. The clone was successfully maintained in B. megaterium PV586, indicating that it is the pBM600 replicon. The presence of a characteristic plasmid replication motif, including six copies of a 21-bp iteron upstream from BMQ_PBM60001, indicates it is most likely a theta replicon. However, the identified replicon shows no homology with the other four theta plasmid replicons in QM B1551. We note that orthologs of QM B1551 plasmid genes are found not only on the genome of QM B1551 but also on DSM319 (Fig. 1; see also Table S2 in the supplemental material). This observation indicates that strain DSM319 is potentially capable of acquisition and maintenance of plasmids.
Evidence for the exchange of genes between plasmids and chromosomes.
We have observed extensive gene exchange between QM B1551 plasmids and the main chromosomes of both strains (Fig. 1, circles 10 and 11). Of the QM B1551 499 protein-coding plasmid genes, 104 have homologs (BSR, >0.4) on either the QM B1551 or DSM319 chromosome (see Table S2). Some genes share more than 90% sequence identity over the full peptide length with their chromosomal homologs. Plasmid genes with homologs on the chromosomes are found on all plasmids except pBM100 (5.4 kb). The majority of genes are encoded on pBM400, pBM500, and pBM700, while only a few genes are found on pBM200, pBM300, and pBM600 (see Table S2). These genes can be classified into three groups. There are 19 genes present on QM B1551 plasmids that have homologs on the QM B1551 chromosome but not on the DSM319 chromosome. Conversely, 32 plasmid genes have homologs on the DSM319 chromosome but not the QM B1551 chromosome. The remaining 53 genes have homologs on both the QM B1551 and DSM319 chromosomes (see Table S2). These findings demonstrate that some of the plasmid-borne capabilities of QM B1551 are also found in the plasmidless DSM319 strain. The fact that the number of genes shared between the plasmids and QM B1551 chromosome is smaller than the number shared with the DSM319 chromosome suggests that the plasmidless state of DSM319 might be the result of a recent event. The homologous genes are found scattered in different locations, both on the plasmids and on the DSM319 and QM B1551 chromosomes. Several of the genes are found in clusters of 2 to 11 genes and appear to have been cotransferred, as they have the same relative positions and orientations on both the plasmid and chromosomes with conserved intergenic nucleotide homology. This set of genes encodes a variety of physiological functions, such as metabolism, transport, regulation, sporulation, and germination, and include 45 conserved hypothetical CDS with no assigned function.
Spore coat biosynthesis.
We detected major differences in the genes involved in spore coat structure and its biosynthesis between B. megaterium and B. subtilis strain 168 (see Table S3 in the supplemental material). More than half of the B. subtilis genes involved in spore coat formation are missing and could not be identified in either B. megaterium strain, which suggests that B. megaterium possesses a different spore coat structure than B. subtilis (23).
Natural competence genes of B. megaterium.
Transformation competence has been well-studied in the Gram-positive model organism B. subtilis 168 (16). To date, no conditions have been found that induce natural competence in B. megaterium. B. megaterium genomes contain 33 competence orthologs to genes known to mediate competence in B. subtilis (see Table S4 in the supplemental material) (28) and that may allow B. megaterium to incorporate foreign genetic material. The genome data revealed that B. megaterium lacks the comQXPA gene cluster and two other competence genes, comFB and comS, that are present in B. subtilis. Having detected almost all known genes for competence in other bacilli (see Table S4), we speculate that competence would be easy to engineer in the well-studied B. megaterium strains. Furthermore, it may occur naturally in some strains of the species.
Flagellar genes of B. megaterium.
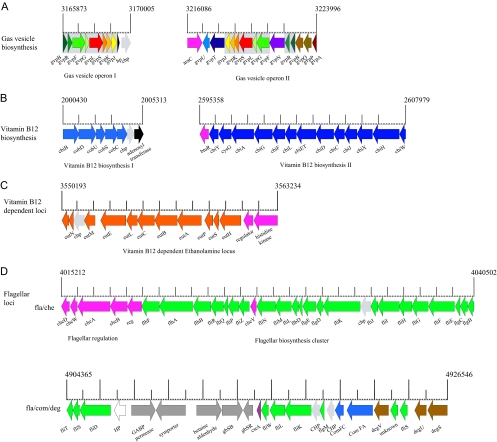
B. megaterium QM B1551 is motile but requires an oxygen-permeable coverslip for sustained motility (P. S. Vary, unpublished data). The flagellar genes of both QM B1551 and DSM319 form two clusters that are almost identical to those found in B. subtilis (Fig. 1 and 5D). Cluster one corresponds to the fla/che 26-kb operon in B. subtilis (42). In this cluster, only CheC, a signal-terminating phosphatase important in chemotaxis protein methylation, is missing from the operon and not found elsewhere in B. megaterium. The second and larger cluster is more complex (Fig. 5D). The fliT, fliS, and fliD genes match those of B. subtilis, but flaG is not found in B. megaterium. Moreover, the B. subtilis gene hag, which codes for flagellin, is not part of the cluster but is found almost 2 Mb away (BMQ_1093), not near any other flagellar genes except an adjacent short flagellin domain gene (BMQ_1092), which is presumably a gene fragment that may have been created during the transposition events that moved the hag gene to its new location. In its place in the cluster are six genes that apparently mediate unrelated metabolic functions (BMQ_5103 to BMQ_5108). These genes code for a GABA permease and associated symporter, a betaine aldehyde dehydrogenase, an alcohol dehydrogenase, and a GbsR transcriptional regulator, as well as a hypothetical protein. The syntenic genes consist of flagellar genes (fliWLKS), two competence-associated genes (comFA and comFC), and the degS-degU genes, which code for a two-component system that regulates the transition from exponential to stationary growth. We note here that DegU has also been shown to regulate motility via σD in B. subtilis (36).
Fig. 5.
Genome features of B. megaterium QM B1551. (A) Gas vesicle-forming proteins. The B. megaterium QM B1551 chromosome features two unique gas vesicle protein-forming loci (gvp) not found in any of the related species or genera. DSM319 has only operon II. The scale (in bp) indicates the genomic location of the gas vesicle loci, which are composed of 8 and 14 genes (gvpBRFGLSKJ and gvpAPQBRNFGLSKJTU). A possible araC regulator is also present. The loci are syntenic and share 8 homologous genes, with the notable exception of gvpN. The absence of colocalizing mobile genetic loci and average GC content suggest a duplication event in strain QM B1551 rather than acquisition via horizontal gene transfer. Homologous genes have been given identical colors and shaded in gray. (B) Vitamin B12 biosynthesis. Both B. megaterium strains encode three phylogenetically unrelated loci that are associated with vitamin B12 biosynthesis; the two multigene loci are shown here. The scale (in bp) indicates the genomic location of these loci in strain QM B1551. Regulatory genes are colored magenta. (C) Vitamin B12-dependent loci. Several enzymes with dependencies on vitamin B12 have been identified in the B. megaterium genome, such as ethanolamine ammonia-lyase (EutBC) and methylmalonyl-CoA mutase (MutAB). The genes eutCBA are also found at Mb 2.52 to 2.53 in both strains. (D) Flagellar biosynthesis. The two flagellar biosynthesis clusters, fla/che and fla/com/deg in QM B1551, are shown, with the upstream regulatory genes colored magenta. The fla/com/deg cluster contains flagellar, competence, and the extracellular enzyme regulatory genes degU and degS.
Metabolic and physiologic capabilities.
The genomic sequences were used to investigate the underlying molecular nature of some of the unique physiological and metabolic features of B. megaterium. The transporter inventory of both B. megaterium strains and potential substrates (http://www.membranetransport.org/index.html) has been analyzed using TransportDB (50).
B. megaterium carries genes for the glyoxylate pathway. In keeping with its known phenotype, B. megaterium uses the Embden-Meyerhof pathway for glycolysis, followed by a classical Krebs cycle. In contrast to other bacilli, B. megaterium contains an intact glyoxylate pathway (see Fig. S2A in the supplemental material). From this starting point, the B. megaterium genome contains genes for the synthesis of all 20 proteinogenic amino acids, all nucleotides and cofactors used by other enzymes, and all necessary fatty acids. Genes for the transporters and special enzymes needed to utilize a wide variety of carbon sources have been found. Aerobic respiration involves three membrane-bound dehydrogenases to transfer electrons from NADH, glycerol-3-phosphate, and succinate to menaquinone (see Fig. S2B). Menaquinone is oxidized by two menaquinone oxidases of the cytochrome bd type, by a cytochrome aa3-type oxidase, which is not found in other bacilli, or by the cytochrome bc1 complex, followed by cytochrome c and cytochrome c oxidase. B. megaterium also contains a fourth oxidase with high sequence similarity to E. coli cytochrome o ubiquinol oxidase, which is not found in other bacilli. Under anaerobic conditions, B. megaterium has all the genes required to perform a mixed acid fermentation that produces lactate, 2,3-butanediol, and acetate, as previously described for B. subtilis (see Fig. S2C) (41, 47). Unlike most other bacilli, B. megaterium is not capable of using nitrate as an electron acceptor, since it lacks a membrane-bound nitrate reductase of the Nar type. Further details about B. megaterium energy metabolism are found in Fig. S2 of the supplemental material.
Strain-specific carbon source utilization.
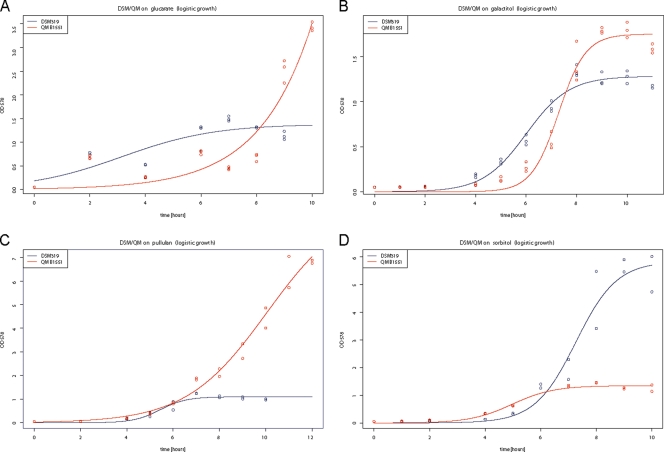
Predictions for the metabolic capabilities of each strain were deduced from in silico comparative genome analysis and validated by growth experiments in vivo (Fig. 6). For strain QM B1551, several unique glucarate-degrading enzymes were found in an operon-like structure, including glucarate permease (BMQ_1892), glucarate dehydratase (BMQ_1893), and the degradative enzymes galactarate dehydratase (BMQ_1888) and 5-dehydro-4-deoxyglucarate dehydratase (BMQ_1890). The final product of this glucarate degradation process is 2,5-dioxopentanoate, which is discharged into the citrate cycle via 2-oxoglutarate. Furthermore, a complete galactitol-specific phosphotransferase system (PTS) is found to be unique in B. megaterium QM B1551 (BMQ_3201 to BMQ_3204). The genes for the corresponding enzymes galactitol-1-phosphate 5-dehydrogenase (BMQ_3200) and an alcohol dehydrogenase (BMQ_3199) are also part of this genomic island. We demonstrated experimentally that B. megaterium QM B1551 (in contrast to strain DSM319) can utilize both glucarate and galactitol as carbon sources (Fig. 6A and B). Additionally, growth experiments on pullulan showed that B. megaterium QM B1551 grows more effectively on pullulan than does DSM319 (Fig. 6C), although both strains are equipped with the same chromosomal pullulanases, AmyX (BMQ_4837) and PulA (BMQ_2037). Strain DSM319 encodes a cellulase-like glycosyl hydrolase (BMD_1113), an additional citrate two-component system (BMD_2292/2293), part of a sucrose-specific PTS (BMD_3721), sorbitol dehydrogenase GutB (BMD_3588) with its transcriptional activator GutR (BMD_3589), and a probable sorbitol transporter (BMD_3587). The sorbitol uptake system proved to be very effective in DSM319 (Fig. 6D), with optical growth densities of 6, while that for QM B1551 was only 1.2.
Fig. 6.
Growth of B. megaterium strains DSM319 (blue) and QM B1551 (red) on different nutrients. Strain-specific gene differences deduced from the analysis of the isolate-specific genes were validated by growth on different nutrients: (A) glucarate, (B) galactitol, (C) pullulan, (D) sorbitol. The optical density was recorded over a time course of up to 13 h. The experimental data were fitted to logistic or exponential growth curves.
Osmotic adjustment and compatible solute accumulation.
In its varied habitats, B. megaterium is exposed to increased osmolarity that triggers water efflux from the cell. It therefore has to adjust the osmotic potential of its cytoplasm to prevent dehydration. The physiology and genetics of osmotic adjustment to high-osmolarity surroundings has been studied intensively in B. subtilis (6). In the initial phase of adjustment to high osmolarity, B. subtilis accumulates large amounts of potassium via the KtrAB and KtrCD systems to curb the efflux of water (22). Both Ktr potassium uptake systems are also present in B. megaterium (BMQ_3903/3904 and BMQ_1339/1233). In the second phase of osmotic adjustment, B. subtilis synthesizes large quantities of the compatible solute proline and acquires various types of organic osmoprotectants (e.g., glycine betaine) from environmental sources via high-affinity transport systems. Recent studies have shown that B. megaterium DSM32 synthesizes proline when it is challenged by high salinity (9). The in silico analysis of the genome sequences of QM B1551 and DSM319 revealed a complete proline biosynthetic gene cluster (proHJA; BMQ_2287 to BMQ_2289) whose counterparts are osmotically induced in B. subtilis, B. licheniformis, and Halobacillus halophilis (26, 71). B. megaterium may also use an exogenous supply of proline for osmoprotective purposes, since it possesses a homolog (BMQ_1420) of the osmotically induced proline uptake system OpuE from B. subtilis (6). Glycine betaine is a very effective osmoprotectant, and B. subtilis can acquire it either from environmental sources or synthesize it from an exogenous supply of the precursor choline via a two-step oxidation process. Based on homology to B. subtilis, several glycine betaine import systems were identified in B. megaterium. Two ABC transporters of the OpuA type, opuACAB (BMQ_0858 to BMQ_0860) and opuAABC (BMQ_1542 to BMQ_1544), an OpuC-like glycine betaine/choline importer (BMQ_3925/3926), and an OpuD-like BCCT-type transport system (BMQ_1351) are present. In addition, a gene cluster encoding the glycine betaine biosynthetic enzymes GbsA and GbsB (BMQ_5106/5107) are present.
Adaptations to an aquatic lifestyle.
Besides its most common soil habitat, B. megaterium is also found in diverse environments, including rice paddies, dried food, seawater, sediments, fish, and even in bee honey (63). Analysis of B. megaterium genome sequences strongly indicates that it is well-adapted to an aquatic lifestyle, since it possesses genes for the formation of gas vesicles. These intracellular structures are often found in marine microorganisms and function as flotation devices, allowing the bacterium to float up and down by adjusting the gas pressure inside the gas vesicle (60). The genome of B. megaterium QM B1551 has two gas vesicle-forming operons (Fig. 5A). The two gvp loci share eight homologous genes (gvpBRFGLSKJ) and are syntenic except for gvpN, which is found between gvpR and gvpF in the larger operon. The smaller locus (BMQ_3224 to BMQ_3231) is not present in DSM319. The larger operon encompasses 14 genes, gvpAPQBRNFGLSKJTU (BMQ_3290 to BMQ_3303), and an araC-like regulator. This gene cluster was previously discovered in B. megaterium strain VT1660, and its functional expression in E. coli renders this bacterium buoyant (33). It should be noted that genes for gas vesicles have been found in genomes of many microorganisms that do not typically live in aquatic habitats, fostering discussions on the physiological function of these floating devices (60). The proteins involved in gas vesicle formation in B. megaterium show a phylogenetic relationship far outside the Bacillus group and are related to proteins present in aquatic Cyanobacteria but also in members of Archaea. One explanation for the gas vesicles could be related to the ability of the bacterium to leave aquatic regions of low oxygen tension.
Biotechnological use of B. megaterium for protein production.
One major advantage of B. megaterium for industrial protein production is its ability to secrete proteins directly into the growth medium (2, 4, 37, 45). B. megaterium employs Sec and Tat systems for this purpose; the relevant genes were all detected in the genomes. The Sec-dependent secretion system is especially widely used and has been investigated for industrial recombinant protein production. A look at both genome sequences of B. megaterium revealed potential genes for approximately 30 secreted proteases (see Table S5 in the supplemental material). Only a few proteases have experimentally been found to be secreted, including metalloprotease InhA, extracellular protease Vpr, aminopeptidase YwaD, and neutral protease NprM (67). The gene for the major extracellular protease NprM was deleted from production strain MS941 (69), which allowed up to 1.2 g/liter functional proteins to be recovered from the medium (4, 20, 25, 54, 66). These data clearly demonstrate the advantage of B. megaterium strains for recombinant protein production.
Vitamin B12 production.
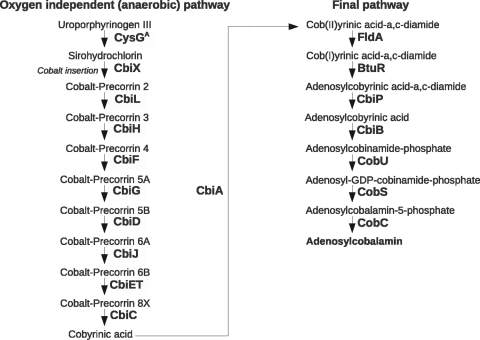
B. megaterium was one of the first biotechnological vitamin B12 producers described for bacteria (61, 64, 70). In agreement with the well-studied biosynthetic pathway in Salmonella enterica, which is known for its ability to synthesize vitamin B12 in the presence and absence of oxygen (5, 8, 49), the genes for oxygen-independent vitamin B12 biosynthesis genes were found in the genomes of B. megaterium. They are organized in two distinct, independent operons (Fig. 5B). The larger operon, cbiWHXJCDETLFGA-cysG-cbiY-btuR, has been thoroughly described (49); the smaller operon, cbiB-cobDUSC, is associated with an uncharacterized ATP:cob(I)alamin adenosyltransferase and codes for the enzymes involved in the final steps of the biosynthetic pathway (Fig. 7). The gene cbiP coding for adenosylcobyrinic acid synthase (EC 6.3.5.10) is isolated in the genome at Mb 2.9 as a single gene with no further B12 biosynthesis context. The genome of both B. megaterium strains now allows the full reconstruction of the vitamin B12 biosynthetic pathway (Fig. 7).
Fig. 7.
Oxygen-independent biosynthesis of adenosylcobalamin. B. megaterium is able to synthesize vitamin B12 in the absence of oxygen due to the presence of the signature gene cbiX, which results in insertion of cobalt at an early stage, in contrast to the oxygen-dependent pathway.
Vitamin B12-dependent enzymes.
Several enzymes with dependencies on vitamin B12 have been identified in silico in the B. megaterium genome. Ethanolamine ammonia-lyase EutBC (EC 4.3.1.7) and methylmalonyl coenzyme A (CoA) mutase MutAB (EC 5.4.99.4) are two that have been shown to contribute to the ability of B. megaterium to degrade lipids. Other than B. megaterium, only Geobacillus kaustophilus and B. halodurans have been found to carry the MutAB genes. EutBC is part of a large B. megaterium species-specific ethanolamine utilization operon, eutHSPABCLEM (BMQ_3678 to BMQ_3686) (Fig. 5C), together with an upstream ethanolamine two-component response regulator system (BMQ_3687/3688) and further uncharacterized genes contributing to ethanolamine utilization (BMQ_3675 to -3677). A second EutBC can also be found as part of a small eutABC-eat cluster (BMQ_2531 to -2534), where Eat is an ethanolamine permease. It is noteworthy that a phage integrase (BMQ_2528) is found three genes downstream, which may indicate a lateral acquisition of this functional unit. Furthermore, the vitamin B12-dependent methionine synthase MetH (BMQ_1293) and a vitamin B12-dependent ribonucleoside-diphosphate reductase (BMQ_4885/4886) were identified in both strains.
Relationship of B. megaterium to other bacilli. (i) Phylogenetic position of B. megaterium.
B. megaterium belongs to a deeply rooted lineage within the genus Bacillus, based on its phenotypic characteristics, genome size, intermediate G+C content, and 16S rRNA phylogeny (Table 1) (1, 46). Whether B. megaterium is more closely related to B. cereus and its relatives or to B. subtilis and its relatives has been difficult to resolve based on 16S rRNA gene phylogeny alone. Trees showing all three possible relationships between these groups have been published recently (27, 44, 68). To address this issue, we performed a phylogenetic analysis on 385 orthologous genes identified using BSR analysis (see Fig. S3 in the supplemental material) (48). A neighbor-joining tree was generated, using Listeria monocytogenes as the outgroup. In this tree B. megaterium, B. cereus, and B. subtilis each anchor clades that are supported in all bootstrap samples. However, the order with which these clades join varies between bootstrap samples, with each of the three possible orders supported by 21 to 51% of the bootstrap samples. Thus, the ambiguous relationship between B. megaterium, B. cereus, and B. subtilis that is seen with 16S rRNA phylogenies is shown to also exist when a large set of orthologous genes is examined, possibly indicating a large amount of lateral gene transfer between these species.
(ii) Genome dynamics in the Bacillus group of organisms.
When orthologous genes were compared between B. megaterium and other Bacillus species, a region of striking synteny was observed neighboring the ori (Fig. 1), from about the 10 o'clock to the 2 o'clock position (Fig. 8; see also Fig. S4 in the supplemental material). The Gram-positive spore former Oceanobacillus iheyensis and Listeria monocytogenes, the causative agent of listeriosis, show similar regions of synteny. Increased phylogenetic distance generally correlated with an increased number of insertions and inversions in the syntenic regions. The region of synteny is dominated by genes involved in sporulation, cell envelope biosynthesis, and the transcription and translation machinery. This phenomenon might be explained by a need for these genes to be readily accessible during germination and sporulation (73).
Fig. 8.
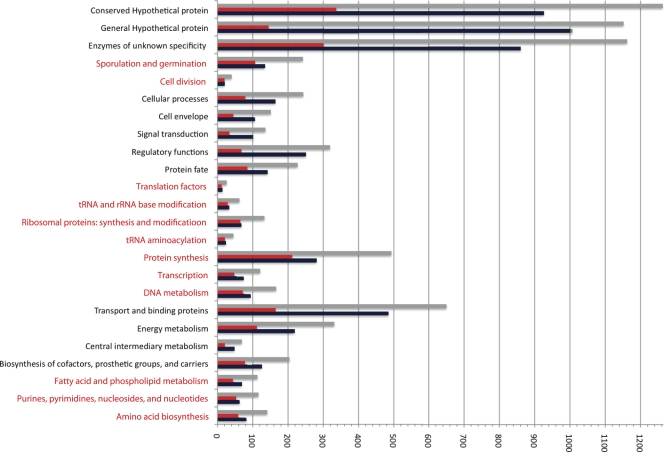
Distinct genomic locations for different functional classes of genes in B. megaterium QM B1551. The number of genes classified by function that were encoded within the conserved syntenic region flanking the ori (red) was compared to the number present in the remainder of the chromosome (green), according to Fig. 1, and to the total chromosomal number (gray). Those functions concentrated in the conserved region that were compared to the remainder of the functions are lettered in red.
The core and pan-genome of the Bacillus group of organisms.
The availability of two complete genomes of B. megaterium (Fig. 1; see also Table S1 in the supplemental material) provides the opportunity to investigate the genomic plasticity and global gene reservoir of the Bacillus group of organisms, the Bacillus pan-genome (39) (Fig. 9). The core genome, genes expected to be present in all genomes, is estimated at 2,009 protein-coding genes (Fig. 9A). We estimated the Bacillus pan-genome to contain 10,534 unique protein-coding genes (Fig. 9C). The pan-genome defines the total number of genes found at least once among all Bacillus genomes. Our estimates of the gene discovery rate indicate that Bacillus has an open pan-genome: on average, 96 new genes would be expected from each new Bacillus genome sequenced (Fig. 9B). These results are comparable to data obtained for E. coli (59) or for Staphylococcus agalacticae and S. pneumoniae (58). As evidenced by the genomic analyses of B. megaterium QM B1551 and DSM319, there is a high degree of genetic diversity even among closely related strains. The gene discovery rate in Bacillus was significantly increased by the fact that each strain had about 300 genes not found in the other strain, with an additional 499 genes found on the QM B1551 plasmids. In a species featuring an open pan-genome, such as E. coli, the predicted pan-genome size is almost 75% larger than the average genome size of the species (59), a finding comparable to our results within the Bacillus group of organisms.
Conclusions and future directions.
The analysis of the genome sequences of the historically and biotechnologically important strains QM B1551 and DSM319 has advanced our knowledge of the metabolic versatility and the evolution of the Gram-positive aerobic spore-forming bacteria. We have identified numerous unique genetic traits not seen in any Bacillus species previously studied, including the presence of gas vesicle proteins, a glyoxylate pathway, and vitamin B12 biosynthesis, and also a lack of homology with the spore coats of B. subtilis. Our data suggest there is considerable DNA exchange between Bacillus and other Gram-positive bacteria (including pathogens), as well as on the intraspecies level between the seven indigenous plasmids and chromosomes. The observed phenomenon of large conserved syntenic regions neighboring the chromosomal ori is key in understanding the lifestyle of B. megaterium and other related spore formers. The availability of the genome sequences of these two B. megaterium strains, the reconstruction of key metabolic and energetic pathways, and the finding that most of the known Bacillus species competence genes are present, together with well-established protocols for genetic modifications, offer a promising future for the further development of B. megaterium as a model organism for systems biotechnology (3, 19, 64).
Supplementary Material
ACKNOWLEDGMENTS
The sequencing of B. megaterium strains QM B1551 and DSM319 was supported with federal funds from the National Science Foundation under NSF contract no. 0802327 and the German Research Foundation under contract no. SFB578.
We thank Vanessa Hering for the growth experiments, Anni Moore and Melvin Duvall for phylogeny discussions, and Stefan Münnich and students of the Bioinformatics classes at NIU for help with the annotation.
We declare that we have no competing financial interests.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Ash C., Farrow J. A. E., Wallbanks S., Collins M. D. 1991. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small subunit ribosomal RNA sequences. Lett. Appl. Microbiol. 13:202–206 [Google Scholar]
- 2. Biedendieck R., et al. 2007. Export, purification, and activities of affinity tagged Lactobacillus reuteri levansucrase produced by Bacillus megaterium. Appl. Microbiol. Biotechnol. 74:1062–1073 [DOI] [PubMed] [Google Scholar]
- 3. Biedendieck R., et al. 2009. Systems biology of recombinant protein production in Bacillus megaterium, p. 133–161 In Scheper T. (ed.), Biosystems engineering, vol. 113, Springer, Berlin, Germany [Google Scholar]
- 4. Biedendieck R., et al. 2007. A sucrose-inducible promoter system for the intra- and extracellular protein production in Bacillus megaterium. J. Biotechnol. 132:426–430 [DOI] [PubMed] [Google Scholar]
- 5. Biedendieck R., et al. 2010. Metabolic engineering of cobalamin (vitamin B12) production in Bacillus megaterium. Microb. Biotechnol. 3:24–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bremer E. 2002. Adaptation to changing osmolarity, p. 385–391 In Sonenshein A. L., Hoch J. A., Losick R.(ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC [Google Scholar]
- 7. Bunk B., Biedendieck R., Jahn D., Vary P. S. 2010. Bacillus megaterium and other bacilli: industrial applications, p. 1–15 Encyclopedia of industrial biotechnology: bioprocess, bioseparation, and cell technology. John Wiley & Sons, Hoboken, NJ [Google Scholar]
- 8. Bunk B., et al. 2010. A short story about a big magic bug. Bioeng. Bugs 1:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bursy J., Pierik A. J., Pica N., Bremer E. 2007. Osmotically induced synthesis of the compatible solute hydroxyectoine is mediated by an evolutionarily conserved ectoine hydroxylase. J. Biol. Chem. 282:31147–31155 [DOI] [PubMed] [Google Scholar]
- 10. Christie G., Gotzke H., Lowe C. R. 2010. Identification of a receptor subunit and putative ligand-binding residues involved in the Bacillus megaterium QM B1551 spore germination response to glucose. J. Bacteriol. 17:4317–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Christie G., Lazarevska M., Lowe C. R. 2008. Functional consequences of amino acid substitutions to GerVB, a component of the Bacillus megaterium spore germinant receptor. J. Bacteriol. 190:2014–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Christie G., Lowe C. R. 2007. Role of chromosomal and plasmid-borne receptor homologues in the response of Bacillus megaterium QM B1551 spores to germinants. J. Bacteriol. 189:4375–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clarke N. A., Cowles P. B. 1952. Studies on the host-virus relationship in a lysogenic strain of Bacillus megaterium. II. The relationship between growth and bacteriophage production in cultures of Bacillus megaterium. J. Bacteriol. 63:177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Bary A. 1884. Vergleichende Morphologie und Biologie der Pilze, Mycetozoen und Bacterien. Wilhelm Engelmann, Leipzig, Germany [Google Scholar]
- 15. Dietrich J. A., et al. 2009. A novel semi-biosynthetic route for artemisinin production using engineered substrate-promiscuous P450(BM3). ACS Chem. Biol. 4:261–267 [DOI] [PubMed] [Google Scholar]
- 16. Dubnau D. 1993. Genetic exchange and homologous recombination, p. 555–584 In Sonenshein A. L., Hoch J. A., Losick R. (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, DC [Google Scholar]
- 17. English J. D., Vary P. S. 1986. Isolation of recombination-defective and UV-sensitive mutants of Bacillus megaterium. J. Bacteriol. 165:155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Foerster H. F., Foster J. W. 1966. Response of Bacillus spores to combinations of germinative compounds. J. Bacteriol. 91:1168–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fürch T., et al. 2007. Effect of different carbon sources on central metabolic fluxes and the recombinant production of a hydrolase from Thermobifida fusca in Bacillus megaterium. J. Biotechnol. 132:385–394 [DOI] [PubMed] [Google Scholar]
- 20. Gamer M., Frode D., Biedendieck R., Stammen S., Jahn D. 2009. A T7 RNA polymerase-dependent gene expression system for Bacillus megaterium. Appl. Microbiol. Biotechnol. 82:1195–1203 [DOI] [PubMed] [Google Scholar]
- 21. He J.-S., Fulco A. J. 1991. A barbiturate-regulated protein binding to a common sequence in the cytochrome P450 genes of rodents and bacteria. J. Biol. Chem. 266:7864–7869 [PubMed] [Google Scholar]
- 22. Holtmann G., Bakker E. P., Uozumi N., Bremer E. 2003. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J. Bacteriol. 185:1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Imamura D., Kuwana R., Takamatsu H., Watabe K. 2010. Localization of proteins to different layers and regions of Bacillus subtilis spore coats. J. Bacteriol. 192:518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kieselburg M. K., Weickert M., Vary P. S. 1984. Analysis of resident and transformant plasmids in Bacillus megaterium. Biotechnology 2:254–259 [Google Scholar]
- 25. Kim J. Y. 2003. Overproduction and secretion of Bacillus circulans endo-beta-1,3-1,4-glucanase gene (bglBC1) in B. subtilis and B. megaterium. Biotechnol. Lett. 25:1445–1449 [DOI] [PubMed] [Google Scholar]
- 26. Kocher S., Averhoff B., Muller V. 1 March 2011. Development of a genetic system for the moderately halophilic bacterium Halobacillus halophilus: generation and characterization of mutants defect in the production of the compatible solute proline. Environ. Microbiol. doi:10.1111/j.1462-2920.2011.02437.x [DOI] [PubMed] [Google Scholar]
- 27. Kolsto A. B., Tourasse N. J., Okstad O. A. 2009. What sets Bacillus anthracis apart from other Bacillus species? Annu. Rev. Microbiol. 63:451–476 [DOI] [PubMed] [Google Scholar]
- 28. Kramer N., Hahn J., Dubnau D. 2007. Multiple interactions among the competence proteins of Bacillus subtilis. Mol. Microbiol. 65:454–464 [DOI] [PubMed] [Google Scholar]
- 29. Kunnimalaiyaan M., Stevenson D. M., Zhou Y., Vary P. S. 2001. Analysis of the replicon region and identification of an rRNA operon on pBM400 of Bacillus megaterium QM B1551. Mol. Microbiol. 39:1010–1021 [DOI] [PubMed] [Google Scholar]
- 30. Kunnimalaiyaan M., Vary P. S. 2005. Molecular characterization of plasmid pBM300 from Bacillus megaterium QM B1551. Appl. Environ. Microbiol. 71:3068–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lach D. A., Sharma V. K., Vary P. S. 1990. Isolation and characterization of a unique division mutant of Bacillus megaterium. J. Gen. Microbiol. 136:545–553 [DOI] [PubMed] [Google Scholar]
- 32. Lemoigne M., Lenoel C. P., Croson M. 1950. Assimilation of acetylacetic acid and beta-hydroxybutyric acid by B. megatherium. Ann. Inst. Pasteur (Paris) 78:705–710(In French.) [PubMed] [Google Scholar]
- 33. Li N., Cannon M. C. 1998. Gas vesicle genes identified in Bacillus megaterium and functional expression in Escherichia coli. J. Bacteriol. 180:2450–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ludwig W., Schleifer K.-H., Whitman W. B. 2009. Revised road map to the phylum Firmicutes, p. 1–14 In Vos P. D., et al.(ed.), Bergey's manual of systematic bacteriology, vol. 3, 2nd ed. Springer, New York, NY [Google Scholar]
- 35. Lwoff A., Siminovitch L., Kjeldgaard N. 1950. Induction of the production of bacteriophages in lysogenic bacteria. Ann. Inst. Pasteur (Paris) 79:815–859(In French.) [PubMed] [Google Scholar]
- 36. Mader U., et al. 2002. Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics. Mol. Genet. Genomics 268:455–467 [DOI] [PubMed] [Google Scholar]
- 37. Malten M., et al. 2006. A Bacillus megaterium plasmid system for the production, export, and one-step purification of affinity-tagged heterologous levansucrase from growth medium. Appl. Environ. Microbiol. 72:1677–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Malten M., Hollmann R., Deckwer W. D., Jahn D. 2005. Production and secretion of recombinant Leuconostoc mesenteroides dextransucrase DsrS in Bacillus megaterium. Biotechnol. Bioeng. 89:206–218 [DOI] [PubMed] [Google Scholar]
- 39. Medini D., Donati C., Tettelin H., Masignani V., Rappuoli R. 2005. The microbial pan-genome. Curr. Opin. Genet. Dev. 15:589–594 [DOI] [PubMed] [Google Scholar]
- 40. Nahrstedt H., Meinhardt F. 2004. Structural and functional characterization of the Bacillus megaterium uvrBA locus and generation of UV-sensitive mutants. Appl. Microbiol. Biotechnol. 65:193–199 [DOI] [PubMed] [Google Scholar]
- 41. Nicholson W. L. 2008. The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanediol dehydrogenase. Appl. Environ. Microbiol. 74:6832–6838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ordal G. W., Marquez-Magana L., Chamberlin M. J. 1993. Motility and chemotaxis, p. 765–784 In Sonenshein A. L., Hoch J. A., Losick R. (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, DC [Google Scholar]
- 43. Panbangred W., Weeradechapon K., Udomvaraphant S., Fujiyama K., Meevootisom V. 2000. High expression of the penicillin G acylase gene (pac) from Bacillus megaterium UN1 in its own pac minus mutant. J. Appl. Microbiol. 89:152–157 [DOI] [PubMed] [Google Scholar]
- 44. Porwal S., Lal S., Cheema S., Kalia V. C. 2009. Phylogeny in aid of the present and novel microbial lineages: diversity in Bacillus. PLoS One 4:e4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Priest F. G. 1977. Extracellular enzyme synthesis in the genus Bacillus. Bacteriol. Rev. 41:711–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Priest F. G., Goodfellow M., Todd C. 1988. A numerical classification of the genus Bacillus. J. Gen. Microbiol. 134:1847–1882 [DOI] [PubMed] [Google Scholar]
- 47. Ramos H. C., et al. 2000. Fermentative metabolism of Bacillus subtilis: physiology and regulation of gene expression. J. Bacteriol. 182:3072–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rasko D. A., Myers G. S., Ravel J. 2005. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Raux E., Lanois A., Rambach A., Warren M. J., Thermes C. 1998. Cobalamin (vitamin B12) biosynthesis: functional characterization of the Bacillus megaterium cbi genes required to convert uroporphyrinogen III into cobyrinic acid a,c-diamide. Biochem. J. 335:167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ren Q., Chen K., Paulsen I. T. 2007. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 35:D274–D279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rossler D., et al. 1991. Phylogenetic diversity in the genus Bacillus as seen by 16S rRNA sequencing studies. Syst. Appl. Microbiol. 14:266–269 [DOI] [PubMed] [Google Scholar]
- 52. Scholle M. D., White C. A., Kunnimalaiyaan M., Vary P. S. 2003. Sequencing and characterization of pBM400 from Bacillus megaterium QM B1551. Appl. Environ. Microbiol. 69:6888–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stahl U., Esser K. 1983. Plasmid heterogeneity in various strains of Bacillus megaterium. Eur. J. Appl. Microbiol. Biotechnol. 17:248–251 [Google Scholar]
- 54. Stammen S., et al. 2010. High-yield intra- and extracellular protein production using Bacillus megaterium. Appl. Environ. Microbiol. 76:4037–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stevenson D. M., Lach D., Vary P. S. 1993. A gene required for germination in Bacillus megaterium is plasmid-borne, p. 197–207 In Balla E., Berencsie G. (ed.), DNA transfer and gene expression in microorganisms. Intercept, Budapest, Hungary [Google Scholar]
- 56. Sussman M. D., Vary P. S., Hartman C., Setlow P. 1988. Integration and mapping of Bacillus megaterium genes which code for small, acid-soluble spore proteins and their protease. J. Bacteriol. 170:4942–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tao Y.-P., Vary P. S. 1991. Isolation and characterization of sporulation lacZ fusion mutants of Bacillus megaterium. J. Gen. Microbiol. 137:797–806 [Google Scholar]
- 58. Tettelin H., et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Touchon M., et al. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. van Keulen G., Hopwood D. A., Dijkhuizen L., Sawers R. G. 2005. Gas vesicles in actinomycetes: old buoys in novel habitats? Trends Microbiol. 13:350–354 [DOI] [PubMed] [Google Scholar]
- 61. Vary P. S. 1992. Development of genetic engineering in Bacillus megaterium, p. 251–310 In Doi R., McGloughlin M. (ed.), Biology of bacilli: application to industry. Butterworths-Heinemann, Boston, MA: [PubMed] [Google Scholar]
- 62. Vary P. S. 1993. The genetic map of Bacillus megaterium, p. 475–487 In Sonenshein A. L., Hoch J. A., Losick R. (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, DC [Google Scholar]
- 63. Vary P. S. 1994. Prime time for Bacillus megaterium. Microbiology 140:1001–1013 [DOI] [PubMed] [Google Scholar]
- 64. Vary P. S., et al. 2007. Bacillus megaterium: from simple soil bacterium to industrial protein production host. Appl. Microbiol. Biotechnol. 76:957–967 [DOI] [PubMed] [Google Scholar]
- 65. Vary P. S., Garbe J. C., Franzen M., Frampton E. W. 1982. Genetics of leucine biosynthesis in Bacillus megaterium. J. Bacteriol. 149:1112–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. von Tersch M. A., Robbins H. L. 1990. Efficient cloning in Bacillus megaterium: comparison to Bacillus subtilis and Escherichia coli cloning hosts. FEMS Microbiol. Lett. 70:305–310 [DOI] [PubMed] [Google Scholar]
- 67. Wang W., Sun J., Hollmann R., Zeng A.-P., Deckwer W.-D. 2006. Proteomic characterization of transient expression and secretion of a stress-related metalloprotease in high cell density culture of Bacillus megaterium. J. Biotechnol. 126:313–324 [DOI] [PubMed] [Google Scholar]
- 68. Wang W., Sun M. 2009. Phylogenetic relationships between Bacillus species and related genera inferred from 16S rDNA sequences. Braz. J. Microbiol. 40:505–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wittchen K. D., Meinhardt F. 1995. Inactivation of the major extracellular protease from Bacillus megaterium DSM319 by gene replacement. Appl. Microbiol. Biotechnol. 42:871–877 [DOI] [PubMed] [Google Scholar]
- 70. Wolf J. B., Brey R. N. 1986. Isolation and genetic characterizations of Bacillus megaterium cobalamin biosynthesis-deficient mutants. J. Bacteriol. 166:51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wood J. M., et al. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130:437–460 [DOI] [PubMed] [Google Scholar]
- 72. Yang Y., et al. 2006. High yield recombinant penicillin G amidase production and export into the growth medium using Bacillus megaterium. Microb. Cell Fact. 5:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zupancic M. L., Tran H., Hofmeister A. E. 2001. Chromosomal organization governs the timing of cell type-specific gene expression required for spore formation in Bacillus subtilis. Mol. Microbiol. 39:1471–1481 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.